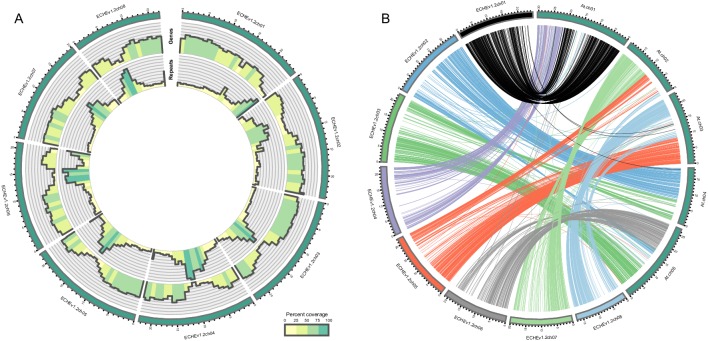
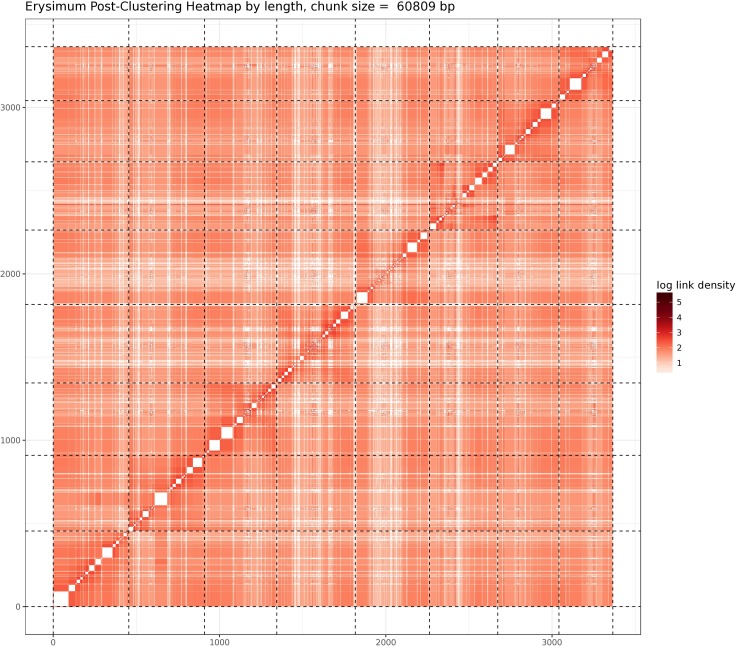
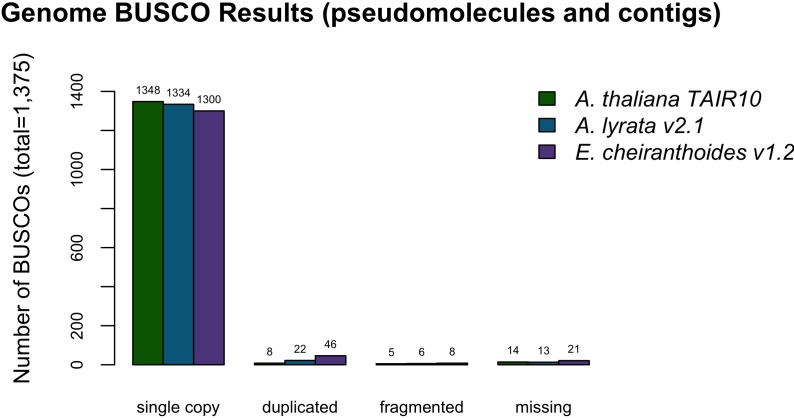
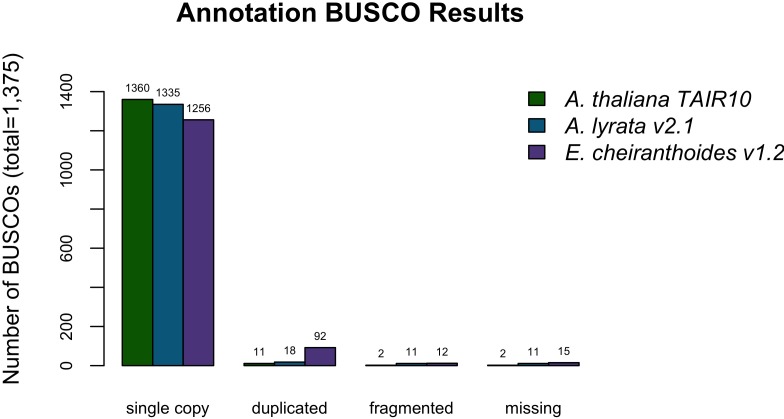
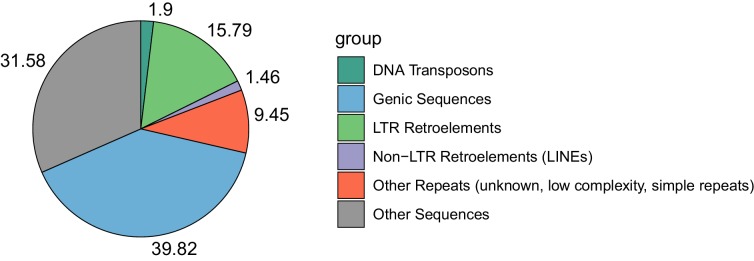
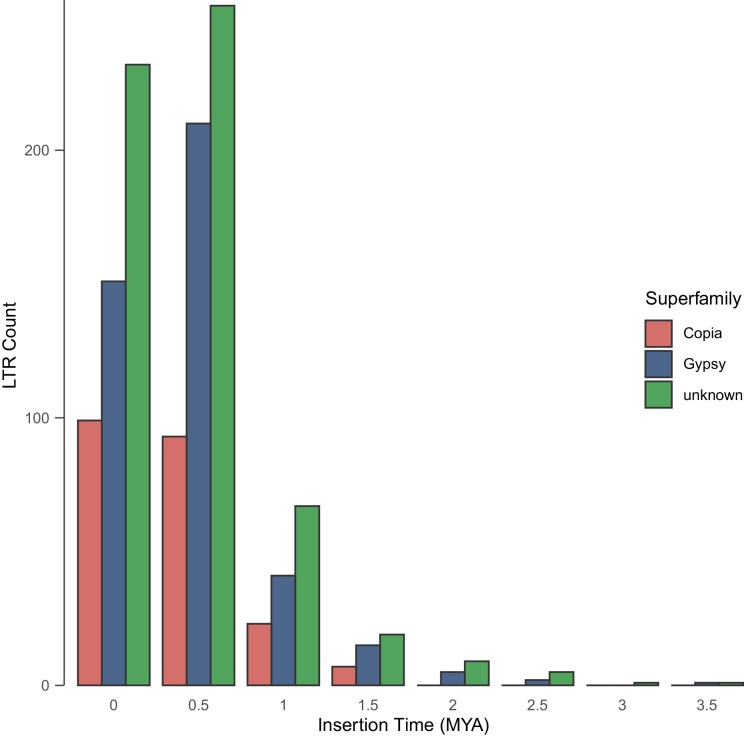
Figure 2. Visualization of the E.cheiranthoides genome assembly.
(A) Circos plot of the E. cheiranthoides genome with gene densities (outer circle) and repeat densities (inner circle) shown as histogram tracks. Densities are calculated as percentages for 1 Mb windows. (B) Synteny plot of E. cheiranthoides and A. thaliana. Lines between chromosomes connect aligned sequences between the two genomes.