Abstract
Although dispersal is generally viewed as a crucial determinant for the fitness of any organism, our understanding of its role in the persistence and spread of plant populations remains incomplete. Generalizing and predicting dispersal processes are challenging due to context dependence of seed dispersal, environmental heterogeneity and interdependent processes occurring over multiple spatial and temporal scales. Current population models often use simple phenomenological descriptions of dispersal processes, limiting their ability to examine the role of population persistence and spread, especially under global change. To move seed dispersal ecology forward, we need to evaluate the impact of any single seed dispersal event within the full spatial and temporal context of a plant’s life history and environmental variability that ultimately influences a population’s ability to persist and spread. In this perspective, we provide guidance on integrating empirical and theoretical approaches that account for the context dependency of seed dispersal to improve our ability to generalize and predict the consequences of dispersal, and its anthropogenic alteration, across systems. We synthesize suitable theoretical frameworks for this work and discuss concepts, approaches and available data from diverse subdisciplines to help operationalize concepts, highlight recent breakthroughs across research areas and discuss ongoing challenges and open questions. We address knowledge gaps in the movement ecology of seeds and the integration of dispersal and demography that could benefit from such a synthesis. With an interdisciplinary perspective, we will be able to better understand how global change will impact seed dispersal processes, and potential cascading effects on plant population persistence, spread and biodiversity.
Keywords: Analytical models, demography, global change, individual-based models, long-distance seed dispersal, population models, seed dispersal
Although dispersal is generally viewed as a crucial determinant for the fitness of any organism, our understanding of its role in the persistence and spread of plant populations remains incomplete. In this perspective, we provide guidance on integrating empirical and theoretical approaches that account for the context dependency of seed dispersal to improve our ability to generalize and predict the consequences of dispersal, and its anthropogenic alteration, across systems. With an interdisciplinary perspective, we will be able to better understand how global change will impact seed dispersal processes, and potential cascading effects on plant population persistence, spread and biodiversity.
Introduction
Dispersal influences individual fitness (Saastamoinen et al. 2018), population persistence (Kendrick et al. 2012) and biodiversity across scales (Vellend 2010), as well as a population’s ability to track shifting habitats, deal with large-scale environmental variability and adapt to novel environments in response to global change (Zhou and Kot 2011; Clobert et al. 2012; Travis et al. 2013). Global change, including climate change, habitat fragmentation and overharvesting, affects the ecology and evolution of dispersal, in turn altering the ability of species to move or adapt to global change events (Travis et al. 2013). For sessile organisms such as plants, dispersal of propagules—defined as the movement away from the parent location—may be the sole opportunity to escape changes in local environmental conditions. Ecological understanding of dispersal has progressed by describing patterns of dispersal and the conditions under which they arise (Nathan and Muller-Landau 2000; Nathan et al. 2012), advancing dispersal theory for populations and communities (Levin et al. 2003; Levine and Murrell 2003) and determining the effectiveness of seed dispersal (Schupp 1993; Schupp et al. 2010, 2017). Nevertheless, the role of seed dispersal in the long-term spatial dynamics of plant populations remains poorly understood. The complexity and context dependence of seed dispersal ecology challenges our ability to generalize across different systems and predict responses of plant diversity to global change. To move towards the predictive understanding necessary to inform conservation strategies requires a systematic examination of dispersal mechanisms and their influence on the persistence and spread of populations.
Seed dispersal ecology is complex and context-dependent (Schupp 2007; see Figure 1 in Beckman et al. 2020). Plants exhibit a diverse array of strategies to disperse their propagules using biotic and abiotic vectors. The majority of plants are dispersed by animals (56 %; Aslan et al. 2013), including mammals, birds, reptiles and ants; some self-disperse, such as through ballistic action, and the rest are dispersed by abiotic means, including wind, water and gravity. Dispersal vectors affect seed viability and the temporal and spatial patterns of seed rain, which influences the ‘seedscape’, i.e. the abiotic and biotic environments surrounding a seed that influence later recruitment stages (Beckman and Rogers 2013). The pattern of seed deposition determines a plant’s interactions with neighbours competing for limiting resources, the likelihood of mortality due to natural enemies, the possibility of avoiding catastrophic losses due to disasters and the potential of reaching microsites suitable for survival, growth and future reproduction (Howe and Smallwood 1982; Schupp and Fuentes 1995; Nathan and Muller-Landau 2000; Beckman and Rogers 2013). For most plants, mortality is highest during the early stages of the life cycle, and the vast majority of seeds do not lead to a reproductive adult (Terborgh et al. 2014). Ecological processes from seed production to recruitment thus determine gene flow and the colonization of new areas, ultimately influencing the spatial distribution of species, community diversity and ecosystem functioning.
Our incomplete understanding of seed dispersal’s role in plant populations stems from seed dispersal ecology being largely based on short-term, local-scale empirical studies for a small number of species, on the one hand, and, on the other hand, theoretical dispersal models that often make simplified assumptions, bringing into question their suitability for making quantitative and system-specific predictions. These barriers exist for several reasons. First, seed dispersal is only one process in the chain of events within a plant’s life cycle (from flower to reproductive adult), and it interacts with several other processes over multiple spatiotemporal scales. Consequently, it is difficult to quantify the demographic importance of dispersal relative to processes affecting survival and growth at later life-history stages. Second, options for controlled experiments are limited because of the difficulty of manipulating dispersal at the spatial, temporal or organizational scales relevant to assess its complete demographic impact (e.g. Augspurger and Kitajima 1992; Coulson et al. 2001; Poulsen et al. 2012). Third, uncovering spatial processes from available observational data on spatial patterns of plant recruitment necessitates the collection of detailed field data to isolate different processes that result in similar patterns (e.g. Wiegand et al. 2009). Fourth, analysing mathematical or simulation models based on realistic assumptions of processes occurring across multiple spatiotemporal and organizational scales and in heterogeneous environments requires mathematical and statistical rigor within an interdisciplinary context (e.g. Harsch et al. 2014). To overcome these challenges and improve our ability to understand and predict the contributions of seed dispersal to populations requires a comprehensive framework that quantitatively integrates dispersal and demography. In other words, we need to evaluate the impact of any single seed dispersal event within the full spatial, temporal and environmental context of a plant’s life history to fully understand the contribution of seed dispersal to population dynamics, thereby closing the seed dispersal loop (Wang and Smith 2002).
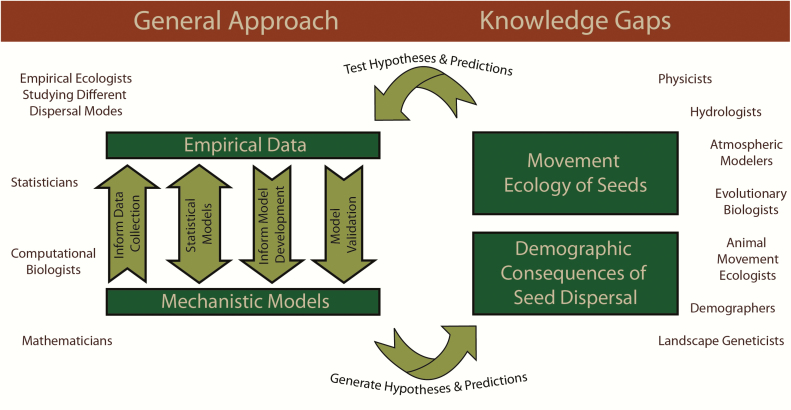
Here, we discuss how the above goal can be reached (Fig. 1). We begin by providing a general perspective on integrating empirical and theoretical methods for addressing the context dependency of seed dispersal to generalize and predict across systems. We then highlight two knowledge gaps that could benefit from such an integrative approach. First, we present advances and challenges in the movement ecology of seeds, considering the multitude of seed dispersal mechanisms and vectors that influence spatial patterns of seed dispersal. Second, we discuss potential pathways for integrating dispersal and demography to reach an improved understanding of population persistence and population spread. Throughout, we demonstrate that advancing the study of seed dispersal and its influence on population dynamics requires increased collaboration among researchers that examine disparate life-history stages of plants from a variety of disciplinary, geographic and organismal perspectives. Such studies will be even more powerful if they take advantage of advances in empirical, statistical, computational and mathematical methods, in tandem with global initiatives and standardized experiments over large geographic extents. We propose promising multidisciplinary and interdisciplinary advances, including opportunities to apply existing frameworks and approaches from other disciplines to advance seed dispersal ecology (Fig. 2). We synthesize suitable theoretical frameworks for this work and discuss concepts, approaches and available data from diverse subdisciplines to help operationalize concepts, highlight recent breakthroughs across research areas and discuss ongoing challenges and open questions. We end with specific strategies to guide future research.
Figure 1.
To advance current knowledge gaps in seed dispersal ecology requires interdisciplinary collaboration in which researchers simultaneously and iteratively collect empirical data and develop mechanistic models that are integrated with statistics.
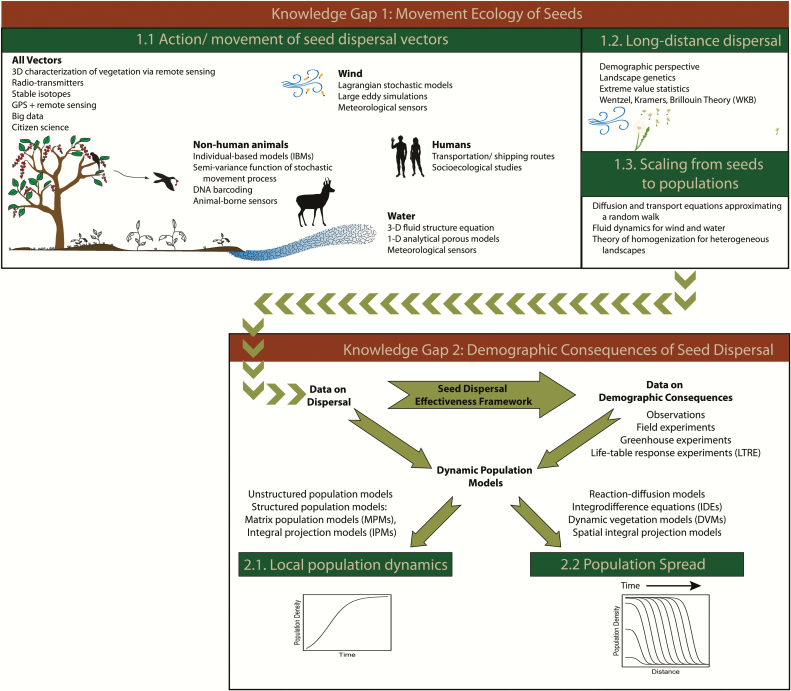
Figure 2.
Examples of the differing empirical and modelling approaches used to quantify dispersal and estimate the impacts of dispersal. We suggest that studies combining multiple approaches are likely to provide greater insight into dispersal dynamics.
A general approach for studying context dependence of seed dispersal
The large number of processes and agents that constitute dispersal (see Figure 1 in Beckman et al. 2020) create a distinct paradox: to predict the consequences of dispersal, we need to simultaneously reduce complexity to generalize across systems and embrace complexity to be able to make system-specific predictions (see also Evans et al. 2013). Reducing complexity can (i) aid in scaling across ecological organizational levels, (ii) reduce the need for data that may not be logistically feasible to collect and (iii) increase the efficiency of the computational models necessary for answering pressing conservation and management issues. By contrast, embracing complexity can (i) provide quantitative predictions for specific conservation and management issues and (ii) allow for a more faithful representation of a particular ecological system. The approach a researcher uses depends on their aim, that is, generalization across systems or specific forecasts, the question of interest and knowledge about the system, including available data.
How to reduce complexity?
To move towards a more fundamental understanding of seed dispersal, we need to know when and how we can generalize dispersal and its impacts on populations. This requires both advances in the theory of dispersal ecology and standardized empirical methods to test and inform theory. Theory can take the form of conceptual, statistical, simulation or mathematical models and allows us to clearly formulate our assumptions and the expected first principles underlying observed patterns while necessarily simplifying the system of interest (Marquet et al. 2014). Theory and standardized data collection will aid us in finding differences and commonalities across systems and can help determine if, when, and how we can scale from local empirical studies to predict qualitative or quantitative responses to global change at larger temporal, spatial or organizational scales. Building and collaborating with international networks of researchers (Frugivory and Seed Dispersal: e.g. Estrada and Fleming 1986; Levey et al. 2002; Hardesty 2007, CoDisperse: e.g. Beckman et al. 2020) we can integrate theory with data from existing studies, long-term data sets and future data collection initiatives developed by an interdisciplinary network of researchers to answer the most pressing questions in seed dispersal ecology.
How to embrace complexity?
To enhance system-specific predictions, we need to address uncertainty, boost simulation capacity and collect relevant ecological and natural history data. Systems-based approaches can be used to understand a system as a whole and to incorporate the complexity of ecosystems as well as uncertainty related to data, model structure and model selection (e.g. Hartig et al. 2012; Milner-Gulland and Shea 2017). We can include mechanistic representations of reproduction, dispersal, growth and survival that allow predictions of dynamic responses to future global change and novel conditions, without assuming static relationships under current environmental conditions. Connecting these models to data requires statistical advances, such as Bayesian Inference or Approximate Bayesian Computation (ABC; Hartig et al. 2011, 2012), that incorporate heterogeneous data into process-based models to reduce uncertainty and test model output with data. Additionally, development of systems-based approaches to study seed dispersal requires computational advances to deal with multi-scale problems, mathematical advances that can approximate complexity and reduce computational expenses, and integration of empirical data across systems and subdisciplines that study the movement of seeds, their corresponding vectors (e.g. wind, water, animals, etc.) and the fitness contributions of seed dispersal.
