Abstract
Cytoplasmic dynein is an AAA+ motor that drives the transport of many intracellular cargoes towards the minus-end of microtubules. Previous in vitro studies characterized isolated dynein as an exceptionally weak motor that moves slowly and diffuses on a microtubule. Recent studies altered this view by demonstrating that dynein remains in an autoinhibited conformation on its own, and processive motility is activated when it forms a ternary complex with dynactin and a cargo adaptor. This complex assembles more efficiently in the presence of Lis1, providing an explanation for why Lis1 is a required cofactor for most cytoplasmic dynein-driven processes in cells. This review describes how dynein motility is activated and regulated by cargo adaptors and accessory proteins.
Keywords: intracellular transport, cytoskeleton, motor proteins, dynein, dynactin, Lis1, cargo adaptors
Cytoplasmic Dynein
In eukaryotes, intracellular cargoes such as organelles, membrane-bound vesicles, and misfolded proteins are trafficked throughout the cytoplasm by molecular motors that walk along actin filaments and microtubules (MTs). While myosin motors use actin as a substrate for motility, kinesins and dyneins walk along MTs. Cytoplasmic dynein is the principle motor responsible for the transport of cargoes towards the minus-end of MTs [1]. Cytoplasmic dynein also drives retrograde transport in neurons [2] and plays critical roles in cell division [3,4]. Mutations that lead to defects in cytoplasmic dynein motility are linked to a variety of human neurological pathologies including spinal muscular atrophy [5], Charcot-Marie Tooth disease [6], cortical development malformations [7,8], and neurodegenerative diseases [9,10]. Other dyneins are localized to cilia, where dynein-2 drives intraflagellar transport (IFT) in a retrograde direction in cilia, and inner and outer arm dyneins power ciliary beating [11]. This review will focus on the mechanism and regulation of cytoplasmic dynein (referred to as dynein hereafter). The functions of dynein-2 and axonemal dyneins are described in recent reviews [11–13].
Because of the complexity and large size of dynein’s structure, the mechanism and regulation of dynein motility have only recently begun to emerge. While studies of Saccharomyces cerevisiae dynein found that the dynein heavy chain (DHC) exhibits processive motility in vitro in the absence of any accessory proteins and cofactors [14], this has not held true for mammalian homologs [15,16]. Recent in vitro reconstitution studies revealed that isolated mammalian dynein remains in an inactive conformation and motility is activated when dynein and its cofactor dynactin form a complex with a coiled-coil adaptor protein (discussed below). These adaptors physically tether dynein to specific intracellular cargoes [17]. Activation of dynein motility is also regulated by accessory proteins that bind directly to the dynein motor domain (discussed below). This review will discuss how components of the dynein transport machinery activate dynein motility and regulate its stepping and force generation along MTs.
An Overview of Dynein and its Regulators
Structure of dynein and dynactin
Mammalian dynein is a large 1.5 MDa complex composed of a homodimer of DHCs and several smaller non-catalytic subunits. The DHC contains a C-terminal motor domain (see Glossary) and an N-terminal tail domain. The motor domain comprises a ring (15 nm diameter) of six AAA+ modules (AAA1–6) [18–20]. In contrast to kinesin, whose MT interface is located on the surface of the ATPase core [21,22], dynein’s MT-binding domain (MTBD) is separated from the AAA+ ring by a coiled-coil stalk [23,24]. The tail domain is involved in the dimerization of the DHCs and contacts the AAA+ ring through a linker region [25,26]. The tail also binds to an intermediate chain (IC) and light-intermediate chain (LIC). Additionally, the unstructured N-terminal region of the IC recruits three light-chain dimers Robl/LC7, Tctex, and LC8 [Fig. 1A] [26–28].
Figure 1. Activation of the dynein transport machinery.
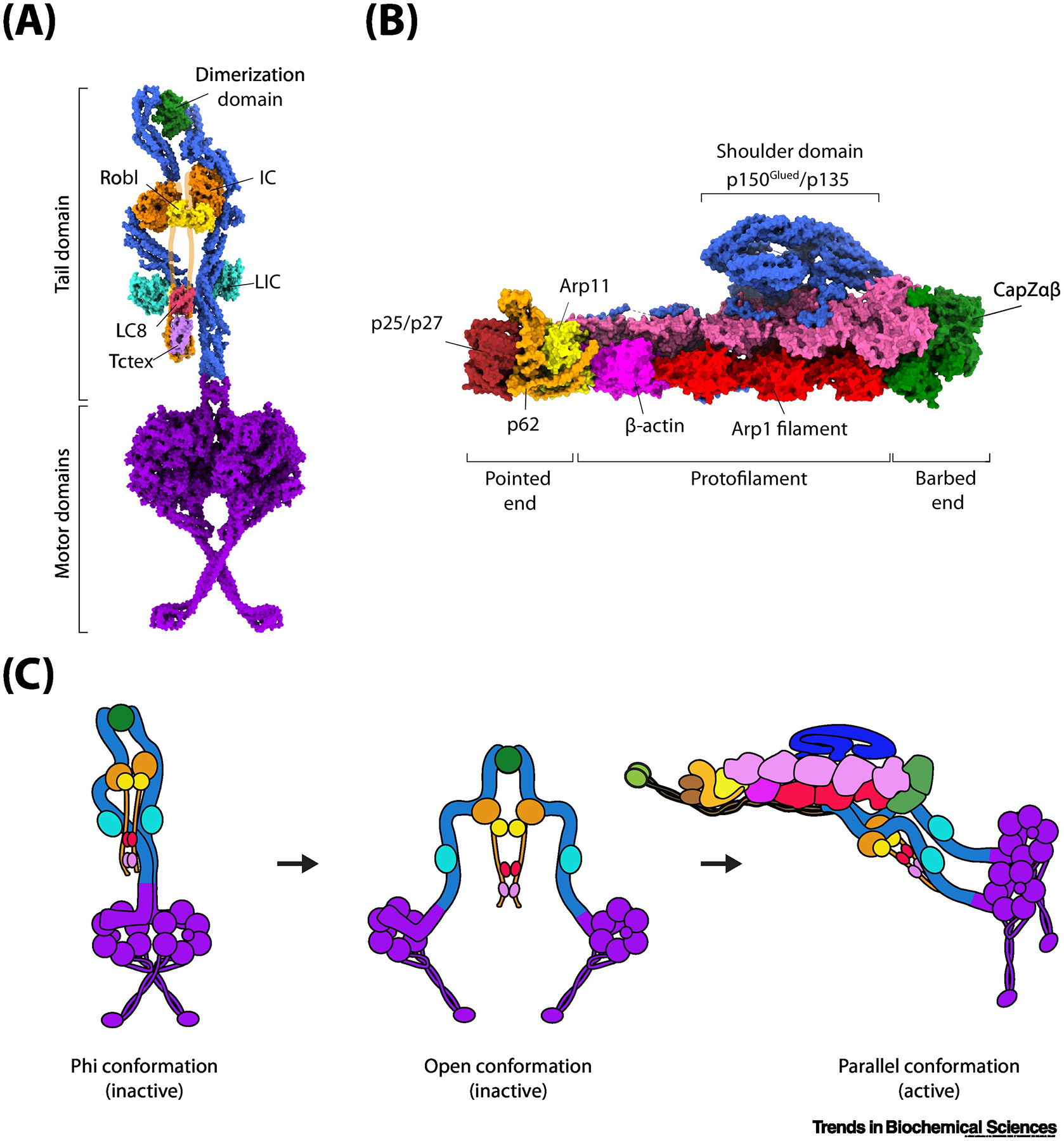
(A) The architecture of the dynein complex in the autoinhibited (phi) conformation (PDB accession: 5NVU). The N-terminal dimerization domain links the two DHCs (blue) together. The intermediate chains (ICs) are held together through an N-terminal unstructured region (purported location shown in orange outline) through homodimers of Robl, LC8, and Tctex. The light intermediate chains (LICs, cyan) bind mid-way along the tail domain. The motor domains (purple) form a stacked structure with the coiled-coil stalks crossed. (B) The architecture of the dynactin complex (PDB accession: 5ADX). The two Arp1 filaments (red and magenta) form the core of the dynactin structure. The barbed end is capped by CapZαβ (green) and the pointed end is capped by p25/p27 (dark red), p62 (orange), and Arp11 (yellow). Atop the Arp1 filaments sits a large shoulder-like domain consisting of p150Glued/p135 (blue). p150Glued also consists of a large coiled-coil projection terminating with the CAP-Gly domain (not shown). (C) Isolated dynein forms an autoinhibited phi conformation through self-dimerization of the motor domains and only weakly interacts with a MT. Disruption of the phi interaction sites separates the two motor domains, but dynein is unable to move processively in this open conformation because motor domains point towards each other. Binding to dynactin and a cargo adaptor orients the motor domains into a parallel conformation for processive movement along MTs.
