Abstract
Nitrogen use efficiency (NUE) is the efficiency with which plants acquire and use nitrogen. Plants have high-affinity nitrate transport systems, which involve certain nitrate transporter (NRT) genes. However, limited data are available on the contribution of the NRT2/3 gene family in barley nitrate transport. In the present study, ten putative NRT2 and three putative NRT3 genes were identified using bioinformatics methods. All the HvNRT2/3 genes were located on chromosomes 3H, 5H, 6H or 7H. Remarkably, the presence of tandem repeats indicated that duplication events contributed to the expansion of the NRT2 gene family in barley. In addition, the HvNRT2/3 genes displayed various expression patterns at selected developmental stages and were induced in the roots by both low and high nitrogen levels. Furthermore, the overexpression of HvNRT2.1 improved the yield related traits in Arabidopsis. Taken together, the data generated in the present study will be useful for genome-wide analyses to determine the precise role of the HvNRT2/3 genes during barley development, with the ultimate goal of improving NUE and crop production.
Introduction
Nitrogen, as a function of nutrient availability, plays an essential role in controlling plant growth and development [1]. Limited nitrogen supplies decrease crop production. However, the excessive use of nitrogen fertilizers results in severe pollution and environmental deterioration [2]. Therefore, it is important to improve nitrogen use efficiency for cereal production with a low nitrogen supply [3].
Improving nitrogen use efficiency (NUE) in plants requires a more complete understanding of the transport of NO3- from the soil to the plant and within the plant itself. To date, two kinetically distinct nitrate uptake systems have been identified by physiological and molecular studies in plant roots, including the low-affinity transport system (LATS), which is encoded by the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER (NRT1/PTR) family (NPF), and the high-affinity transport system (HATS), which is encoded by the nitrate transporter 2 (NRT2) family [4–8]. The functions of some NRT genes have been analyzed in plants. In Arabidopsis, AtNRT1.1 (CHL1), a dual-affinity nitrate transporter, functions as high-affinity and low-affinity nitrate transporters, when it is phosphorylated and dephosphorylated, respectively [9]. OsNRT1.1B (OsNPF6.5) is a putative homolog of CHL1, and it contributes to grain yield and NUE in rice [10]. The overexpression of OsNRT1.1A (OsNPF6.3) greatly improves N utilization and grain yield, as well as shortens the maturation time [11]. There is a lesser number of NRT2 genes than NRT1, and they are expressed predominantly in the root [8, 12]. In rice, OsNRT2.3 is transcribed into two spliced isoforms (OsNRT2.3a and OsNRT2.3b), and the overexpression of OsNRT2.3b leads to an improvement in grain yield and NUE by 40% [13]. However, when OsNRT2.3a expression is decreased, xylem loading of nitrate is impaired, and there is decreased plant growth at low nitrate levels [14]. This indicates that both alternative splices execute different functions in rice development.
NRT3 (also named NAR2) proteins are partner proteins that interact with most NRT2 proteins and contribute to high-affinity nitrate uptake [15]. In Atnrt3.1 mutant plant, the expression levels of AtNRT1.1 and AtNRT2.1 are reduced in response to nitrate induction, and constitutive high-affinity influx and high-affinity nitrate-inducible influx are dramatically decreased [15]. Similarly, OsNRT2.1, OsNRT2.2 and OsNRT2.3a are also synchronously suppressed in Osnar2.1 mutant, while high- and low-affinity nitrate transport is greatly impaired. A further analysis revealed that OsNAR2.1 not only interacts with OsNRT2.1/OsNRT2.2 but also with OsNRT2.3a [16]. Thus, the NRT2 genes combine with the NRT3 genes to enable the plant to cope with variable nitrate supplies.
In barley, four HvNRT2 (HvNRT2.1-2.4) and three HvNRT3 (HvNAR2.1-2.3) genes were isolated from the roots. The expression pattern of the HvNRT2 genes was also characterized under various nitrogen sources, and a highly specific interaction is suggested between HvNRT2.1 and HvNAR2.3 by using 15N-enriched nitrate uptake into Xenopus oocytes. However, until now, none of the HvNRT2/3 gene family have been described at the barley genome level and their functions are still unclear [17–19]. In the present study, we identified HvNRT2 and HvNRT3 genes in barley using bioinformatics and constructed a phylogenetic tree. Then, the expression patterns of these genes in response to low and high nitrate levels were analyzed. In addition, the HvNRT2.1 gene was overexpressed to determine its function in Arabidopsis. These results will be useful in further investigations into the functions of the NRT family in high-affinity nitrate uptake in plants.
Materials and methods
Plant materials
Barley cv. Morex was used in the present study. The seeds were surface-sterilized and germinated on wet filter paper at 25°C for 3 days. The germinated seeds were transferred into 60-well plastic containers (25 L) with aerated and nitrogen-free one-tenth-strength modified Johnson’s solution for 4 days [20]. The plants were then supplied with 0.2 mM NO3- (Low nitrogen, LN) or 5 mM NO3- (High nitrogen, HN). The plants were grown in a growth chamber at 25°C/22°C (day/night) under a 16-h/8-h light/dark cycle and 70% relative humidity. At 0 h, 1 h, 3 h, 6 h, 12 h, 24 h, 48 h, and 72 h after the treatment, the total roots were harvested from ten plants and were immediately frozen in liquid nitrogen for RNA extraction, with three biological replicates at each time point. For each replicate, the RNA from four plants of each genotype was analyzed. The Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used as the wild type. The plants were grown in a greenhouse in soil at 20°C under long-day conditions (16 h light/8 h dark). For the in vitro seedling assays, the seeds were surface sterilized and cold treated at 4°C for 3 days in the dark and were then exposed to white light. The plants were grown at 20°C on horizontal or vertical plates containing Murashige and Skoog (MS) medium, 3% sucrose, and 0.9% agarose (Merck).
