Abstract
Background and aims
The β-catenin signaling pathway is one of the most commonly deregulated pathways in cancer cells. Amino-acid substitutions within armadillo repeats 5 and 6 (K335, W383, and N387) of β-catenin are found in several tumor types, including liver tumors. We investigated the mechanisms by which these substitutions increase signaling and the effects on liver carcinogenesis in mice.
Methods
Plasmids encoding tagged full-length β-catenin (CTNNB1) or β-catenin with the K335I or N387K substitutions, along with MET, were injected into tails of FVB/N mice. Tumor growth was monitored, and livers were collected and analyzed by histology, immunohistochemistry, and quantitative reverse-transcription PCR. Tagged full-length and mutant forms of β-catenin were expressed in HEK293, HCT116, and SNU449 cells, which were analyzed by immunoblots and immunoprecipitation. A panel of β-catenin variants and cell lines with knock-in mutations were analyzed for differences in N-terminal phosphorylation, half-life, and association with other proteins in the signaling pathway.
Results
Mice injected with plasmids encoding K335I or N387K β-catenin and MET developed larger, more advanced, tumors than mice injected with plasmids encoding wild-type β-catenin and MET. K335I and N387K β-catenin bound APC with lower affinity than wild-type β-catenin, but still interacted with scaffold protein AXIN1 and in the nucleus with TCF7L2. This interaction resulted in increased transcription of genes regulated by β-catenin. Studies of protein structures supported the observed changes in relative binding affinities.
Conclusion
Expression of β-catenin with mutations in armadillo repeats 5 and 6, along with MET, promotes formation of liver tumors in mice. In contrast to N-terminal mutations in β-catenin that directly impair its phosphorylation by GSK3 or binding to BTRC, the K335I or N387K substitutions increase signaling via reduced binding to APC. However, these mutant forms of β-catenin still interact with the TCF family of transcription factors in the nucleus. These findings reveal how these amino acid substitutions increase β-catenin signaling in cancer cells.
Keywords: hepatocellular carcinoma, hepatocarcinogenesis, gene regulation, mouse model
INTRODUCTION
The β-catenin signaling pathway is one of the most commonly deregulated pathways among cancers.1 In normal cells, β-catenin signaling is maintained at low levels by a destruction complex consisting of the APC tumor suppressor, scaffold proteins AXIN1, AXIN2, and the kinases GSK3 and CK1α. In this complex, CK1α initiates β-catenin (CTNNB1) phosphorylation at S45, followed by sequential GSK3 phosphorylation at T41, S37 and finally S33. Next, the phosphorylated 32DpSGXXpS37 motif promotes binding of the ubiquitin ligase β-TrCP (BTRC), which targets β-catenin for proteasomal degradation. When cells are exposed to Wnt ligands, this β-catenin breakdown complex is temporarily inhibited, leading to the stabilization of β-catenin. As a result, it translocates into the nucleus and associates with members of the TCF/LEF family of transcription factors, thus regulating the expression of specific downstream Wnt/β-catenin target genes.2,3
In several tumor types this pathway is constitutively activated through mutational (in)activation of one of the core elements of the destruction complex. In colorectal cancers predominantly ‘loss-of-function’ mutations are observed in the APC gene. For these mutations it has become well-accepted that they are selected on providing a “just-right” level of β-catenin signaling that is optimal for tumor initiation and progression.4 Other tumor types, such as hepatocellular and endometrial carcinomas,5–7 mainly acquire oncogenic β-catenin (CTNNB1) mutations at the N-terminal S/T phosphorylation residues, making the protein more resistant to proteolytic degradation. Recently, Rebouissou and co-workers showed that also for these CTNNB1 mutations a clear genotype-phenotype correlation exists in liver cancer.8 In-frame exon3 deletions that remove the entire N-terminal phosphorylation domain and D32-S37 amino-acid alterations directly affecting the β-TrCP recognition motif, lead to highly active β-catenin variants. T41 mutations were associated with moderate activity, while S45 mutations showed a weak but clear activation of the pathway. The reduced activity of the latter two mutations is most likely the result of residual phosphorylation of the β-TrCP interaction domain, leading to some breakdown of these mutants.9 In this liver cancer study also two more recently recognized mutational CTNNB1 hotspots were included, that is K335I and N387K, which are located in armadillo repeat domains 5 and 6 of the β-catenin protein, respectively. Currently, according to the COSMIC website,10 mutations at these residues have been observed in more than 100 individual tumors, especially those of the liver, colon and kidney (Figure 1, Supplemental Table S1). They lead to a weak but significant enhancement of β-catenin signaling.8,11 However, the mechanism leading to their increased activity is still unknown.12 Here, by exploring potential mechanisms that may explain their increased signaling propensity, we identify a novel mechanism of enhanced β-catenin signaling.
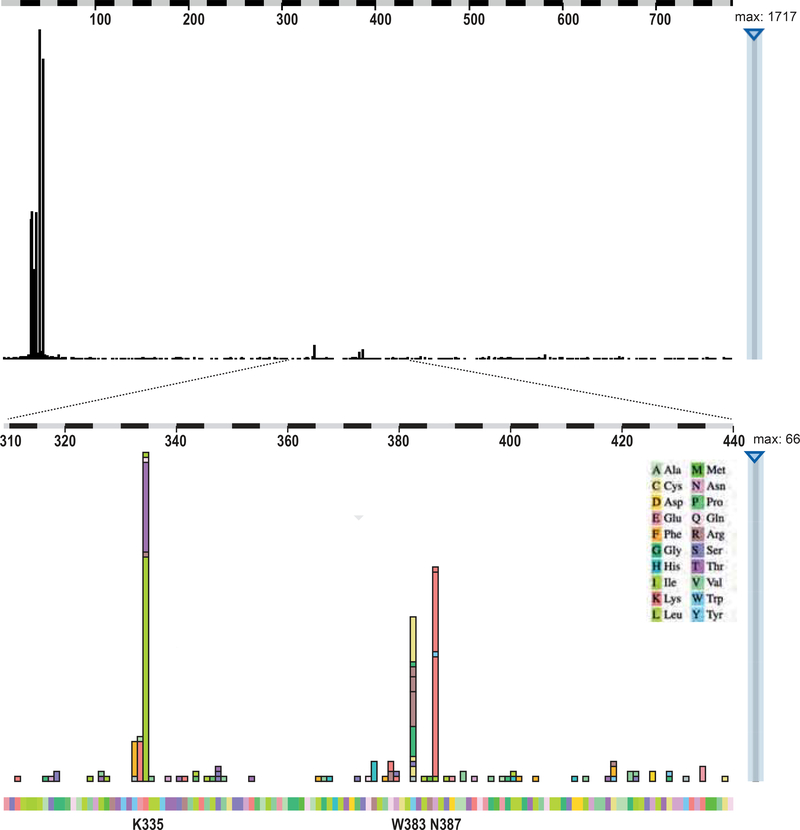
Figure 1. Oncogenic hotspot mutations observed in armadillo repeats 5 and 6 of β-catenin.
Three amino acids are commonly mutated in cancers, i.e. K335, W383, and N387. Top panel shows overview of all reported amino acid substitutions along the β-catenin protein, while bottom panel shows mutations restricted to amino acids 310–440. Details such as involved cancer types can be explored interactively on the COSMIC website, but are also presented in Supplemental Table S1. The following alterations have been observed at least 4 times: Y333F 7x; E334K 8x; K335T 18x; K335I 46x; R376H 4x; W383C 12x; W383R 12x; W383G 7x; N387K 40x.
METHODS
COSMIC Database analysis
To obtain the cancer-related CTNNB1 mutations depicted in Supplemental Figure S1, we analyzed the COSMIC website (https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=CTNNB1) filtering it on amino acids 310–440. Data were updated until January 2019.