Confronting complexity with models and data
We believe that a promising approach to confront the complexity and context dependency of seed dispersal is to allow for feedbacks between empirical observations and the exploration of dynamics by simultaneously and iteratively collecting data and developing models. This approach would allow for data to inform the development and refinement of model assumptions, parameters and structure, and for models to elucidate mechanisms driving empirical patterns. By collecting data on dispersal processes simultaneously with model development, we can use models to develop hypotheses and predictions that can be tested empirically, and with an iterative approach, we can refine models based on empirical results to develop and test new hypotheses and predictions. Proper incorporation of stochasticity can help determine the limits to prediction as well as experimental challenges. In addition, models can be developed based on the results from manipulative experiments and project the consequences of dispersal for higher organizational levels (e.g. populations, communities) or over a larger spatial and temporal extent than is possible with manipulative experiments alone. Results of these models can be compared to observational data to help discern whether and how dispersal processes lead to empirical patterns observed over larger spatiotemporal scales. Finally, mechanistic models can predict responses to different scenarios of novel conditions anticipated from global change models. Mechanistic models range from analytical models to complicated simulation models (Box 1; Dieckmann et al. 2000; Jongejans et al. 2008). In addition, phenomenological models can be useful in describing dispersal patterns (e.g. Bullock et al. 2017) and approximating mechanistic models of dispersal for inclusion in process-based models. Data collection efforts can include synthesis of existing knowledge or collection of data from manipulative experiments and observations from the field, greenhouse and laboratory. The most appropriate modeling approach depends on research questions, assumptions and type of data available (Box 1).
Box 1. Overview of models.
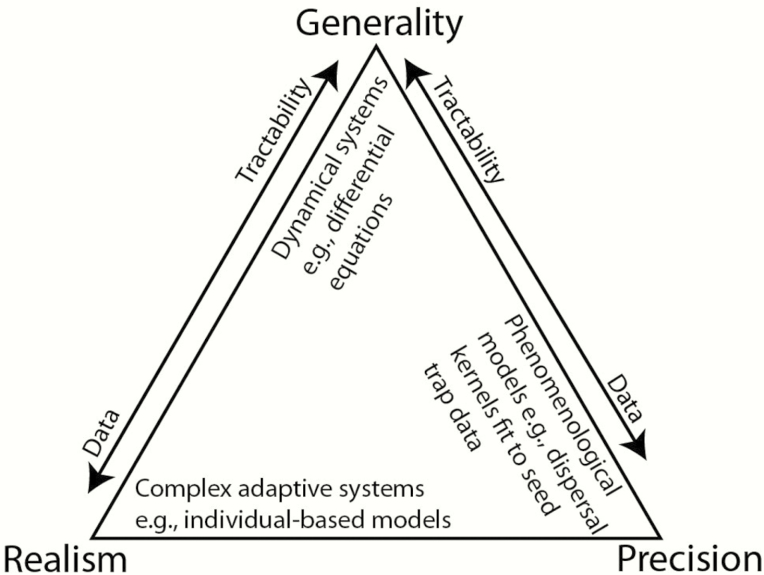
Developing and evaluating process-based models requires empirical studies to identify the processes to be included (model structure), the descriptions used for those processes (model selection) and data on parameters (Grimm and Railsback 2011). Based on the purpose of the model, researchers will need to decide how to balance generality, realism and precision (Fig. 3).
Figure 3.
Trade-offs in model building as discussed by Levins (1966): the goals of models are to maximize generality, realism and precision but trade-offs exist such that only two of these three goals can be captured. While there is philosophical doubt on whether these trade-offs exist (Evans 2012), maximizing all three goals will likely result in a model that is intractable and impossible to analyse (Silverman 2018).
Analytical mathematical models offer conceptual insights on the qualitative behaviour of the system by using simplifying assumptions that allow the general contribution of different processes and parameters to be evaluated. This can be particularly helpful when data are limited (Bullock et al. 2012). Analytical models can also facilitate scaling from individual seeds to populations by approximating computationally expensive simulations while retaining key dispersal mechanisms (e.g. Travis et al. 2011). More complicated models that are fine-tuned for a specific system are thought to have greater predictive power (Evans et al. 2013), though this requires further investigation as adding more complicated model structure increases uncertainty (Sun et al. 2016). Simulation models, such as individual-based models (also known as agent-based models), are becoming more sophisticated as computing power increases and can be quite useful for suggesting how individual-level processes give rise to complex population-level phenomena. However, complicated simulations trade analytical tractability, computational inexpensiveness and fewer data requirements for direct incorporation of natural complexity, real-world variability and uncertainty (Fig. 3).
Further assumptions to consider during model development are whether, and how, to incorporate time, space, stochasticity and individuals. Does the question of interest involve static relationships or changes over time (i.e. static vs. dynamic models)? If researchers are interested in changes over time, do the entities in the model experience time continuously (e.g. overlapping generations) or discretely (e.g. seasonality), and what temporal scales are relevant? Is space important; should it be continuous or discrete, and what spatial scales are relevant? How important is it to consider deterministic vs. stochastic model versions? Can the system be modelled assuming large population sizes or are interactions among discrete individuals important to consider? Other questions to consider involve the detail of processes to be included. For example, does dispersal need to be represented by detailed movement pathways or are phenomenological dispersal patterns sufficient? What is the importance of demographic variation? How important are interactions with mutualists (e.g. mycorrhizae) and antagonists (e.g. competitors, natural enemies) at the site of deposition?
In summary, seed dispersal is a complex and context-dependent process, but we assert that the seed dispersal loop can be closed and the contribution of seed dispersal to plant population dynamics can be quantified from multidisciplinary and interdisciplinary perspectives. We can achieve this by synthesizing recent advances in analytical mathematical models, computational simulation models, statistics, data synthesis and coordinated data collection on dispersal and recruitment processes. Such an integration will ultimately help balance necessary complexity with tractability.
Next, we discuss advances and challenges in confronting this context dependency with data and models in the context of two knowledge gaps: (i) mechanisms underlying the movement ecology of seeds and resulting dispersal patterns and (ii) demographic consequences of this movement.
Gap 1: Understanding the movement ecology of seeds
This first knowledge gap focuses on improving our mechanistic understanding of the movement of individual seeds in order to generalize dispersal mechanisms and patterns across systems and to predict dispersal under novel conditions. Studies uncovering spatial patterns of seed dispersal have tended to focus on population-level patterns (e.g. Eulerian methods), but are becoming increasingly mechanistic by focusing on the movement of individual seeds (e.g. Langrarian approaches; Turchin 1998). To describe population-level spatial patterns of seed dispersal, ecologists have estimated dispersal kernels (probability density function of dispersal distances; Nathan et al. 2012) by combining seed traps with inverse modeling (Nathan and Muller-Landau 2000) and incorporating genetic information from seeds and parents (e.g. Hardesty 2007; Jones and Muller-Landau 2008). While these analyses increase our understanding of the variation in seed dispersal patterns, dispersal kernels tend to be phenomenological (Nathan et al. 2012; but see Katul et al. 2005; Codling et al. 2008 for examples of mechanistically derived dispersal kernels) and therefore have limited capability for forecasting changes in dispersal itself under novel conditions resulting from global change. In addition, these phenomenological models tend to describe the spatial patterns of seeds arising from the final outcome of dispersal and not the process of dispersal, while a mechanistic understanding can only be achieved by partitioning the contributions of different dispersal vectors (see Rogers et al. 2019 for solutions to this issue).
A more mechanistic understanding of the movement of individual seeds requires explicitly quantifying the action and movement of different seed dispersal vectors and their interactions with plants. A challenge will be measuring the extent of long-distance dispersal (LDD), rare events that are particularly difficult to study but likely critical to the establishment of new populations, colonization after disturbance and rapid plant migration in response to climate change (Nathan 2006). Finally, we need to be able to scale up movements of individual seeds to effectively generalize and predict spatial patterns that emerge at the population level. Development of models informed by empirical data will help us incorporate the necessary level of complexity for dispersal vectors and their interactions with plants, measure the extent of LDD and scale from the movement of individual seeds to describe population-level spatial patterns.
Action/movement of seed dispersal vectors
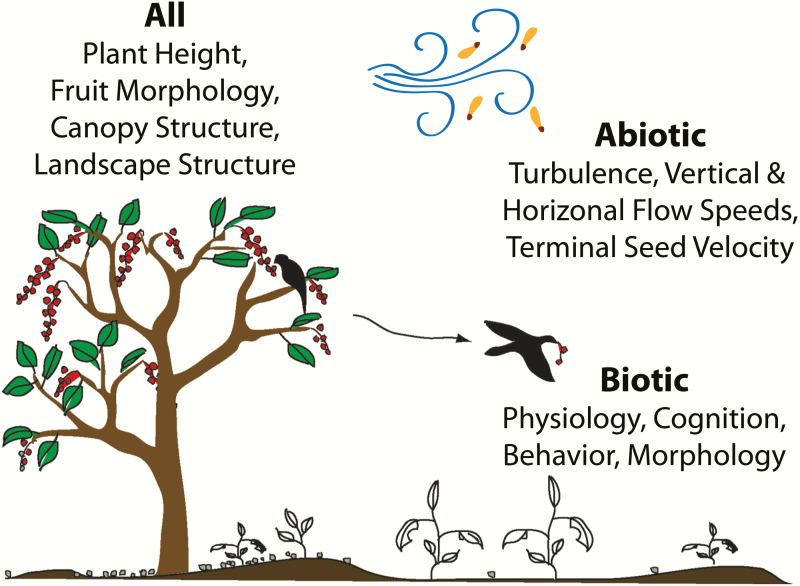
Across species, plants are dispersed by a range of dispersal vectors; even an individual seed may be dispersed by a suite of abiotic and biotic means. These vectors have different consequences for seed dispersal patterns and require a range of empirical and mathematical methods to uncover and describe associated processes (Fig. 4). Investigating all the actions, movements and processes influencing the journey of a seed at the plant, population or species level is daunting but a mechanistic understanding is possible by integrating empirical and theoretical approaches. One approach is to describe functional groups to generalize across species as discussed by Aslan et al. (2019). Another is to draw general lessons from analysis of total dispersal kernels for key species (Rogers et al. 2019). Here, we highlight data-driven quantitative approaches that enable researchers to describe these complex processes and advance a mechanistic understanding of different vectors, focusing on wind, water and animals, including humans.
Figure 4.
Examples of processes influencing abiotically and biotically dispersed seeds.
For abiotically dispersed plant species, we can gain an increased mechanistic understanding of dispersal processes from physics and hydrology. The physics of the transport of propagules—such as spores, pollen and seeds—due to the action of wind or water is a vast field in its own right (e.g. Okubo and Levin 1989; Isard and Gage 2001; Katul et al. 2005; Nathan et al. 2011b; Aylor 2017). For wind dispersal, a typical seed dispersal event first involves release of a seed from the plant canopy. For plants in the herbaceous layer, seeds are generally dispersed above the canopy, and any seeds released under the canopy settle immediately due to very low wind speeds (Soons et al. 2004). For trees and shrubs, the seed will experience dispersal within the canopy due to canopy-scale turbulence, sometimes followed by escape from the canopy and transport via the surface layer or even higher levels of the atmospheric boundary layer, before being deposited (Augspurger 1986). Each of these steps involves turbulence and advective flows with different properties. Hence, one of the challenges is to connect dispersal processes that dominate at different scales (Pauchard and Shea 2006). A variety of numerical simulation methods have been developed, including Lagrangian stochastic models (Katul et al. 2005; Kuparinen 2006; Aylor 2017) and large eddy simulations (Chamecki et al. 2009; Nathan et al. 2011b). These mechanistic models, varying in levels of complexity, have given us insights on the importance of seed abscission, canopy structure, plant height and land surface heterogeneity on LDD through effects on turbulence and wind speed, but additional advances in theory are required to generalize across systems (Nathan et al. 2011b). For seed dispersal by water, obtaining a fine-scale resolution of flow requires numerically solving 3-D fluid-structure interaction equations, which is extremely expensive computationally. In other scenarios, 1-D analytical porous models may suffice to resolve flow through vegetative beds including sea grasses, reefs and macrophytes (e.g. Brinkman 1949; Strickland et al. 2017). These modeling approaches for abiotically dispersed plant species can be further developed with advances in data collection. For example, remote sensing now enables 3-D characterizations of vegetation (e.g. Lefsky et al. 2002; Eitel et al. 2016), and meteorological sensors (FLUXNET; Baldocchi et al. 2001) allow monitoring speeds and turbulence of wind and water at high spatial and temporal resolutions.
The dispersal of a seed by an animal depends on the vector’s life-history strategy, local abundances and distributions of dispersers and fruiting trees, landscape structure, and individual characteristics of the animal and fruit themselves (Nathan et al. 2008; Cortes and Uriarte 2012; Schupp et al. 2019; Snell et al. 2019). Spatially explicit individual-based models (IBMs) can integrate data on dispersal processes, such as gut retention time, animal movement and number of seeds dispersed, to determine the spatial locations of seeds and contribution to long-distance seed dispersal. For example, Kleyheeg et al. (2017) predicted seed dispersal patterns by the mallard (Anas platyrhynchos), an important dispersal vector of wetland plants, by using a spatially explicit, mechanistic simulation model developed from high-resolution data on gut-passage times and landscape-scale movements of the mallard. Pires et al. (2018) estimated that LDD reduced by at least two-thirds following extinctions of mammals in specific Pleistocene assemblages using a mechanistic simulation model incorporating seed ingestion, gut retention, animal movement and seed deposition. Animal movement relevant to seed dispersal can occur across multiple spatiotemporal scales; for example, an animal may forage at fine spatial and temporal scales but search for foraging sites at long distances. The movement path of an individual arises from an animal’s internal state, navigation capacity, motion capacity and the environment (Nathan et al. 2008). Multiple behaviours of animal movement are quantified using observational data on the locations of individual animals collected at predetermined fixed time intervals, and some of the derived quantities used to describe movement are sensitive to the choice of sampling rate. These sampling rates should be guided by the research question and the movement process under investigation. Fleming et al. (2014) recently developed an approach using a semi-variance function of a stochastic movement process that enables identification of multiple modes of animal movement that vary across spatiotemporal timescales (e.g. foraging, simple random search and home range) and provides a solution to the sampling rate problem. Using this approach, they were able to incorporate foraging behaviour into existing animal movement models. Finally, understanding the preference and avoidance of certain habitats within the landscape by animal seed dispersers will be necessary for determining subsequent growth and survival of plants after deposition. For example, Kleyheeg et al. (2017) found that landscape configuration governs mallard movements, and transport of seeds to core areas may help maintain connectivity of wetland plant populations.