Sequence database searches
The amino acid sequences of the NRT2 and NRT3 genes generated from Arabidopsis [15, 21], rice [22], and maize [23, 24] were used as query sequences. The barley sequence data were sourced from the Morex (http://webblast.ipk-gatersleben.de/barley/) [25], Gramene (http://ensembl.gramene.org/Hordeum_vulgare/Info/Index), and NCBI databases (http://www.ncbi.nlm.nih.gov/). BLAST programs (TBLASTN and BLASTN) were available for the IPK barley genome database and the NCBI barley EST database. Multiple database searches were performed to collect all members of the barley NRT2 and NRT3 genes. Domain searches (PF07690.14 and PF16974.3) were performed using SignalP (http://www.cbs.dtu.dk/services/SignalP/) [26], HMMSCAN (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) [27], and SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/) [28], with the default cutoff parameters. The isoelectric points and protein molecular weights were obtained with the help of the proteomics and sequence analysis tools on the ExPASy proteomics server (http://expasy.org/) [29]. The names of the HvNRT2 genes were designated according to their ascending order in barley chromosomes. However, the names of the HvNRT3 genes were given according to the definition provided by Tong et al. [30].
Chromosomal location, gene structure, and duplication events of the NRT2/3 genes
The chromosomal locations were retrieved from the IPK database (http://webblast.ipk-gatersleben.de/barley/). All the genes were mapped to the chromosomes with MapDraw software [31]. The exon/intron structures were constructed using GSDS (http://gsds.cbi.pku.edu.cn/) [32]. Tandem duplication genes were identified manually if they were within the 10 predicted genes or within 30 kb of each other on the physical barley map [33]. Segmental duplications were identified by BLASTP in ten predicted proteins upstream and downstream of each of the HvNRT2/3 genes [34].
Phylogenetic tree analysis
Full-length amino acid sequences of the HvNRT2/3 genes identified in barley were aligned using the Clustal X 1.83 program with the default pairwise and multiple alignment parameters. The phylogenetic tree was constructed based on this alignment using the neighbor joining (NJ) method in MEGA version 6 with the following parameters: Poisson correction; pairwise deletion; uniform rates; and bootstrap (1000 replicates) [35]. Conserved motifs were investigated by multiple alignment analyses using MEME version 3.0 [36].
NRT2 and NRT3 gene expression analyses
Gene expression data from eight tissues of the cultivar ‘Morex’ were obtained from the barley genome database (http://apex.ipk-gatersleben.de/apex/f?p=284:10:6281639160219::NO). Deep RNA sequencing (RNA-seq) was carried out on fifteen Morex tissues from almost all stages of the barley life cycle, which was comprised 4-day embryos derived from a germinated seed, the roots and shoots from a 10 cm seedling (10 cm shoot stage), inflorescences (5 cm and 1-1.5 cm), developing grains at 5 and 15 days after pollination (DAP), an etiolated seedling (10 DAP), the epidermal strips and roots (28 DAP), rachis of inflorescences (35 DAP), lemma, lodicule, and palea (42 DAP), developing tillers at the 3rd internode (42 DAP), and senescing leaves (56 DAP) [25]. The expression patterns are presented as heat maps in green/yellow/red coding, which reflects the FPKM (Fragments Per Kilobase of transcript per Million mapped reads), with red indicating a high expression level, yellow indicating a moderate expression level, and green indicating a low expression level.
Isolation of total RNA and quantitative real-time PCR
Total RNA was isolated using an RNA extraction kit (TRIzol reagent, Invitrogen, USA), and the isolated RNA was incubated with RNase-free DNase I (TaKaRa, Japan) to remove any contaminating DNA. The RNA quality and yield were analyzed by agarose gel electrophoresis and a NanoDrop 1000 Spectrophotometer V 3.7. First strand cDNA was generated from 2 μg of total RNA with M-MLV reverse transcriptase (TaKaRa, Japan) using random primers. The specific primers used for the quantitative real-time PCR analysis are listed in S1 Table. The reactions were carried out in 20 μl reaction systems containing 10 mM Tris-HCl (pH 8.5), 50 mM KCl, 2 mM MgCl2, 0.4 μl DMSO, 200 mM dNTPs, 10 pmol specific PCR primers, 1 U Taq DNA polymerase, and 0.5 μl SYBR GREEN I fluorescence dye. Quantitative real-time PCR was performed using a ViiA™ 7 Real-Time PCR System (Applied Biosystems, USA). The running protocol was as follows: 94°C for 3 min, followed by 40 cycles at 94°C for 30 s; 58°C for 30 s; 72°C for 30 s; and a final extension of 72°C for 5 min. The amplification of HvActin (Accession number HORVU1Hr1G002840) was employed as an internal standard. All the reactions were run in triplicate. The Ct values were determined by ViiA™ 7 software using the default settings. The relative expression levels of the target genes were determined using the 2-ΔΔCt method [37]. For each sample, the PCR was performed with three biological replicates.
Generation of Arabidopsis transgenic plants
The full-length coding sequence of HvNRT2.1 was cloned into the Gateway entry vector pDNOR221 (S1 Table). After confirmation by DNA sequencing, the HvNRT2.1 fragments were recombined into the pB2GW7 destination vector, which expresses under the control of the CaMV35S promoter. The resulting construct was introduced into the Agrobacterium tumefaciens strain GV3101 and was used to transform the Arabidopsis plants using the floral dip method. The transgenic plants were selected by herbicide treatment, and three representative T3 homozygous lines were obtained for the phenotypic analysis. The statistical analysis of the differences between the wide type and transgenic plants was performed by using a Student’s t-test.
Silique and seed size measurements
Mature siliques were harvested, and their lengths were recorded using vernier calipers. The seed size measurement was performed as described previously [38]. Briefly, the siliques were harvested once they were dry. The seeds were allowed to dry completely in centrifuge tubes for at least seven days before the measurement. Next, the seeds were spread and taped onto a piece of white paper, so that none of the seeds were touching each other. Using a flatbed scanner (Canon CanoScan 9000F), images of each accession was obtained at a resolution of 300 dpi with transmitted light. The “particle analysis” feature of ImageJ software was used to measure the seed length, width, and area. Three biological replicates were undertaken with 10 plants per transformed plant.