Plasmids and construction
N-terminal FLAG-tagged β-catenin variants were constructed by using the pcDNA-5′UT-FLAG vector as previously described.13 Briefly, the constructs of wild-type (WT), S33Y and exon3 deletion were generated by using the Gibson assembly method (NEB). Based on the WT backbone, all other variants (G34V, S37F, T41A, S45P, Y333F, E334K, K335I, K335T, R376H, W383G, W383R, N387K and R582W) were constructed by using Q5 site-directed mutagenesis (NEB). The GFP-APC (1199–2167) plasmid was generated by cloning a 2.9 kb EcoRI fragment of mouse Apc into pEGFP-C1. It encodes all 20 amino acid β-catenin binding repeats and all AXIN-binding domains of APC. pcDNA3.1 myc-mβTrCP WD1–7 (241–569) was a gift from Michael Ruppert (Addgene #62977).14 pCS2-MT mouse Axin (Axin MTFu1) was a gift from Frank Costantini (Addgene #21287) and encodes an N-terminal 6xMyc-tagged mouse AXIN1 variant of 832 aa.15 The pT3-EF1α-β-catenin-K335I and pT3-EF1α-β-catenin-N387K constructs were generated by subcloning a Bsu36I-EcorI fragment in the pT3-EF1α-β-catenin-WT plasmid (with N-terminal Myc-tag). pT3-EF1α-c-Met and pCMV/sleeping beauty transposase (pCMV/SB) plasmids have been described previously.16
Mice and hydrodynamic tail injection
Hydrodynamic tail vein injection was performed basically as previously described.17–19 Briefly, 20 μg of either pT3-EF1α-β-catenin-WT, K335I or N387K construct was mixed with 20 μg pT3- EF1α-c-Met and 1.6 μg pCMV/SB in 2 ml saline. This solution was injected in the lateral tail vein of 6–8 week wild-type FVB/N mice purchased from The Jackson Laboratory (Sacramento, CA). Mice were monitored via abdominal palpation every three days. The mice were euthanized and were considered to be “dead” when they developed palpable abdominal mass based on the IACUC protocol. Mice were housed and fed in accordance with protocols approved by the Committee for Animal Research at the University of California, San Francisco (San Francisco, CA). Livers and tumors were analyzed basically as previously described (see Supplemental Methods).17–19
CRISPR/Cas9 mediated knock-in of CTNNB1 mutations
Previously, we generated SNU449 clones in which the AXIN1 p.R712* mutation was successfully repaired using CRISPR/Cas9 technology (manuscript submitted). Clone-20 thereof and HEK293 cells were used to knock-in β-catenin S37F and K335I mutations as described in Supplemental Methods.
Half-life determination
N-terminal FLAG-tagged β-catenin variant plasmids (200ng) were co-transfected into HEK293 cells with pEGFP-C1 (50ng) using FuGENE® HD. The pEGFP-C1 plasmid generates a highly stable GFP protein serving as loading control. After 24 hours, the cells were treated with cycloheximide (50 μg/ml) for 0, 30, 60, 90, 120, and 150 minutes. Next, cells were washed with cold PBS two times and lysed in 2x Laemmli sample buffer (120 mM Tris-Cl pH 6.8, 20% glycerol, 4% SDS) with 0.1 M DTT, and heated for 5 min at 95°C.
Immunoprecipitation
For the immunoprecipitation (IP) assays using expression plasmids, HEK293 or HCT116 cells were seeded in 6-well plates before transfection. When reaching 60–80% confluence, 1μg N-terminal FLAG-tagged β-catenin variant plasmids were co-transfected with either 1μg of APC-, AXIN- or βTrCP-expression plasmids using FuGENE® HD. After 24 hours, FLAG-tagged β-catenin was IPed using ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich). Knock-in cell lines were seeded in 15cm round plates, lysed, incubated with primary antibody, which was IPed using Pierce Protein A/G Magnetic beads (Thermo Scientific™). Details of the IP protocols can be found in Supplemental Methods.
Western blotting, antibodies and quantification
Fluorescent western blotting and quantitative analysis were performed basically as previously described.20 Primary antibodies are listed in Supplemental Methods. As secondary antibodies we used anti-rabbit or anti-mouse IRDye-conjugated antibodies (LI-COR Biosciences, Lincoln, USA). Protein intensity was detected with the Odyssey 3.0 Infrared Imaging System and analyzed by Image Studio Lite Ver5.2. For APC western blot analysis we used Immobilon ECL Ultra HRP substrate (MerckMillipore). Membranes for ECL detection were blocked and incubated using Immobilon Block-PO reagent (MerckMillipore).
β-catenin reporter assays
HEK293 cells were seeded in 24-well plates to reach approximately 50–70% confluency on the day of transfection. Each well was transfected with 5 ng of the β-catenin expression vectors, 250 ng Topflash or Fopflash, and 10 ng CMV-Renilla using Fugene HD (Promega). For the knock-in HEK293 and SNU449 cells we used the more sensitive WRE/MRE reporters, using 12-well and 6-well plates, respectively. After two days, luciferase activities were measured in a LumiStar Optima luminescence counter (BMG LabTech, Offenburg, Germany) and normalized for transfection efficiency by using the Dual Luciferase Reporter Assay system (Promega) according to the manufacturer’s instruction. Transfections were performed in triplicate and the mean and standard error were calculated for each condition. The β-catenin reporter activities are shown as TOP/FOPflash or WRE/MRE ratios.
Structure analysis
Structures were analyzed and figures were created using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. Overlays were created by superimposing β-catenin (chain A) from the β-catenin-TCF complex (1G3J.pdb), β-catenin-E-cadherin complex (1I7W.pdb) or the β-catenin-AXIN complex (1Q27) onto chain A of the β-catenin-phosphorylated 20AA APC complex (1TH1.pdb).
Statistical method
Expression differences tested by qRT-PCR and luciferase assays were tested by unpaired t-test. Statistical differences are depicted as follows: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. All statistical analyses were performed using GraphPad Prism 6.
RESULTS
K335I and N387K β-catenin variants are potent inducers of hepatocellular carcinoma formation in mice
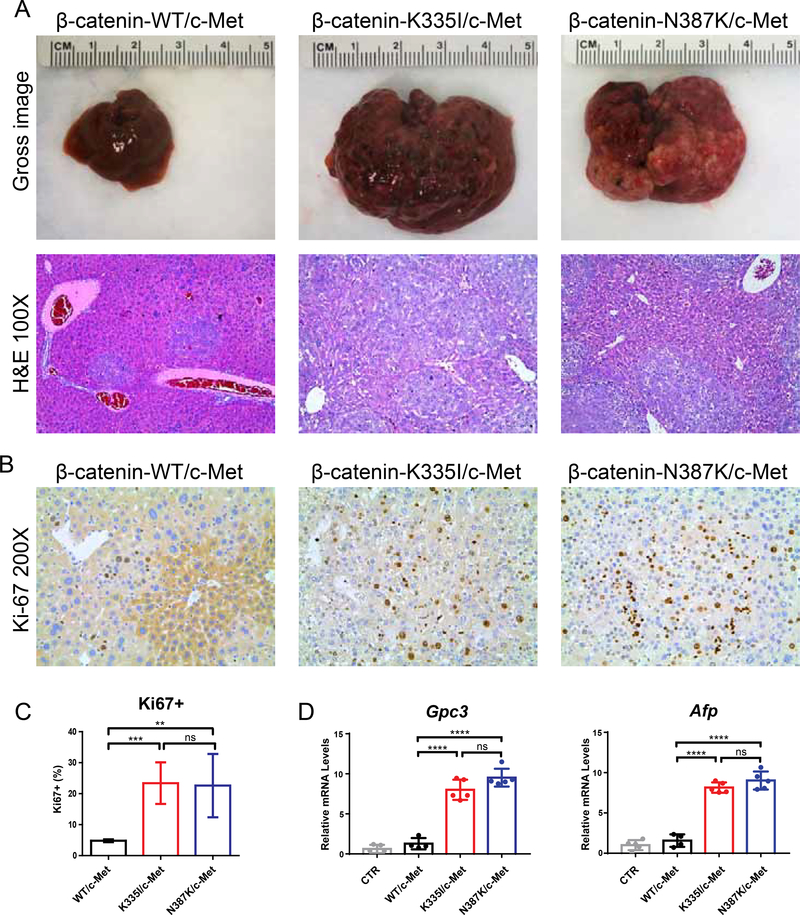
To directly demonstrate the oncogenic potential of the β-catenin K335I and N387K variants, we hydrodynamically injected them in the tail vein of mice.21 Wild-type β-catenin was taken along as control. All variants were co-transfected with c-Met as previous work demonstrated that oncogenic β-catenin alone is insufficient for HCC development.16,17 In strong contrast to the mice injected with wild-type β-catenin, all K335I/N387K injected mice became moribund 6–7 weeks after injection showing a massive increase in liver weight and tumor burden (Figure 2A). Histologically, we observed the presence of well to moderately-differentiated HCC with solid or macro-trabecular growth patterns (Figure 2A, Supplemental Figure S1), in accordance with previous observations.16,17 Lesions are positive for the hepatocyte marker HNF4α, while negative for the biliary marker CK19 (Supplemental Figure S2). In wild-type injected mice only occasional HCC-like foci were observed. This marked difference in oncogenic potential was confirmed by immunohistochemical analysis of the proliferation marker Ki-67 (Figure 2B,C) and qRT-PCR analysis showing an elevated expression of the HCC markers Gpc3 and Afp (Figure 2D).