To better understand the mechanisms of seed dispersal in socioecological systems, we need to consider both accidental and deliberate seed dispersal by humans (e.g. Wichmann et al. 2009; Taylor et al. 2012), which can occur over great distances that are potentially global in scale (Bullock et al. 2018). Methods are being developed to quantify and model seed dispersal by humans. Relevant advances in invasion biology include genetic analysis to identify seed sources (e.g. Eriksen et al. 2014), transportation/shipping route mapping (e.g. Miller and Ruiz 2014; Chapman et al. 2017) and socioecological studies of human behaviours and movements (e.g. Wilson et al. 2016). From the results of these studies, some of these interactions may be generalizable and predictable (e.g. based on plant traits; Bullock et al. 2018). For example, vehicles were observed to disperse seeds in a directional manner in Berlin, in which seed traps near outbound lanes tended to have native seeds and exotic non-crop seeds, while inbound lanes tended to have exotic crops (von der Lippe and Kowarik 2008).
Recent empirical advances can aid a mechanistic understanding of seed dispersal and the development of mechanistic models described above (e.g. Nathan et al. 2011b; Cortes and Uriarte 2012). These advances include detailed data on seed movement (e.g. stable isotopes: Carlo et al. 2009; radio transmitters: Hirsch et al. 2012; DNA barcoding: González-Varo et al. 2014), animal movement (e.g. Movebank: Kranstauber et al. 2011; integrating GPS tracking with remote sensing: Kays et al. 2015; animal-borne sensors: Wilmers et al. 2015) and the abiotic environment (Baldocchi et al. 2001; Davies and Asner 2014). For example, using telemetric thread tags (Hirsch et al. 2012), Jansen et al. (2012) found that secondary dispersers have a greater role in LDD than previously thought. In addition, future research can link trait data (e.g. TRY: Kattge et al. 2011; D3: Hintze et al. 2013; KEW: Royal Botanic Gardens Kew 2016) to dispersal processes to help reduce complexity of interactions and models (Aslan et al. 2019). The quantity of data relevant for dispersal is increasing into the realm of big data (Allan et al. 2018), and rapid access is eased through curated repositories. These repositories can be used to improve our ability to incorporate intraspecific variation in seed dispersal (Schupp et al. 2019), which can have important consequences for plant populations, communities and evolution (Snell et al. 2019; Schreiber and Beckman 2020). While current advances allow us to study dispersal vector characteristics at very fine spatial and temporal resolution, the question remains whether this also captures variation among and within populations and species.
Long-distance dispersal
Long-distance dispersal—often a rare event—is critical to the spread of populations (Ferrandino 1993; Kot et al. 1996; Hastings et al. 2005). Advances have been made in operationalizing the concept of LDD, but challenges remain concerning how to measure these rare events. Jordano (2017) recently introduced operative definitions of LDD using a demographic perspective in which propagules can contribute to LDD by expanding a species’ range when they colonize new areas outside of the source population or disperse away from close relatives outside of the genetic neighbourhood in which parents mate. He identified three types of LDD: (i) LDD within the genetic neighbourhood, (ii) short-distance dispersal outside of the genetic neighbourhood and (iii) strict-sense LDD (i.e. LDD outside the genetic neighbourhood). The question remains: once operationalized, how do we measure such rare events? This remains a major challenge in empirical ecology, but perspectives from population genetics, statistics, and physics are improving our ability to empirically measure the importance of LDD in gene flow and species distributions.
Genetic analysis of populations can link individuals to their source populations and has been a useful tool for understanding the importance of rare, long-distance events in colonizing new areas. Using genetic information from 25 species, Alsos and colleagues found that multiple dispersal events from several source regions contributed to post-glacial colonization of five islands in the Arctic; source regions were 280 to >3000 km away and were frequently not the closest ones—suggesting a greater role for deterministic rather than stochastic factors resulting in LDD (Alsos et al. 2007, 2015). Landscape genetics reveal that multiple LDD events were also responsible for mountain hemlock colonization on Alaska’s Kenai Peninsula (USA) following Pleistocene glaciation (Johnson et al. 2017). Using molecular techniques, observations of fruit consumption, and data from seed traps and faeces, Jordano et al. (2007) were able to quantify the contribution of different dispersal vectors to LDD for the mahaleb cherry (Prunus mahaleb) and found that LDD of this plant species is driven by a small subset of large frugivores. Citizen science projects can also shed light on the extent and magnitude of LDD events. For example, in Sweden, Auffret and Cousins (2013) found that humans dispersed meadow species, especially those with hooked or appendaged seeds and persistent seed banks, from 1.3 to 110 km.
Extreme value theory, introduced by Gumbel (1958), can be applied to dispersal distances obtained from molecular tools, tracking dispersal vector movement and censored data (e.g. the maximum observed distance moved from a fruiting tree) to estimate the frequency and extent of rare dispersal events. This statistical technique has been widely used in other disciplines such as climatology, hydrology, engineering, insurance, finance and, more recently, ecology to estimate the frequency and extent of rare events, for example, the return interval of large floods (e.g. Gaines and Denny 1993; Gutschick and BassiriRad 2003). Extreme value statistics have been applied to the study of plant dispersal only very recently, likely due to limited data in the past (García et al. 2017). Using data on seed dispersal distances obtained from genetic analyses of the vertebrate-dispersed mahaleb cherry, García et al. (2017) found that seeds could be dispersed outside of the focal population with low, but non-zero probability (Pr(X ≥ 1 km) = 0.10, Pr(X ≥ 5 km) = 4 × 10−4 and Pr(X ≥ 10 km) = 7 × 10−5). Extreme value statistics can give insight into invasions, the loss of dispersal services or the likelihood of populations tracking suitable habitat (García et al. 2017), and may therefore be useful for generalizing across systems. However, these methods are phenomenological models fit to existing data and assume stationarity, while extreme value statistics are likely to shift under global change.
Physics can contribute insights on measuring rare events mechanistically, which can help in predicting LDD under global change. In nature, there are two types of rare events: (i) discrete and uncorrelated, such as an unusually long pause between two consecutive events in a Poisson process, and (ii) a sequence of cumulative rare events. Long-distance dispersal by animals is most likely of the first type. Long-distance dispersal by wind is most likely of the second type, as an unlikely sequence of turbulent events sustain the seed in the air for an unlikely long period of time (all the while being pushed by wind in the direction parallel to the ground). Physicists have developed a powerful approach for understanding the statistics and dynamics of a sequence of cumulative rare events. A rare chain of events connects the initial and final state. In the case of a LDD event, the initial and final states would be the source and destination locations of a propagule. An event would be considered rare, if the dispersal event was much longer than a typical dispersal event (quantified, for instance, by the standard deviation of dispersal distances). The key insight behind this approach is that a very unlikely chain of events (e.g. a dispersal event that is much longer than a typical dispersal event) unfolds along an essentially deterministic realization—the least unlikely out of all unlikely realizations connecting these initial and final states. The more unlikely the chain of events, the more it will be dominated by this least unlikely realization (or chain of events). Deviations around this ‘optimal path’ quickly decrease in probability, even though the probability of this least unlikely path is also small (given some fixed waiting time). The Wentzel, Kramers, Brillouin (WKB) theory—originally developed for calculating the rates of (rare) tunnelling events in Quantum Mechanics—has found applications in fields as diverse as population biology and epidemiology in recent years (Ovaskainen and Meerson 2010). This WKB theory is the method by which an optimal path (or trajectory) can be found, and the probability of a rare event can be evaluated. Given the properties of noise (e.g. its correlation function), the method gives a certain cost function that measures the relative probability of any one path. Minimizing this cost function over the functional space of paths gives the optimal path, and the cost function along the optimal path gives the dominant contribution to the probability of a rare event. Attempts are currently under way to adapt this theory to hydrodynamics (Laurie and Bouchet 2015; Bouchet et al. 2018), overcoming challenges imposed by the high dimensionality of dynamics involved. Ovaskainen and Meerson (2010) provide both an accessible exposition of this technique and its biological applications, and a clear explanation of challenges of applying these ideas in complex situations.
Scaling from individual seed movements to population-level patterns
Both generalizing and predicting population-level patterns of seed dispersal from the movements of individual seeds require relevant advances in data, mathematics and computation. First, it requires a detailed understanding of the mechanisms of the focal system as introduced above and the natural histories of the relevant players. Second, it requires mathematical and computational advances that efficiently scale from interactions occurring over short time scales (on the order of minutes) and spatial scales (on the order of mm–cm) to patterns emerging over months to years across landscapes and regions. Modeling population-level patterns of the movement of individual seeds in response to the action, behaviour and movement of dispersal vectors can be computationally expensive and require extensive data for calibration. There are only a few plant species for which complex, mechanistic models have been developed and for which enough data exist to parameterize these models to predict population-level spatial patterns of seed dispersal from individual seed movements (Nathan et al. 2011b; Cortes and Uriarte 2012). These models are better-developed for ballistic- and wind-dispersed plants than animal-dispersed plants (Nathan et al. 2011b; Cortes and Uriarte 2012), in large part because of the complexity of animal behaviour and movement (Zwolak 2018). However, the field of animal movement ecology has advanced tremendously over the last decades with tracking and analytical methods constantly improving (Börger 2016). Collaborating more closely with animal movement researchers opens up new opportunities for developing improved models of animal-dispersed plants.
To scale from individual movements to population-level patterns, we can approximate complex mechanistic models with models that make simplifying assumptions. One multi-scale mathematical approach is to begin with random walks of individuals and use various approximations to arrive at diffusion or transport equations that describe the collective movement of individuals (Turchin 1998; Codling et al. 2008). Coupled with functions of seed retention time (Bullock et al. 2011), these approximations can describe seed dispersal by animals and give important insight into how variability in retention times (in animal guts or externally) influences LDD (Guttal et al. 2011). A fluid dynamics approach used by physicists, hydrologists and atmospheric modellers can help overcome challenges in scaling from the local interactions of a seed with the physical dynamics of wind and water to large-scale dispersal patterns. Under certain assumptions, the effect of wind on the dispersal kernel and LDD can be approximated using analytical mathematical results, such as the Wald analytical LDD model (e.g. Katul et al. 2005). The empirical, analytical and numerical methods for the treatment of abiotic dispersal in heterogeneous landscapes require further development in future research (Brinkman 1949; Bohrer et al. 2008; Nathan et al. 2011b; Katul and Poggi 2012; Trakhtenbrot et al. 2014). Advances have been made on this front by Powell and Zimmermann (2004), who developed an analytical solution to approximate the migration of plants dispersed by animals based on the theory of homogenization, which could be extended to abiotically dispersed plants. Through this technique, Powell and Zimmermann (2004) incorporated caching activity by harvester ants for wild ginger (Asarum canadense), by Blue Jays (Cyanocitta cristata) for oaks (Quercus) and by Clark’s nutcrackers (Nucifraga columbiana) for whitebark pine (Pinus albicaulis), and were able to predict migration rates of trees that matched the paleo-record, except in the case of the Holocene migration of wild ginger.
Future research can advance mechanistic dispersal kernels and predictive seed dispersal ecology. First, dispersal kernels can better incorporate interactions between dispersal vectors and individual seeds, which tend to occur across multiple spatial and temporal scales. For example, different dispersal mechanisms can be incorporated mathematically into the dispersal kernel, which gives the long-term limit after all seeds land, to evaluate the effects of different vectors quantitatively using methods from applied mathematics (Rogers et al. 2019). Second, dispersal kernels should account for non-stationarity in driving factors and depend on the environment. That way, kernels can change with time, space and shifts in the environment, important for predicting dispersal in novel landscapes. Standardized data initiatives provide a valuable means for evaluating the magnitude and causes of non-stationarity across space and time. Third, integrating multiple dispersal vectors, non-stationary dispersal kernels, and improved and standardized dispersal vector monitoring and data collection with process-based models will allow predicting the spatiotemporal distribution of seeds of entire populations across the landscape.
Gap 2: Understanding the demographic consequences of seed dispersal
So far, we have discussed how to understand seed dispersal as a process. To understand the importance of seed dispersal for the dynamics of a population over multiple generations, we need to understand how this process interacts with stages across a plant’s entire life cycle, from seed production through juvenile and adult survival and growth. Prediction of the demographic consequences of seed dispersal remains a large challenge due to the context dependence of seed dispersal (Schupp et al. 2010), heterogeneity of the environment (Nathan et al. 2011a), the long lifespans of many adult plants and interdependent processes occurring over multiple spatial and temporal scales (Mokany et al. 2014). A promising path forward is integrating the Seed Dispersal Effectiveness Framework (Schupp 1993; Schupp et al. 2010) with advances in mathematical and computational methods (e.g. Godinez-Alvarez and Jordano 2007; Cortes and Uriarte 2012). The Seed Dispersal Effectiveness Framework—an important progression in embracing the context dependence of seed dispersal and moving towards an ability to generalize across systems—provides a roadmap for evaluating the contribution of each dispersal vector to the production of a new adult by evaluating the quantity of seeds dispersed and quality of seed dispersal in different contexts (Schupp 1993; Schupp et al. 2010). This information can be incorporated into process-based dynamic models of populations to examine the influence of dispersal compared to other life-history stages on the growth and spread of populations over multiple generations as discussed below.
Local population dynamics
To evaluate the role of seed dispersal in population dynamics, we need to explicitly integrate over critical determinants of seed dispersal effectiveness (Schupp et al. 2010), including pre-dispersal, dispersal and post-dispersal processes operating across different life stages. Hitherto, the Seed Dispersal Effectiveness Framework has mostly been applied to single species (but see Donoso et al. 2016; Fricke et al. 2018), but Aslan et al. (2019) outline an approach to generalize across functional groups. Such attempts at generalizing seed dispersal effectiveness across species and systems are necessary because empirical data for operationalizing the Seed Dispersal Effectiveness Framework are still scarce (but see Simmons et al. 2018). For example, data on dispersal and its delayed consequences for plant survival and growth are limited in temporal scale, following seeds for only a few years (Clark et al. 1999a; Howe and Miriti 2004), and are highly species-specific, with data amount and quality varying widely among plant species and interacting species that influence plant dispersal, growth and survival (Agrawal et al. 2007). Additionally, for long-lived plants such as trees, we are limited to collecting data on the early stages of recruitment. As a result, it is unclear how variation in dispersal and heterogeneity in the seedscape across space and time will influence later stages of recruitment. Population models constitute an alternative approach, and parameterizing population models with observational and experimental data on the effectiveness of different dispersal vectors and their deposition in varying seedscapes helps elucidate the role of dispersal in the demographic process (e.g. Brodie et al. 2009b). Local population dynamics can be modelled to assume a range of biological complexity (reviewed in Jongejans et al. 2008). Such models include unstructured population models (e.g. exponential growth), structured population models that include stages or ages (e.g. matrix population models [MPMs]; Caswell 2001), spatially explicit IBMs (e.g. Adler and Muller-Landau 2005; Beckman et al. 2012) and dynamic vegetation models (DVMs; Snell et al. 2014).