Nitrate and nitrogen content measurements
To determine the tissue nitrate content at maturity, Col-0 and transgenic plants were weighed and boiled in distilled water for 5 min, and the nitrate content was determined by the Cataldo method [39]. Whole plants were dried to a constant weight at 80°C and were ground in a Cyclotec 1093 sample mill (Hoganas City, Sweden) before being sieved through a 0.5 mm screen. The total nitrogen content in the plants was quantified according to the Kjeldahl method by using FOSS Kjeltec TM 2300 (Foss Analytical AB, Sweden) [40]. The statistical analysis of the differences in aerial part traits between the Col-0 and transgenic plants was performed using a Student’s t-test.
Results
Identification of NRT2/3 genes in the barley genome
A total of ten putative HvNRT2 and three putative HvNRT3 genes from the entire barley genome were identified (Table 1). The HvNRT2 genes encoded proteins ranging from 483 (HvNRT2.10) to 514 (HvNRT2.2) amino acids, with protein masses that ranged from 50.40 kD to 55.38 kD and protein pI values ranging from 7.01 (HvNRT2.10) to 8.98 (HvNRT2.1) (Table 1). However, the HvNRT3 genes encoded low molecular weight proteins compared to the HvNRT2 genes, with the protein masses ranging from 21.13 kD to 21.27 kD. These were also basic proteins (the pI values ranged from 9.00 to 9.23). A further analysis revealed that HvNRT3.2 was located on the end of 5HL, HvNRT2.2-9 was on the end of 6HS, HvNRT3.1 on the centromeric regions of 6H, and the remaining genes were distributed on the long arm of chromosomes 3, 6, and 7 (Fig 1). Remarkably, the duplication event analysis indicated that 8 (8/10, 80%) of the HvNRT2 genes were tandem repeated (Fig 1). The tandem duplicated genes contained two clusters, and each cluster contained four genes (HvNRT2.2-5 and HvNRT2.6-9 gene pairs). The gene structural analyses showed that the HvNRT2 genes contained one exon. However, the HvNRT3 genes shared three exons (S1 Fig).
Table 1. The information of HvNRT2/3 genes in barley.
| Gene Name | Gene ID | Chr. | Physical Position on Barley Genome (start position (bp)-end position (bp)) | Coding Sequence Length (bp) | Amino Acid Length (aa) | Mass (Da) | pI |
|---|---|---|---|---|---|---|---|
| HvNRT2.1 | HORVU3Hr1G066090.1 | 3 | 503310429-503312717 | 1542 | 514 | 55376.14 | 8.98 |
| HvNRT2.2 | HORVU6Hr1G005570.1 | 6 | 12363356-12364512 | 1521 | 507 | 54703.55 | 8.39 |
| HvNRT2.3 | HORVU6Hr1G005580.1 | 6 | 12371132-12373090 | 1521 | 507 | 54673.52 | 8.39 |
| HvNRT2.4 | HORVU6Hr1G005590.1 | 6 | 12378589-12380579 | 1521 | 507 | 54627.44 | 8.39 |
| HvNRT2.5 | HORVU6Hr1G005600.2 | 6 | 12385842-12387748 | 1527 | 509 | 54974.94 | 8.53 |
| HvNRT2.6 | HORVU6Hr1G005720.1 | 6 | 12654106-12655479 | 1524 | 508 | 54658.86 | 8.67 |
| HvNRT2.7 | HORVU6Hr1G005770.1 | 6 | 12754340-12756226 | 1527 | 509 | 54553.54 | 8.14 |
| HvNRT2.8 | HORVU6Hr1G005780.1 | 6 | 12765305-12766893 | 1521 | 507 | 54367.15 | 7.9 |
| HvNRT2.9 | HORVU6Hr1G005930.1 | 6 | 13075246-13077142 | 1524 | 508 | 54636.74 | 8.17 |
| HvNRT2.10 | HORVU7Hr1G098550.4 | 7 | 598494068-598496471 | 1449 | 483 | 50402.35 | 7.01 |
| HvNAR3.1 | MLOC_3053.1 | 6 | 268,053,711-268,054,907 | 591 | 197 | 21129.3 | 9.11 |
| HvNAR3.2 | HORVU5Hr1G115500.3 | 5 | 646684461-646686179 | 777 | 259 | 21268.37 | 9 |
| HvNAR3.3 | HORVU6Hr1G053710.1 | 6 | 336809019-336810586 | 594 | 198 | 21137.32 | 9.23 |
Fig 1. The chromosomal location of the HvNRT2/3 genes in the barley physical map.
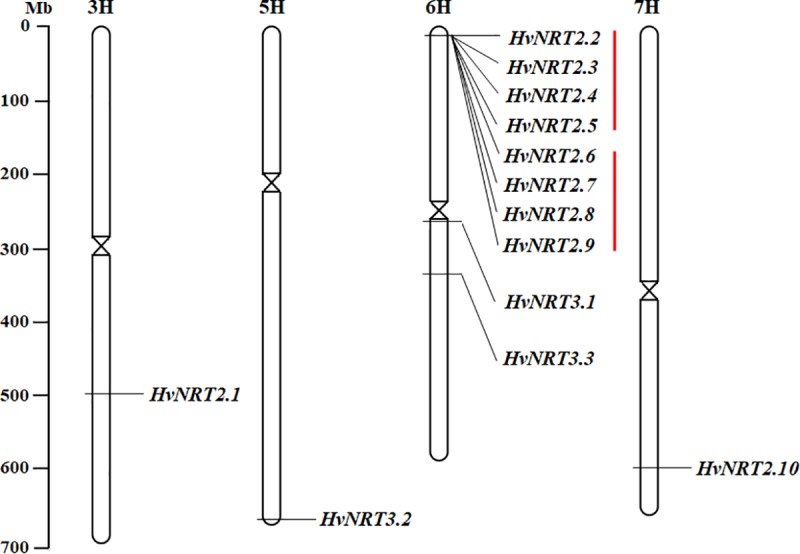
The red lines represent tandemly duplicated gene pairs.
Conserved amino acid sequence and phylogenetic tree for the HvNRT2/3 proteins in plants
Bioinformatic methods were employed to analyze the HvNRT2 and HvNRT3 proteins in the present study, revealing that an MFS domain and an NAR domain were present in the HvNRT2 and HvNRT3 proteins, respectively (S2 Fig). The alignment and comparison of the HvNRT2 full length protein sequences illustrated that the proteins had ten (HvNRT2.10), eleven (HvNRT2.1/6/8), or twelve (HvNRT2.2/3/4/5/7/9) (S2 Fig) transmembrane domains, while only one transmembrane region was found in the HvNRT3 proteins.
