Manogepix is a broad-spectrum antifungal agent that inhibits glycosylphosphatidylinositol (GPI) anchor biosynthesis. Using whole-genome sequencing, we characterized two efflux-mediated mechanisms in the fungal pathogens Candida albicans and Candida parapsilosis that resulted in decreased manogepix susceptibility. In C. albicans, a gain-of-function mutation in the transcription factor gene ZCF29 activated expression of ATP-binding cassette transporter genes CDR11 and SNQ2.
KEYWORDS: APX001, fosmanogepix, manogepix, APX001A, Gwt1, antifungal, GPI anchor, glycosylphosphatidylinositol, antifungal therapy, efflux
ABSTRACT
Manogepix is a broad-spectrum antifungal agent that inhibits glycosylphosphatidylinositol (GPI) anchor biosynthesis. Using whole-genome sequencing, we characterized two efflux-mediated mechanisms in the fungal pathogens Candida albicans and Candida parapsilosis that resulted in decreased manogepix susceptibility. In C. albicans, a gain-of-function mutation in the transcription factor gene ZCF29 activated expression of ATP-binding cassette transporter genes CDR11 and SNQ2. In C. parapsilosis, a mitochondrial deletion activated expression of the major facilitator superfamily transporter gene MDR1.
INTRODUCTION
Invasive fungal infections cause significant mortality and morbidity in humans, killing >1.5 million people annually (1, 2). A rapidly growing immunocompromised population is at particular risk, including those undergoing chemotherapy or solid organ transplantation or those infected with HIV (3–5). Current antifungal treatments are limited to three major classes of drugs: azoles, polyenes, and echinocandins (6). Issues of safety, tolerability, and the evolution of fungal drug resistance necessitate the development of antifungals with new mechanisms of action.
Fosmanogepix (formerly APX001, formerly E1211) is a novel intravenous (i.v.) and orally available N-phosphonooxymethyl prodrug that is currently in clinical development for the treatment of life-threatening invasive fungal infections that are often resistant to standard-of-care antifungal therapy (ClinicalTrials identifier NCT03604705) (7, 8). Fosmanogepix is converted by systemic phosphatases to the active moiety, manogepix (MGX; APX001A, formerly E1210) (9). MGX targets the essential fungal acyltransferase Gwt1 (10), blocking inositol acylation of glycosylphosphatidylinositol (GPI) anchors and trafficking of GPI-anchored proteins from the endoplasmic reticulum (ER) (11, 12). GPI anchors are attached to proteins in the ER and mediate their trafficking and attachment to the cell surface (13). MGX does not inhibit the mammalian Gwt1 homolog, PIGW (12). Gwt1 inhibition halts fungal growth, activates unfolded protein stress responses, and alters the composition of the fungal cell wall to expose immunostimulatory β-(1→3)-glucans (14). MGX has activity against the major fungal pathogens Candida albicans (15), Candida auris (16), Cryptococcus neoformans (17), and Aspergillus fumigatus (18), as well as less common pathogens, including Fusarium and Scedosporium (19).
To further explore the therapeutic potential of fosmanogepix, it is important to understand the potential for evolution of drug resistance. Spontaneous and serial passage experiments revealed that the GWT1 missense mutations V162A (heterozygous) and V163A in C. albicans and Candida glabrata, respectively, demonstrated 16- and 32-fold increases in MGX MIC values (20). These mutations are hypothesized to impede drug binding to the target and differ from essential catalytic residues in Gwt1 (10, 21). Off-target mutations driving increases in MGX MIC values are largely unknown, although mutations in EMP24 suppress toxicity of the Gwt1 inhibitor gepinacin (14). Emp24 facilitates quality control of GPI assembly by directing mature GPI-anchored proteins from the ER to the Golgi complex (22). Emp24 loss of function is predicted to release immature GPI-anchored proteins that accumulate during Gwt1 inhibition (23).
Spontaneous mutants of C. albicans (strain 5-3) and C. parapsilosis (strain 5-2) were identified that demonstrated decreased susceptibility to MGX and fluconazole (FLC) (20). The C. albicans mutant demonstrated 4-fold and 2-fold increases in the MICs of MGX and FLC, respectively, versus MICS of the isogenic wild-type strain, while the C. parapsilosis mutant demonstrated 8-fold and 4-fold increased MICs of MGX and FLC, respectively, versus MICs of the isogenic wild-type strain (20) (Table 1). In vitro susceptibility assays were performed as described in CLSI M27-A3, except that the dilution scheme consisted of 2-fold serial dilutions from 5 μM to 0.0049 μM (1.792 μg/ml to 0.00175 μg/ml). MIC values were determined at 50% growth inhibition relative to that of drug-free controls at 48 h (24).