Figure 2. K335I and N387K β-catenin variants are potent inducers of HCC development in mice.
(A) Hydrodynamic tail vein injection of β-catenin-K335I and N387K variants resulted in a strong increase of liver weights within 6–7 weeks, in contrast to wild-type β-catenin injected animals (N=5 each). All variants were co-transfected with c-Met. H&E staining revealed multiple large foci of well-differentiated HCC with solid or macro-trabecular growth patterns. (B) IHC analysis for the proliferation marker Ki67, and (C) quantification of five representative fields. (D) Expression of HCC markers Gpc3 and Afp was evaluated by qRT-PCR in normal (CTR) and transfected livers. All data are presented as mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001; ns, non-significant.
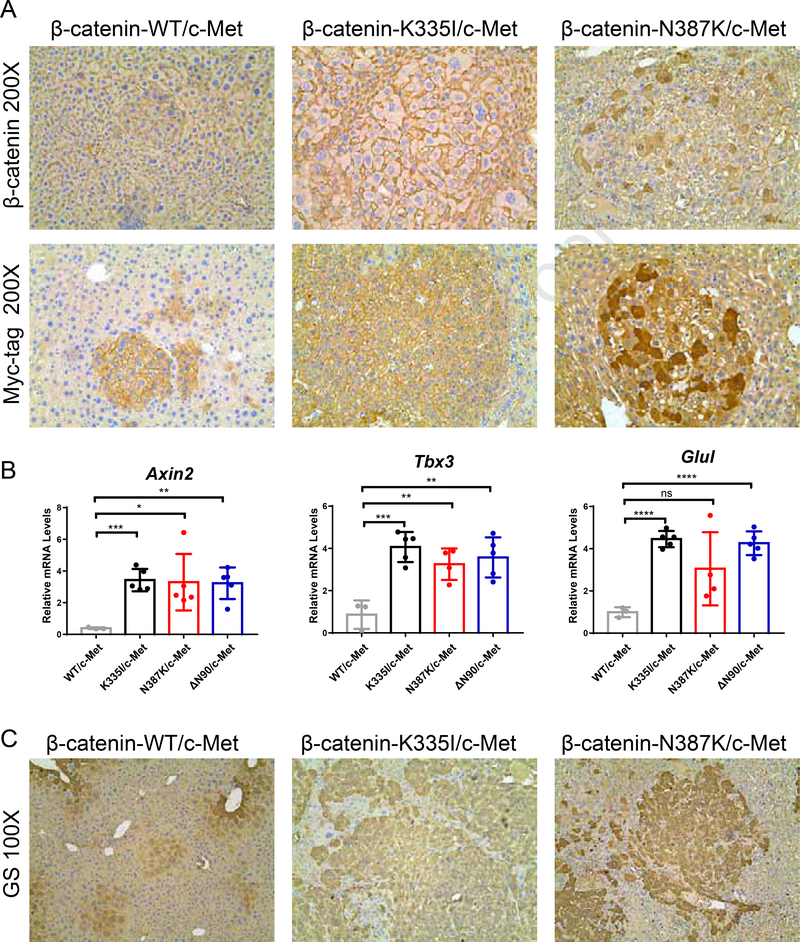
Staining for β-catenin showed a predominant membranous location (Figure 3A). Occasionally a weak cytoplasmic or nuclear staining was observed, especially for the N387K variant. This result was confirmed with an antibody detecting the Myc-tag present in the construct (Figure 3A). The lack of a prominent nuclear accumulation is in line with previous observations for the “weak” β-catenin S45Y variant.17 Nevertheless, we observed a strong increase in the (liver-specific) β-catenin target genes Axin2, Tbx3 and Glul (Figure 3B), the latter also confirmed by immunohistochemical staining for Glutamine Synthetase (GS) (Figure 3C).
Figure 3. Immunohistochemical analysis of β-catenin patterns and target gene expression.
(A) IHC for β-catenin and associated Myc-tag reveals a predominant membranous location for transfected wild-type, K335I, and N387K variants. Occasionally a stronger cytoplasmic staining is observed, especially for the N387K variant. (B) qRT-PCR analysis of β-catenin target genes Axin2, Tbx3, and Glul. RNA isolated from previously injected β-catenin-Δ90N/c-Met animals was used as positive control.17 (C) IHC confirmation of Glutamine Synthethase (GS) expression.
Taken together, these results show that both the K335I and N387K variants compared with wild-type β-catenin, possess a strong potential to induce liver tumor formation in mice, associated with a clear induction of target gene expression.
Half-life time of β-catenin mutants correlates with their activation potential
The signaling activity of β-catenin is strongly regulated by proteolytic degradation. Therefore, we analyzed the half-life times of various commonly observed β-catenin variants in comparison with wild-type protein (Supplemental Figure S3). Interestingly, K335I and N387K variants show a similar protein stability as wild-type β-catenin, with half-life times between 65–100 minutes. The S45P mutation increases the half-life up to 110 minutes, while mutations affecting the D32-S37 β-TrCP binding motif and the EX3-del variant, extend it to more than 2 hours. Thus, these analyses show that protein stability largely correlates with the reported signaling activities, that is, mutants with weak enhanced signaling capabilities show a turnover comparable to the wild-type protein, whereas more active signaling variants show a clearly increased half-life.
K335 and N387 variants are not affected in N-terminal phosphorylation and β-TrCP binding
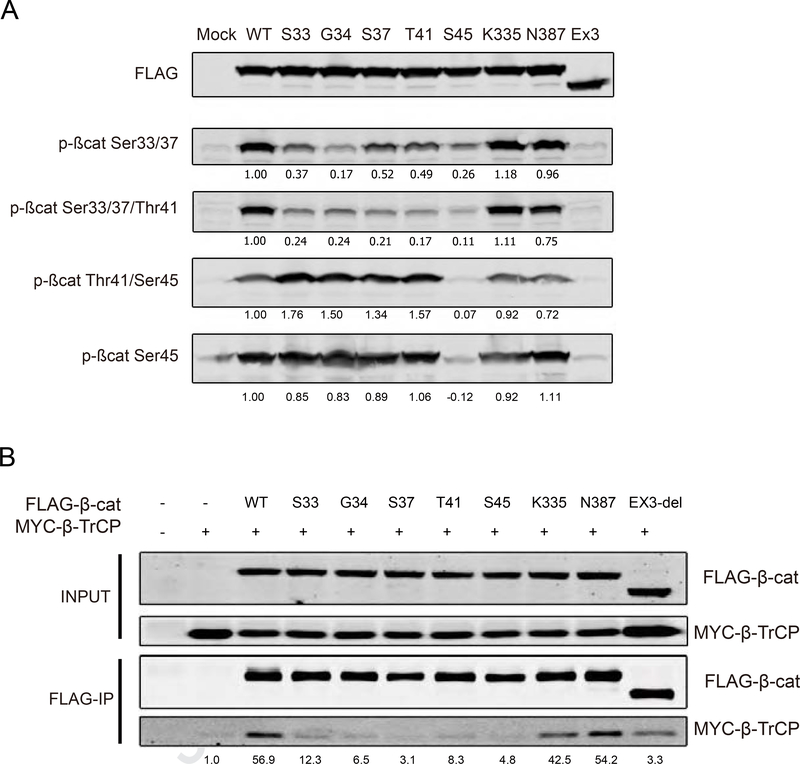
β-catenin protein stability is largely regulated by sequential N-terminal S/T phosphorylation. Hence, we determined the phosphorylation status of β-catenin variants transfected in HEK293 cells (Figure 4A). The EX3-del variant lacking all S/T residues cannot be recognized by any of the phospho-antibodies. All other variants are readily phosphorylated at S45 comparable to the wild-type protein, except for the S45P mutant. In contrast, the GSK3 mediated phosphorylation at residues T41, S37 and S33 is reduced in all S33 to S45 mutants. In accordance with previous reports, we still observed some residual S/T phosphorylation in all these variants.9 Importantly, in this over-expression assay the K335 and N387 mutants showed no obvious deviation from the phosphorylation pattern observed for the wild-type protein.