According to the phylogenetic tree of the NRT2 and NRT3 proteins in barley, rice, maize, and Arabidopsis, the NRT2 proteins could clearly be divided into three distinct clusters (Ⅰ, Ⅱ, and Ⅲ). A total of 18 NRT2 genes belonged to cluster Ⅰ, including five NRT2 genes derived from dicotyledonous Arabidopsis and 13 NRT2 genes from monocotyledonous plants (8, 2, and 3 genes from barley, rice, and maize, respectively). AtNRT2.5 and AtNRT2.7 represented clusters Ⅱ and Ⅲ, containing four and three genes, according to the phylogenetic tree, respectively (Fig 2A). In the NRT3 phylogenetic tree, the proteins derived from the dicotyledonous and monocotyledonous plants clustered together (Fig 2B), indicating an evolutionary dichotomy of the NRT2/3 genes between the dicot and monocot plants. In addition, the number of NRT2 and NRT3 genes in barley was larger than in other plants.
Fig 2. Phylogenetic analysis of the NRT2/3 proteins in Arabidopsis, maize, rice, and barley.
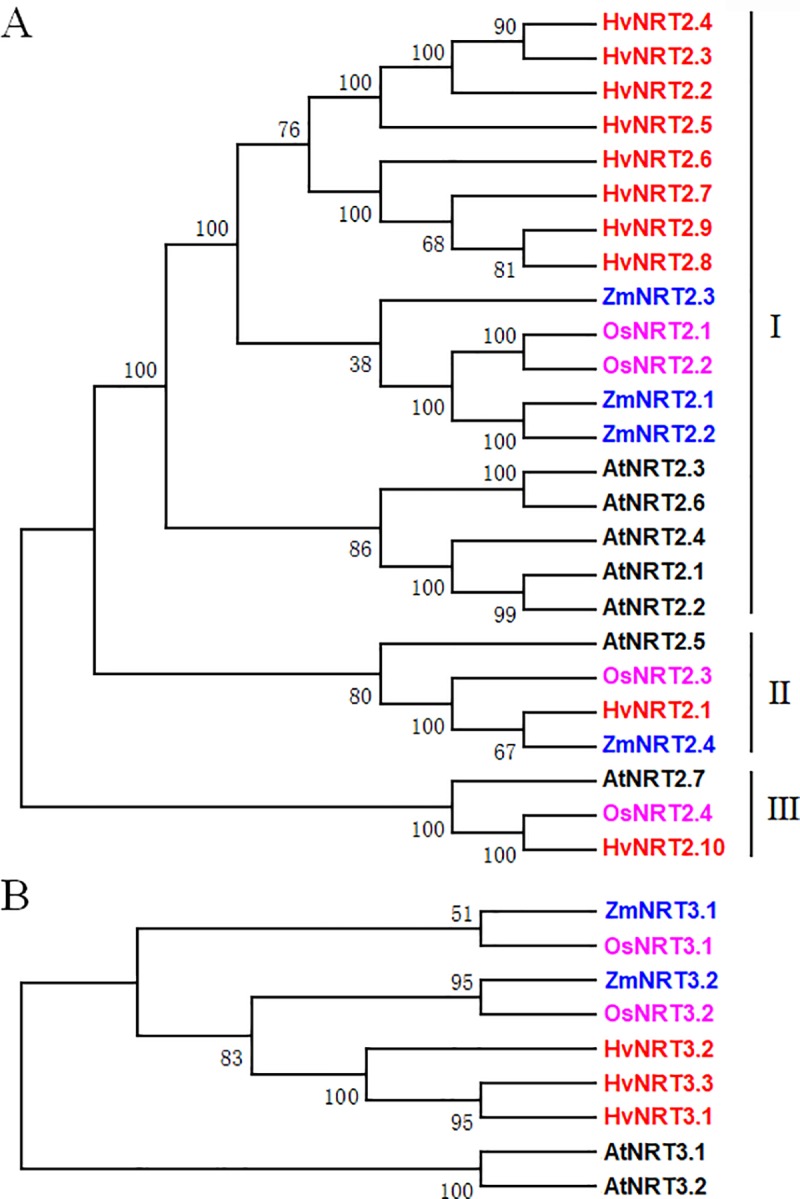
A, the phylogenetic analysis of the NRT2 genes; B, the phylogenetic analysis of the NRT3 genes. NRT2 gene sequence of Arabidopsis, maize, and rice generated from the report by Plett et al. [20].
HvNRT2/3 gene expression patterns at different developmental stages
An exploration of the expression patterns of genes contributes to our understanding of their molecular function [41]. Thus, we searched the deep RNA sequencing (RNA-seq) data from the cultivar ‘Morex’ [25, 42]. A heat map of the FPKM signal values for the 13 HvNRT2/3 genes in the selected Morex tissues is presented in Fig 3. All of the HvNRT2/3 genes displayed relatively high expression levels in the 4-day embryos dissected from germinating seeds (except HvNRT2.10) and in roots (10 cm shoot stage and 28 DAP). Conversely, a limited expression of the HvNRT2/3 genes was observed in the developing inflorescences (0.5-1.5 cm) and grains (5 and 15 DAP). In addition, HvNRT2.10 had a higher expression level than other genes in the shoots (10 cm shoot stage), while HvNRT3.2 had a relatively high expression level in the senescing leaves. Remarkably, HvNRT3.2 showed an exceptionally high expression level compared to the other genes in the developing tillers (the third internode at 42 DAP), etiolated seedlings (10 DAP), lemmas (42 DAP), lodicules (42 DAP), palea (42 DAP), epidermis (28 DAP), and rachises (35 DAP). This expression pattern analysis may contribute to our understanding of the function of the HvNRT2/3 genes in barley.
Fig 3. Expression profile analysis of the HvNRT2/3 genes in barley.