TABLE 1.
C. albicans 5-3 and C. parapsilosis 5-2 demonstrate elevated MICs to MGX and FLC, while C. albicans 5-3 and C. albicans ZCF29/ZCF29W986L are resistant to beauvericin
| Background | Strain | Manogepix |
Fluconazole |
Beauvericin |
|||
|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | Fold change vs WT | MIC (μg/ml) | Fold change vs WT | MIC (μg/ml) | Fold change vs WT | ||
| C. albicans ATCC 90028 | WT | 0.0035 | 0.125 | 3.125 | |||
| 5-3 | 0.014 | 4 | 0.5 | 4 | 100 | 32 | |
| C. parapsilosis ATCC 22019 | WT | 0.007 | 2 | 25 | |||
| 5-2 | 0.056 | 8 | 4 | 2 | 25 | 1 | |
| C. albicans LC191 | WT | 0.0035 | 0.125 | 3.125 | |||
| ZCF29/ZCF29W986L | 0.028 | 8 | 0.25 | 2 | 100 | 32 | |
These two strains were not mutated in the GWT1 gene; thus, we hypothesized that elevated MIC values for MGX and FLC could result from enhanced expression of multidrug efflux pumps, affecting the susceptibility to the structurally and mechanistically distinct antifungal agents. Although drug efflux has been frequently associated with antifungal resistance (25), it has not been described for MGX. To assess efflux in the strains with elevated MGX MIC values, we incubated cultures with Nile red, which accumulates in lipid membranes and is actively extruded from cells by efflux (26). Nile red is a substrate for the ATP-binding cassette (ABC) transporters Cdr1 and Cdr2, and the major facilitator superfamily transporter Mdr1 (26), which are the efflux pumps most frequently resulting in azole resistance in Candida (25). For these experiments, unit equivalents of an optical density at 600 nm (OD600) of 1 of log-phase cultures were washed 2 times in 1 ml buffer A (20 mM Na-HEPES, 150 mM NaCl, pH 7.5), resuspended in 1 ml buffer A, and incubated at 30°C for 2 h. Nile red was added to 7 μM and incubated for 1 h. Stained cells were washed 2 times in buffer A, and then efflux was initiated by addition of glucose to 1% (wt/vol). Nile red fluorescence was determined by flow cytometry after 30 min (Beckman CytoFLEX, phycoerythrin (PE) filter A01-1-0052; analysis with CytExpert 2.3) and visualized by fluorescence microscopy on a Zeiss AxioObserver.Z1 (Chroma Tech ET Cy5 filter). Both C. albicans and C. parapsilosis MGX mutants with elevated MIC values accumulated ∼60% less Nile red than their parental strains (Fig. 1A and B), implicating drug efflux in the decreased susceptibility to MGX.
FIG 1.