An extremely powerful set of analytical tools have been developed for both MPMs and integral projection models (IPMs) to predict population growth rate, stable stage distribution and sensitivity to small perturbations in the model parameters (Caswell 2001; Ellner and Rees 2006). These analytical models rely on the law of large numbers, and thus model mean populations that encounter each other in proportion to their average abundance (i.e. a mean-field assumption), generally assume homogeneous environments, and provide asymptotic results. Such structured population models are useful to examine different hypotheses of how present conditions influence populations by examining population projections (Caswell 2001). We can use these models to analyse the population growth rate and its sensitivity under different dispersal scenarios (e.g. no dispersal, one dispersal vector, a community of dispersal vectors), and this has improved our understanding of whether and under what conditions dispersal is important for a particular species and our ability to predict the consequences of shifting the community of dispersal vectors. For example, Godinez-Alvarez and Jordano (2007) proposed integrating the Seed Dispersal Effectiveness Framework with MPMs to evaluate the influence of dispersal vectors on the dynamics of plant populations. By building the projection matrix based on the quantity and quality of dispersal by one bat and three bird seed dispersers, Godinez-Alvarez and Jordano (2007) found that dispersal vector identity influenced population growth rates of the cactus Neobuxbaumia tetetzo. In MPMs, plants are categorized in discrete stages by size or life-history stage (e.g. seed, seedling, juvenile, adult), which is more appropriate for plants as they can remain in the same stage for multiple years and/or have unknown or difficult-to-measure ages. Integral projection models can accommodate both discrete and continuous descriptions of fecundity, survival and growth based on size and age (Easterling et al. 2000), which is especially important for long-lived species as individual variation within stages can influence population dynamics (Zuidema et al. 2010). The dynamics of transients, important in the conservation and management of populations, can also be analysed (Caswell 2006). For example, Elwood et al. (2018) showed that scatter-hoarders can have significant effects on both short- and long-term population dynamics of American Chestnut (Castanea dentata). An important advance in analysing dynamics of structured population models is the development of tools to examine consequences of random variation in vital rates (i.e. stochastic demography; Boyce et al. 2006). This stochastic variation affects estimates of population growth, persistence and resilience compared to deterministic versions of structured population models (Boyce et al. 2006). As anthropogenic pressures can increase or decrease environmental variability, the implications of this variation for demography should be carefully considered (e.g. Snell et al. 2019). In addition, future research can explicitly include post-dispersal mechanisms, such as competition, mortality due to natural enemies and microsite suitability for growth (Howe and Miriti 2004) into these suite of population models to increase their capability of predicting dynamics in response to novel conditions.
To explicitly incorporate post-dispersal processes, researchers could use a systems approach to examine the influence of dispersal by animals on local plant dynamics. Cortes and Uriarte (2012) proposed integrating the Seed Dispersal Effectiveness Framework with the movement ecology paradigm developed by Nathan and colleagues (2008) that combines internal states, motion and navigation capacities of individuals with external factors to study movement. This could be done with IBMs (Grimm and Railsback 2005) or DVMs (Snell et al. 2014). For example, Loayza and Knight (2010) used an IBM parameterized by field studies on seed dispersal movement and the quantitative and qualitative components of seed dispersal effectiveness for two bird dispersers of the tree Guettarda viburnoides in a forest-savanna mosaic in Bolivia. Their model predicted that dispersal by Purplish Jays (Cyanocorax cyanomelas; pulp consumers which frequently dropped seeds) increased population growth due to a positive impact of seed handling and an increased likelihood of reaching suitable habitat (woody patches), whereas dispersal by Chestnut-eared Aracaris (Pteroglossus castanotis; ‘legitimate’ seed dispersers that swallow the fruit whole and pass the endocarp intact) decreased population growth, due to dispersal to unsuitable habitats (forest islands). Dynamic vegetation models include demographic, ecological and physiological processes as well as biotic interactions (i.e. competition) and range from models that simulate forest dynamics through growth and mortality of individual trees to models that simulate biogeochemical cycles and vegetation distributions through plant functional types (Snell et al. 2014). However, only a few DVMs currently include seed dispersal (e.g. Sato et al. 2007; Snell 2014; Snell and Cowling 2015; Lehsten et al. 2019), and none yet includes the level of detail outlined here. Dynamic vegetation models with more realistic seed dispersal processes can capture interactions in novel non-analogue environments, useful for predicting population dynamics when interspecific interactions and demographic processes shift.
Data requirements for population models can come from long-term observational studies, manipulative or accidental experiments (e.g. systems that have lost dispersers as a result of global change; HilleRisLambers et al. 2013), or combinations thereof. Data on the dispersal process as discussed above, including the action/activity, occurrence, abundance and movement patterns of dispersal vectors, inform potential deposition sites of different dispersal vectors. Long-term data from censusing give information on survival, growth and recruitment through time and space (i.e. the quality of seed dispersal), necessary for long-lived species. Field and greenhouse experiments can provide detailed information on the suitability of deposition sites for plant recruitment. Seed addition experiments in different habitats can be used to quantify how the action/movement of different seed dispersal vectors influence recruitment in different microsite conditions (Turnbull et al. 2000; Clark et al. 2007). To evaluate the influence of interspecific interactions within deposition sites, controlled greenhouse and field studies can exclude mycorrhizal and nurse–plant associations to measure the effect of these changes on plant growth and nutrient exchange (van der Heijden 2010) or impose or simulate herbivory, seed predation, pathogen attack or parasitism to measure growth rates and other fitness correlates in the presence of natural enemies (e.g. Agrawal 1999). Accidental experiments enable researchers to functionally manipulate dispersal or realistically simulate its absence and evaluate the impact across all life-history stages, not just those that are most tractable for experiments (e.g. seeds, small seedlings). For example, Brodie et al. (2009a) predicted a decline in population growth rate of the canopy tree Choerospondias axillairs in over-hunted forests using a combination of accidental experiments, manipulative seed germination experiments, and population matrix models. How population growth rates differ depending on the quality of seed dispersal across different life-history stages and habitats can be investigated within population models using life table response experiment (LTRE) analyses (Caswell 2001). A LTRE (term introduced by Caswell 1989) compares vital rates under different experimental or observational conditions. A LTRE analysis examines differences in a demographic summary statistic derived from these vitals rates, such as population growth rate, across the study conditions. This is done by decomposing differences in the demographic summary statistic into contributions from the differences in vital rates across study conditions (Caswell 1989, 2001). For example, Loayza and Knight (2010) used a LTRE analysis to compare population growth rates of the tree G. viburnoides between two habitats in which seeds are deposited by different bird seed dispersers. They decomposed the difference in predicted population growth due to contributions from the differences in seedling growth, small tree growth and adult tree fecundity (Loayza and Knight 2010).
Population spread
Dispersal and population spread are at the centre of a fundamental question in global change biology and invasion ecology (Clark et al. 1998; Pauchard and Shea 2006; Jongejans et al. 2008, 2011; Lockwood et al. 2013): if habitats change due to habitat destruction or climate change, will seed dispersal and population growth allow the plant population to track its suitable habitat? Or if a plant species’ seeds are transported into a novel habitat, will seed dispersal and population growth allow the species to naturalize or even become invasive? Information on dispersal processes and demographic transitions from the Seed Dispersal Effectiveness Framework can aid the development of models to predict the spread of populations invading new areas and evaluate the relative importance of seed dispersal. Analytical approaches used to model population spread include reaction-diffusion models that combine continuous time population models with diffusion (i.e. population-level approximation of random walks as discussed in Scaling from individual seed movements to population-level patterns), which are widely and successfully used in spatial ecology (Okubo and Levin 2002; Cantrell and Cosner 2004). Their discrete time analogues, integrodifference equations (IDEs), offer several appealing features for modeling plant populations. Integrodifference equations can incorporate discrete stage structure (Neubert and Caswell 2000) and more closely represent seasonality in natural systems. They also offer greater flexibility in describing dispersal events via redistribution kernels (or probability density functions for seed shadows) (Kot and Schaefer 1986). As discussed in the previous section, systems approaches can also be used to model population spread rates, which assume discrete interacting individuals. Santini et al. (2016) found that IBMs predicted slower spread rates of mammals compared to an IDE, most likely due to the inherent stochasticity in IBMs. A functional perspective of seed dispersal effectiveness (Aslan et al. 2019) could help the incorporation of dispersal into DVMs to simulate range shifts of plants (Snell et al. 2014). For an overview of the types of models that integrate dispersal and demography, see Jongejans et al. (2008).
Integrodifference equations (IDEs) have been used to examine the spread of invading organisms (Kot et al. 1996; Hastings et al. 2005; Skarpaas and Shea 2007) and the influence of climate change on shifts in species ranges (Zhou and Kot 2011; Harsch et al. 2014). Exponentially bounded kernels result in constant speed of population spread/invasion in integrodifference equations. However, fat-tailed kernels, such as the bivariate version of Student’s t-distribution that fits many dispersal vector–plant combinations (Clark et al. 1999b), may lead to accelerating invasion speeds (Kot et al. 1996). Clark et al. (2001) developed an alternative approach to estimate finite spread rates using the expected velocity for the location of the furthest-forward individual. Using this method, they found slower spread rates than predicted by analytical models, and these slower rates were in line with paleorecords (Clark et al. 2001). Mechanistic models for wind-dispersed species that incorporate dispersal and demography have been used to determine causes of variation and predict spread rates in response to climate-mediated changes in dispersal (Nathan et al. 2011a; Bullock et al. 2012; Teller et al. 2016). Life table response experiment analyses (introduced in the previous section) of IDEs can determine the contributions of differences in demography and dispersal to differences in spread rates across populations as was done for both inter- and intraspecific bird populations by Caswell et al. (2003) and different management scenarios of the annual herb Rhinanthus minor (Bullock et al. 2008). By integrating seed dispersal effectiveness with LTRE analysis, researchers can examine how different dispersers influence population spread rates through their effects on demography and dispersal or how changes in vital rates and seed dispersal due to global change could influence population spread rates. Recent advances provide new opportunities to understand the influence of dispersal processes on population spread. Mathematicians have developed promising approaches to incorporate individual variation (spatial IPMs; Jongejans et al. 2011), fragmented landscapes (i.e. reaction-diffusion models: Maciel and Lutscher 2013; integrodifference equations: Gilbert et al. 2014), stochasticity (Caswell et al. 2011) and temporally variable environments (Caswell et al. 2011; Schreiber and Ryan 2011; Ellner and Schreiber 2012). A good description of a variety of methods for calculating discrete-time invasion rates from data is available in Lewis et al. (2006).
Empirical advances to measure spread (e.g. remote sensing via unmanned aerial vehicles and telemetry) can be combined with models to elucidate important dispersal vectors. For example, Vellend et al. (2006) estimated migration rates for Trillium grandiflorum using an IDE parameterized with data on deer movements from telemetry and gut passage to describe dispersal and demographic transitions under different levels of herbivory, and these estimates were much faster than previous estimates based on ant dispersal. In addition, vehicles can disperse seeds long distances and facilitate the spread of invasive species that can disrupt land management and ecosystem function of natural plant communities. For example, vehicles aided the spread of cheatgrass, which has overtaken sagebrush in the western arid regions of the USA (Strickland et al. 2015)—and this has implications for cattle grazing and water storage. Future research should further develop approaches to determine if and when it is necessary to consider LDD in the context of population spread (Kot et al. 1996). For determining spread rates of populations, Neubert and Caswell (2000) suggested that data on the distances dispersed by seeds are more important than knowing the proportion of seeds dispersed at long distances—as long as this proportion is small—and that it is more feasible to measure the distance travelled by LDD vectors than the proportion dispersed by each vector that results in different dispersal kernels.
Recommendations for future research
Moving towards a mechanistic and predictive understanding of the movement of seeds and the demographic consequences of this movement requires collaboration across a large group of scientists working at different scales, in different bioregions, using a wide arsenal of tools. At present, seed dispersal research is carried out by researchers from an array of subdisciplines with diverse but poorly aligned goals and approaches. Disparate literature bodies investigate seed dispersal from ecological, mathematical, theoretical, computational, statistical, genetic, physical and evolutionary angles. While each subdiscipline can contribute insights into particular aspects of seed dispersal, no single disciplinary method or conceptual framework can independently close the loop on seed dispersal and its contributions to plant populations. We provide recommendations for future research focusing on strategies to accommodate diverse but potentially limited data.
Collate existing, disparate data sets
The highly context-dependent nature of empirical data and limited knowledge of dispersal and its consequences for plant fitness impede our ability to generalize and predict response of plants under global change. However, there is a wealth of available knowledge that has not yet been synthesized for analysis. Rich new data sets are currently emerging in ecology as a result of advances in remote sensing data (Kerr and Ostrovsky 2003; Pettorelli et al. 2014), environmental sensor data (Rundel et al. 2009; Wilmers et al. 2015), long-term data from research sites such as Long-term Ecological Research (LTER) and Forest Global Earth Observatory (ForestGEO; Anderson-Teixeira et al. 2015), emergence of new large collaborative networks (e.g. Templ et al. 2018; USA National Phenology Network 2018) and globally distributed experiments (e.g. Nutrient Network; Borer et al. 2014), and increased digital availability of data. Data sources include publicly available data sets, including data on dispersal distances (e.g. Tamme et al. 2014; Bullock et al. 2017), traits (e.g. Kleyer et al. 2008; Kattge et al. 2011; Royal Botanic Gardens Kew 2016), networks (e.g. Pigot et al. 2016), demography (e.g. Salguero-Gómez et al. 2015), plant phylogenies (useful for generalization based on cross-species comparisons and understanding the evolutionary implications of seed dispersal; Zanne et al. 2013) and species distributions (e.g. Enquist et al. 2016), and unpublished data sets (seed dispersal networks, dispersal kernels, spatial dispersal data, movement data, etc.). There is also an abundance of existing data within the gray and white literature, including data from conservation areas and government organizations, in a variety of languages. Automated text analysis as used in the social sciences (e.g. Wilkerson and Casas 2017) can identify documents with relevant data in multiple languages. Currently, a repository for data on dispersal processes is lacking and requires appropriate cyberinfrastructure to assimilate large quantities of disparate data into models. Existing cyberinfrastructure, such as the National Science Foundation-funded CyVerse developed for the life sciences, is one option. CyVerse allows for flexible storage of heterogeneous data and is able to interface with existing repositories that house relevant data (Goff et al. 2011; Merchant et al. 2016). The time is ripe for creating a repository for dispersal data for synthesis and analysis. These data can be linked to existing available data sets to close the two knowledge gaps discussed above and improve our ability to generalize across systems and predict outcomes for specific systems.