Figure 4. K335 and N387 variants are not affected in N-terminal phosphorylation and β-TrCP binding.
(A) FLAG-tagged β-catenin variants were transfected in HEK293 and subjected to western blotting with phospho-specific β-catenin antibodies (Table 3 in supplemental methods). Endogenous β-catenin is visible in the mock and Ex3-del transfected lanes, which was subtracted from the transfected variants. Signal of wild-type (WT) protein was set to 1, to which all other values were normalized. K335I and N387K variants are similarly phosphorylated as wild-type protein. (B) FLAG-tagged β-catenin variants were co-transfected with Myc-tagged β-TrCP into HEK293 cells. Two hours prior to FLAG-IP, cells were treated with the proteasome inhibitor MG132. The amount of co-IPed β-TrCP was determined after normalization with β-TrCP input signal. Signal of IP obtained with Myc-tagged β-TrCP alone was set to 1. K335I and N387K variants are not affected in their association with β-TrCP.
Next, we determined the binding capacity of β-TrCP to each variant in HEK293 cells. Our immunoprecipitation (IP) protocol resulted in a weak unspecific binding of Myc-tagged β-TrCP in the negative control (lane 2 of Figure 4B). For the S33-S45 mutants similar or at most slightly increased levels of co-IPed β-TrCP were observed, whereas both K335 and N387 mutants showed equal binding capabilities comparable to the wild-type protein. Overall, these analyses show that K335I and N387K mutant proteins are not seriously affected in N-terminal phosphorylation and binding to β-TrCP.
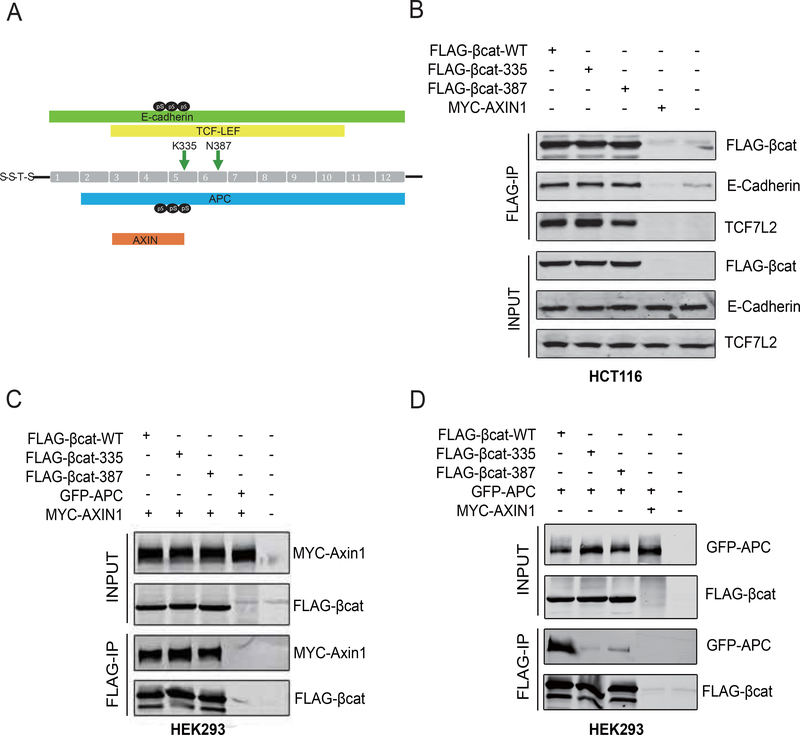
K335 and N387 variants show reduced binding to the APC protein
As protein half-life, N-terminal phosphorylation and β-TrCP binding are not clearly affected for the K335I and N387K variants, we sought for an alternative explanation explaining their increased signaling behavior. Both residues are located in armadillo repeats 5 and 6 of β-catenin in the center of the protein. In total 12 armadillo repeats (residues 141–664) form a superhelix with a positively charged groove, which can associate with a large number of proteins.2 We focused on well-established binding partners at the cell membrane, nucleus and within the destruction complex, that is E-cadherin, TCF7L2 (also known as TCF4), AXIN1, and APC. Except for AXIN1, these have all been shown to bind an extended area encompassing both K335 and N387 residues (Figure 5A). Within epithelial cells, most β-catenin is captured at the cell membrane through interactions with cadherins. Hence, a reduced binding affinity between β-catenin and E-cadherin is expected to increase the signaling pool of β-catenin, possibly leading to enhanced nuclear signaling. However, immunoprecipitation of wild-type, K335I and N387K variants from HCT116 cells showed no altered binding to E-cadherin (Figure 5B). Alterations in binding to TCF7L2 within the nucleus are also expected to affect activation of β-catenin target genes, but again we failed to show any difference between the tested variants (Figure 5B). Likewise, binding of co-transfected MYC-tagged AXIN1 was not affected by both mutations (Figure 5C). In contrast, binding of a co-transfected GFP-tagged APC fragment was strongly impaired for the K335I and N387K mutants (Figure 5D). Taken together, these IP experiments show that compared to wild-type β-catenin, both mutants associate less strongly with APC, one of the core proteins of the destruction complex.
Figure 5. K335I and N387K variants selectively show reduced binding to APC.
(A) Schematic representation of the reported binding domains of APC, AXIN, TCF/LEF and E-cadherin to the armadillo repeat region of β-catenin. Position of K335I and N387K mutations are indicated by arrows. pS indicates Serine phosphorylation present at binding interface of APC or E-cadherin. (B) FLAG-tagged WT, K335I and N387K β-catenin variants were transfected in HCT116 cells. Myc-tagged-AXIN1 and non-transfected cells were used as negative controls. Following FLAG-IP, endogenous E-cadherin and TCF7L2 were shown to bind equally to wild-type β-catenin and both variants. (C) The same β-catenin variants were co-transfected with Myc-tagged-AXIN1 into HEK293 cells. All variants bind equally to AXIN1. (D) Co-transfection with GFP-APC in HEK293 cells. GFP-APC expresses a GFP-tagged murine APC fragment (aa1199–2167) encoding all the 20 amino-acid β-catenin-binding repeats and AXIN-binding domains. Both the K335I and N387K variants associate much weaker to GFP-APC. In the latter two experiments co-transfection of GFP-APC and Myc-AXIN1 was used as negative IP-control. All transfections experiments were performed 3 times with identical results.
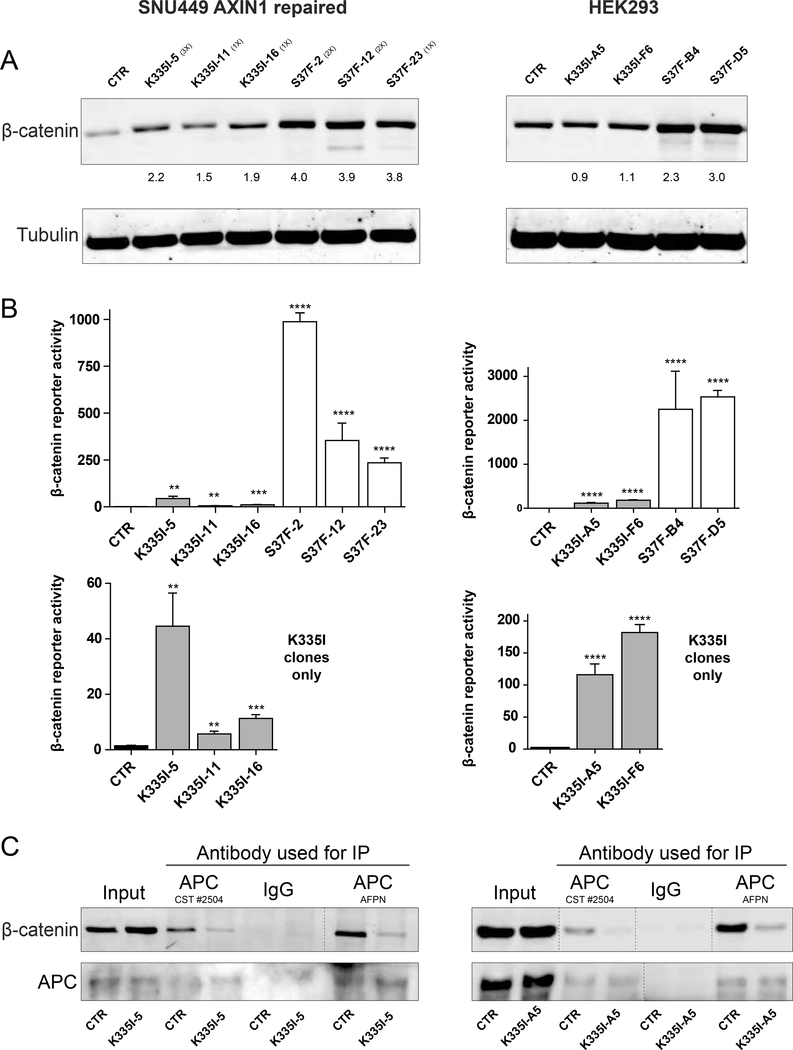
Endogenous expression in cell lines confirms increased signaling and reduced affinity for APC
As overexpression may affect the stoichiometry of the β-catenin breakdown complex, we wished to confirm our results at endogenous expression levels. Previously, we generated SNU449 liver cancer cells in which the AXIN1 p.R712* mutation was successfully repaired using CRISPR/Cas9 technology (manuscript submitted), so that no known β-catenin activating mutations remain. In these and HEK293 cells we successfully knocked-in the K335I mutation. For direct comparison we also generated cells carrying the “strong” S37F mutation (see Supplemental Table S2 for clone details). Attempts to generate N387K knock-in cell lines failed. Western blot analysis of SNU449 mutant cells revealed a modest increase in total β-catenin levels, especially for the S37F mutant cells (Figure 6A), while in HEK293 this was only the case for S37F clones. All K335I clones showed a significant increase in β-catenin reporter activity (Figure 6B), although not as pronounced when compared with the S37F variant. These results were confirmed with AXIN2 qRT-PCR (Supplemental Figure S4). Immunoprecipitation of endogenous APC using two independent antibodies confirmed the reduced binding of the K335I variant (Figure 6C). These results confirm the increased signaling propensity of the armadillo variants through reduced APC binding also at endogenous levels.