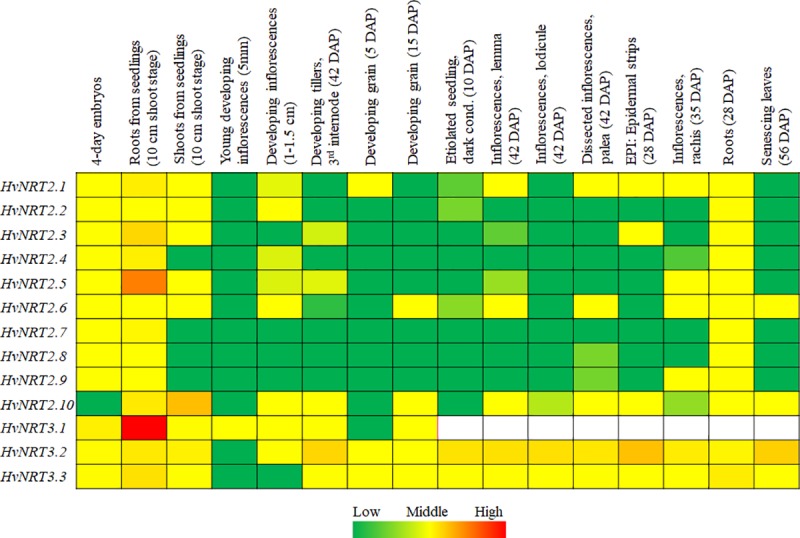
The expression patterns are presented as heat maps in green/yellow/red coding, which reflects the FPKM, with red indicating high expression levels, yellow indicating moderate expression levels, and green indicating low expression levels. The data on the expression of the HvNRT3.1 gene were only gathered from eight selected tissues [39]. The default values are shown in white.
HvNRT2/3 gene expression analysis under low and high nitrogen levels
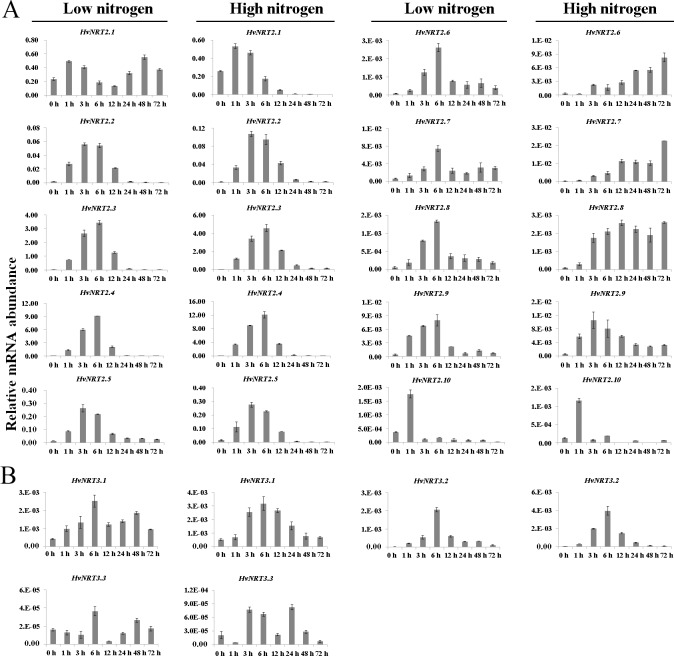
The expression pattern of the HvNRT2 and HvNRT3 genes in barley roots was investigated using quantitative real-time PCR. As shown in Fig 4, when the barley roots were exposed to low (0.2 mM) and high (5 mM) nitrogen levels, the number of transcripts of seven HvNRT2 and two HvNRT3 genes increased, with peaks at 1 h (HvNRT2.1/10 genes), 3 h (HvNRT2.2/5 genes), and 6 h (HvNRT2.3/4/9 and HvNRT3.1/2 genes), before gradually decreasing. However, the mRNA levels of HvNRT2.6/7/8 rapidly increased and peaked at 6 h under the low nitrogen conditions and at 72 h under the high nitrogen conditions. In addition, the expression of HvNRT3.3 displayed two peaks at 3 h and 24 h and responded more quickly to high rather than low nitrogen conditions.
Fig 4. Expression patterns of the HvNRT2/3 genes in the roots pretreated with low and high nitrogen levels.
A, expression patterns of the HvNRT2 genes; B, expression patterns of the HvNRT3 genes.
Morphological characterization of the HvNRT2.1 overexpression plants
In order to investigate the functional roles of the barley NRT genes, HvNRT2.1, which has a high homology to OsNRT2.3, and was overexpressed in Arabidopsis. In the transgenic lines, HvNRT2.1 was introduced into the genomic DNA and was expressed at the transcription level in three independent overexpression plants (S3 Fig). Phenotypic changes were observed between the Col-0 and overexpression plants at the mature stage (Fig 5). There were no obvious differences in the plant height, dry weight, and number of siliques per plant between the Col-0 and transgenic lines (Fig 6A, 6B and 6D). However, 7.9%, 18.5%, and 23.3% increments in the length of siliques, yield per plant and grain number per siliques, respectively, were observed in the transgenic lines compared with Col-0 (Fig 6C, 6E and 6F). The size of the seeds, including the seed length, width, and thousand grain weight, was higher in the three independent HvNRT2.1 transgenic lines compared to the wild type (Fig 6G, 6H and 6I). The seed length and width significantly increased by 11.5% and 24.5% compared to the wild type, respectively. The thousand grain weight dramatically increased by 37.0%, suggesting that the barley HvNRT2.1 gene positively regulated seed development. In addition, the seeds from the overexpression plant accumulated significantly more nitrate and nitrogen, and these increased by 47.2% and 41.4%, respectively, compared with the wide type for the mature plants (Fig 7).
Fig 5. Phenotype of the overexpression and Col-0 plants.