Use novel statistical techniques to integrate disparate data with process-based models
Differences in model structure and parameterization based on limited data can create large uncertainties in model predictions (Hartig et al. 2012) and necessitates systematic examination of dispersal mechanisms as well as high-resolution data (Cortes and Uriarte 2012; Mokany et al. 2014). We can take advantage of systematic reviews and statistical approaches, such as meta-analyses (e.g. Markl et al. 2012), inverse modeling (e.g. Ribbens et al. 1994) and imputation methods (e.g. Santini et al. 2016), to integrate the growing body of available data with process-based models. Systematic reviews and meta-analyses can help identify processes that require model development, as well as parameter ranges for these models. Recent statistical advances in merging process-based models with Bayesian or approximate Bayesian methods can reduce uncertainty by incorporating different types of data (Hartig et al. 2011, 2012), facilitating identification of relevant processes by better utilizing existing data, a major advantage of modern statistics and computing that has not yet been exploited. For example, approximate Bayesian approaches (e.g. ABC) enable one to infer parameters from a variety of process-based models including stochastic individual-based simulation models, which cannot be informed by statistical theory such as maximum likelihood or Bayesian methods because their likelihood functions cannot be explicitly calculated. In addition, new methods are continuing to be developed to accommodate sparse data and fill gaps in trait data (e.g. Swenson 2014; Schrodt et al. 2015; Santini et al. 2016).
Scale from the movement of individual seeds to population-level patterns of dispersal and recruitment using analytical approximations
Using analytical models developed from empirical data, we can explore alternative hypotheses regarding dispersal that can be tested in the field, make broadly applicable predictions that can be evaluated across systems and explore sensitivity to parameters (important when data are limited; Bullock et al. 2012). Results from these empirical studies enable the refinement of theoretical models. In cases where it would be infeasible or unethical to use empirical experimentation at the scales necessary to explore population dynamics, models can be used to evaluate competing hypotheses. Approximations require less data for parameterization and are efficient, and thus can help inform pressing management issues (Travis et al. 2011). Finally, these approximations can be included as submodels of more complex simulation models to reduce their complexity and data requirements and predict consequences of dispersal at larger organization, spatial or temporal scales.
Conduct sensitivity analyses of models to determine sufficiency of available data
Developing process-based models can guide effective data collection by determining the sensitivity of models to variation in parameters or structure (Milner-Gulland and Shea 2017). For example, parameters that are identified as being disproportionally important for determining plant responses will require more detailed data collection (e.g. Nathan et al. 2011b; Mokany et al. 2014). We can examine whether missing data or poorly parameterized values influence model output or produce contradictory patterns. Models can guide the choice of empirical sampling designs and appropriate statistical models by evaluating the sensitivity of results to different sampling designs and statistical models (virtual ecologist approach; Skarpaas et al. 2005; Zurell et al. 2010). Finally, this process will guide the development of methods and protocols for standardized data collection that can be included in both existing and new long-term studies. Standardized data collection efforts informed by theory will facilitate cross-site comparisons in both data analysis and model outputs, can help evaluate model predictions and will facilitate the investigation of future questions in seed dispersal ecology.
Create coordinated research networks and standardized data collection protocols to fill remaining data gaps
We encourage researchers to coordinate research activities and utilize a variety of empirical methods (e.g. censuses, seed traps, genetics, radio-tracking, remote sensing, etc.) to study a diversity of seed dispersal vectors and plant growth forms (woody plants, herbaceous plants, grasses, etc.) building upon existing standardized data collection protocols and global networks (e.g. Borer et al. 2014; Anderson-Teixeira et al. 2015; Saatkamp et al. 2019). A summary of data needs as identified by the participants of the Seed Dispersal Workshop is provided in Supporting Information —File S1. Empirical ecologists are able to generate important case study data on local processes occurring over short time periods that can serve as model systems for testing theory. We can use theory to examine whether and how information from case studies can be generalized across systems and extrapolated to larger organizational, spatial and temporal scales. In addition, coordinating and standardizing data collections can help overcome shortcomings in empirical studies to increase the number of focal species and the spatial and temporal scope. Often empirical ecologists are geographically scattered, and researchers working in tropical vs. temperate systems or Old World vs. New World systems are largely segregated—publishing in different journals and attending different conferences. Therefore, increasing international collaborations and global integration across regions will be necessary to enable generalization to ecosystems worldwide. Based on participant experiences described at the Seed Dispersal Workshop, there seems to be little communication among researchers studying abiotic vs. biotic dispersal vectors or among researchers working on biotic dispersal vectors, that is researchers working on endozoochoric, epizoochoric and seed-caching organisms. Linking these perspectives may advance our understanding of the importance of different dispersal vectors. In addition, we propose that closer collaborations among ecologists, mathematicians, hydrologists, atmospheric modellers and physicists exploring the movement of animals, water and wind will bring important insights to these efforts.
Predict consequences of dispersal over larger organization and spatiotemporal scales
System-specific forecasts will require the development and application of novel analytical and efficient computational methods for models. Computational models based on dispersal theory and parameterized with system-specific data hold promise for evaluating the importance of dispersal within ecosystems. Such generalizations may elucidate the qualitative and quantitative effects of species-specific dispersal kernels and disperser loss on plant populations. Collaboration and information sharing between empiricists, mathematicians, modellers and theoreticians may help address this challenge, by directing empirical data collection to efficiently address model parameter needs and by helping ecological modellers to incorporate relevant variables as they develop increasingly mechanistic models. These models can be evaluated with future empirical studies.
Conclusions
To tackle the complexity and context dependency of seed dispersal, we urge a better integration of empirical and theoretical approaches. This requires enhanced communication and collaboration across researchers in different disciplines, across geographic locations, and studying different aspects of plant life histories and environmental conditions that influence dispersal and demography. Existing models need to be further developed and refined to evaluate the role of dispersal on population persistence and spread; better predict extinction risk of species; and evaluate conservation and management strategies. Synthesis of data on dispersal processes, seed dispersal effectiveness across multiple life-history stages and demography represents an opportunity to develop theory for generalization across systems and to identify relevant processes that require model development and data collection for system-specific predictions.
Supporting Information
The following additional information is available in the online version of this article—
File S1. Summary of data needs.
Sources of Funding
Ideas for this manuscript initiated during the Seed Dispersal Workshop held in May 2016 at the Socio-Environmental Synthesis Center in Annapolis, MD and supported by the US National Science Foundation Grant DEB-1548194 to N.G.B. and the National Socio-Environmental Synthesis Center under the US National Science Foundation Grant DBI-1052875. D.Z. received funding from the Swiss National Science Foundation (SNF, grant: PZ00P3_168136/1) and from the German Science Foundation (DFG, grant: ZU 361/1-1).
Contributions by the Authors
N.G.B. led the development of the concepts, writing and revising of the manuscript with input from C.E.A. and H.S.R. All authors contributed to the development of concepts and are listed in order of contribution and alphabetical order within each level of contribution.
Conflict of Interest
None declared.
Acknowledgements
We thank the staff of the National Socio-Environmental Synthesis Center for logistical support for the workshop. We would also like to thank Anny Chung, Sarah Bogen, Binod Borah, Elsa Jos, Eric Sodja, Cole Carlson and Justin Tirrell for a helpful brainstorming session on figures and Kim McConkey and two anonymous reviewers for their constructive feedback.
Literature Cited
- Adler FR, Muller-Landau HC. 2005. When do localized natural enemies increase species richness? Ecology Letters 8:438–447. [Google Scholar]
- Agrawal AA. 1999. Induced responses to herbivory in wild radish: effects on several herbivores and plant fitness. Ecology 80:1713–1723. [Google Scholar]
- Agrawal AA, Ackerly DD, Adler F, Arnold AE, Cáceres C, Doak DF, Post E, Hudson PJ, Maron J, Mooney KA, Power M, Schemske D, Stachowicz J, Strauss S, Turner MG, Werner E. 2007. Filling key gaps in population and community ecology. Frontiers in Ecology and the Environment 5:145–152. [Google Scholar]
- Allan BM, Nimmo DG, Ierodiaconou D, VanDerWal J, Koh LP, Ritchie EG. 2018. Futurecasting ecological research: the rise of technoecology. Ecosphere 9:e02163. [Google Scholar]
- Alsos IG, Ehrich D, Eidesen PB, Solstad H, Westergaard KB, Schonswetter P, Tribsch A, Birkeland S, Elven R, Brochmann C. 2015. Long-distance plant dispersal to North Atlantic islands: colonization routes and founder effect. AoB Plants 7:plv036; doi: 10.1093/aobpla/plv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsos IG, Eidesen PB, Ehrich D, Skrede I, Westergaard K, Jacobsen GH, Landvik JY, Taberlet P, Brochmann C. 2007. Frequent long-distance plant colonization in the changing Arctic. Science 316:1606–1609. [DOI] [PubMed] [Google Scholar]
- Anderson-Teixeira KJ, Davies SJ, Bennett AC, Gonzalez-Akre EB, Muller-Landau HC, Wright SJ, Abu Salim K, Almeyda Zambrano AM, Alonso A, Baltzer JL, Basset Y, Bourg NA, Broadbent EN, Brockelman WY, Bunyavejchewin S, Burslem DF, Butt N, Cao M, Cardenas D, Chuyong GB, Clay K, Cordell S, Dattaraja HS, Deng X, Detto M, Du X, Duque A, Erikson DL, Ewango CE, Fischer GA, Fletcher C, Foster RB, Giardina CP, Gilbert GS, Gunatilleke N, Gunatilleke S, Hao Z, Hargrove WW, Hart TB, Hau BC, He F, Hoffman FM, Howe RW, Hubbell SP, Inman-Narahari FM, Jansen PA, Jiang M, Johnson DJ, Kanzaki M, Kassim AR, Kenfack D, Kibet S, Kinnaird MF, Korte L, Kral K, Kumar J, Larson AJ, Li Y, Li X, Liu S, Lum SK, Lutz JA, Ma K, Maddalena DM, Makana JR, Malhi Y, Marthews T, Mat Serudin R, McMahon SM, McShea WJ, Memiaghe HR, Mi X, Mizuno T, Morecroft M, Myers JA, Novotny V, de Oliveira AA, Ong PS, Orwig DA, Ostertag R, den Ouden J, Parker GG, Phillips RP, Sack L, Sainge MN, Sang W, Sri-Ngernyuang K, Sukumar R, Sun IF, Sungpalee W, Suresh HS, Tan S, Thomas SC, Thomas DW, Thompson J, Turner BL, Uriarte M, Valencia R, Vallejo MI, Vicentini A, Vrška T, Wang X, Wang X, Weiblen G, Wolf A, Xu H, Yap S, Zimmerman J. 2015. CTFS-ForestGEO: a worldwide network monitoring forests in an era of global change. Global Change Biology 21:528–549. [DOI] [PubMed] [Google Scholar]
- Aslan CE, Beckman NG, Rogers HS, Bronstein JL, Zurell D, Hartig F, Shea K, Pejchar L, Neubert MG, Poulsen JR, HillRisLambers J, Miriti MN, Loiselle BA, Effiom EO, Zambrano J, Schupp EW, Pufal G, Johnson J, Bullock JM, Brodie JF, Bruna EM, Cantrell RS, Decker R, Fricke EC, Gurski K, Hastings A, Kogan O, Powell JA, Razafindratsima OH, Sandor M, Schreiber SJ, Snell RS, Strickland C, Zhou Y. 2019. Employing plant functional groups to advance seed dispersal ecology and conservation. AoB Plants 11:plz006; doi: 10.1093/aobpla/plz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan CE, Zavaleta ES, Tershy B, Croll D. 2013. Mutualism disruption threatens global plant biodiversity: a systematic review. PLoS One 8:e66993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffret AG, Cousins SA. 2013. Humans as long-distance dispersers of rural plant communities. PLoS One 8:e62763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augspurger CK. 1986. Morphology and dispersal potential of wind-dispersed diaspores of Neotropical trees. American Journal of Botany 73:353–363. [Google Scholar]