Figure 6. Endogenous expression in cell lines confirms increased signaling and reduced affinity for APC.
(A) Introduction of the K335I and S37F mutation in SNU449 cells leads to a modest increase in total β-catenin protein levels. In HEK293 cells this is restricted to S37F clones. Numbers in brackets refer to the number of CTNNB1 alleles expressing that variant. HEK293 clones are homozygous in all cases. (B) β-catenin luciferase reporter assay of all available clones. WRE/MRE ratios are shown. Data are presented as mean ± SD. **,P<0.01; ***, P<0.001; ****, P<0.0001. Bottom half shows the K335I clones in comparison with the parental line. (C) IP of endogenous APC reveals reduced binding of K335I. Two independent APC antibodies were used. Vertical dashed lines indicate removed irrelevant lanes.
Protein structures of β-catenin complexes provide a rationale for reduced binding of mutant β-catenin selectively to the APC protein
To identify potential mechanisms underlying the differential binding of AXIN1, APC, TCF7L2 and E-Cadherin to both mutants, we analyzed available protein structures of β-catenin in complex with fragments of these proteins (Supplemental Figure S5A).22–25 In all complexes, β-catenin-K355 forms a hydrogen bond with a backbone carbonyl oxygen of the bound partner protein (Supplemental Figure S5B–D). In addition, K335 forms specific interactions with sidechains from the bound proteins, which are different for each partner and depend on its phosphorylation status. The seven 20-amino acid repeats (AAR) of APC contain a conserved SxxxSLSSL motif that is phosphorylated on multiple Serine residues by GSK3/CK1 when APC is associated with AXIN, which strongly increases the binding to β-catenin.25–27 Interestingly, in the β-catenin-APC complex the positively charged K335 sidechain forms ionic interactions and hydrogen bonds with two negatively charged phosphorylated Serines (pS1504 and pS1507) in the APC 20-AAR motif (Supplemental Figure S5B), all of which are lost when mutated to Isoleucine or Threonine. In the β-catenin-TCF complex, no Serines are available within this region for phosphorylation, instead K335 forms a single ionic interaction with the negatively charged sidechain of D40 (Supplemental Figure S5C). E-cadherin carries a phosphorylated Serine (S692) at the corresponding position of APC-Ser1507 and forms an ionic bond with K335, however it carries an apolar A688 at the position corresponding to pS1504 in APC (Supplemental Figure S5D). In AXIN only hydrophobic residues are present at this interface (not shown), which form weak van-der-Waals contacts with the polar amine of K335, but might form more favorable contacts when the Lysine is mutated to hydrophobic Isoleucine. Summarized, K335 forms more energetically favorable interactions with APC than with any of the other proteins, explaining the observed relatively large effect of K355 mutation on APC binding specifically.
The N387K mutation will influence hydrogen bond formation between β-catenin and the peptide backbone of APC, LEF/TCF and E-cadherin, and is expected to reduce the strength of the interaction in all complexes (Supplemental Figures S5E and S6A/S6B). In this case the structural analysis provided no direct clues explaining the reduced binding of specifically APC to this variant.
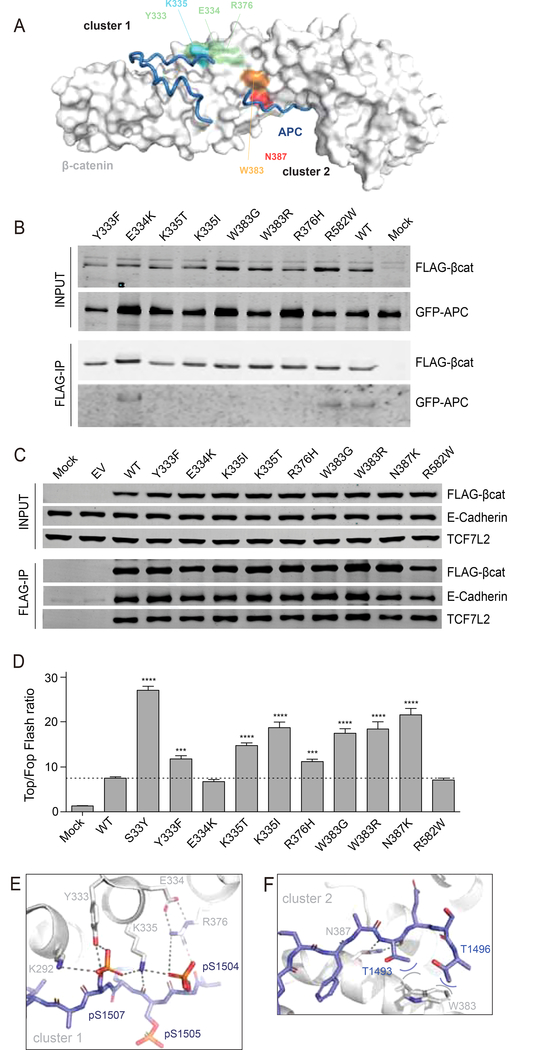
Selective loss of APC binding is a common feature of armadillo repeat 5 and 6 mutations
Currently, more than 150 individual neoplasias have been identified carrying amino acid alterations in repeats 5/6 of β-catenin (Figure 1, Supplemental Table S1). Besides the K335 and N387 amino acid alterations, residues Y333, E334, R376 and especially W383 are also mutated in neoplasias regularly. Within the β-catenin structure these residues are organized in two clusters in close proximity of each other (Figure 7A). We generated expression plasmids for these variants and tested them for binding to APC, TCF7L2 and E-cadherin. We also took along one commonly observed R582W variant (n=14) located towards the end of the armadillo repeats. As can be seen in Figure 7B/C most of these newly generated mutants show loss of APC binding, while all retain binding to TCF7L2 and E-cadherin. The only exceptions are the E334K and R582W variants that behave identical to wild-type protein with respect to APC binding.
Figure 7. Increased signaling and selective reduction of APC binding for most armadillo repeat 5/6 variants.