Dynein motility is powered by ATP hydrolysis of the AAA1 site [23], coupled with conformational changes in the linker domain [29,30]. In the absence of a nucleotide in AAA1, dynein is strongly bound to the MT and the linker is in a straight conformation. ATP binding to AAA1 triggers MT release by altering the registry of the coiled-coil stalk [31,32] and forces the linker into a bent conformation referred to as the priming stroke [33,34]. The priming stroke provides a net bias towards the minus-end as dynein takes a step [35]. After ATP hydrolysis, the motor rebinds to the MT. MT binding accelerates phosphate release, and the linker reverts back to its straight conformation in the ADP-bound state [1,23]. This step acts as the force-generating power stroke of dynein [36].
Dynactin is a large 1.1 MDa multi-subunit complex that is required for nearly all dynein-driven processes in cells. The complex consists of 23 individual subunits that are assembled around a filament consisting of the actin-related protein 1 (Arp1) [25] [Fig. 1B]. The filament has a polarity similar to actin. The pointed end of the filament is capped by Arp11, p25/27, and p62, while the barbed end binds to the CapZαβ heterodimer. On top of the dynactin filament sits the p150Glued subunit, which forms a large shoulder-like projection and contains an extended region consisting of three coiled-coiled domains [30,37] and an N-terminal Cap-Gly domain that binds to MTs [38,39]. The Cap-Gly domain regulates recruitment of dynactin to the MT by binding to plus-end tracking proteins (+TIPs) such as CLIP170 and EB1 [40–43] or through post-translational modifications present on the tubulin C-terminal tail [44,45].
Cargo adaptors activate dynein for processive motility
Until recently, most of our knowledge of how dynein steps along the MT and responds to external forces came from the in vitro studies of S. cerevisiae dynein. These studies have shown that DHC is a highly processive motor in the absence of associated chains and other cofactors such as dynactin [14]. Unlike kinesin and myosin V, which have a coordinated hand-over-hand stepping mechanism [46,47], dynein moves along MTs through uncoordinated stepping behavior between its two motor domains, with a variable step-size and a high frequency of sideways and backward steps [48–50]. S. cerevisiae dynein motility stalls when subjected to 3–4 pN resistive forces [50–52], suggesting that it produces forces comparable to 6 pN stall force of plus-end-directed kinesin [53].
Initial in vitro studies on isolated mammalian dynein deviated significantly from the studies on S. cerevisiae dynein, as this motor has a very low affinity to MTs and exhibits little to no processive motility [15,16]. Surprisingly, dynein also had a low affinity for its cofactor dynactin, and the addition of dynactin had a minor effect on dynein motility [54,55]. Because mammalian dynein appeared to be active in MT gliding assays, in which many surface-immobilized motors glide MTs, it was initially proposed that individual dyneins are not sufficient to drive robust motility, but multiple dyneins need to form a team to be able to perform a wide variety of functions inside cells [15,56,57].
Studies in mammalian cells showed that dynein binds to intracellular cargos through adaptor proteins that contain a long coiled-coil. This was first demonstrated by activation of dynein-driven transport of Golgi-associated vesicles by an N-terminal coiled-coil of Bicaudal D homologue 2 (BicD2N) [58,59], suggesting that these adaptors are required to activate dynein motility. In vitro reconstitution experiments of recombinant dynein, dynactin, and a cargo adaptor [60,61] have verified this prediction. These studies have shown that dynein remains in an autoinhibited conformation on its own and the addition of dynactin has little to no effect on motility. Remarkably, the addition of BicD2N was sufficient to assemble a stable complex with dynein and dynactin. This complex exhibited robust processive movement at speeds similar to dynein-driven cargos in cells [60,61]. Several other cargo adaptors have been shown to activate dynein motility in a similar manner through in vitro reconstitution experiments, including Hook3 [61,62], NIN [17], NINL [17], CRACR2a [63], Rab45 [63], and BicD family-like cargo adaptor 1 (BicDL1) [64]. In vitro studies in crude cell lysates have identified additional dynein-activators, such as Hook1 [65], Rab11-FIP3 [61], and Spindly [61], but these have not yet been tested to activate dynein with pure components.
Studies in live cells identified additional candidate adaptors for dynein, such as TRAK and HAP1 [66–69]. TRAK1 and TRAK2 associates with the Mitochondrial Rho GTPase 1 (Miro1) on the outer mitochondrial membrane [70–73] and serves as a cargo adaptor that links dynein and kinesin-1 to mitochondria. TRAK1 and TRAK2 each contain a domain homologous to BiCDL-1, further suggesting that these adaptors activate dynein [66,74]. Another candidate adaptor is the huntingtin-associated protein 1 (HAP1), which is involved in the transport of autophagosomes [75–78]. Together with huntingtin, HAP1 recruits both dynein and kinesin in order to regulate the dynamics of various organelles in neurons [79]. Additionally, the MT-binding protein NuMA has been implicated as a a cargo adaptor through its interaction with dynein and dynactin [4]. NuMA is essential for bipolar mitotic spindle assembly and maintenance, and is relocated to spindle poles by dynein during cell division [80]. NuMA contains a Spindly-like motif, which is essential for dynein recruitment, suggesting that it functions similarly to known cargo adaptors [81]. It remains to be demonstrated whether these adaptors activate dynein-dynactin motility through in vitro reconstitution experiments. Table 1 summarizes the cargo adaptors that link dynein to specific cargos.
Table 1.
The list of cargo adaptors that recruit dynein to intracellular cargos.
| Adaptor | Intracellular Cargo | Known binding partners (not including dynein and dynactin) | Evidence of dynein interaction | Demonstration of dynein activation | Refs |
|---|---|---|---|---|---|
| BicD1 | Golgi-derived vesicles, cytoplasmic vesicles | Rab6a, Rab6b | Proteomics | N/A | [17] |
| BicD2 | Golgi-derived vesicles, cytoplasmic vesicles | Rab6a, KIF5B | Co-IP, In vitro reconstitution | In vitro reconstitution | [60,61] |
| BicDL-1 | Secretory vesicles | KIF1C, Rab6a, Rab6b | Co-IP, In vitro reconstitution | In vitro reconstitution | [64] |
| HOOK1 | Late endosomes | FTS, AKTIP | Co-IP | Cell lysate isolation | [65,154] |
| HOOK2 | Golgi-derived vesicles | FTS, AKTIP | Co-IP | N/A | [155] |
| HOOK3 | Secretory vesicles | KIF1C | Co-IP, In-vitro reconstitution | In vitro reconstitution | [61,62] |
| SPDL1 | Kinetochore | KNTC1, ZW10 | Co-IP | Cell lysate isolation | [61,82] |
| Rab11FIP3 | Recycling endosomes | Rab11a | Co-IP | Cell lysate isolation | [61] |
| NINL | Centrosome | APC | Co-IP, In-vitro reconstitution | In vitro reconstitution | [17] |
| NIN | Centrosome | GSK3β | Co-IP, In-vitro reconstitution | In vitro reconstitution | [17] |
| CRACR2a | Endosomes | ORAI1, STIM1 | Co-IP, In-vitro reconstitution | In vitro reconstitution | [63] |
| Rab45 | Endosomes | N/A | Co-IP, In-vitro reconstitution | In vitro reconstitution | [63] |
| CCDC88A | Endosomes | DISC1, AKT1 | Co-IP | N/A | [17] |
| CCDC88B | Lytic granules | DOCK8, HSPA5 | N/A | N/A | [17] |
| CCDC88C | N/A | DVL1 | Co-IP | N/A | [17] |
| NUMA1 | Microtubules | LGN, RAE1, SPAG5 | IP | N/A | [80] |
| TRAK1 | Mitochondria | KIF5B, Miro | Co-IP | N/A | [66] |
| TRAK2 | Mitochondria, Peroxisomes | KIF5A, KIF5C | Co-IP | N/A | [66] |
| HAP1 | Autophagosomes, BDNF-containing vesicles, | HTT, KIF5B | Indirect IP (interaction with dynactin) | N/A | [75] |
| RILP | Late endosomes, lysosomes | Rab7, Rab36 | Co-IP | N/A | [156] |
| JIP3 | APP vesicles, lysosomes, JNK | APP | Indirect IP | N/A | [157] |
Cargo adaptors do not contain a well conserved sequence. However, they all contain long coil-coil regions and typically share several conserved structural features such as the CC1-box and Hook domain which both bind to the light intermediate chain [62,82,83], and the Spindly motif that binds to the pointed end of dynactin [82].