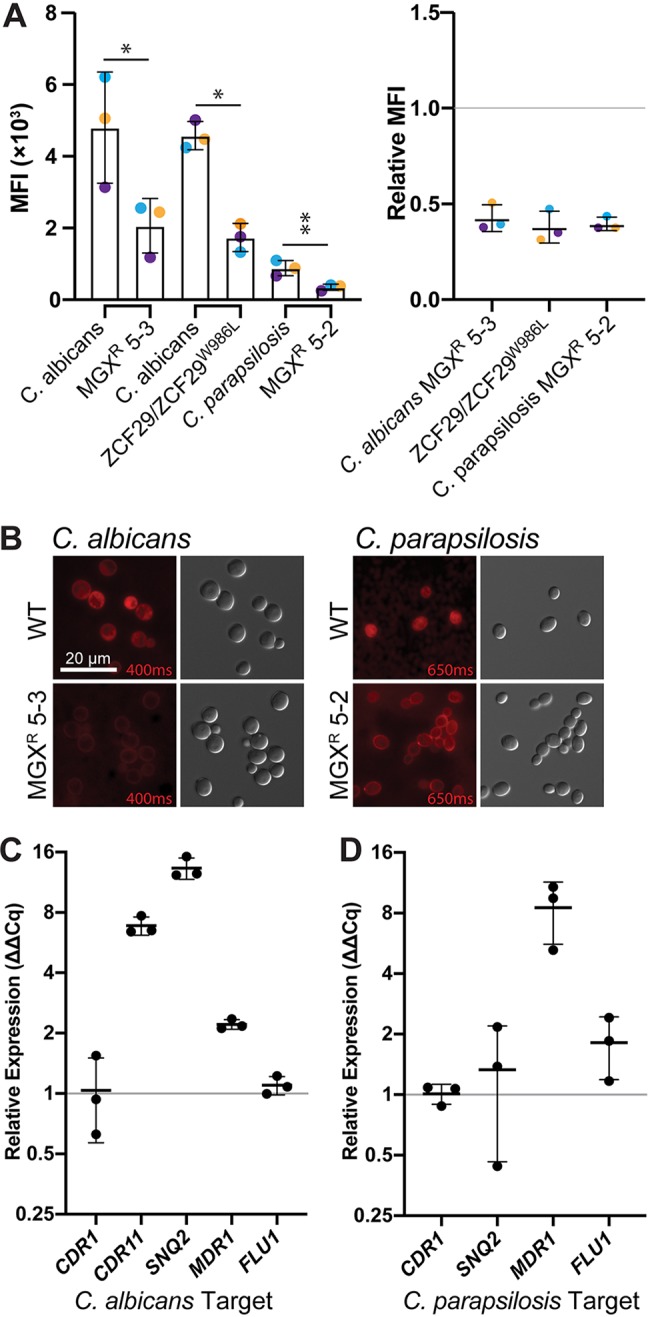
Drug efflux is activated in mutants of C. albicans and C. parapsilosis with reduced susceptibility to MGX. (A) C. albicans 5-3 and C. parapsilosis 5-2 mutants have reduced accumulation of the general efflux pump substrate Nile red. Nile red fluorescence was monitored by flow cytometry. (Left) Median fluorescence intensity (MFI; PE) ± standard deviation (SD) measured in 3 independent experiments (10,000 events/sample). (Right) Ratios of median fluorescence intensity for indicated mutant-wild type pair. Differences between groups were determined by ratio paired t test. *, P ≤ 0.05; **, P ≤ 0.005. Colored points indicate experimental replicates. (B) Representative micrographs of C. albicans and C. parapsilosis wild-type strains and mutants with decreased MGX susceptibility stained with Nile red, prepared the same as for those in panel A. Exposure times (milliseconds) are indicated in red. (C) Relative transcript levels of CDR11, SNQ2, and MDR1 but not CDR1 or FLU1 are upregulated in C. albicans MGXr 5-3. RT-qPCR data are mean fold changes ± SDs from 3 biological replicates assayed in technical triplicates, normalized to ACT1 and GPD1. (D) Transcript levels of MDR1 but not CDR1, SNQ2, or FLU1 are upregulated in C. parapsilosis MGXr 5-2. Experiments were performed the same as for those in panel C and normalized to ACT1.
To identify mutations responsible for activation of drug efflux, we sequenced the genomes of the MGX mutants using the Illumina MiSeq platform (Genewiz). Adaptor sequences and low-quality reads were removed using Trimmomatic v0.39 (27). Paired reads were assembled to Candida Genome Database (28) C_albicans_SC5314_A21 (29) and C_parapsilosis_CDC317 (30) by using Bowtie2 v2.3.5.1 (31) (Table 2). Missense single nucleotide variants (SNVs) between parental and mutant assemblies were detected using Mutect v1.1.7 (32) and SnpEff v2.6.3 (33) and validated by Sanger sequencing. Loss of heterozygosity or aneuploidy was not detected by the Yeast Mapping Analysis Pipeline (34) and CNV-seq (35).
TABLE 2.