(A) Surface representation of β-catenin in grey with bound APC as tube representation in blue. K335 in cluster-1 is colored light-blue, with surrounding residues Y333, E334 and R376 in light-green. N387 in cluster-2 is colored red, while the other commonly mutated W383 is colored orange. (B) Co-transfection of indicated FLAG-tagged β-catenin variants with GFP-APC in HEK293 cells. Following FLAG-IP, most of the commonly observed armadillo repeat 5/6 variants associate much weaker to GFP-APC. (C) FLAG-tagged β-catenin variants were transfected in HCT116 cells. Following FLAG-IP all variants are shown to bind equally to endogenous TCF7L2 and E-cadherin. (D) A Top-Fopflash β-catenin reporter assay in HEK293 cells shows that all armadillo repeat 5/6 variants that bind weaker to APC show a 1.5–3 fold increased signaling compared with wild-type β-catenin. Top-Fopflash ratios are shown. Experiments were performed in triplo and repeated twice. Data are presented as mean ± SD. ***, P<0.001; ****, P<0.0001. (E) Details of the cluster-1 complex, with β-catenin as grey cartoon and APC as blue stick representation. A six-residue ion-pair network is formed between E334-R376;R376-pS1504;pS1504-K335;K335-pS1507;pS1507-K292. In fact, the network is even bigger and extends beyond K292 (not shown). Hydrogen bonds are indicated with dashed lines. (F) Details of the cluster-2 complex. W383 packs against the hydrophobic methyl-groups (blue carbons) of the sidechains of T1493 and T1496 and so forms favorable van-der-Waals interactions (curved lines).
Next, we performed β-catenin reporter assays for all the armadillo repeat 5/6 variants taking S33Y and wild-type β-catenin along as controls (Figure 7D). All variants showing a reduced binding to APC, yield a 1.5–3 fold higher induction of reporter activity compared with wild-type, while the E334K and R582W variants behave similar to wild-type.
Importantly, analysis of the available protein structures shows that all additional mutated residues associated with increased signaling (Y333, R376, and W383), form direct interactions with APC (Figure 7E/F). In fact, binding of cluster-1 residues (Figure 7A/E) to APC creates an extensive network in which oppositely charged groups alternate and form multiple ionic interactions and hydrogen bonds with neighboring residues. Any mutation in this network will affect several highly optimized interactions, explaining the relatively large effect on APC binding that we observed. E334 does not form direct interactions with APC, possibly explaining its lack of increased signaling when mutated, although it may be involved in proper orienting the R376 residue.
In TCF and E-cadherin, the highly cooperative cluster-1 network is much reduced in size due to replacement of crucial Serine sidechains (that are phosphorylated in APC) with hydrophobic residues (V44 in TCF and A688 in E-cadherin; Supplemental Figure S6C/S6D). These apolar residues cannot favorably interact with the polar sidechains in cluster-1, which means that the contribution of this cluster to the stability of TCF and cadherin complexes with β-catenin is weaker than for APC, explaining why cluster-1 mutations have a relatively smaller effect on their binding.
In cluster-2 (Figure 7A/F), the sidechain of W383 forms van-der-Waals contacts with two Threonine residues on APC, forming a small hydrophobic core that contributes to complex-stability. Mutation of W383 will disrupt this energetically favorable region, resulting in loss of APC binding. TCF residues do not interact with the sidechain of W383 in β-catenin, and its mutation will not affect stability of the TCF complex (Supplemental Figure S6E). The situation is less clear in E-cadherin, in which favorable interactions between W383 from β-catenin and Y681 from E-cadherin are formed (Supplemental Figure S6F).
Armadillo repeat 5 and 6 mutations co-occur with other β-catenin enhancing mutations in colorectal, but not in hepatocellular carcinomas
Mutations in the repeats 5 and 6 region of β-catenin have been mainly reported in liver, colon and kidney tumors,5,8,28–31 and anecdotally in several other tumor types. The stronger activating β-catenin mutations encompassing exon3 are known to occur in a mutually exclusive fashion with disease-causing APC mutations,32 showing that either mutation is sufficient to drive tumorigenesis. When we analyzed colorectal cancers carrying armadillo repeat 5/6 mutations for concomitant defects in other genes linked to β-catenin signaling, these were observed in 11 out of 13 cancers (Supplemental Table S3). Seven out of 13 cancers carried APC mutations, but interestingly 3 out of 7 express a truncated APC protein that is expected to retain a significant level of β-catenin regulation, that is 3 or more of the 20 AARs.4 Three other cancers carry truncating AXIN2 or RNF43 mutations that are generally also considered to be weak signaling activators. A similar analysis of 20 hepatocellular carcinomas identified no accompanying β-catenin signaling activating, except for a single truncating ZNRF3 mutation (Supplemental Table S4). Thus, in contrast to liver cancers, the armadillo hotspot mutations observed in colorectal cancers are often accompanied by other “weak” mutations that may synergistically enhance β-catenin signaling to levels supporting tumor growth.
DISCUSSION
The β-catenin signaling pathway is one of the most commonly deregulated pathways among cancers.1 In colorectal cancers this is predominantly accomplished by inactivating APC mutations, while e.g. in liver cancers aberrant activation has been mainly attributed to activating mutations in the CTNNB1 gene (20–25%).4,7,33–35 The exon3 related β-catenin mutations acquire enhanced signaling activities by interfering with proper N-terminal phosphorylation and subsequent proteolytic degradation. More recently, mutations in the armadillo repeat domains 5 and 6 of β-catenin have been identified in a substantial number of cancers, especially those of the liver. Here, we provide direct evidence that these mutants are potent inducers of HCC development in mice. In addition, we uncover a novel mode of β-catenin activation for these mutants, i.e., they reduce the binding to APC, while simultaneously retaining binding to TCF/LEF. The latter is important to allow sufficient enhancement of β-catenin target genes driving tumor formation. A weaker binding to APC is expected to increase the signaling pool of β-catenin, enhancing the expression of β-catenin target genes driving tumor formation. If such a mutation would however simultaneously reduce binding to the essential nuclear TCF/LEF transcriptional co-factors, the overall activation of target genes would not increase or even be reduced compared to wild-type β-catenin. In accordance, mutation of K312 and K435 residues is rarely observed, as these so-called “charged buttons” of β-catenin are essential for binding of most proteins to the armadillo superhelix by forming an ionic bond to conserved glutamate residues.22,36
Mapping the mutated residues on the protein structure of β-catenin, two clusters emerge that appear critical binding domains with APC (Figure 7A). Cluster-1 is centered around K335 and further involves residues Y333 and R376. Previously, these have all been identified to directly interact with phosphorylated 20AA repeats of APC, but also to some extent with cadherins and TCF/LEF.25 However, the IP experiments show that their mutation selectively results in loss of APC binding, while leaving binding to TCF and E-cadherin unaffected. For the K335 residue itself, the structural analyses provide a rationale as it forms more energetically favorable interactions with APC than with any of the other proteins. Likewise, the structural analyses reveal that Y333 is not critical for TCF binding, while R376 is not important for cadherin binding. It is interesting to consider that an extensive ion-pair network is formed upon β-catenin-APC complex formation, while much smaller, fragmented networks are present in the TCF and E-cadherin complexes. Ion-pair networks are cooperative entitiesfrom which removal of a single residue through mutation will have a relatively large effect on the stability of this region.37–39 This may explain why the mutation of K335 has a much larger effect on complex formation with APC (in which it is a central residue forming multiple ionic bonds) than on complex formation with TCF and E-cadherin (in which the Lysine is a peripheral residue in the smaller network).
Cluster-2 involves residues W383 and N387. Previously, the W383 residue was already identified as important for APC binding as a W383A mutant failed to bind APC, while retaining LEF/TCF7L2 association,40 which is in accordance with the structural analyses because TCF does not interact with W383. For the commonly mutated N387 residue the structural analysis provided no direct clues explaining the reduced binding of specifically APC. Nevertheless, our IP and structural analyses provide clear evidence that both clusters are more relevant for APC binding than for TCF and cadherins.
Despite two decades of research the exact mechanism through which APC functions in the destruction complex is still not fully understood. Several models not necessarily excluding each other have been proposed.41,42 One model proposes that APC is involved in sequestering β-catenin within the cytoplasm, thereby preventing it from entering the nucleus.43,44 Other models suggest that APC protects S/T phosphorylated β-catenin from dephosphorylation by PP2A, that it may be required to interface β-catenin with the ubiquitin and proteasome machinery, or that it is required to displace AXIN from phosphorylated β-catenin to allow a new cycle of β-catenin breakdown.24,45,46 Whichever model will turn out to be true, in all cases a weaker association of mutant β-catenin with APC will make the process less efficient and effectively lead to more signaling.
How to reconcile the weak APC/mutant β-catenin interaction with the rather modest increased signaling activity associated with these variants? Current models of APC’s contribution to the destruction complex propose that direct binding of β-catenin to APC is not absolutely required for a proper level of β-catenin turnover, as long as APC is present within the destruction complex, for example through binding to AXIN.41,42,44,47 This partial functional redundancy between APC and AXIN is most likely also responsible for keeping the signaling activity of the armadillo repeats 5/6 mutants in check, although it will be less efficient when compared with wild-type β-catenin.