Activation of Mammalian Cytoplasmic Dynein: A Structural Perspective
The autoinhibited conformation
Early observations that mammalian dynein was predominantly non-processive in the absence of dynactin or a cargo adaptor suggested that dynein is autoinhibited when not transporting cargo [15], similar to kinesin-1 [84–86]. Low-resolution electron microscopy (EM) studies showed that autoinhibition occurs via self-dimerization of the dynein motor domains. This conformation was nicknamed the phi-particle, due to its similarity to the Greek letter phi (Φ) [15,87]. High-resolution cryo-EM structures of dynein-1 [26] and dynein-2 [88,89] revealed that self-dimerization is mediated primarily by previously uncharacterized interactions between the linker domains, linker-AAA5, and the AAA4-AAA5 [Fig. 1A]. The phi dynein is unable to walk processively along MTs because hydrophobic interactions between the crossed coil-coil stalk regions prevent both monomers to simultaneously interact with the MT. In addition, the registry of the coiled-coils stalks and the nucleotide-binding site of AAA1 are trapped in a low-affinity MT-binding state [33].
Assembly of the active dynein-dynactin transport complex
Disruption of the self-dimerization interface with site-specific point mutations creates an open conformation with higher MT-binding affinity [26]. However, the motor domains remain in an antiparallel conformation and do not move processively. In the presence of a cargo adaptor, binding of dynein to dynactin forces the tail domains to point in the same direction. This results in parallel conformation of the motor domains [Fig. 1C] [37,64,90], which is ideal for MT binding and processive movement. It remains unclear why the heads cannot form a parallel orientation in the absence of dynactin and a cargo adaptor. One hypothesis is that, when not bound to dynactin, the dimerization domain prefers an antiparallel conformation. Alternatively, dimerization of the light chains at the unstructured regions of the intermediate chain may restrict conformational freedom of the motor domains by stabilizing the tail domains into an antiparallel orientation [Fig. 1C; middle panel].
Regulation of dynein stoichiometry by cargo adaptors
Recent EM studies showed that the coiled-coil region of a cargo adaptor extends along a lower exposed region of the dynactin filament to recruit a second dynein motor [64,90]. The two dyneins are packed closely together with their motor domains positioned side-by-side [Fig. 2A]. Their tails make extensive intermolecular contacts, which may reinforce a parallel orientation between the DHCs to favor processive motility.
Figure 2. Recruitment of two dyneins to dynactin increases the force generation and velocity of the complex.
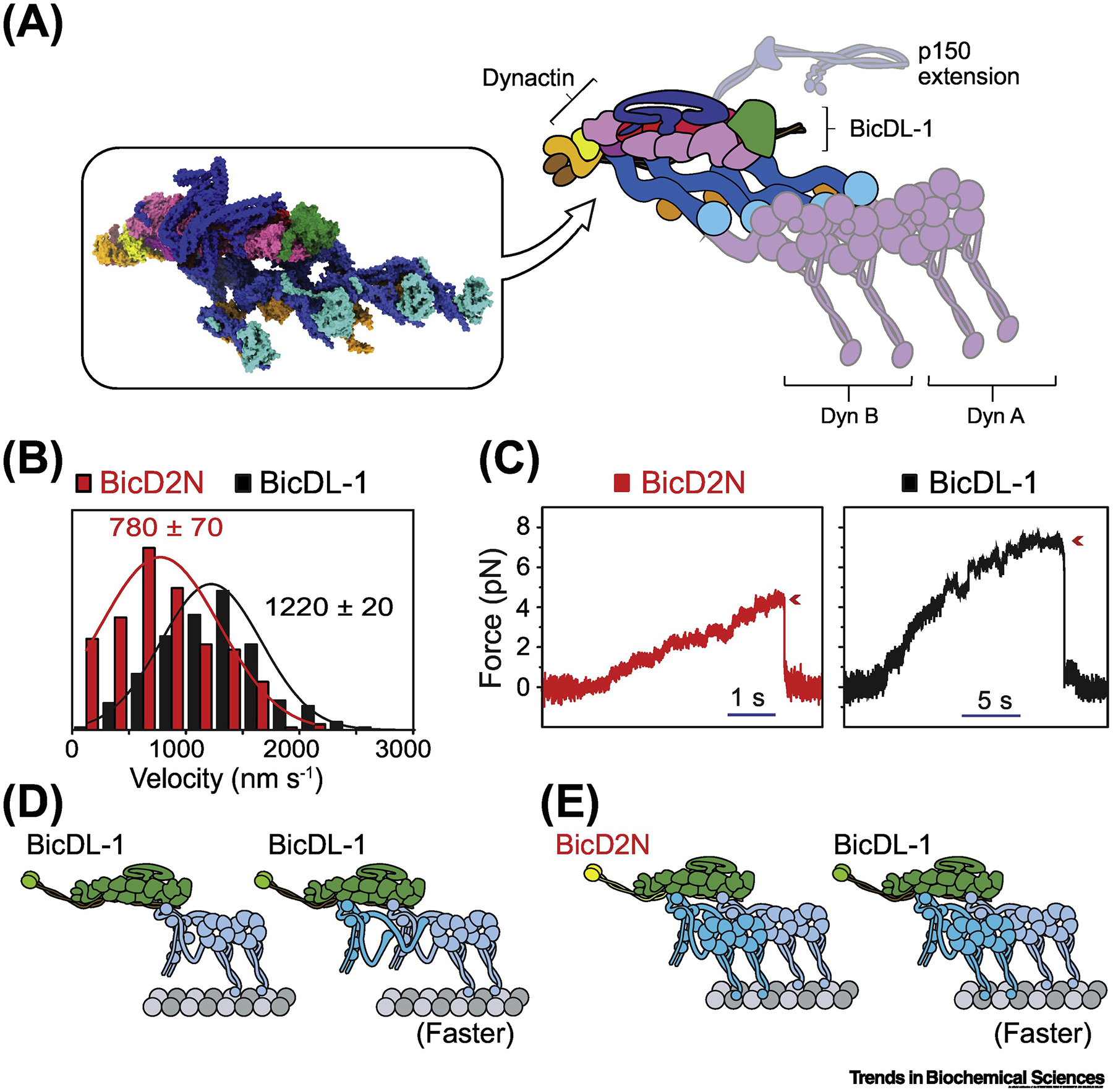
(A) (Left box) The architecture of two dynein tail domains bound to dynactin and the cargo adaptor BicDL-1 (PDB accession: 6F1T). (Right) Schematic representation of the structure with the purported positions of the dynein motor domains and p150 extension. Dynein A binds to the center of the dynactin filament, while dynein B binds dynactin closer towards the barbed end. (B) Complexes assembled with BicDL-1 moves faster than complexes assembled with the BicD2N adaptor. The mean velocities are calculated from Gaussian fits (solid curves, mean ± s.e.m.) (modified from [93]). (C) Complexes assembled with the BicDL-1 adaptor stall at higher forces than complexes assembled with BicD2N (modified from [93]). (D) Recruitment of two dyneins to dynactin by BicDL-1 results in faster movement. Motor domains of the second dynein are dispensable for increased speed. (E) Complexes assembled with BicDL-1 move faster than complexes assembled with BicD2N when recruiting the same number of dynein motors.