Fungal strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| C. albicans | ||
| ATCC 90028 | Wild type | ATCC |
| ATCC 90028 5-3 | ZCF29W986L/ZCF29 | 20 |
| CaLC191 (DAY185) | URA3/ura3::imm434 HIS1/his1::hisG ARG4/arg4::hisG | 45 |
| CaLC3815 | URA3/ura3::imm434 HIS1/his1::hisG ARG4/arg4::hisG ZCF29W986L/ZCF29 | 36 |
| C. parapsilosis | ||
| ATCC 22019 | Wild type | ATCC |
| ATCC 22019 5-2 | capafmp06Δ3780-5672 | 20 |
In C. albicans MGXr 5-3, a single heterozygous SNV resulted in a W986L substitution in the Zn(II)2Cys6 transcription factor Zcf29. This gain-of-function mutation was previously identified in beauvericin-resistant C. albicans (36). Consistent with this connection, C. albicans MGXr 5-3 demonstrated a 32-fold increase in the MIC value versus that in the wild-type (WT) strain to beauvericin, and an engineered ZCF29/ZCF29W986L C. albicans mutant demonstrated an 8-fold increase in the MGX MIC value versus that in the WT strain (Table 1). Transcript levels for efflux pump genes were assessed using reverse transcriptase quantitative real-time PCR (RT-qPCR). RNA was extracted from 10 ml log-phase yeast extract-peptone-dextrose (YPD) cultures grown at 30°C by using a Qiagen RNeasy minikit. RNA was treated with Qiagen RNase-free DNase and reverse transcribed using Bio-Rad iScript cDNA synthesis kit. qPCR was performed using Thermo Fisher Scientific SYBR green master mix and oligonucleotide primers described in Table S2 in the supplemental material. Data were analyzed using Bio-Rad CFX Manager 3.1. Transcript levels of efflux genes CDR11, SNQ2, and MDR1 were increased in MGXr 5-3 by 6.8, 13.3, and 2.2-fold, respectively (Fig. 1C), consistent with transcriptional profiling of C. albicans ZCF29/ZCF29W986L (36). Transcript levels of CDR1 and FLU1 were unchanged (Fig. 1C).
No missense SNVs were detected in C. parapsilosis MGXr 5-2; however, a deletion was detected in the mitochondrial chromosome from bases 15430 to 17322. The boundaries of this deletion were defined using MitoDel v3.0 (37), which identified 2,087 reads across this junction. This deletion interrupts CAPAFMP06 (COX1), which encodes cytochrome c oxidase, and CAPAFMP06.3/CAPAFMP06.4, intronic open reading frames (ORFs) to COX1 that encode enzymes with predicted roles in mRNA splicing (38). Consistent with a respiratory defect, C. parapsilosis MGXr 5-2 formed petite colonies on YP glucose-containing medium and did not grow on YP medium containing the nonfermentable carbon source glycerol (Fig. 2A). Furthermore, when subcultured in glycerol-supplemented synthetic complete medium for 2 h and then stained with 10 nM mitochondrial membrane potential-dependent stain MitoTracker Red CMXRos (Invitrogen) for 30 min, C. parapsilosis MGXr 5-2 showed ∼60% reduced staining relative to that of the parental strain. MitoTracker Red fluorescence was quantified by flow cytometry (Fig. 2B) (Beckman CytoFLEX, DsRed filter A01-1-0053) and visualized by fluorescence microscopy (Fig. 2C) on a Zeiss AxioObserver.Z1 (Chroma Tech ET HQ DsRed filter).
FIG 2.