Mutations in this region of β-catenin have been mainly reported in liver cancers, Wilms tumors and those of the large intestine,5,8,28–31 and anecdotally in several other tumor types, but their true frequency is probably underestimated as in the past CTNNB1 mutation analysis was largely restricted to the exon3 hotspot. Therefore, whole-exome or genome mutational analyses that are becoming more routine procedures for tumor investigations nowadays, are likely to uncover more frequently these specific mutations. Given their weak activation nature, these mutations are more likely going to be identified in tumor types that require a minimal to moderate activation of the pathway to support their growth.4,48,49 In that respect, their presence in colorectal cancer is somewhat surprising as these cancers generally select for higher β-catenin signaling levels.4 Our analysis shows however that in colorectal cancer the armadillo hotspot mutations are often accompanied by other (weak) mutations that synergistically may enhance β-catenin signaling to levels supporting tumor growth. A similar scenario may be at work for Wilms tumors, in which the armadillo repeat 5/6 mutations often associate with those of WTX (official name AMER1),31 also leading to a modest signaling enhancement. In liver cancers however, the weak signaling associated with these armadillo repeat variants appears to be sufficient to support tumor growth as they are not clearly associated with defects in other Wnt/β-catenin signaling related components.
This latter observation is also in line with our tumor induction experiments in mice and the analysis of SNU449 and HEK293 cell lines with knock-in mutations. In a direct comparison, endogenous levels of the K335I mutant are clearly more potent inducers of β-catenin signaling than wild-type β-catenin, but rather weak in comparison with the “strong” S37F mutation. Nevertheless, within 6–7 weeks after injection they lead to a massive HCC formation, which is comparable in time-frame with the S45Y and Δ90N-β-catenin variants.17 Immunohistochemical analysis demonstrated however no clear nuclear accumulation for the K335I and N387K variants. Similar observations have been made for K335/N387 mutant human liver tumors, which also lack a clear nuclear β-catenin accumulation associated with a weak activation of target genes.8 Combined these results show that a strong nuclear presence is no prerequisite for an efficient liver cancer formation. In support, we have recently shown that liver specific loss of AXIN1 requires β-catenin to induce hepatocarcinogenesis, which was however associated with only a weak activation of β-catenin target genes.18 Buchert et al. have shown that late-onset hepatocellular tumors were present in all mice carrying a hypomorphic APC mutation associated with just a modest increase in β-catenin signaling, while tumor formation was absent or largely prevented with slightly increased or decreased signaling.48 All these examples show that only modest increases in β-catenin signaling may nevertheless be relevant for hepatocellular tumorigenesis.
In conclusion, our molecular and structural analyses uncover a novel mutational mechanism of enhanced β-catenin signaling. In contrast to the N-terminal mutations in β-catenin that directly impair its phosphorylation by GSK3/CK1α or binding to β-TrCP, the hotspot mutations within armadillo repeats 5 and 6 lead to enhanced signaling through reduced binding to APC, while simultaneously retaining the interaction with their nuclear TCF/LEF transcriptional co-factors. The latter is important to allow sufficient enhancement of β-catenin target genes driving tumor formation. A second difference with the N-terminal mutants appears to be that they often co-occur with other mutations affecting β-catenin signaling, at least in colorectal and Wilms tumors.
Supplementary Material
Table 1.
List of primary antibodies used for immunohistochemistry
| Antibody | Catalogue Number | Company | Species | Dilution |
|---|---|---|---|---|
| β-Catenin | BD 610153 | BD Biosciences | Mouse | 1 200 |
| Glutamine Synthetase | BD 610518 | BD Biosciences | Mouse | 1 500 |
| Maine Medical Center Research | ||||
| Myc-tag | Vli01 | Institute | Rabbit | 1 150 |
| Ki-67 | MA5–14520 | Thermo Fisher scientific | Rabbit | 1 150 |
| HNF4a | ab181604 | Abcam | Rabbit | 1 2000 |
| CK19 | ab133496 | Abcam | Rabbit | 1 500 |
Table 2.
List of primer pairs used in qRT-PCR analysis
| Gene name | Forward | Reverse |
|---|---|---|
| 18s rRNA | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT |
| Mouse-Axin2 | GCTCCAGAAGATCACAAAGAGC | AGCTTTGAGCCTTCAGCAT C |
| Mouse-Glul | CAGGCTGCCATACCAACTTCA | TCCTCAATGCACTTCAGACCAT |
| Mouse-Tbx3 | CAGGCAGCCTTCAACTGCTT | GGACACAGATCTTTGAGGTT GGA |
| Mouse-Afp | TCTGCTGGCACGCAAGAAG | TCGGCAGGTTCTGGAAACTG |
| Mouse-Gpc3 | CAGCCCGGACTCAAATGGG | CAGCCGTGCTGTTAGTTGGTA |
| Human-AXIN2 | CAAACTTTCGCCAACCGTGGTTG | GGTGCAAAGACATAGCCAGAACC |
Table 3.
List of antibodies used for western blotting
| Antibody | Catalogue Number | Company | Species | Dilution |
|---|---|---|---|---|
| FLAG | F3165 | Sigma-Aldrich | Mouse | 1 1000 |
| pβ-catenin (S33/37) | 2009 | Cell Signaling Technology | Rabbit | 1 1000 |
| pβ-catenin (S33/37/T41) | 9561 | Cell Signaling Technology | Rabbit | 1 1000 |
| pβ-catenin (T41/S45) | 9565 | Cell Signaling Technology | Rabbit | 1 1000 |
| pβ-catenin (S45) | 9564 | Cell Signaling Technology | Rabbit | 1 1000 |
| TCF7L2 | 05–511 | Merck | Mouse | 1 1000 |
| E-cadherin | 3195 | Cell Signaling Technology | Rabbit | 1 1000 |
| Myc-tag | Ab9106 | Abcam | Rabbit | 1 1000 |
| GFP | A-11122 | Thermo Fisher scientific | Rabbit | 1 1000 |
| APC | MABC202 (FE9) | Sigma-Aldrich | Mouse |
Antibody specificities of phospho-specific β-catenin antibodies as provided by Cell Signaling Technology are as follows p-βcat Ser33/37 recognizes β-catenin when phosphorylated at Ser33 R Ser37, or both; p-βcat Ser33/37/Thr41 recognizes any phosphorylation at Ser33/37/Thr41 except for pSer33 alone; p-βcat Thr41/Ser45 recognizes pThr41 R pSer45, or both.
Table 4.
sgRNA and ss DN sequences used to introduce the S37F and K335I mutations into cell lines. The lowercase nucleotides are silent mutations in the ss DNs that will alter the gRNA recognition sequence to prevent re-cutting. nderlined and bold A will introduce the S37F and K335I mutation.
| sgRNA S37F | gCT GGACT CTGGAATCCATT C |
| sgRNA K335I | GATACTTACGAAAAACTACTG |
| ss DN S37F | CCACTCATACAGGACTTGGGAGGTATCCACATCCTCTTCCTCAGGATTGCCTTTACCACTCAGAGAAGGAGCTGTGGTAGTGGCACCAAAgTGaATTCCAGAGTCCAGGTAAGACTGTTGCTGCCAG |
| ss DN K335I | ATAGATACATATACTTACCAGCTTCTACAATAGCCGGCTTATTACTAGAGCAGACAGATAGCACCTTCAGCAGCTTGTGGTCCAtAGaAGTATTTCGTAAGTATAGGTCCTCATTATATTTAC |
Table 5.
List of antibodies used for immunoprecipitation
| Antibody | Catalogue Number | Company | Species |
|---|---|---|---|
| β-catenin | 8480 | Cell Signaling Technology | Rabbit |
| APC | 2504 | Cell Signaling Technology | Rabbit |
| IgG | 2729 | Cell Signaling Technology | Rabbit |
| APC | AFPN | homemade4 | Rabbit |
References:
Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8 2281-2308.
Richardson CD, Ray GJ, DeWitt MA, et al. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 2016;34 339-44.
Harayama T, Riezman H. Detection of genome-edited mutant clones by a simple competition-based PCR method. PLoS ne 2017;12 e0179165.
Smits R, Kielman MF, Breukel C, et al. Apc1638T a mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev 1999;13 1309-21.