Single-molecule studies revealed that complexes that recruit a second dynein dimer move faster and produce substantially increased forces [Figures 2B, C]. Intriguingly, recruitment of two dyneins to a native scaffold (i.e. dynactin and a cargo adaptor) results in faster movement (1.2 μm/s) than complexes with single dynein (0.8 μm/s) [Fig. 2B] [64]. This is not consistent with previous in vitro studies, which observed little to no change when multiple processive motors are assembled to an artificial DNA scaffold [91,92].
Several models have been proposed to explain the increased speed of dynein supercomplexes. For example, side-by-side positioning of the motors may restrict dynein’s intrinsic backward and sideways stepping behavior, thus allowing the complex to take a more direct route along the MT. However, stepping measurements showed that recruitment of second dynein does not alter the frequency of sideways and backward steps, but increases the ATPase activity and the stepping rate [93]. A recent single molecule study suggested that inter-dimer contacts that occur through the adjacent tail domains increase the stepping rate of the AAA+ domains [Figures 2D, E] [93]. It remains to be determined whether these interactions allosterically regulate the motor domains or produce long-range structural rearrangements to increase motor speed. In addition, complexes bound to BicD2 or BicDL-1 exhibit different velocities even when recruiting the same number of motors [Fig. 2D] [64,93]. Therefore, cargo adaptors not only activate the dynein-dynactin complex for processive motility but also regulate dynein’s velocity and force-production.
Mammalian dynein-dynactin produces substantial forces
In contrast to S. cerevisiae dynein, early optical trapping measurements found that isolated mammalian dynein produced low forces (0.5–1 pN) in vitro [94–97]. This seemingly contrasted with the robust and efficient transport of dynein-driven cargos in cells [38,98]. It has been proposed that dynein forms large teams to generate higher forces required to transport large intracellular cargos. The primary evidence in support of this model comes from optical trapping of beads nonspecifically adsorbed to native mammalian dynein purified from brain extract, which observed a ~ 1–2 pN periodicity of peak forces. In vivo optical trapping measurements of motile phagocytosed latex beads and purified early/late phagosomes observed stall forces up to nearly 20 pN [56,57,99,100]. It has been proposed that the peak periodicity represents the force production of single or a pair of dynein motors and that large forces (12–20 pN) are generated by more than ten dyneins simultaneously engaging with the MT.
Because initial in vitro studies of mammalian dynein used inactive protein, models proposed to explain mammalian dynein behavior differed significantly from those for S. cerevisiae dynein. Recent advances in generating recombinant mammalian dynein have allowed for the precise delineation of the motor’s force generation in the presence and absence of cargo adaptors. In line with previous in vitro measurements, recombinant dynein alone produces ~ 1–2 pN force, yet exhibits primarily diffusive and non-processive motility [101]. In the presence of cargo adaptors, the complexes that primarily recruit a single dynein stall at forces similar to that of S. cerevisiae dynein (3.5 pN) [101]. Recruitment of second dynein nearly doubles the force generation (5–7 pN, [Fig. 2C]) [64], consistent with previous reports that multiple dyneins can collectively generate higher mechanical forces than a single motor [56,57]. These studies suggest that retrograde cargos are driven by a single or a few dynein-dynactin complexes [102], rather than a large ensemble of weak motors.
The underlying reason for the controversy between in vitro and in vivo force measurements of dynein remains to be elucidated. One major difference between these studies is the absence of clear stalling events of intracellular cargos. It is possible that peak forces measured in vivo do not represent the maximum force generation of the motor complex while providing information about the number of motor complexes carrying a cargo. Alternatively, the presence of other motors, cargo adaptors and regulatory complexes on a cargo affects dynein force generation in complex regulatory pathways.
Lis1 Serves as an Assembly Factor of Dynein-Dynactin
Lis1 and NudE/NudEL are the only known regulatory proteins that interact directly with the dynein motor domain. Lis1 was initially identified as the causative gene associated with lissencephaly-1 (smooth brain disorder) [103]. Studies in live cells showed that Lis1 is a cofactor required for dynein’s cellular functions, including recruitment of dynein to the plus-end of MTs, proper positioning of the spindle positioning, and anchoring of dynein to the cell cortex [43,104–107]. These studies also led to the discovery of NudE/NudEL, which are homologous proteins that bind to Lis1 and participate in dynein regulatory functions [108–112].
The mechanism by which Lis1/NudEL regulates dynein remained enigmatic. In vivo studies suggested that Lis1 is required for targeting dynein to the MTs, and it dissociates from dynein to initiate transport [105–107]. However, in vitro studies on isolated S. cerevisiae and mammalian dynein initially reported contradictory observations that Lis1 stimulates dynein activity [113,114], or exerts an inhibitory effect that is relieved upon binding of NudEL [115]. Recent structural and single-molecule studies on S. cerevisiae dynein have elucidated a more complex mode of regulation [116]. Specifically, Lis1 binding near the AAA3 site increases the MT binding affinity and displaces the linker domain from its docking site at AAA5, thereby inhibiting or reducing the speed of dynein motility [117]. Depending on the nucleotide state of the AAA3 site, the second β-propeller domain of a Lis1 dimer binds to the coiled-coil stalk and reduces the MT affinity of dynein, thereby increasing the speed of the complex [116].
Recent studies showed that Lis1 increases the frequency and velocity of active mammalian dynein-dynactin in vitro [43,114,118]. These studies proposed that Lis1’s main role is to promote the formation of dynein/dynactin complexes containing two dynein dimers [Fig. 3] [119,120]. According to this model, Lis1 binds to dynein in the open conformation and prevents switching back to the phi conformation. Therefore, Lis1 increases the efficiency of the first dynein’s ability to assemble a complex with dynactin and a cargo adaptor, and increases the likelihood of complexes to recruit two dyneins [119,120]. In support of this model, human Lis1 binds to dynein at a region of AAA3/4 that is not accessible in the phi particle and has a higher affinity for dynein mutants that are predominantly in the open state [120]. Lis1 binding may also promote a parallel orientation of the motor domains [120], but this model needs to be further tested by cryoEM. Interestingly, Lis1 does not colocalize with moving cargoes [121,122], consistent with the observation that Lis1 dissociates from dynein-dynactin during or after the complex formation in vitro [119,120]. The S. cerevisiae homolog Pac1 was also found to promote the active conformation of dynein, suggesting that this mechanism is conserved in eukaryotes [123]. Consistent with these results, the requirement of Lis1 and NudE in HookA-mediated dynein activation in A. nidulans can be bypassed by expressing a dynein mutant incapable of forming the phi conformation [124].
Figure 3. Lis1 promotes the assembly of two dynein dimers onto dynactin.
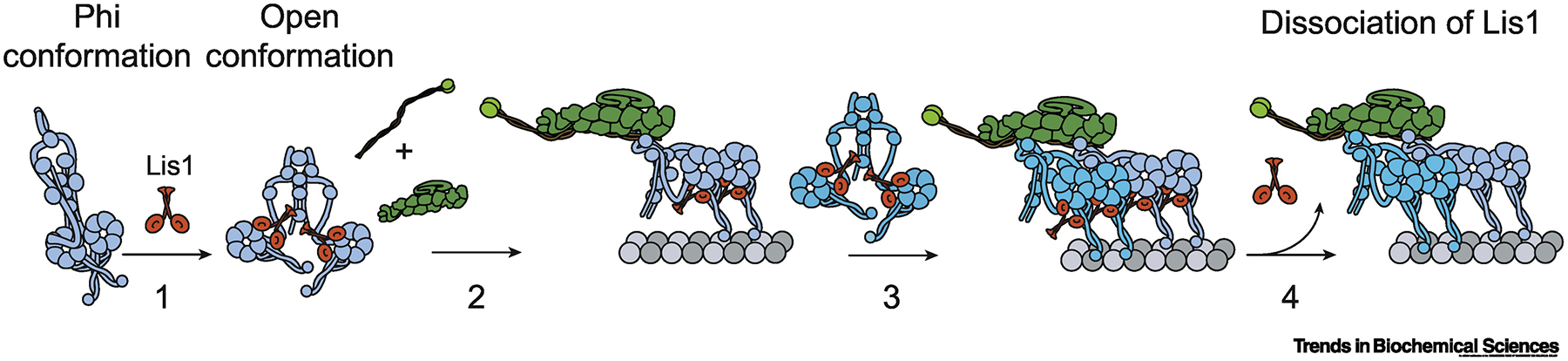
(1) Lis1 preferentially binds to and stabilizes dynein when it is in the ‘open’ conformation. (2) Dynein has a higher affinity for dynactin in the open conformation. (3) Lis1 recruits a second open dynein to dynactin. (4) Lis1 dissociates from most of the activated two dynein complexes after the assembly.