- Augspurger CK, Kitajima K. 1992. Experimental studies of seedling recruitment from contrasting seed distributions. Ecology 73:1270–1284. [Google Scholar]
- Aylor DE. 2017. Aerial dispersal of pollen and spores. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Baldocchi D, Falge E, Gu L, Olson R, Hollinger D, Running S, Anthoni P, Bernhofer C, Davis K, Evans R, Fuentes J, Goldstein A, Katul G, Law B, Lee X, Malhi Y, Meyers T, Munger W, Oechel W, Paw U KT, Pilegaard K, Schmid HP, Valentini R, Verma S, Vesala T, Wilson K, Wofsy S. 2001. FLUXNET: a new tool to study the temporal and spatial variability of ecosystem-scale carbon dioxide, water vapor, and energy flux densities. Bulletin of the American Meteorological Society 82:2415–2434. [Google Scholar]
- Beckman NG, Aslan CE, Rogers HR. 2020. Introduction to the special issue: the role of seed dispersal in plant populations: perspectives and advances in a changing world. AoB Plants. 12:plaa010; doi: 10.1093/aobpla/plaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman NG, Neuhauser C, Muller-Landau HC. 2012. The interacting effects of clumped seed dispersal and distance- and density-dependent mortality on seedling recruitment patterns. Journal of Ecology 100:862–873. [Google Scholar]
- Beckman NG, Rogers HS. 2013. Consequences of seed dispersal for plant recruitment in tropical forests: interactions within the seedscape. Biotropica 45:666–681. [Google Scholar]
- Bohrer G, Katul GG, Nathan R, Walko RL, Avissar R. 2008. Effects of canopy heterogeneity, seed abscission and inertia on wind-driven dispersal kernels of tree seeds. Journal of Ecology 96:569–580. [Google Scholar]
- Borer ET, Harpole WS, Adler PB, Lind EM, Orrock JL, Seabloom EW, Smith MD. 2014. Finding generality in ecology: a model for globally distributed experiments. Methods in Ecology and Evolution 5:65–73. [Google Scholar]
- Börger L. 2016. EDITORIAL: stuck in motion? Reconnecting questions and tools in movement ecology. The Journal of Animal Ecology 85:5–10. [DOI] [PubMed] [Google Scholar]
- Bouchet F, Marston JB, Tangarife T. 2018. Fluctuations and large deviations of Reynolds stresses in zonal jet dynamics. Physics of Fluids 30:015110. [Google Scholar]
- Boyce MS, Haridas CV, Lee CT; The NSDWG 2006. Demography in an increasingly variable world. Trends in Ecology & Evolution 21:141–148. [DOI] [PubMed] [Google Scholar]
- Brinkman HC. 1949. A calculation of the viscous force exerted by a flowing fluid on a dense swarm of particles. Applied Scientific Research 1:27–34. [Google Scholar]
- Brodie JF, Helmy OE, Brockelman WY, Maron JL. 2009a. Bushmeat poaching reduces the seed dispersal and population growth rate of a mammal-dispersed tree. Ecological Applications 19:854–863. [DOI] [PubMed] [Google Scholar]
- Brodie JF, Helmy OE, Brockelman WY, Maron JL. 2009b. Functional differences within a guild of tropical mammalian frugivores. Ecology 90:688–698. [DOI] [PubMed] [Google Scholar]
- Bullock JM, Bonte D, Pufal G, da Silva Carvalho C, Chapman DS, García C, García D, Matthysen E, Delgado MM. 2018. Human-mediated dispersal and the rewiring of spatial networks. Trends in Ecology & Evolution 33:958–970. [DOI] [PubMed] [Google Scholar]
- Bullock JM, Galsworthy SJ, Manzano P, Poschlod P, Eichberg C, Walker K, Wichmann MC. 2011. Process-based functions for seed retention on animals: a test of improved descriptions of dispersal using multiple data sets. Oikos 120:1201–1208. [Google Scholar]
- Bullock JM, González LM, Tamme R, Götzenberger L, White SM, Pärtel M, Hooftman DAP. 2017. A synthesis of empirical plant dispersal kernels. Journal of Ecology 105:6–19. [Google Scholar]
- Bullock JM, Pywell RF, Coulson-Phillips SJ. 2008. Managing plant population spread: prediction and analysis using a simple model. Ecological Applications 18:945–953. [DOI] [PubMed] [Google Scholar]
- Bullock JM, White SM, Prudhomme C, Tansey C, Perea R, Hooftman DAP. 2012. Modelling spread of British wind-dispersed plants under future wind speeds in a changing climate. Journal of Ecology 100:104–115. [Google Scholar]
- Cantrell RS, Cosner C. 2004. Diffusion and ecological processes: modern perspectives. New York: John Wiley & Sons. [Google Scholar]
- Carlo T, Tewksbury J, Martínez del Río C. 2009. Tracking the fate of seedlings with a stable isotope. Ecology 91:3516–3525. [DOI] [PubMed] [Google Scholar]
- Caswell H. 1989. Analysis of life table response experiments I. Decomposition of effects on population growth rate. Ecological Modelling 46:221–237. [Google Scholar]
- Caswell H. 2001. Matrix population models. Construction, analysis, and interpretation, 2nd edn. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Caswell H. 2006. Sensitivity analysis of transient population dynamics. Ecology Letters 10:1–15. [DOI] [PubMed] [Google Scholar]
- Caswell H, Hunter CM, Neubert MG. 2011. Demography and dispersal: invasion speeds and sensitivity analysis in periodic and stochastic environments. Theoretical Ecology 4:407–421. [Google Scholar]
- Caswell H, Lensink R, Neubert MG. 2003. Demography and dispersal: life table response experiments for invasion speed. Ecology 84:1968–1978. [Google Scholar]
- Chamecki M, Meneveau C, Parlange MB. 2009. Large eddy simulation of pollen transport in the atmospheric boundary layer. Journal of Aerosol Science 40:241–255. [Google Scholar]
- Chapman D, Purse BV, Roy HE, Bullock JM. 2017. Global trade networks determine the distribution of invasive non-native species. Global Ecology and Biogeography 26:907–917. [Google Scholar]
- Clark JS, Beckage B, Camill P, Cleveland B, HillRisLambers J, Lichter J, McLachlan J, Mohan J, Wyckoff P. 1999a. Interpreting recruitment limitation in forests. American Journal of Botany 86:1–16. [PubMed] [Google Scholar]
- Clark JS, Fastie C, Hurtt G, Jackson ST, Johnson C, King GA, Lewis M, Lynch J, Pacala S, Prentice C, Schupp EW, Webb T, Wyckoff P. 1998. Reid’s paradox of rapid plant migration - dispersal theory and interpretation of paleoecological records. BioScience 48:13–24. [Google Scholar]
- Clark JS, Lewis M, Horvath L. 2001. Invasion by extremes: population spread with variation in dispersal and reproduction. The American Naturalist 157:537–554. [DOI] [PubMed] [Google Scholar]
- Clark CJ, Poulsen JR, Levey DJ, Osenberg CW. 2007. Are plant populations seed limited? A critique and meta-analysis of seed addition experiments. The American Naturalist 170:128–142. [DOI] [PubMed] [Google Scholar]
- Clark JS, Silman M, Kern R, Macklin E, Hille Ris Lambers J. 1999b. Seed dispersal near and far: patterns across temperate and tropical forests. Ecology 80:1475–1494. [Google Scholar]
- Clobert J, Baguette M, Benton T, Bullock J, eds. 2012. Dispersal ecology and evolution. Oxford, UK: Oxford University of Press. [Google Scholar]
- Codling EA, Plank MJ, Benhamou S. 2008. Random walk models in biology. Journal of the Royal Society, Interface 5:813–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Uriarte M. 2012. Integrating frugivore behavior and animal movement: A review of the evidence and implication for scaling seed dispersal. Biological Reviews 88:255–272. [DOI] [PubMed] [Google Scholar]
- Coulson SJ, Bullock JM, Stevenson MJ, Pywell RF. 2001. Colonization of grassland by sown species: dispersal versus microsite limitation in responses to management. Journal of Applied Ecology 38:204–216. [Google Scholar]
- Davies AB, Asner GP. 2014. Advances in animal ecology from 3D-LiDAR ecosystem mapping. Trends in Ecology & Evolution 29:681–691. [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Law R, Metz JA, eds. 2000. The geometry of ecological interactions: simplifying spatial complexity. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Donoso I, García D, Rodríguez-Pérez J, Martínez D. 2016. Incorporating seed fate into plant–frugivore networks increases interaction diversity across plant regeneration stages. Oikos 125:1762–1771. [Google Scholar]
- Easterling MR, Ellner SP, Dixon PM. 2000. Size-specific sensitivity: applying a new structured population model. Ecology 81:694–708. [Google Scholar]
- Eitel JUH, Höfle B, Vierling LA, Abellán A, Asner GP, Deems JS, Glennie CL, Joerg PC, LeWinter AL, Magney TS, Mandlburger G, Morton DC, Müller J, Vierling KT. 2016. Beyond 3-D: the new spectrum of lidar applications for earth and ecological sciences. Remote Sensing of Environment 186:372–392. [Google Scholar]
- Ellner SP, Rees M. 2006. Integral projection models for species with complex demography. The American Naturalist 167:410–428. [DOI] [PubMed] [Google Scholar]
- Ellner SP, Schreiber SJ. 2012. Temporally variable dispersal and demography can accelerate the spread of invading species. Theoretical Population Biology 82:283–298. [DOI] [PubMed] [Google Scholar]
- Elwood EC, Lichti NI, Fitzsimmons SF, Dalgleish HJ. 2018. Scatterhoarders drive long- and short-term population dynamics of a nut-producing tree, while pre-dispersal seed predators and herbivores have little effect. Journal of Ecology 106:1191–1203. [Google Scholar]
- Enquist BJ, Condit R, Peet RK, Schildhauer M, Thiers BM. 2016. Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Preprints 4:e2615v2. [Google Scholar]
- Eriksen RL, Hierro JL, Eren Ö, Andonian K, Török K, Becerra PI, Montesinos D, Khetsuriani L, Diaconu A, Kesseli R. 2014. Dispersal pathways and genetic differentiation among worldwide populations of the invasive weed Centaurea solstitialis L. (Asteraceae). PLoS One 9:e114786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada A, Fleming TH, eds. 1986. Frugivores and seed dispersal. Dordrecht, The Netherlands: Springer. [Google Scholar]
- Evans MR. 2012. Modelling ecological systems in a changing world. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MR, Grimm V, Johst K, Knuutila T, de Langhe R, Lessells CM, Merz M, O’Malley MA, Orzack SH, Weisberg M, Wilkinson DJ, Wolkenhauer O, Benton TG. 2013. Do simple models lead to generality in ecology? Trends in Ecology & Evolution 28:578–583. [DOI] [PubMed] [Google Scholar]
- Ferrandino FJ. 1993. Dispersive epidemic waves: I. Focus expansion within a linear planting. Phytopathology 83:795–802. [Google Scholar]
- Fleming CH, Calabrese JM, Mueller T, Olson KA, Leimgruber P, Fagan WF. 2014. From fine-scale foraging to home ranges: a semivariance approach to identifying movement modes across spatiotemporal scales. The American Naturalist 183:E154–E167. [DOI] [PubMed] [Google Scholar]
- Fricke EC, Bender J, Rehm EM, Rogers HS. 2018. Functional outcomes of mutualistic network interactions: a community-scale study of frugivore gut passage on germination. Journal of Ecology 107:757–767. [Google Scholar]
- Gaines SD, Denny MW. 1993. The largest, smallest, highest, lowest, longest, and shortest: extremes in ecology. Ecology 74:1677–1692. [Google Scholar]
- García C, Borda-de-Água L, Rees M. 2017. Extended dispersal kernels in a changing world: insights from statistics of extremes. Journal of Ecology 105:63–74. [Google Scholar]
- Gilbert MA, White SM, Bullock JM, Gaffney EA. 2014. Spreading speeds for stage structured plant populations in fragmented landscapes. Journal of Theoretical Biology 349:135–149. [DOI] [PubMed] [Google Scholar]
- Godinez-Alvarez H, Jordano P. 2007. An empirical approach to analysing the demographic consequences of seed dispersal by frugivores. In: Dennis A, Schupp E, Green R, Westcott DA, eds. Seed dispersal: theory and its application in a changing world. Wallingford, UK: CAB Iternational, 391–406. [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, Muir A, Merchant N, Lowry S, Mock S, Helmke M, Kubach A, Narro M, Hopkins N, Micklos D, Hilgert U, Gonzales M, Jordan C, Skidmore E, Dooley R, Cazes J, McLay R, Lu Z, Pasternak S, Koesterke L, Piel WH, Grene R, Noutsos C, Gendler K, Feng X, Tang C, Lent M, Kim SJ, Kvilekval K, Manjunath BS, Tannen V, Stamatakis A, Sanderson M, Welch SM, Cranston KA, Soltis P, Soltis D, O’Meara B, Ane C, Brutnell T, Kleibenstein DJ, White JW, Leebens-Mack J, Donoghue MJ, Spalding EP, Vision TJ, Myers CR, Lowenthal D, Enquist BJ, Boyle B, Akoglu A, Andrews G, Ram S, Ware D, Stein L, Stanzione D. 2011. The iPlant collaborative: cyberinfrastructure for plant biology. Frontiers in Plant Science 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Varo JP, Arroyo JM, Jordano P, Gilbert M. 2014. Who dispersed the seeds? The use of DNA barcoding in frugivory and seed dispersal studies. Methods in Ecology and Evolution 5:806–814. [Google Scholar]
- Grimm V, Railsback SF. 2005. Individual-based modeling and ecology. Princeton, NJ: Princeton University Press. [Google Scholar]
- Grimm V, Railsback SF. 2011. Pattern-oriented modelling: a ‘multi-scope’ for predictive systems ecology. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbel E. 1958. Statistics of extremes. New York: Columbia University Press. [Google Scholar]
- Gutschick VP, BassiriRad H. 2003. Extreme events as shaping physiology, ecology, and evolution of plants: toward a unified definition and evaluation of their consequences. New Phytologist 160:21–42. [DOI] [PubMed] [Google Scholar]
- Guttal V, Bartumeus F, Hartvigsen G, Nevai AL. 2011. Retention time variability as a mechanism for animal mediated long-distance dispersal. PLoS One 6:e28447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty BD. 2007. How far do offspring recruit from parent plants? A molecular approach to understanding effective dispersal. In: Dennis A, Schupp E, Green R, Westcott DA, eds. Seed dispersal: theory and its application in a changing world. Wallingford, UK: CAB International, 277–299. [Google Scholar]