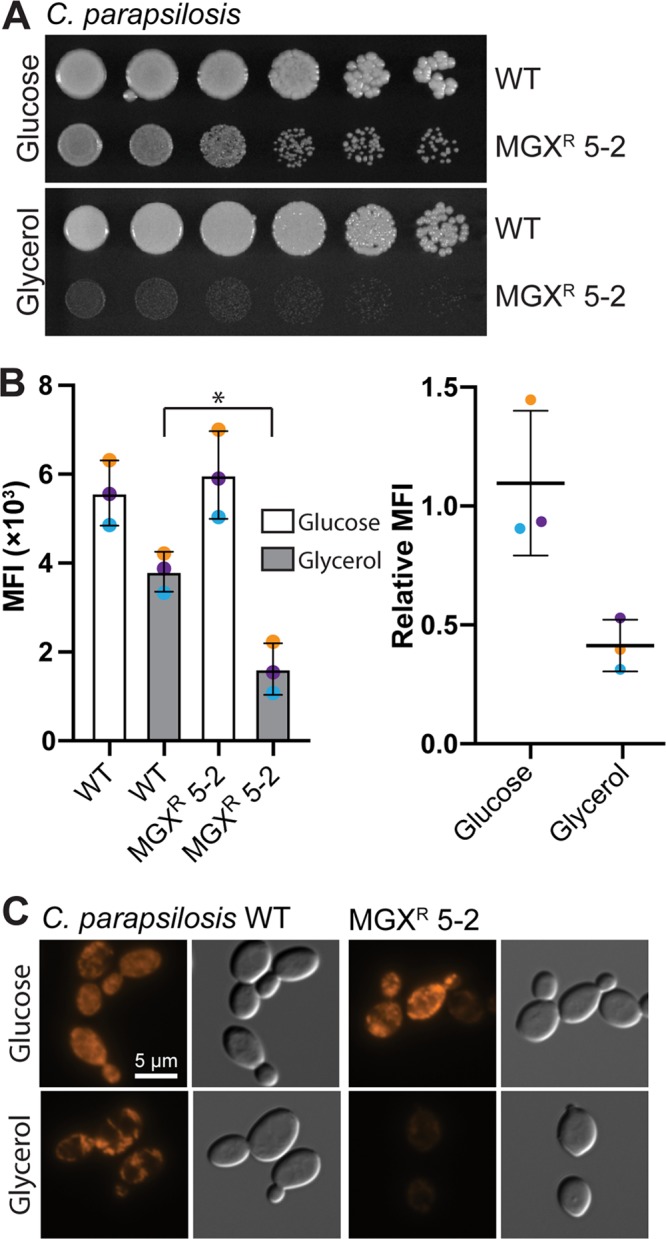
C. parapsilosis MGXr 5-2 has a defect in mitochondrial function. (A) C. parapsilosis MGXr 5-2 forms petite colonies on YPD agar and does not grow on YP-glycerol agar; 10-fold dilutions of stationary-phase cultures of C. parapsilosis were spotted on YP agar containing 2% (wt/vol) d-glucose or glycerol and then photographed after 48 h of growth at 30°C. (B) C. parapsilosis MGXr 5-2 has reduced mitochondrial membrane potential when subcultured in medium containing 2% (wt/vol) glycerol. MitoTracker Red CMXRos fluorescence was monitored by flow cytometry. Data are median fluorescence intensities (DsRed) ± SDs from 3 independent experiments (10,000 events/sample). Differences between groups were determined by ratio paired t test. *, P ≤ 0.05. (C) Representative micrographs of MitoTracker Red-stained cells prepared the same as for those in panel B.
Respiratory competence is linked to drug susceptibility in diverse fungal pathogens and is often driven by efflux pump overexpression (39). Indeed, transcript levels of MDR1 were 8.5-fold upregulated in C. parapsilosis MGXr 5-2 (Fig. 1C), consistent with a previously described petite mutant of C. albicans with decreased susceptibility to FLC (40, 41). In some Saccharomyces cerevisiae and C. glabrata petite mutants, the Pdr1/Pdr3 transcription factors induce expression of ABC transporter genes PDR5 (ortholog of CDR1), SNQ2, and YOR1 (42–44). This is not the case in C. parapsilosis MGXr 5-2, as transcript levels of CDR1 and SNQ2 were unchanged (Fig. 1C).
In conclusion, we have identified two efflux-mediated mechanisms conferring reduced susceptibility to MGX in two Candida species. In C. albicans, a gain-of-function mutation in the transcription factor gene ZCF29 activated expression of ABC transporter genes CDR11 and SNQ2. In C. parapsilosis, a mitochondrial deletion activated expression of the major facilitator superfamily (MFS) transporter gene MDR1. The MIC of MGX was at maximum 0.0056 μg/ml, suggesting that these individual mutations may not result in clinically significant resistance. Additionally, loss of mitochondrial function is expected to impair virulence, as observed with some C. albicans petite mutants (41).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided by Amplyx. S.D.L. was supported by an NSERC postdoctoral fellowship (516840-2018).