Intracellular Cargo Transport: Integrating Information from in vitro Studies
The ability to reconstitute active dynein-dynactin complexes and in-depth structural and biophysical studies in vitro have provided unprecedented insight into the activation and regulation of dynein motility. However, how dynein is recruited to specific cargos and its motility is coordinated with plus-end-directed kinesin to determine which direction the cargo moves remain unclear. Recent studies in live cells have provided fresh insight into these questions.
Dynein-mediated intracellular transport
Despite the key role that cargo adaptors play in activating dynein motility, it remains to be demonstrated if cargo adaptors serve as a general mechanism for recruiting the active motor complex to its cargoes. Although numerous cargo adaptors have been shown to activate dynein, only a subset of adaptors have been shown to interact directly with intracellular cargos [Fig. 4]. The strongest evidence in support of this model is based on experiments with the BicD/BicDL and HOOK family of cargo adaptors. Both BicD2 and BicDL-1 have been shown to regulate Golgi vesicle transport in mammalian cells by interacting through their C-terminal regions with the Rab6 family of small GTPases [8,58,125–127]. BicD1 also functions in the Golgi transport pathway [58], but it has not yet shown to be a bona-fide cargo adaptor in vitro. Additionally, BicD2, along with its Drosophila homologue BicD, interact with the RNA-binding protein Egalitarian to facilitate long-range mRNA transport [128–130]. Several in vitro reconstitution studies established that Egalitarian and an mRNA are required to disrupt an autoinhibited BicD/BicD2 conformation in order enable formation of the activated dynein complex [129,130]. Within the HOOK family, Hook1 and Hook3 have both been shown to regulate endosomal trafficking in several endocytic pathways [131–133]. While Hook3 has been shown to activate dynein through in vitro reconstitution experiments, Hook1’s purported role as a dynein activator has been demonstrated in cell lysate [134].
Figure 4. Cargo adaptors link dynein to intracellular cargos.
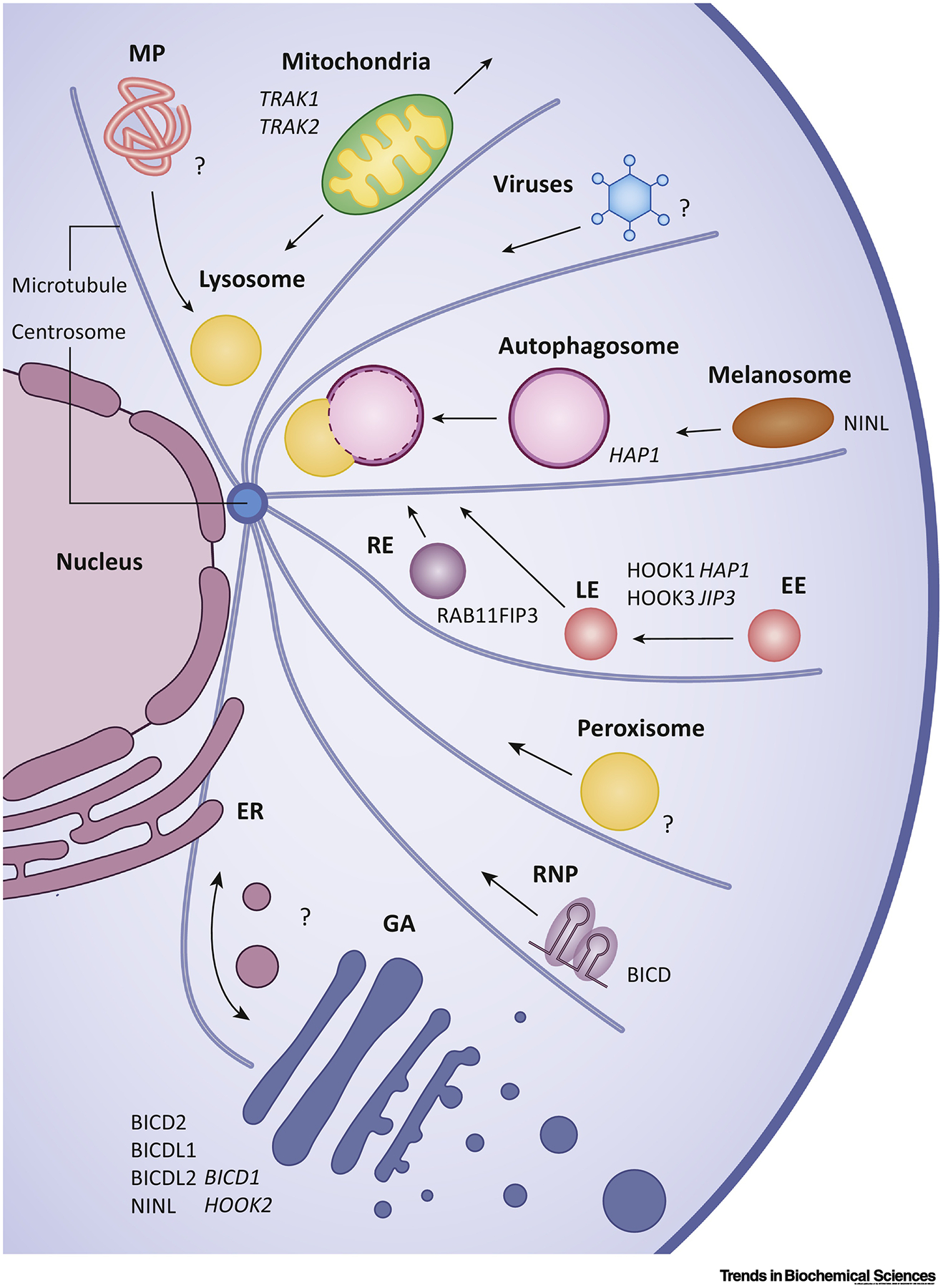
Schematic representation of the intracellular cargoes that dynein transports within the cell. Cargo adaptors that are confirmed through in vitro reconstitution to activate dynein are shown in regular font, while hypothesized adaptors are italicized. Question marks indicate cargos where the identity of the adaptor remains unknown. Arrows indicate direction of intracellular cargo transport. Abbreviations: MP, misfolded proteins; RE, recycling endosome; LE, late endosome; EE, early endosome; GA, Golgi apparatus; ER, endoplasmic reticulum; RNP, ribonucleic particle; TRAK1, trafficking kinesin-binding protein 1 (also known as OIP-106); TRAK2, trafficking kinesin-binding protein 2 (also known as GRIF-1); HAP1, huntingtin-associated protein 1; JIP3, JNK-interacting protein 1; RAB11FIP3, Rab11 family-interacting protein 3; BICD, Bicaudal D; BICDl/2, Bicaudal D homologue 1/2; BICDLl/2, BICD family-like protein 1/2; NINL, Ninein-like protein.
The physiological significance for recruiting two dyneins to dynactin remains unclear. One possibility is that cargo adaptors control the number of dyneins recruited to dynactin to regulate dynein-mediated transport [64]. In vitro reconstitution studies showed that BicDL-1 recruits two dyneins, while BiCD2N primarily recruits a single dynein [64]. Complexes that recruit two dyneins move faster, travel longer distances on a MT, and produce substantially increased forces [64], and this may lead to more efficient transport in the retrograde direction. Consistent with this idea, overexpression of BicDL-1 relative to BicD2 increases the mean velocity of Rab6 vesicles in the retrograde direction [60], whereas reduction of BicDL-1 expression relative to BicD2 leads to increased anterograde transport of Rab6 secretory vesicles [126]. Alternatively, full-length BicD2 also recruits two dyneins to dynactin in the presence of Lis1. While these complexes produce higher forces, they still move slower than complexes assembled with BicDL-1 [64,93]. It remains to be established whether the differences in motility and force generation of dynein-dynactin assembled with BicDL-1 can account for the shift of balance of bidirectional transport in dynein’s favor, relative to complexes assembled with BicD2N.