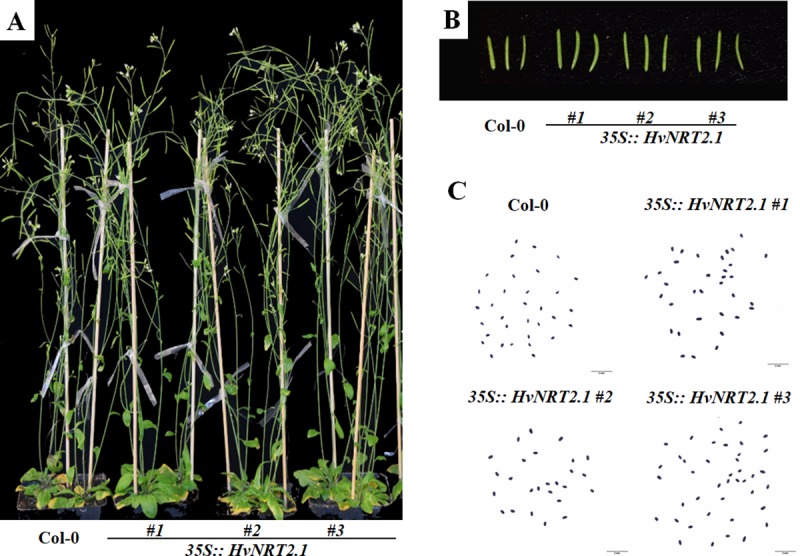
A, Whole plant; B, Silique; C, Seed.
Fig 6. Phenotype analysis of the Col-0 and overexpression plants.
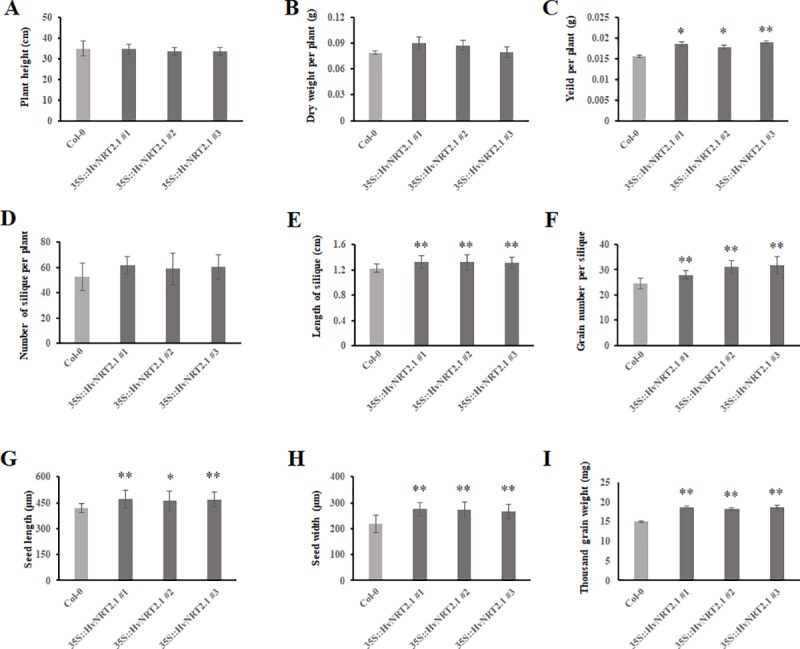
A, plant height; B, dry weight; C, yield per plant; D, number of silique per plant; E, length of silique; F, grain number per plant; G, seed length; H, seed width; I, thousand grain weight. ∗p < 0.05; ∗∗p < 0.01.
Fig 7. Nitrate and nitrogen content in the Col-0 and overexpression seeds.
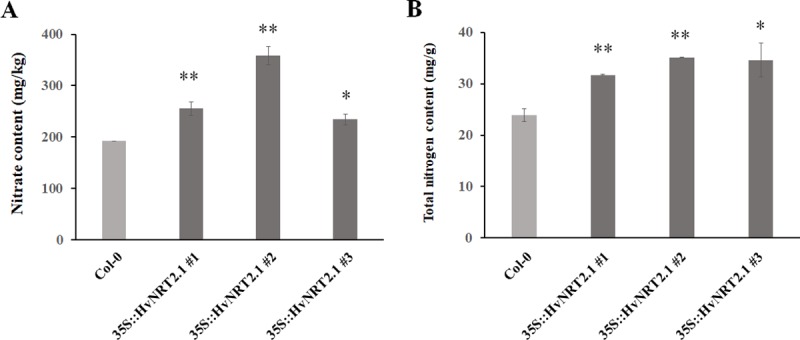
A, nitrate content; B, nitrogen content. ∗p < 0.05; ∗∗p < 0.01.
Discussion
Both NRT2 and NRT3 proteins are involved in high-affinity nitrate uptake in plants [15]. Due to the sequencing of plant genomes, the NRT2/3 genes have been widely analyzed in plants. For example, a total of 4, 7, and 4 NRT2 genes have been analyzed in rice, Arabidopsis, and maize, respectively, as well as two NRT3 genes [8, 15, 23–24, 43]. Although several HvNRT2 and HvNRT3 genes have been cloned in barley [30, 44], there is no information about barley HvNRT2/3 genes at the genome level. The present study identified ten putative NRT2 and three putative NRT3 genes that encode components of the high-affinity nitrate transport system (HATS) in barley. The number of NRT2/3 genes in barley is greater than those in rice, Arabidopsis, and maize. A duplication event analysis indicated that tandem duplication played an important role in the expansion of the NRT2 genes and may have contributed to the difference in the number of NRT genes in barley, rice, Arabidopsis, and maize [45]. In addition, SignalP and SMART predicted that three HvNRT3 proteins had signal peptides with 21 (HvNRT3.2/3) and 25 (HvNRT3.1) amino acids (S2 Table), which was similar to that of the AtNRT3 genes in Arabidopsis.
Phylogenetic analyses and evolutionary relationships are used to investigate the functions of genes in plants [46]. In rice, OsNRT2.1 and OsNRT2.2 share identical amino acid sequences and are clustered together with other NRT2 genes. Rice is a monocotyledon plant known to have undergone tandem duplication [8]. Interestingly, similar events were detected at the end of 6HS, where the HvNRT2.2/3/4/5 and HvNRT2.6/7/8/9 genes were clustered. Both HvNRT2.1 and HvNRT2.10 were closely related to OsNRT2.3 and OsNRT2.4 based on the phylogenetic tree. It is worth noting that barley has one exon in common with rice (except OsNRT2.4) but not Arabidopsis, suggesting that these NRT2 genes diverged before the monocotyledon-dicotyledon separation in higher plants [8, 12]. The structure of the NRT3 genes with two exons is conserved in plants, while divergence is observed in the protein homology between monocotyledon and dicotyledon plants [8, 15].