L.E.C. and L.W. are cofounders and shareholders in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics. L.E.C. is a consultant for Boragen, a small-molecule development company focused on leveraging the unique chemical properties of boron chemistry for crop protection and animal health. M.K. is an employee of Amplyx. K.J.S. was an employee of Amplyx and is now an independent consultant at Hearts Consulting Group. S.D.L. has no conflicts.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low C-Y, Rotstein C. 2011. Emerging fungal infections in immunocompromised patients. F1000 Med Rep 3:14. doi: 10.3410/M3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 5.Enoch DA, Ludlam HA, Brown NM. 2006. Invasive fungal infections: a review of epidemiology and management options. J Med Microbiol 55:809–818. doi: 10.1099/jmm.0.46548-0. [DOI] [PubMed] [Google Scholar]
- 6.Robbins N, Wright GD, Cowen LE. 2016. Antifungal drugs: the current armamentarium and development of new agents. Microbiol Spectr 4:903–922. doi: 10.1128/microbiolspec.FUNK-0002-2016. [DOI] [PubMed] [Google Scholar]
- 7.Hodges MR, Ople E, Shaw KJ. 2017. First-in-human study to assess safety, tolerability and pharmacokinetics of APX001 administered by intravenous infusion to healthy subjects, poster 1840. IDweek 2017, San Diego, CA. [Google Scholar]
- 8.Hodges MR, Ople E, Shaw KJ. 2017. Phase 1 study to assess safety, tolerability and pharmacokinetics of single and multiple oral doses of APX001 and to investigate the effect of food on APX001 bioavailability, poster 1860. IDweek 2017, San Diego, CA. [Google Scholar]
- 9.Hata K, Horii T. 2011. In vitro and in vivo antifungal activities of E1211, a water-soluble prodrug of E1210, abstr F1-1377. Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL. [Google Scholar]
- 10.Umemura M, Okamoto M, Nakayama K-I, Sagane K, Tsukahara K, Hata K, Jigami Y. 2003. GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J Biol Chem 278:23639–23647. doi: 10.1074/jbc.M301044200. [DOI] [PubMed] [Google Scholar]
- 11.Tsukahara K, Hata K, Nakamoto K, Sagane K, Watanabe N-A, Kuromitsu J, Kai J, Tsuchiya M, Ohba F, Jigami Y, Yoshimatsu K, Nagasu T. 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol Microbiol 48:1029–1042. doi: 10.1046/j.1365-2958.2003.03481.x. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe N-A, Miyazaki M, Horii T, Sagane K, Tsukahara K, Hata K. 2012. E1210, a new broad-spectrum antifungal, suppresses Candida albicans hyphal growth through inhibition of glycosylphosphatidylinositol biosynthesis. Antimicrob Agents Chemother 56:960–971. doi: 10.1128/AAC.00731-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittet M, Conzelmann A. 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 14.McLellan CA, Whitesell L, King OD, Lancaster AK, Mazitschek R, Lindquist S. 2012. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem Biol 7:1520–1528. doi: 10.1021/cb300235m. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Lee MH, Paderu P, Lee A, Jimenez-Ortigosa C, Park S, Mansbach RS, Shaw KJ, Perlin DS. 2018. Significantly improved pharmacokinetics enhances in vivo efficacy of APX001 against echinocandin- and multidrug-resistant Candida isolates in a mouse model of invasive candidiasis. Antimicrob Agents Chemother 62:e00425-18. doi: 10.1128/AAC.00425-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hager CL, Larkin EL, Long L, Zohra Abidi F, Shaw KJ, Ghannoum MA. 2018. In vitro and in vivo evaluation of the antifungal activity of APX001A/APX001 against Candida auris. Antimicrob Agents Chemother 62:e02319-17. doi: 10.1128/AAC.02319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw KJ, Schell WA, Covel J, Duboc G, Giamberardino C, Kapoor M, Moloney M, Soltow QA, Tenor JL, Toffaletti DL, Trzoss M, Webb P, Perfect JR. 2018. In vitro and in vivo evaluation of APX001A/APX001 and other Gwt1 inhibitors against Cryptococcus. Antimicrob Agents Chemother 62:e00523-18. doi: 10.1128/AAC.00523-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebremariam T, Alkhazraji S, Alqarihi A, Jeon HH, Gu Y, Kapoor M, Shaw KJ, Ibrahim AS. 2018. APX001 is effective in the treatment of murine invasive pulmonary aspergillosis. Antimicrob Agents Chemother 63:e01713-18. doi: 10.1128/AAC.01713-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castanheira M, Duncanson FP, Diekema DJ, Guarro J, Jones RN, Pfaller MA. 2012. Activities of E1210 and comparator agents tested by CLSI and EUCAST broth microdilution methods against Fusarium and Scedosporium species identified using molecular methods. Antimicrob Agents Chemother 56:352–357. doi: 10.1128/AAC.