Coordination of bidirectional transport
How does the cell determine which direction the cargo moves? Most cargos simultaneously recruit plus- and minus-end directed motors. These opposite polarity motors compete to determine which direction the cargo moves, but they also need each other to return back to their origin of transport. Inhibition of kinesin not only decreases anterograde transport, but retrograde transport is also similarly reduced, usually within minutes [135]. Similarly, dynein inhibition leads to a significant reduction in anterograde transport [136–138].
There is a range of cargoes that exhibit unidirectional movement without frequent pausing and backward movement, indicating that tight coordination exists between motors of opposite polarity [43,139]. For example, intraflagellar transport (IFT) is driven by kinesin-2 in the anterograde direction, and dynein-2 in the retrograde direction. When kinesin-2 drives anterograde IFT, dynein-2 is autoinhibited and facing away from the MT [140]. As a result, transport of IFT trains along the length of cilia is often smooth, without frequent pauses and reversals [141,142]. Once IFT trains arrive at the ciliary tip, kinesin-II is either inactivated or decoupled from IFT cargoes before retrograde transport occurs [140,143]. It has been reported that this ‘hand-off’ process is regulated by post-translational modifications, but the precise mechanism of IFT turnover remains to be elucidated [144]. It is possible that IFT trains spontaneously disassemble once they arrive at the ciliary tip and reassemble in a manner different from their assembly at the ciliary base, such that dyneins now face towards the MT, while the kinesin binding site is either blocked or facing away from the MT in retrograde trains. Alternatively, motors may be regulated by signaling proteins that interact with tip tracking proteins at the MT plus-end [145,146].
Unlike IFT, most cargoes exhibit pauses and reversals as they are transported in neurons [135,147–150]. It has been proposed that these reversals may be due to a mechanical tug-of-war between opposite polarity motors. However, in vitro tug-of-war experiments fail to exhibit frequent reversals and often resulted in slow motility along the direction of a winning motor [56,57]. This is in contrast with intracellular cargos that usually move at near the full speed of the winning motor in between frequent reversals [138,152]. Therefore, it is unlikely that kinesin and dynein continuously compete in a tug-of-war during intracellular transport. Instead, opposite polarity motors on a cargo may be regulated in trans to prevent a continuous tug-of-war and slowdown of motility. This regulatory mechanism may shut off transiently for a tug-of-war between kinesin and dynein, which would lead to a pause. It may also or switch the active motor carrying the cargo, which would lead to a reversal of cargo transport.
Several studies suggested that cargo adaptors play a major role in regulating the directionality of intracellular cargoes. Recent studies found that Hook3 simultaneously binds dynein-dynactin and kinesin-3 [151]. Unlike previous tug-of-war assays that used artificial scaffolds, these complexes moved at full speed in the direction of a winning motor [151]. However, the kine sin-3-Hook3-dynein complex did not exhibit pausing or reversals. It is possible that the reversals are not frequent enough to observe at a limited length of MTs used in in vitro assays (~ 10 μm). Alternatively, switching between anterograde and retrograde transport could be regulated by other factors missing from these in vitro experiments [153]. Future structural and biophysical experiments will be required to assess whether cargo adaptors directly determine which direction the cargo moves by controlling the activity of dynein and kinesin motors.
Concluding Remarks and Future Perspectives
The past decade has witnessed a dramatic advancement in our understanding of the mechanism and regulation of dynein motors. High-resolution EM structures have revealed the structure of isolated dynein and the detailed structural changes that occur upon binding to dynactin and a cargo adaptor that activate dynein for processive motility. Single-molecule experiments revealed how cargo adaptors regulate motility and force-production of mammalian dynein. Despite these advances, we still do not have a comprehensive list of cargo adaptors that specifically recruit dynein to hundreds of cargoes inside cells. Of particular interest will be to understand how cargo adaptors that bind both kinesin and dynactin regulate the net directionality of the complex. It will also be important to determine whether other cofactors contribute to regulating directionality (see Outstanding Questions). In vitro reconstitution of MT motors, cargo adaptors, and their regulatory components will play an especially important role in this effort. Furthermore, integration of these in vitro reconstitution conditions with detailed biophysical and structural experiments will allow us to dissect the complex mechanistic features of the MT transport machinery. Ultimately, these approaches will advance the field towards understanding the mechanism and regulation of bidirectional cargo transport along MTs.
Outstanding Questions.
In addition to already identified cargo adaptors, what other adaptors activate dynein for processive motility? There is a growing list of ‘candidate’ adaptors obtained from studies in live cells and these need to be verified in vitro.
How do cargos recruit kinesin and dynein motors and regulate their activity to move in a particular direction along MTs?
What are the molecular cues that govern transient reversals and pauses of intracellular cargos?
Highlights.
Dynein remains in an inactive conformation on its own, and motility is activated when it forms a complex with dynactin and a cargo adaptor.
Previous studies identified isolated mammalian dynein as an exceptionally weak motor that diffuses on an MT and produces low forces.
In vitro reconstitution of the mammalian dynein-dynactin complex recently altered this view, and revealed that dynein is a robust motor that walks rapidly and produces forces comparable to plus-end-directed kinesin.
CryoEM and single-molecule studies showed that cargo adaptors recruit two dynein motors to dynactin for faster movement and higher force production.
Lis1, the only accessory factor that binds directly to dynein’s motor domain, promotes the formation of fully activated dynein/dynactin complexes containing two dynein dimers, and then dissociates from dynein leading to rapid motility.
A subset of cargo adaptors that simultaneously recruit kinesin and dynein to the cargo, may coordinate kinesin-dynein activity to determine which direction the cargo moves.
Acknowledgments
We sincerely apologize to all those colleagues whose important work was not cited in this paper due to space limitations. This work was supported by a grant from the National Institutes of Health (RO1-GM116204) to A.Y.