Acknowledgments
Grant support: This research was financially supported by a China Scholarship Council PhD fellowship to Pengyu Liu (File NO. 2014 0822 0029), Shan Li (File NO. 2014 0806 0053) and Manning Qian (File NO. 2018 0832 0464); as well as NIH grant (R01CA204586) to Xin Chen. Joyce Lebbink is supported by the gravitation program (024.001.028) from the Netherlands Organization for Scientific Research (NWO).
List of abbreviations
- APC
adenomatous polyposis coli
- AXIN1/2
axis inhibition protein 1/2
- CK1α
casein kinase 1α
- COSMIC
Catalogue Of Somatic Mutations In Cancer
- CTNNB1
β-catenin (cadherin-associated protein) beta 1
- DMEM
Dulbecco’s Modified Eagle Medium
- DTT
Dithiothreitol
- GFP
Green fluorescent protein
- GLUL
glutamine synthase
- GSK3
Glycogen synthase kinase 3
- HCC
hepatocellular carcinoma IP immunoprecipitation
- LGR5
Leucine-rich repeat-containing Gprotein coupled receptor 5
- PBS
Phosphate buffered saline
- SDS
Sodium dodecyl sulfate
- TCF
T-cell factor/lymphoid enhancer-binding factor
- TCGA
The Cancer Genome Atlas
- β-TrCP
Beta-transducin repeats-containing proteins
- WT
wild-type
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J 2012;31:2714–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017;169:985–999. [DOI] [PubMed] [Google Scholar]
- 4.Albuquerque C, Bakker ER, van Veelen W, et al. Colorectal cancers choosing sides. Biochim Biophys Acta Rev Cancer 2011;1816:219–231. [DOI] [PubMed] [Google Scholar]
- 5.Pilati C, Letouze E, Nault JC, et al. Genomic Profiling of Hepatocellular Adenomas Reveals Recurrent FRK-Activating Mutations and the Mechanisms of Malignant Transformation. Cancer Cell 2014;25:428–41. [DOI] [PubMed] [Google Scholar]
- 6.Network CGAR, Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Network CGAR. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–1341 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebouissou S, Franconi A, Calderaro J, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different β-catenin activity associated with liver tumor progression. Hepatology 2016;64:2047–2061. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Vogelstein B, Kinzler KW. Phosphorylation of beta-catenin at S33, S37, or T41 can occur in the absence of phosphorylation at T45 in colon cancer cells. Cancer Res 2003;63:5234–5. [PubMed] [Google Scholar]
- 10.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li CM, Kim CE, Margolin AA, et al. CTNNB1 mutations and overexpression of Wnt/beta-catenin target genes in WT1-mutant Wilms’ tumors. Am J Pathol 2004;165:1943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colnot S Focusing on beta-catenin activating mutations to refine liver tumor profiling. Hepatology 2016;64:1850–1852. [DOI] [PubMed] [Google Scholar]
- 13.Dubbink HJ, Hollink I, Avenca Valente C, et al. A novel tissue-based β-catenin gene and immunohistochemical analysis to exclude familial adenomatous polyposis among children with hepatoblastoma tumors. Pediatr Blood Cancer 2018;65:e26991. [DOI] [PubMed] [Google Scholar]
- 14.Deng W, Vanderbilt DB, Lin CC, et al. SOX9 inhibits beta-TrCP-mediated protein degradation to promote nuclear GLI1 expression and cancer stem cell properties. J Cell Sci 2015;128:1123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng L, Fagotto F, Zhang T, et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 1997;90:181–92. [DOI] [PubMed] [Google Scholar]
- 16.Tao J, Xu E, Zhao Y, et al. Modeling a human hepatocellular carcinoma subset in mice through coexpression of met and point-mutant beta-catenin. Hepatology 2016;64:1587–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao Y, Xu M, Tao J, et al. Oncogenic potential of N-terminal deletion and S45Y mutant beta-catenin in promoting hepatocellular carcinoma development in mice. BMC Cancer 2018;18:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao Y, Wang J, Karagoz E, et al. Axis inhibition protein 1 (Axin1) Deletion-Induced Hepatocarcinogenesis Requires Intact beta-Catenin but Not Notch Cascade in Mice. Hepatology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adebayo Michael AO, Ko S, Tao J, et al. Inhibiting Glutamine-Dependent mTORC1 Activation Ameliorates Liver Cancers Driven by beta-Catenin Mutations. Cell Metab 2019;29:1135–1150 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Xu L, Liu P, et al. Blocking Wnt Secretion Reduces Growth of Hepatocellular Carcinoma Cell Lines Mostly Independent of beta-Catenin Signaling. Neoplasia 2016;18:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol 2014;184:912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham TA, Weaver C, Mao F, et al. Crystal structure of a beta-catenin/Tcf complex. Cell 2000;103:885–96. [DOI] [PubMed] [Google Scholar]
- 23.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 2001;105:391–402. [DOI] [PubMed] [Google Scholar]
- 24.Xing Y, Clements WK, Kimelman D, et al. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev 2003;17:2753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing Y, Clements WK, Le Trong I, et al. Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function. Mol Cell 2004;15:523–33. [DOI] [PubMed] [Google Scholar]
- 26.Rubinfeld B, Albert I, Porfiri E, et al. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 1996;272:1023–6. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Xing Y, Hinds TR, et al. The third 20 amino acid repeat is the tightest binding site of APC for beta-catenin. J Mol Biol 2006;360:133–44. [DOI] [PubMed] [Google Scholar]
- 28.Gadd S, Huff V, Walz AL, et al. A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet 2017;49:1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33:125–136 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perotti D, Hohenstein P, Bongarzone I, et al. Is Wilms tumor a candidate neoplasia for treatment with WNT/beta-catenin pathway modulators? Mol Cancer Ther 2013;12:2619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin PJ, Kinzler KW, Sparks AB. beta-Catenin Mutations: Insights into the APC Pathway and the Power of Genetics. Cancer Res 2016;76:5587–5589. [DOI] [PubMed] [Google Scholar]
- 33.Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226–1239 e4. [DOI] [PubMed] [Google Scholar]
- 34.Dahmani R, Just PA, Perret C. The Wnt/beta-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2011;35:709–13. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Pan Q, Fuhler GM, et al. Action and function of Wnt/beta-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol 2017;52:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci 2007;120:3337–44. [DOI] [PubMed] [Google Scholar]
- 37.Lebbink JH, Knapp S, van der Oost J, et al. Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface. J Mol Biol 1999;289:357–69. [DOI] [PubMed] [Google Scholar]
- 38.Lebbink JH, Consalvi V, Chiaraluce R, et al. Structural and thermodynamic studies on a salt-bridge triad in the NADP-binding domain of glutamate dehydrogenase from Thermotoga maritima: cooperativity and electrostatic contribution to stability. Biochemistry 2002;41:15524–35. [DOI] [PubMed] [Google Scholar]
- 39.Karshikoff A, Nilsson L, Ladenstein R. Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J 2015;282:3899–917. [DOI] [PubMed] [Google Scholar]
- 40.von Kries JP, Winbeck G, Asbrand C, et al. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat Struct Biol 2000;7:800–7. [DOI] [PubMed] [Google Scholar]
- 41.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol 2013;5:a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kappel EC, Maurice MM. Molecular regulation and pharmacological targeting of the beta-catenin destruction complex. Br J Pharmacol 2017;174:4575–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha NC, Tonozuka T, Stamos JL, et al. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell 2004;15:511–21. [DOI] [PubMed] [Google Scholar]
- 44.Roberts DM, Pronobis MI, Poulton JS, et al. Deconstructing the β-catenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Mol Biol Cell 2011;22:1845–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y, Fu C, Ishikawa S, et al. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell 2008;32:652–61. [DOI] [PubMed] [Google Scholar]
- 46.Pronobis MI, Rusan NM, Peifer M. A novel GSK3-regulated APC:Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient betacatenin destruction. Elife 2015;4:e08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pronobis MI, Deuitch N, Posham V, et al. Reconstituting regulation of the canonical Wnt pathway by engineering a minimal beta-catenin destruction machine. Mol Biol Cell 2017;28:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchert M, Athineos D, Abud HE, et al. Genetic dissection of differential signaling threshold requirements for the Wnt/beta-catenin pathway in vivo. PLoS Genet 2010;6:e1000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakker ER, Hoekstra E, Franken PF, et al. beta-Catenin signaling dosage dictates tissue-specific tumor predisposition in Apc-driven cancer. Oncogene 2013;32:4579–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.