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor M, Moloney M, Soltow QA, Pillar CM, Shaw KJ. 2019. Evaluation of resistance development to the Gwt1 inhibitor manogepix (APX001A) in Candida species. Antimicrob Agents Chemother 64:e01387-19. doi: 10.1128/AAC.01387-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagane K, Umemura M, Ogawa-Mitsuhashi K, Tsukahara K, Yoko-O T, Jigami Y. 2011. Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p. J Biol Chem 286:14649–14658. doi: 10.1074/jbc.M110.193490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. 2011. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol Biol Cell 22:2924–2936. doi: 10.1091/mbc.E11-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann PA, McLellan CA, Koseoglu S, Si Q, Kuzmin E, Flattery A, Harris G, Sher X, Murgolo N, Wang H, Devito K, de Pedro N, Genilloud O, Kahn JN, Jiang B, Costanzo M, Boone C, Garlisi CG, Lindquist S, Roemer T. 2015. Chemical genomics-based antifungal drug discovery: targeting glycosylphosphatidylinositol (GPI) precursor biosynthesis. ACS Infect Dis 1:59–72. doi: 10.1021/id5000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Revie NM, Iyer KR, Robbins N, Cowen LE. 2018. Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol 45:70–76. doi: 10.1016/j.mib.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394:87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. 2017. The Candida Genome Database (CGD): incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45:D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissman J, Muzzey D, Schwartz K, Weissman JS, Sherlock G. 2013. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol 14:R97. doi: 10.1186/gb-2013-14-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guida A, Lindstädt C, Maguire SL, Ding C, Higgins DG, Corton NJ, Berriman M, Butler G. 2011. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics 12:628. doi: 10.1186/1471-2164-12-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbey DA, Funt J, Lurie-Weinberger MN, Thompson DA, Regev A, Myers CL, Berman J. 2014. YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Med 6:1–16. doi: 10.1186/s13073-014-0100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie C, Tammi MT. 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shekhar-Guturja T, Tebung WA, Mount H, Liu N, Köhler JR, Whiteway M, Cowen LE. 2016. Beauvericin potentiates azole activity via inhibition of multidrug efflux, blocks Candida albicans morphogenesis, and is effluxed via Yor1 and circuitry controlled by Zcf29. Antimicrob Agents Chemother 60:7468–7480. doi: 10.1128/AAC.01959-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosworth CM, Grandhi S, Gould MP, LaFramboise T. 2017. Detection and quantification of mitochondrial DNA deletions from next-generation sequence data. BMC Bioinformatics 18:407–436. doi: 10.1186/s12859-017-1821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nosek J, Novotna M, Hlavatovicova Z, Ussery DW, Fajkus J, Tomaska L. 2004. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol Genet Genomics 272:173–180. doi: 10.1007/s00438-004-1046-0. [DOI] [PubMed] [Google Scholar]
- 39.Shingu-Vazquez M, Traven A. 2011. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell 10:1376–1383. doi: 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S, Clancy CJ, Nguyen KT, Clapp W, Nguyen MH. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob Agents Chemother 51:1855–1858. doi: 10.1128/AAC.00182-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng S, Clancy CJ, Zhang Z, Hao B, Wang W, Iczkowski KA, Pfaller MA, Nguyen MH. 2007. Uncoupling of oxidative phosphorylation enables Candida albicans to resist killing by phagocytes and persist in tissue. Cell Microbiol 9:492–501. doi: 10.1111/j.1462-5822.2006.00805.x. [DOI] [PubMed] [Google Scholar]
- 42.Moye-Rowley WS. 2005. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354:15–21. doi: 10.1016/j.gene.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Gulshan K, Schmidt JA, Shahi P, Moye-Rowley WS. 2008. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol Cell Biol 28:5851–5864. doi: 10.1128/MCB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brun S, Bergès T, Poupard P, Vauzelle-Moreau C, Renier G, Chabasse D, Bouchara J-P. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob Agents Chemother 48:1788–1796. doi: 10.1128/aac.48.5.1788-1796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blankenship JR, Heitman J. 2005. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect Immun 73:5767–5774. doi: 10.1128/IAI.73.9.5767-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