Glossary:
- AAA+ proteins
Diverse protein family of ring-shaped P-loop NTPases
- Bicaudal D homologue 2
a coiled-coil adaptor that links dynein to Rab6A vesicles
- BicDL-1
a coiled-coil adaptor that links dynein to Rab6A vesicles
- HOOK3
a coiled-coil adaptor that links dynein to Golgi apparatus membranes
- Linker domain
a mechanical element that drives dynein stepping towards the minus-end
- Lis1
a dimeric regulatory protein that binds directly to the dynein motor domain
- Motor domain
a catalytic AAA+-ring that powers dynein motility
- Nudel
a coiled-coil protein that binds to Lis1
- p150Glued
the largest subunit of dynactin that interacts with MTs
- Phi particle
the inactive conformation of dynein formed by the self-dimerization of DHCs
- Processivity
the ability of a motor to take many steps before dissociating from its substrate
- Tail domain
an extended region of the DHC that interacts with the IC, LICS, and LCs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roberts AJ et al. (2013) Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol 14, 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirokawa N et al. (2010) Molecular motors in neurons: Transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 3.Heald R et al. (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425 [DOI] [PubMed] [Google Scholar]
- 4.Merdes A et al. (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458 [DOI] [PubMed] [Google Scholar]
- 5.Baloh R et al. (2012) Mutations in the Tail Domain of DYNC1H1 Cause Dominant Spinal Muscular Atrophy. Neurology 78, 1714–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weedon MN et al. (2011) Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant Axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet 89, 308–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirier K et al. (2013) Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet 45, 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipka J et al. (2013) Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem. Soc. Trans 41, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 9.Willemsen MH et al. (2012) Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet 49, 179–183 [DOI] [PubMed] [Google Scholar]
- 10.Hafezparast M et al. (2003) Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science 300, 808–812 [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa H and Marshall WF (2011) Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell Biol 12, 222–234 [DOI] [PubMed] [Google Scholar]
- 12.Viswanadha R et al. (2017) Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol 9:a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi D and Takeda H (2012) Ciliary motility: The components and cytoplasmic preassembly mechanisms of the axonemal dyneins. Differentiation 83, S23–S29 [DOI] [PubMed] [Google Scholar]
- 14.Reck-Peterson SL et al. (2006) Single-Molecule Analysis of Dynein Processivity and Stepping Behavior. Cell 126, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torisawa T et al. (2014) Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat. Cell Biol 16, 1118–1124 [DOI] [PubMed] [Google Scholar]
- 16.Trokter M et al. (2012) Reconstitution of the human cytoplasmic dynein complex. Proc. Natl. Acad. Sci 109, 20895–20900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redwine WB et al. (2017) The human cytoplasmic dynein interactome reveals novel activators of motility. Elife 6:e28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon T et al. (2011) X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol 18, 638–642 [DOI] [PubMed] [Google Scholar]
- 19.Kon T et al. (2012) The 2.8 Å crystal structure of the dynein motor domain. Nature 484, 345–350 [DOI] [PubMed] [Google Scholar]
- 20.Carter AP et al. (2011) Crystal Structure of the Dynein Motor Domain. Science 331, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woehlke G et al. (1997) Microtubule interaction site of the kinesin motor. Cell 90, 207–216 [DOI] [PubMed] [Google Scholar]
- 22.Sindelar CV and Downing KH (2007) The beginning of kinesin’s force-generating cycle visualized at 9-Å resolution. J. Cell Biol 177, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts AJ et al. (2009) AAA+ Ring and Linker Swing Mechanism in the Dynein Motor. Cell 136, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter AP (2013) Crystal clear insights into how the dynein motor moves. J. Cell Sci 126, 705–713 [DOI] [PubMed] [Google Scholar]
- 25.Urnavicius L et al. (2015) The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K et al. (2017) Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell 169, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J et al. (2005) Solution structure of isoform 1 of Roadblock/LC7, a light chain in the dynein complex. J. Mol. Biol 354, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 28.Williams JC et al. (2007) Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. U. S. A 104, 10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cianfrocco MA et al. (2015) Mechanism and Regulation of Cytoplasmic Dynein. Annu. Rev. Cell Dev. Biol 31, 83–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter AP et al. (2016) How dynein and dynactin transport cargos: A structural perspective. Curr. Opin. Struct. Biol 37, 62–70 [DOI] [PubMed] [Google Scholar]
- 31.Kon T et al. (2009) Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol 16, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbons IR et al. (2005) The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem 280, 23960–23965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt H et al. (2015) Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter AP et al. (2008) Structure and Functional Role of Dynein’s Microtubule-Binding Domain. Science 322, 1691–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Can S et al. (2019) Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess SA et al. (2003) Dynein structure and power stroke. Nature 421, 715–718 [DOI] [PubMed] [Google Scholar]
- 37.Chowdhury S et al. (2015) Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol 22, 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moughamian AJ and Holzbaur ELF (2012) Dynactin Is Required for Transport Initiation from the Distal Axon. Neuron 74, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayloo S et al. (2014) Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat. Commun 5:4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puls I et al. (2003) Mutant dynactin in motor neuron disease. Nat. Genet 33, 455–456 [DOI] [PubMed] [Google Scholar]
- 41.Steinmetz MO and Akhmanova A (2008) Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci 33, 535–545 [DOI] [PubMed] [Google Scholar]
- 42.Weisbrich A et al. (2007) Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol 14, 959–967 [DOI] [PubMed] [Google Scholar]
- 43.Jha R et al. (2017) Combinatorial regulation of the balance between dynein microtubule end accumulation and initiation of directed motility. EMBO J. 36, 3387–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenney RJ et al. (2016) Tyrosination of α-tubulin controls the initiation of processive dynein–dynactin motility. EMBO J. 35, 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbosa DJ et al. (2017) Dynactin binding to tyrosinated microtubules promotes centrosome centration in C. elegans by enhancing dynein-mediated organelle transport. PLoS Genet. 13(7):e1006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yildiz A et al. (2004) Kinesin Walks Hand-Over-Hand. Science 303, 676–678 [DOI] [PubMed] [Google Scholar]
- 47.Yildiz A et al. (2003) Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 48.DeWitt MA et al. (2012) Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science 335, 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu W et al. (2012) Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol 19, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belyy V et al. (2014) Cytoplasmic dynein transports cargos via load-sharing between the heads. Nat. Commun 5:5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao L et al. (2019) Molecular mechanism of cytoplasmic dynein tension sensing. Nat. Commun 10:3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicholas MP et al. (2015) Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proc. Natl. Acad. Sci. U. S. A 112, 6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visscher K et al. (1999) Single kinesin molecules studied with a molecular force clamp. Nature 400, 184–189 [DOI] [PubMed] [Google Scholar]
- 54.King SJ and Schroer TA (2000) Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol 2, 20–24 [DOI] [PubMed] [Google Scholar]
- 55.Kardon JR et al. (2009) Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc. Natl. Acad. Sci. U. S. A 106, 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rai AK et al. (2013) Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 [DOI] [PubMed] [Google Scholar]
- 57.Rai A et al. (2016) Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell 164, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matanis T et al. (2002) Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol 4, 986–992 [DOI] [PubMed] [Google Scholar]
- 59.Splinter D et al. (2012) BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell 23, 4226–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlager MA et al. (2014) In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenney RJ et al. (2014) Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroeder CM and Vale RD (2016) Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J. Cell Biol 214, 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y et al. (2019) CRACR2a is a calcium-activated dynein adaptor protein that regulates endocytic traffic. J. Cell Biol 218, 1619–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urnavicius L et al. (2018) Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olenick MA et al. (2016) Hook adaptors induce unidirectional processive motility by enhancing the Dynein-Dynactin interaction. J. Biol. Chem 291, 18239–18251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Spronsen M et al. (2013) TRAK/Milton Motor-Adaptor Proteins Steer Mitochondrial Trafficking to Axons and Dendrites. Neuron 77, 485–502 [DOI] [PubMed] [Google Scholar]
- 67.Engelender S et al. (1997) Huntingtin-associated protein 1 (HAP1) interacts with the p150 Glued subunit of dynactin. Hum. Mol. Genet 6, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 68.McGuire JR et al. (2006) Interaction of Huntingtin-associated protein-1 with kinesin light chain: Implications in intracellular trafficking in neurons. J. Biol. Chem 281, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 69.Li SH et al. (1998) Interaction of Huntingtin-associated protein with dynactin P150(Glued). J. Neurosci 18, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X et al. (2011) PINK1 and Parkin target miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fransson Å et al. (2006) The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun 344, 500–510 [DOI] [PubMed] [Google Scholar]
- 72.Wang X and Schwarz TL (2009) The Mechanism of Ca2+-Dependent Regulation of Kinesin-Mediated Mitochondrial Motility. Cell 136, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalinski AL et al. (2019) Deacetylation of Miro1 by HDAC6 blocks mitochondrial transport and mediates axon growth inhibition. J. Cell Biol 218, 1871–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brickley K and Stephenson FA (2011) Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J. Biol. Chem 286, 18079–18092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li XJ and Li SH (2005) HAP1 and intracellular trafficking. Trends Pharmacol. Sci 26, 1–3 [DOI] [PubMed] [Google Scholar]
- 76.Ross CA et al. (2003) A huntingtin-associated protein enriched in brain with implications for pathology. Nature 378, 398–402 [DOI] [PubMed] [Google Scholar]