- Harsch MA, Zhou Y, HilleRisLambers J, Kot M. 2014. Keeping pace with climate change: stage-structured moving-habitat models. The American Naturalist 184:25–37. [DOI] [PubMed] [Google Scholar]
- Hartig F, Calabrese JM, Reineking B, Wiegand T, Huth A. 2011. Statistical inference for stochastic simulation models–theory and application. Ecology Letters 14:816–827. [DOI] [PubMed] [Google Scholar]
- Hartig F, Dyke J, Hickler T, Higgins SI, O’Hara RB, Scheiter S, Huth A. 2012. Connecting dynamic vegetation models to data - an inverse perspective. Journal of Biogeography 39:2240–2252. [Google Scholar]
- Hastings A, Cuddington K, Davies KF, Dugaw CJ, Elmendorf S, Freestone A, Harrison S, Holland M, Lambrinos J, Malvadkar U, Melbourne BA, Moore K, Taylor C, Thomson D. 2005. The spatial spread of invasions: new developments in theory and evidence. Ecology Letters 8:91–101. [Google Scholar]
- HilleRisLambers J, Ettinger AK, Ford KR, Haak DC, Horwith M, Miner BE, Rogers HS, Sheldon KS, Tewksbury JJ, Waters SM, Yang S. 2013. Accidental experiments: ecological and evolutionary insights and opportunities derived from global change. Oikos 122:1649–1661. [Google Scholar]
- Hintze C, Heydel F, Hoppe C, Cunze S, König A, Tackenberg O. 2013. D3: the dispersal and diaspore database - baseline data and statistics on seed dispersal. Perspectives in Plant Ecology Evolution and Systematics 15:180–192. [Google Scholar]
- Hirsch BT, Kays R, Jansen PA. 2012. A telemetric thread tag for tracking seed dispersal by scatter-hoarding rodents. Plant Ecology 213:933–943. [Google Scholar]
- Howe HF, Miriti M. 2004. When seed dispersal matters. BioScience 54:651–660. [Google Scholar]
- Howe HF, Smallwood J. 1982. The ecology of seed dispersal. Annual Review of Ecology and Systematics 13:201–228. [Google Scholar]
- Isard SA, Gage SH. 2001. Flow of life in the atmosphere: an airscape approach to invasive organisms. East Lansing, MI: Michigan State University Press. [Google Scholar]
- Jansen PA, Hirsch BT, Emsens WJ, Zamora-Gutierrez V, Wikelski M, Kays R. 2012. Thieving rodents as substitute dispersers of megafaunal seeds. Proceedings of the National Academy of Sciences of the United States of America 109:12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JS, Gaddis KD, Cairns DM, Konganti K, Krutovsky KV. 2017. Landscape genomic insights into the historic migration of mountain hemlock in response to Holocene climate change. American Journal of Botany 104:439–450. [DOI] [PubMed] [Google Scholar]
- Jones FA, Muller-Landau HC. 2008. Measuring long-distance seed dispersal in complex natural environments: an evaluation and integration of classical and genetic methods. Journal of Ecology 96:642–652. [Google Scholar]
- Jongejans E, Shea K, Skarpaas O, Kelly D, Ellner SP. 2011. Importance of individual and environmental variation for invasive species spread: a spatial integral projection model. Ecology 92:86–97. [DOI] [PubMed] [Google Scholar]
- Jongejans E, Skarpaas O, Shea K. 2008. Dispersal, demography and spatial population models for conservation and control management. Perspectives in Plant Ecology Evolution and Systematics 9:153–170. [Google Scholar]
- Jordano P. 2017. What is long-distance dispersal? And a taxonomy of dispersal events. Journal of Ecology 105:75–84. [Google Scholar]
- Jordano P, Garcia C, Godoy JA, García-Castaño JL. 2007. Differential contribution of frugivores to complex seed dispersal patterns. Proceedings of the National Academy of Sciences of the United States of America 104:3278–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattge J, Díaz S, Lavorel S, Prentice IC, Leadley P, Bönisch G, Garnier E, Westoby M, Reich PB, Wright IJ, Cornelissen JHC, Violle C, Harrison SP, Van Bodegom PM, Reichstein M, Enquist BJ, Soudzilovskaia NA, Ackerly DD, Anand M, Atkin O, Bahn M, Baker TR, Baldocchi D, Bekker R, Blanco CC, Blonder B, Bond WJ, Bradstock R, Bunker DE, Casanoves F, Cavender-Bares J, Chambers JQ, Chapin Iii FS, Chave J, Coomes D, Cornwell WK, Craine JM, Dobrin BH, Duarte L, Durka W, Elser J, Esser G, Estiarte M, Fagan WF, Fang J, Fernández-Méndez F, Fidelis A, Finegan B, Flores O, Ford H, Frank D, Freschet GT, Fyllas NM, Gallagher RV, Green WA, Gutierrez AG, Hickler T, Higgins SI, Hodgson JG, Jalili A, Jansen S, Joly CA, Kerkhoff AJ, Kirkup D, Kitajima K, Kleyer M, Klotz S, Knops JMH, Kramer K, Kühn I, Kurokawa H, Laughlin D, Lee TD, Leishman M, Lens F, Lenz T, Lewis SL, Lloyd J, Llusiá J, Louault F, Ma S, Mahecha MD, Manning P, Massad T, Medlyn BE, Messier J, Moles AT, Müller SC, Nadrowski K, Naeem S, Niinemets Ü, Nöllert S, Nüske A, Ogaya R, Oleksyn J, Onipchenko VG, Onoda Y, Ordoñez J, Overbeck G, Ozinga WA, Patiño S, Paula S, Pausas JG, Peñuelas J, Phillips OL, Pillar V, Poorter H, Poorter L, Poschlod P, Prinzing A, Proulx R, Rammig A, Reinsch S, Reu B, Sack L, Salgado-Negret B, Sardans J, Shiodera S, Shipley B, Siefert A, Sosinski E, Soussana JF, Swaine E, Swenson N, Thompson K, Thornton P, Waldram M, Weiher E, White M, White S, Wright SJ, Yguel B, Zaehle S, Zanne AE, Wirth C. 2011. TRY - a global database of plant traits. Global Change Biology 17:2905–2935. [Google Scholar]
- Katul GG, Poggi D. 2012. The effects of gentle topographic variation on dispersal kernels of inertial particles. Geophysical Research Letters 39:L03401. [Google Scholar]
- Katul GG, Porporato A, Nathan R, Siqueira M, Soons MB, Poggi D, Horn HS, Levin SA. 2005. Mechanistic analytical models for long-distance seed dispersal by wind. The American Naturalist 166:368–381. [DOI] [PubMed] [Google Scholar]
- Kays R, Crofoot MC, Jetz W, Wikelski M. 2015. Terrestrial animal tracking as an eye on life and planet. Science 348:aaa2478. [DOI] [PubMed] [Google Scholar]
- Kendrick GA, Waycott M, Carruthers TJB, Cambridge ML, Hovey R, Krauss SL, Lavery PS, Les DH, Lowe RJ, Vidal OMi, Ooi JLS, Orth RJ, Rivers DO, Ruiz-Montoya L, Sinclair EA, Statton J, van Dijk JK, Verduin JJ. 2012. The central role of dispersal in the maintenance and persistence of seagrass populations. BioScience 62:56–65. [Google Scholar]
- Kerr JT, Ostrovsky M. 2003. From space to species: ecological applications for remote sensing. Trends in Ecology & Evolution 18:299–305. [Google Scholar]
- Kleyer M, Bekker RM, Knevel IC, Bakker JP, Thompson K, Sonnenschein M, Poschlod P, Van Groenendael JM, Klimes L, Klimesovâ J, Klotz S, Rusch GM, Hermy M, Adriaens D, Boedeltje G, Bossuyt B, Dannemann A, Endels P, Götzenberger L, Hodgson JG, Jackel A-K, Kühn I, Kunzmann D, Ozinga WA, Römermann C, Stadler M, Schlegelmilch J, Steendam HJ, Tackenberg O, Wilmann B, Cornelissen JHC, Erickson O, Garnier E, Peco B. 2008. The LEDA traitbase: a database of life-history traits of Northwest European flora. Journal of Ecology 96:1266–1274. [Google Scholar]
- Kleyheeg E, Treep J, Jager M, Nolet BA, Soons MB. 2017. Seed dispersal distributions resulting from landscape-dependent daily movement behaviour of a key vector species, Anas platyrhynchos. Journal of Ecology 105:1279–1289. [Google Scholar]
- Kot M, Lewis M, van den Driessche P. 1996. Dispersal data and the spread of invading organisms. Ecology 77:2027–2042. [Google Scholar]
- Kot M, Schaefer HM. 1986. Discrete-time growth-dispersal models. Mathematical Biosciences 80:109–136. [Google Scholar]
- Kranstauber B, Cameron A, Weinzerl R, Fountain T, Tilak S, Wikelski M, Kays R. 2011. The Movebank data model for animal tracking. Environmental Modelling & Software 26:834–835. [Google Scholar]
- Kuparinen A. 2006. Mechanistic models for wind dispersal. Trends in Plant Science 11:296–301. [DOI] [PubMed] [Google Scholar]
- Laurie J, Bouchet F. 2015. Computation of rare transitions in barotropic quasi-geostropic equations. New Journal of Physics 17:015009–015033. [Google Scholar]
- Lefsky MA, Cohen WB, Parker GG, Harding DJ. 2002. Lidar remote sensing for ecosystem studies. BioScience 52:19–30. [Google Scholar]
- Lehsten V, Mischurow M, Lindström E, Lehsten D, Lischke H. 2019. LPJ-GM 1.0: simulating migration efficiently in a dynamic vegetation model. Geoscientific Model Development Discussion 12:893–908. [Google Scholar]
- Levey DJ, Silva W, Galetti M, eds. 2002. Seed dispersal and frugivory: ecology, evolution, and conservation. New York: CAB International. [Google Scholar]
- Levin SA, Muller-Landau HC, Nathan R, Chave J. 2003. The ecology and evolution of seed dispersal: a theoretical perspective. Annual Review of Ecology, Evolution, and Systematics 34:575–604. [Google Scholar]
- Levine J, Murrell D. 2003. The community-level consequences of seed dispersal patterns. Annual Review of Ecology, Evolution, and Systematics 34:549–574. [Google Scholar]
- Levins R. 1966. The strategy of model building in population biology. American Scientist 54:421–431. [Google Scholar]
- Lewis MA, Neubert MG, Caswell H, Clark JS, Shea K. 2006. A guide to calculating discrete-time invasion rates from data. In: Cadotte MW, McMahon SM, Fukami T, eds. Conceptual ecology and invasion biology: reciprocal approaches to nature. Dordrecht, The Netherlands: Springer, 169–192. [Google Scholar]
- Loayza AP, Knight T. 2010. Seed dispersal by pulp consumers, not “legitimate” seed dispersers, increases Guettarda viburnoides population growth. Ecology 91:2684–2695. [DOI] [PubMed] [Google Scholar]
- Lockwood JL, Hoopes MF, Marchetti MP. 2013. Invasion ecology. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Maciel GA, Lutscher F. 2013. How individual movement response to habitat edges affects population persistence and spatial spread. The American Naturalist 182:42–52. [DOI] [PubMed] [Google Scholar]
- Markl JS, Schleuning M, Forget PM, Jordano P, Lambert JE, Traveset A, Wright SJ, Böhning-Gaese K. 2012. Meta-analysis of the effects of human disturbance on seed dispersal by animals. Conservation Biology: the Journal of the Society for Conservation Biology 26:1072–1081. [DOI] [PubMed] [Google Scholar]
- Marquet PA, Allen AP, Brown JH, Dunne JA, Enquist BJ, Gillooly JF, Gowaty PA, Green JL, Harte J, Hubbell SP, O’Dwyer J, Okie JG, Ostling A, Ritchie M, Storch D, West GB. 2014. On theory in ecology. BioScience 64:701–710. [Google Scholar]
- Merchant N, Lyons E, Goff S, Vaughn M, Ware D, Micklos D, Antin P. 2016. The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biology 14:e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AW, Ruiz GM. 2014. Arctic shipping and marine invaders. Nature Climate Change 4:413–416. [Google Scholar]
- Milner-Gulland EJ, Shea K. 2017. Embracing uncertainty in applied ecology. The Journal of Applied Ecology 54:2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokany K, Prasad S, Westcott DA. 2014. Loss of frugivore seed dispersal services under climate change. Nature Communications 5:3971. [DOI] [PubMed] [Google Scholar]
- Nathan R. 2006. Long-distance dispersal of plants. Science 313:786–788. [DOI] [PubMed] [Google Scholar]
- Nathan R, Getz WM, Revilla E, Holyoak M, Kadmon R, Saltz D, Smouse PE. 2008. A movement ecology paradigm for unifying oragnismal movement research. Proceedings of the National Academy of Sciences of the United States of America 105:19052–19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R, Horvitz N, He Y, Kuparinen A, Schurr FM, Katul GG. 2011a. Spread of North American wind-dispersed trees in future environments. Ecology Letters 14:211–219. [DOI] [PubMed] [Google Scholar]
- Nathan R, Katul GG, Bohrer G, Kuparinen A, Soons M, Thompson SE, Trakhtenbrot A, Horn H. 2011b. Mechanistic models of seed dispersal by wind. Theoretical Ecology 4:113–132. [Google Scholar]
- Nathan R, Klein E, Robledo-Arnuncio JJ, Revilla E. 2012. Dispersal kernels: a review. In: Clobert J, Baguette M, Benton TG, Bullock JM, eds. Dispersal ecology and evolution. Oxford, UK: Oxford University of Press, 187–210. [Google Scholar]
- Nathan R, Muller-Landau HC. 2000. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends in Ecology & Evolution 15:278–285. [DOI] [PubMed] [Google Scholar]
- Neubert MG, Caswell H. 2000. Demography and dispersal: calculation and sensitivity analysis of invasion speed for structured populations. Ecology 81:1613–1628. [Google Scholar]
- Okubo A, Levin SA. 1989. A theoretical framework for data analysis of wind dispersal of seeds and pollen. Ecology 70:329–338. [Google Scholar]
- Okubo A, Levin SA. 2002. Diffusion and ecological processes: modern perspectives. New York: Springer-Verlag. [Google Scholar]
- Ovaskainen O, Meerson B. 2010. Stochastic models of population extinction. Trends in Ecology & Evolution 25:643–652. [DOI] [PubMed] [Google Scholar]
- Pauchard A, Shea K. 2006. Integrating the study of non-native plant invasions across spatial scales. Biological Invasions 8:399–413. [Google Scholar]
- Pettorelli N, Laurance WF, O’Brien TG, Wegmann M, Nagendra H, Turner W. 2014. Satellite remote sensing for applied ecologists: opportunities and challenges. Journal of Applied Ecology 51:839–848. [Google Scholar]
- Pigot AL, Bregman T, Sheard C, Daly B, Etienne RS, Tobias JA. 2016. Quantifying species contributions to ecosystem processes: a global assessment of functional trait and phylogenetic metrics across avian seed-dispersal networks. Proceedings of the Royal Society B: Biological Sciences 283:20161597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires MM, Guimarães PR, Galetti M, Jordano P. 2018. Pleistocene megafaunal extinctions and the functional loss of long-distance seed-dispersal services. Ecography 41:153–163. [Google Scholar]