Expression analyses of organs is employed to predict the potential role of specific genes in plant growth and development, as well as their response to different environments [47]. NRT2 genes, as members of the high-affinity transport system, combined with NRT3 genes, were detected in the roots with high abundance levels, indicating that both gene families are responsible for induction either at low or high nitrate levels in the root tissue [8, 12, 15]. All of the thirteen genes investigated had high expression levels in the root tissue and displayed obvious changes following low and high nitrogen treatments. Remarkably, HvNRT2.10 is a homologous of AtNRT2.7 and OsNRT2.4, with 45% and 51% amino acid identity, respectively. It has a similar pattern of expression compared to its homologs, with higher expression levels in the shoots than the roots, and is also relatively highly expressed in seeds, which is consistent with the AtNRT2.7 gene [48, 49]. In rice, a decreased expression of OsNRT2.3a is related to the accumulation of significantly higher levels of nitrate and total nitrogen in the root and lower levels in the shoot [14]. However, enhancing the expression of OsNRT2.3b improves the growth, yield, and NUE in rice [13]. The HvNRT2.1 protein had a similar amino acid length and showed 83.3% identity with OsNRT2.3a, but only 78.4% identity with OsNRT2.3b genes, indicating that HvNRT2.1 plays an important role in long-distance nitrate transport, from the roots to shoots, under low nitrate supply levels in barley. These results will be useful in further investigations of HvNRT2.1 and HvNRT2.10 in barley.
NRT3 genes encode a partner protein that interacts with some members of NRT2 in plants [16, 50]. For example, AtNRT2.1 combined with AtNRT3.1 forms a 150-kDa plasma membrane complex. Both genes were thought to constitute the high-affinity nitrate transporter of Arabidopsis roots [50]. OsNRT3.1 (OsNAR2.1) was induced by low and high nitrogen levels, and in vitro experiments demonstrated that OsNAR2.1 interacts with OsNRT2.1 and OsNRT2.2, as well as OsNRT2.3a [16]. Remarkably, eight HvNRT2 genes and two HvNRT3 genes displayed co-expression patterns under low nitrogen conditions. Therefore, the interactions between the HvNRT2 and HvNRT3 genes should be further investigated to clarify the functions of the NRT genes in barley.
Several NRT2 genes have been characterized in plants. For example, an Atnrt2.1-1 mutant was specifically deficient in HATS, and a further analysis showed that when the ATNRT2.1 protein combined with ATNAR2.1, it forms a complex that transports nitrate efficiently [15, 51, 52]. ATNRT2.7, which had a high expression level in the dry seeds, was localized to the vacuolar membrane and displayed a positive regulation of nitrate content in the seeds. In the present study, the overexpression of HvNRT2.1 led to high nitrate and nitrogen contents and improved the yield-related traits of Arabidopsis. However, both a biomass and plant height increase were not observed in the overexpression plants, indicating that the HvNRT2.1 gene plays a specific role in nitrate and total nitrogen accumulation in plants.
Conclusion
In summary, ten putative NRT2 and three putative NRT3 genes were identified from barley sequencing using bioinformatic methods. A duplication event analysis indicated that tandem repeats contributed to the expansion of the NRT2 gene family in barley. A phylogenetic tree revealed that the NRT2/3 proteins displayed a clear divergence between the monocot and dicot plants. In addition, the HvNRT2/3 genes displayed various expression patterns at selected development stages and were induced in the roots under low and high nitrogen levels. Remarkably, HvNRT2.1, as a homologous gene of OsNRT2.3, significantly improved the yield-related traits in Arabidopsis. Thus, the data generated in the present study will be powerful for genome-wide analyses to determine the precise role of the HvNRT2/3 genes during barley development, with the ultimate goal of improving NUE and crop production.
Supporting information
(TIF)
Amino acid sequence alignments of the HvNRT2 (A) and HvNRT3 (B) proteins. The conserved transmembrane sequence regions are indicated by the black lines above the sequences. The red frame indicates the MFS and NAR domains in the HvNRT2 and HvNRT3 proteins, respectively.
(TIF)
A, PCR analysis at the genomic level; B, RT-PCR analysis at the transcription level.
(TIF)
(XLSX)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported from the National Natural Science Foundation of China (31771771, 31401370 and 31571648), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (17KJB210006), National Barley and Highland Barley Industrial Technology Specially Constructive Foundation of China (CARS-05), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci.U.S.A. 2008; 105: 4939–4944. 10.1073/pnas.0800211105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tilman D. Global environmental impacts of agricultural expansion: the need for sustainable and efficient practices. Proc. Natl. Acad. Sci.U.S.A. 1999; 96: 5995–6000. 10.1073/pnas.96.11.5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassman KG, Peng S, Olk D C, Ladha JK, Reichardt W, Dobermann A, et al. Opportunities for increased nitrogen-use efficiency from improved resource management in irrigated rice systems. Field Crops Res. 1998; 56: 7–39. 10.1016/S0378-4290(97)00140-8 [DOI] [Google Scholar]
- 4.Daniel-Vedele F, Filleur S, Caboche M. Nitrate transport: a key step in nitrate assimilation. Curr Opin Plant Biol. 1998; 1: 235–239. 10.1016/s1369-5266(98)80110-6 [DOI] [PubMed] [Google Scholar]
- 5.Galvan A, Fernandez E. Eukaryotic nitrate and nitrite transporters. Cellular and Molecular Life Sciences. 2001; 58: 225–233. 10.1007/PL00000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass ADM, Brito DT, Kaiser BN, Kronzucker HJ, Kumar A, Okamoto M, et al. Nitrogen transport in plants, with an emphasis on the regulations of fluxes to match plant demand. J Plant Nutr Soil Sc. 2001; 164: 199–207. 10.1002/1522-2624(200104)164:23.3.CO;2-B [DOI] [Google Scholar]
- 7.Williams LE, Miller AJ. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Biol. 2001; 52: 659–688. 10.1146/annurev.arplant.52.1.659 [DOI] [PubMed] [Google Scholar]
- 8.Cai C, Wang JY, Zhu YG, Shen QR, Li B, Tong YP, et al. Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol. 2008; 50: 443–451. 10.1111/j.1744-7909.2008.00642.x [DOI] [PubMed] [Google Scholar]
- 9.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009; 138: 1184–1194. 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 10.Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet. 2015; 47: 834 10.1038/ng.3337 [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Hu B, Yuan DY, Liu YQ, Che RH, Hu YC, et al. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell. 2018; 30: 638–651. 10.1105/tpc.17.00809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto M, Vidmar JJ, Glass ADM. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol. 2003; 44: 304–317. 10.1093/pcp/pcg036 [DOI] [PubMed] [Google Scholar]