- 77.McGuire JR et al. (2005) Interaction of Huntingtin-associated Protein-1 with Kinesin Light Chain. J. Biol. Chem 281, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 78.Wu LLY and Zhou XF (2009) Huntingtin associated protein 1 and its functions. Cell Adhes. Migr 3, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wong YC and Holzbaur ELF (2014) The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci 34, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merdes A et al. (2000) Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol 149, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okumura M et al. (2018) Dynein–dynactin–NuMA clusters generate cortical spindle-pulling forces as a multiarm ensemble. Elife 7:e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gama JB et al. (2017) Molecular mechanism of kinetochore dynein recruitment by the Rod/Zw10/Zwilch complex and Spindly. J. Cell Biol 216, 654–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reck-Peterson SL et al. (2018) The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol 19, 382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stock MF et al. (1999) Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J. Biol. Chem 274, 14617–14623 [DOI] [PubMed] [Google Scholar]
- 85.Hackney DD and Stock MF (2000) Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol 2, 257–260 [DOI] [PubMed] [Google Scholar]
- 86.Kaan HYK et al. (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333, 883–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amos LA (1989) Brain dynein crossbridges microtubules into bundles. J. Cell Sci 93, 19–28 [DOI] [PubMed] [Google Scholar]
- 88.Toropova K et al. (2017) Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat. Struct. Mol. Biol 24, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toropova K et al. (2019) Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol 26, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grotjahn DA et al. (2018) Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat. Struct. Mol. Biol 25, 203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Derr ND et al. (2012) Tug-of-War in Motor Protein Ensembles. Science 338, 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furuta K et al. (2013) Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci 110, 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elshenawy MM et al. (2019) Cargo adaptors regulate stepping and force generation of mammalian dynein – dynactin. Nat. Chem. Biol 15, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mallik R et al. (2005) Building complexity: An in vitro study of cytoplasmic dynein with in vivo implications. Curr. Biol 15, 2075–2085 [DOI] [PubMed] [Google Scholar]
- 95.Mallik R et al. (2004) Cytoplasmic dynein functions as a gear in response to load. Nature 427, 649–652 [DOI] [PubMed] [Google Scholar]
- 96.Gennerich A et al. (2007) Force-Induced Bidirectional Stepping of Cytoplasmic Dynein. Cell 131, 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cho C et al. (2008) Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J. Biol. Chem 283, 25839–25845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schlager MA et al. (2014) Bicaudal D Family Adaptor Proteins Control the Velocity of Dynein-Based Movements. Cell Rep. 8, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 99.Leidel C et al. (2012) Measuring molecular motor forces in VIVO: Implications for tug-of-war models of bidirectional transport. Biophys. J 103, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hendricks AG et al. (2012) Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci 109, 18447–18452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Belyy V et al. (2016) The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol 18, 1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cella Zanacchi F et al. (2019) Quantifying Protein Copy Number in Super Resolution Using an Imaging-Invariant Calibration. Biophys. J 116, 2195–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reiner O et al. (1993) Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364, 717–721 [DOI] [PubMed] [Google Scholar]
- 104.Dobyns WB et al. (2011) Lissencephaly. A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA 270, 2838–2842 [DOI] [PubMed] [Google Scholar]
- 105.Faulkner NE et al. (2000) A role for the lissencephaly gene Lis1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol 2, 784–791 [DOI] [PubMed] [Google Scholar]
- 106.Smith DS et al. (2000) Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol 2, 767–775 [DOI] [PubMed] [Google Scholar]
- 107.Tai CY et al. (2002) Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol 156, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Niethammer M et al. (2000) NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711 [DOI] [PubMed] [Google Scholar]
- 109.Liang Y et al. (2004) Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J. Cell Biol 164, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J et al. (2005) NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol 7, 686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shu T et al. (2004) Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 [DOI] [PubMed] [Google Scholar]
- 112.Yan X et al. (2003) Human Nudel and NudE as Regulators of Cytoplasmic Dynein in Poleward Protein Transport along the Mitotic Spindle. Mol. Cell. Biol 23, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mesngon MT (2006) Regulation of Cytoplasmic Dynein ATPase by Lis1. J. Neurosci 26, 2132–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baumbach J et al. (2017) Lissencephaly-1 is a context-dependent regulator of the human dynein complex. Elife 6:e21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yamada M et al. (2008) LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 27, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.DeSantis ME et al. (2017) Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell 170, 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Toropova K et al. (2014) Lis1 regulates dynein by sterically blocking its mechanochemical cycle. Elife 3:e03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gutierrez PA et al. (2017) Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J. Biol. Chem 292, 12245–12255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Elshenawy MM et al. (2019) Lis1 activates dynein motility by pairing it with dynactin. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Htet ZM et al. (2019) Lis1 promotes the formation of maximally activated cytoplasmic dynein-1 complexes. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lenz JH et al. (2006) A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25, 2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Egan MJ et al. (2012) Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol 197, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marzo MG et al. (2019) Pac1 / LIS1 promotes an uninhibited conformation of dynein that coordinates its localization and activity. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qiu R et al. (2019) LIS1 regulates cargo-adapter – mediated activation of dynein by overcoming its autoinhibition in vivo. J. Cell Biol 218, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hoogenraad CC et al. (2001) Mammalian golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20, 4041–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schlager MA et al. (2010) Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 29, 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Short B et al. (2002) The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr. Biol 12, 1792–1795 [DOI] [PubMed] [Google Scholar]
- 128.Dienstbier M et al. (2009) Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sladewski TE et al. (2018) Recruitment of two dyneins to an mRNA-dependent bicaudal D transport complex. Elife 7:e36306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McClintock MA et al. (2018) RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. Elife 7:e36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Luiro K et al. (2004) Interconnections of CLN3, Hook1 and Rab proteins link Batten disease to defects in the endocytic pathway. Hum. Mol. Genet 13, 3017–3027 [DOI] [PubMed] [Google Scholar]
- 132.Walenta JH et al. (2001) The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol 152, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maldonado-Báez L et al. (2013) Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J. Cell Biol 201, 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Olenick MA et al. (2019) Dynein activator Hook1 is required for trafficking of BDNF-signaling endosomes in neurons. J. Cell Biol 218, 220–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ally S et al. (2009) Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol 187, 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martin M et al. (1999) Cytoplasmic Dynein, the Dynactin Complex, and Kinesin Are Interdependent and Essential for Fast Axonal Transport. Mol. Biol. Cell 10, 3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sainath R and Gallo G (2015) The dynein inhibitor Ciliobrevin D inhibits the bidirectional transport of organelles along sensory axons and impairs NGF-mediated regulation of growth cones and axon branches. Dev. Neurobiol 75, 757–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Encalada SE et al. (2011) Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell 144, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hancock WO (2014) Bidirectional cargo transport: Moving beyond tug of war. Nat. Rev. Mol. Cell Biol 15, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jordan MA et al. (2018) The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol 20, 1250–1255 [DOI] [PubMed] [Google Scholar]
- 141.Kozminski KG et al. (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U. S. A 90, 5519–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dentler W (2005) Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol 170, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chien A et al. (2017) Dynamics of the IFT machinery at the ciliary tip. Elife 6:e28606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liang Y et al. (2014) FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell 30, 585–597 [DOI] [PubMed] [Google Scholar]
- 145.Pan J and Snell WJ (2005) Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev. Cell 9, 431–438 [DOI] [PubMed] [Google Scholar]
- 146.Pederson LB et al. (2005) ChlamydomonasIFT172 Is EncodedbyFLA11, Interacts with CrEB1, and Regulates IFT at the Flagellar Tip. Curr. Biol 15, 262–265 [DOI] [PubMed] [Google Scholar]
- 147.Soppina V et al. (2009) Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. U. S. A 106, 19381–19386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deacon SW et al. (2003) Dynactin is required for bidirectional organelle transport. J. Cell Biol 160, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Levi V et al. (2006) Organelle transport along microtubules in Xenopus melanophores: Evidence for cooperation between multiple motors. Biophys. J 90, 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kural C et al. (2005) Kinesin and dynein move a peroxisome in vivo: A tug-of-war or coordinated movement? Science 308, 1469–1472 [DOI] [PubMed] [Google Scholar]
- 151.Kendrick AA et al. (2019) Hook3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein-1 and KIF1C. J. Cell Biol 218, 2982–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hendricks AG et al. (2010) Motor Coordination via a Tug-of-War Mechanism Drives Bidirectional Vesicle Transport. Curr. Biol 20, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaiser F et al. (2004) IIGP, a member of the IFN inducible and microbial defense mediating 47 kDa GTPase family, interacts with microtubule binding protein hook3. J. Cell Sci 117, 1747–1756 [DOI] [PubMed] [Google Scholar]
- 154.Lee IG et al. (2018) A conserved interaction of the dynein light intermediate chain with dynein-dynactin effectors necessary for processivity. Nat. Commun 9:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dwivedi D et al. (2019) The dynein adaptor Hook2 plays essential roles in mitotic progression and cytokinesis. J. Cell Biol 218, 871–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scherer J et al. (2014) PKA-dependent dynein switching from lysosomes to adenovirus: A novel form of host-virus competition. J. Cell Biol 205, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cavalli V et al. (2005) Sunday driver links axonal transport to damage signaling. J. Cell Biol 168, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]