- Poulsen JR, Clark CJ, Bolker BM. 2012. Experimental manipulation of seed shadows of an afrotropical tree determines drivers of recruitment. Ecology 93:500–510. [DOI] [PubMed] [Google Scholar]
- Powell JA, Zimmermann NE. 2004. Multiscale analysis of active seed dispersal contributes to resolving Reid’s paradox. Ecology 85:490–506. [Google Scholar]
- Ribbens E, Silander JA, Pacala SW. 1994. Seedling recruitment in forests: calibrating models to predict patterns of tree seedling dispersion. Ecology 75:1794–1806. [Google Scholar]
- Rogers HS, Beckman NG, Hartig F, Johnson JS, Pufal G, Shea K, Zurell D, Bullock JM, Cantrell RS, Loiselle B, Pejchar L, Razafindratsima OH, Sandor ME, Schupp EW, Strickland WC, Zambrano J. 2019. The total dispersal kernel: a review and future directions. AoB Plants 11:plz042; doi: 10.1093/aobpla/plz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal Botanic Gardens Kew 2016. Seed Information Database (SID). http://data.kew.org/sid/ (August 2016). [Google Scholar]
- Rundel PW, Graham EA, Allen MF, Fisher JC, Harmon TC. 2009. Environmental sensor networks in ecological research. The New Phytologist 182:589–607. [DOI] [PubMed] [Google Scholar]
- Saastamoinen M, Bocedi G, Cote J, Legrand D, Guillaume F, Wheat CW, Fronhofer EA, Garcia C, Henry R, Husby A, Baguette M, Bonte D, Coulon A, Kokko H, Matthysen E, Niitepõld K, Nonaka E, Stevens VM, Travis JMJ, Donohue K, Bullock JM, Del Mar Delgado M. 2018. Genetics of dispersal. Biological Reviews of the Cambridge Philosophical Society 93:574–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatkamp A, Cochrane A, Commander L, Guja LK, Jimenez-Alfaro B, Larson J, Nicotra A, Poschlod P, Silveira FAO, Cross AT, Dalziell EL, Dickie J, Erickson TE, Fidelis A, Fuchs A, Golos PJ, Hope M, Lewandrowski W, Merritt DJ, Miller BP, Miller RG, Offord CA, Ooi MKJ, Satyanti A, Sommerville KD, Tangney R, Tomlinson S, Turner S, Walck JL. 2019. A research agenda for seed-trait functional ecology. The New Phytologist 221:1764–1775. [DOI] [PubMed] [Google Scholar]
- Salguero-Gómez R, Jones OR, Archer CR, Buckley YM, Che-Castaldo J, Caswell H, Hodgson D, Scheuerlein A, Conde DA, Brinks E, de Buhr H, Farack C, Gottschalk F, Hartmann A, Henning A, Hoppe G, Römer G, Runge J, Ruoff T, Wille J, Zeh S, Davison R, Vieregg D, Baudisch A, Altwegg R, Colchero F, Dong M, de Kroon H, Lebreton J-D, Metcalf CJE, Neel MM, Parker IM, Takada T, Valverde T, Vélez-Espino LA, Wardle GM, Franco M, Vaupel JW. 2015. The COMPADRE Plant Matrix Database: an open online repository for plant demography. Journal of Ecology 103:202–218. [Google Scholar]
- Santini L, Cornulier T, Bullock JM, Palmer SCF, White SM, Bocedi G, Rondinini C, Travis JMJ. 2016. Modelling spread rate in terrestrial mammals and the ability to track a shifting climate: a trait space approach. Global Change Biology 22:2415–2424. [DOI] [PubMed] [Google Scholar]
- Sato H, Itoh A, Kohyama T. 2007. SEIB-DGVM: A new dynamic global vegetation model using a spatially explicit individual-based approach. Ecological Modelling 200:279–307. [Google Scholar]
- Schreiber SJ, Ryan ME. 2011. Invasion speeds of structured populations in fluctuating environments. Theoretical Ecology 4:423–434. [Google Scholar]
- Schreiber SJ, Beckman NG. 2020. Individual variation in dispersal and fecundity increases rates of spatial spread. AoB Plants. 12:plaa001; doi: 10.1093/aobpla/plaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodt F, Kattge J, Shan H, Fazayeli F, Joswig J, Banerjee A, Reichstein M, Bönisch G, Díaz S, Dickie J, Gillison A, Karpatne A, Lavorel S, Leadley P, Wirth CB, Wright IJ, Wright SJ, Reich PB. 2015. BHPMF – a hierarchical Bayesian approach to gap-filling and trait prediction for macroecology and functional biogeography. Global Ecology and Biogeography 24:1510–1521. [Google Scholar]
- Schupp EW. 1993. Quantity, quality, and the effectiveness of seed dispersal by animals. Vegetatio 107/108:15–29. [Google Scholar]
- Schupp E. 2007. The suitability of a site for seed dispersal is context-dependent. In: Dennis A, Schupp E, Green R, Westcott DA, eds. Seed dispersal: theory and its application in a changing world. Wallingford, UK: CAB International, 445–462. [Google Scholar]
- Schupp EW, Fuentes M. 1995. Spatial patterns of seed dispersal and the unification of plant-population ecology. Ecoscience 2:267–275. [Google Scholar]
- Schupp EW, Jordano P, Gómez JM. 2010. Seed dispersal effectiveness revisited: a conceptual review. The New Phytologist 188:333–353. [DOI] [PubMed] [Google Scholar]
- Schupp EW, Jordano P, Gómez JM. 2017. A general framework for effectiveness concepts in mutualisms. Ecology Letters 20:577–590. [DOI] [PubMed] [Google Scholar]
- Schupp EW, Zwolak R, Jones LR, Snell RS, Beckman NG, Aslan C, Cavazos BR, Effiom E, Fricke EC, Montaño-Centellas F, Poulsen J, Razafindratsima OH, Sandor ME, Shea K. 2019. Intrinsic and extrinsic drivers of intraspecific variation in seed dispersal are diverse and pervasive. AoB Plants 11:plz067; doi: 10.1093/aobpla/plz067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E. 2018. Modelling in population biology. In: Methodological investigations in agent-based modelling: with applications for the social sciences. Cham, Switzerland: Springer International Publishing, 61–81. [Google Scholar]
- Simmons BI, Sutherland WJ, Dicks LV, Albrecht J, Farwig N, García D, Jordano P, González-Varo JP. 2018. Moving from frugivory to seed dispersal: Incorporating the functional outcomes of interactions in plant-frugivore networks. The Journal of Animal Ecology 87:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarpaas O, Shea K. 2007. Dispersal patterns, dispersal mechanisms and invasion wave speeds for Carduus thistles. American Naturalist 170:421–430. [DOI] [PubMed] [Google Scholar]
- Skarpaas O, Shea K, Bullock JM. 2005. Optimizing dispersal study design by Monte Carlo simulation. Journal of Applied Ecology 42:731–739. [Google Scholar]
- Snell RS. 2014. Simulating long distance seed dispersal in a dynamic vegetation model. Global Ecology and Biogeography 23:89–98. [Google Scholar]
- Snell RS, Beckman NG, Fricke E, Loiselle BA, Carvalho CS, Jones LR, Lichti NI, Lustenhouwer N, Schreiber SJ, Strickland C, Sullivan LL, Cavazos BR, Giladi I, Hastings A, Holbrook KM, Jongejans E, Kogan O, Montaño-Centellas F, Rudolph J, Rogers HS, Zwolak R, Schupp EW. 2019. Consequences of intraspecific variation in seed dispersal for plant demography, communities, evolution, and global change. AoB Plants 11:plz016; doi: 10.1093/aobpla/plz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell RS, Cowling SA. 2015. Consideration of dispersal processes and northern refugia can improve our understanding of past plant migration rates in North America. Journal of Biogeography 42:1677–1688. [Google Scholar]
- Snell RS, Huth A, Nabel JEMS, Bocedi G, Travis JMJ, Gravel D, Bugmann H, Gutiérrez AG, Hickler T, Higgins SI, Reineking B, Scherstjanoi M, Zurbriggen N, Lischke H. 2014. Using dynamic vegetation models to simulate plant range shifts. Ecography 37:1184–1197. [Google Scholar]
- Soons MB, Heil GW, Nathan R, Katul GG. 2004. Determinants of long-distance seed dispersal by wind in grasslands. Ecology 85:3056–3068. [Google Scholar]
- Strickland C, Dangelmayr G, Shipman PD, Kumar S, Stohlgren TJ. 2015. Network spread of invasive species and infectious diseases. Ecological Modelling 309–310:1–9. [Google Scholar]
- Strickland C, Miller L, Santhanakrishnan A, Hamlet C, Battista N, Pasour V. 2017. Three-dimensional low reynolds number flows near biological filtering and protective layers. Fluids 2:62. [Google Scholar]
- Sun Z, Lorscheid I, Millington JD, Lauf S, Magliocca NR, Groeneveld J, Balbi S, Nolzen H, Müller B, Schulze J, Buchmann CM. 2016. Simple or complicated agent-based models? A complicated issue. Environmental Modelling & Software 86:56–67. [Google Scholar]
- Swenson NG. 2014. Phylogenetic imputation of plant functional trait databases. Ecography 37:105–110. [Google Scholar]
- Tamme R, Götzenberger L, Zobel M, Bullock JM, Hooftman DA, Kaasik A, Pärtel M. 2014. Predicting species’ maximum dispersal distances from simple plant traits. Ecology 95:505–513. [DOI] [PubMed] [Google Scholar]
- Taylor K, Brummer T, Taper ML, Wing A, Rew LJ. 2012. Human-mediated long-distance dispersal: an empirical evaluation of seed dispersal by vehicles. Diversity and Distributions 18:942–951. [Google Scholar]
- Teller BJ, Zhang R, Shea K. 2016. Seed release in a changing climate: initiation of movement increases spread of an invasive species under simulated climate warming. Diversity and Distributions 22:708–716. [Google Scholar]
- Templ B, Koch E, Bolmgren K, Ungersböck M, Paul A, Scheifinger H, Rutishauser T, Busto M, Chmielewski FM, Hájková L, Hodzić S, Kaspar F, Pietragalla B, Romero-Fresneda R, Tolvanen A, Vučetič V, Zimmermann K, Zust A. 2018. Pan European Phenological database (PEP725): a single point of access for European data. International Journal of Biometeorology 62:1109–1113. [DOI] [PubMed] [Google Scholar]
- Terborgh J, Zhu K, Alvarez-Loayza P, Cornejo Valverde F. 2014. How many seeds does it take to make a sapling? Ecology 95:991–999. [DOI] [PubMed] [Google Scholar]
- Trakhtenbrot A, Katul GG, Nathan R. 2014. Mechanistic modeling of seed dispersal by wind over hilly terrain. Ecological Modelling 274:29–40. [Google Scholar]
- Travis JMJ, Delgado M, Bocedi G, Baguette M, Bartoń K, Bonte D, Boulangeat I, Hodgson JA, Kubisch A, Penteriani V, Saastamoinen M, Stevens VM, Bullock JM. 2013. Dispersal and species’ responses to climate change. Oikos 122:1532–1540. [Google Scholar]
- Travis JMJ, Harris CM, Park KJ, Bullock JM. 2011. Improving prediction and management of range expansions by combining analytical and individual-based modelling approaches. Methods in Ecology and Evolution 2:477–488. [Google Scholar]
- Turchin P. 1998. Quantitative analysis of movement: measuring and modeling population redistribution in animals and plants. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Turnbull LA, Crawley MJ, Rees M. 2000. Are plant populations seed limited? A review of seed sowing experiments. Oikos 88:225–238. [Google Scholar]
- USA National Phenology Network 2018. Plant and animal phenology data. Tucson, AZ: USA-NPN. [Google Scholar]
- van der Heijden MG. 2010. Mycorrhizal fungi reduce nutrient loss from model grassland ecosystems. Ecology 91:1163–1171. [DOI] [PubMed] [Google Scholar]
- Vellend M. 2010. Conceptual synthesis in community ecology. The Quarterly Review of Biology 85:183–206. [DOI] [PubMed] [Google Scholar]
- Vellend M, Knight TM, Drake JM. 2006. Antagonistic effects of seed dispersal and herbivory on plant migration. Ecology Letters 9:319–326. [DOI] [PubMed] [Google Scholar]
- von der Lippe M, Kowarik I. 2008. Do cities export biodiversity? Traffic as dispersal vector across urban-rural gradients. Diversity and Distributions 14:18–25. [Google Scholar]
- Wang BC, Smith T. 2002. Closing the seed dispersal loop. Trends in Ecology & Evolution 17:379–385. [Google Scholar]
- Wichmann MC, Alexander MJ, Soons M, Galsworthy SJ, Dunne L, Gould R, Fairfax C, Niggemann M, Hails RS, Bullock JM. 2009. Human mediated dispersal of seeds over long distances. Proceedings of the Royal Society B-Biological Sciences 276:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand T, Martínez I, Huth A. 2009. Recruitment in tropical tree species: revealing complex spatial patterns. The American Naturalist 174:E106–E140. [DOI] [PubMed] [Google Scholar]
- Wilkerson J, Casas A. 2017. Large-scale computerized text analysis in political science: opportunities and challenges. Annual Review of Political Science 20:529–544. [Google Scholar]
- Wilmers CC, Nickel B, Bryce CM, Smith JA, Wheat RE, Yovovich V. 2015. The golden age of bio-logging: how animal-borne sensors are advancing the frontiers of ecology. Ecology 96:1741–1753. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Hardisty DJ, Epanchin-Niell RS, Runge MC, Cottingham KL, Urban DL, Maguire LA, Hastings A, Mumby PJ, Peters DP. 2016. A typology of time-scale mismatches and behavioral interventions to diagnose and solve conservation problems. Conservation Biology: the Journal of the Society for Conservation Biology 30:42–49. [DOI] [PubMed] [Google Scholar]
- Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM. 2013. Three keys to the radiation of angiosperms into freezing environments. Nature 506:89. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kot M. 2011. Discrete-time growth-dispersal models with shifting species ranges. Theoretical Ecology 4:13–25. [Google Scholar]
- Zuidema PA, Jongejans E, Chien PD, During HJ, Schieving F. 2010. Integral Projection Models for trees: a new parameterization method and a validation of model output. Journal of Ecology 98:345–355. [Google Scholar]
- Zurell D, Berger U, Cabral JS, Jeltsch F, Meynard CN, Münkemüller T, Nehrbass N, Pagel J, Reineking B, Schröder B, Grimm V. 2010. The virtual ecologist approach: simulating data and observers. Oikos 119:622–635. [Google Scholar]
- Zwolak R. 2018. How intraspecific variation in seed-dispersing animals matters for plants. Biological Reviews of the Cambridge Philosophical Society 93:897–913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.