- 13.Fan XR, Tang Z, Tan YW, Zhang Y, Luo BB, Yang M, et al. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci.U.S.A. 2016; 113: 7118–7123. 10.1073/pnas.1525184113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z, Fan XR, Li Q, Feng HM, Miller AJ, Shen QR, et al. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 2012; 160: 2052–2063. 10.1104/pp.112.204461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, et al. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006; 140: 1036–1046. 10.1104/pp.105.074385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan M, Fan XR, Feng HM, Miller AJ, Shen QR, Xu GH. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011; 34: 1360–1372. 10.1111/j.1365-3040.2011.02335.x [DOI] [PubMed] [Google Scholar]
- 17.Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996; 175: 223–231. 10.1016/0378-1119(96)00154-0 [DOI] [PubMed] [Google Scholar]
- 18.Vidmar JJ, Zhuo D, Siddiqi M Y, Glass ADM. Isolation and characterization of HvNRT2. 3and HvNRT2. 4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiology, 2000; 122: 783–792. 10.1104/pp.122.3.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong YP, Zhou JJ, Li ZS, Miller AJ. A two‐component high‐affinity nitrate uptake system in barley. Plant Journal. 2005; 41: 442–450. 10.1111/j.1365-313X.2004.02310.x [DOI] [PubMed] [Google Scholar]
- 20.Vidmar JJ, Zhuo D, Siddiqi MY, Glass AD. Isolation and characterization of HvNRT2.3 and HvNRT2.4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiol. 2000; 122: 783–792. 10.1104/pp.122.3.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orsel M, Krapp A, Daniel-Vedele F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002; 129: 886–896. 10.1104/pp.005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki R, Hasegawa H. Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breeding Sci. 2006; 56: 295–302. 10.1007/s10265-010-0375-9 [DOI] [Google Scholar]
- 23.Plett D, Toubia J, Garnett T, Tester M. Kaiser B. N, Baumann U. Dichotomy in the NRT gene families of dicots and grass species. PloS One. 2010; 5: e15289 10.1371/journal.pone.0015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorgona A, Lupini A, Mercati F, Di DL, Sunseri F, Abenavoli MR. Nitrate uptake along the maize primary root: an integrated physiological and molecular approach. Plant Cell Environ. 2011; 34: 1127–1140. 10.1111/j.1365-3040.2011.02311.x [DOI] [PubMed] [Google Scholar]
- 25.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017; 544: 427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- 26.Petersen TN, Brunak S, Von HG, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011; 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 27.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F. et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43: W30–W38. 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014; 43: D257–D260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40: W597–603. 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong YP, Zhou JJ, Li ZS, Miller AJ. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005; 41: 442–450. 10.1111/j.1365-313X.2004.02310.x [DOI] [PubMed] [Google Scholar]
- 31.Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan. 2003; 25: 317–321. 10.1016/S0891-0618(02)00103-5 [DOI] [PubMed] [Google Scholar]
- 32.Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015; 31: 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003; 132: 530–543. 10.1104/pp.103.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006; 16: 510–519. 10.1101/gr.4680506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey T, Elkan C, Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Inltman RA, Brutlog D,Karp P, Lathrop R, Searls D, eds, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. American Association for Artificial Intelligence Press, Menlo Park, CA, (1994) pp 28–36. [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2ΔΔCt method. Methods. 2001; 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 38.Herridge RP, Day RC, Baldwin S, Macknight RC. Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods, 2011; 7:1–11. 10.1186/1746-4811-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cataldo DA, Haroon M, Schrader LE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal. 1975; 6: 71–80. 10.1080/00103627509366547 [DOI] [Google Scholar]
- 40.Kjeldahl C. A new method for the determination of nitrogen in organic matter. Z Anal Chem. 1883; 22: 366–382. 10.1007/BF02514058 [DOI] [Google Scholar]
- 41.Mao DH, Chen CY. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. Plos One. 2012; 7: e47275–e47275. 10.1371/journal.pone.0047275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012; 491: 711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- 43.Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F. Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta. 2004; 219: 714–721. 10.1007/s00425-004-1266-x [DOI] [PubMed] [Google Scholar]
- 44.Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996; 175: 223–231. 10.1016/0378-1119(96)00154-0 [DOI] [PubMed] [Google Scholar]
- 45.Forde BG. Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta. 2000; 1465: 219–235. 10.1016/s0005-2736(00)00140-1 [DOI] [PubMed] [Google Scholar]
- 46.Wen Z, Tyerman SD, Dechorgnat J, Ovchinnikova E, Dhugga KS, Kaiser BN. Maize NPF6 proteins are homologs of Arabidopsis CHL1 that are selective for both nitrate and chloride. Plant Cell. 2017; 29: 2581–2596. 10.1105/tpc.16.00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004; 136: 2621–2632. 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, et al. Daniel-Vedele F. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell. 2007; 19: 1590–1602. 10.1105/tpc.107.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei J, Zheng Y, Feng H, Qu H, Fan X, Yamaji N, et al. OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. Journal of experimental botany. 2018; 69: 1095–1107. 10.1093/jxb/erx486 [DOI] [PubMed] [Google Scholar]
- 50.Kotur Z, Mackenzie N, Ramesh S, Tyerman SD, Kaiser BN, Glass AD. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012; 194: 724–731. 10.1111/j.1469-8137.2012.04094.x [DOI] [PubMed] [Google Scholar]
- 51.Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A. Major alterations of the regulation of root NO3- uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 2001; 127: 262–271. 10.1104/pp.127.1.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, et al. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 2006; 142: 1304–1317. 10.1104/pp.106.085209 [DOI] [PMC free article] [PubMed] [Google Scholar]