Abstract
MX2 protein is a dynamin-like GTPase2 that has recently been identified as an interferon-induced restriction factor of HIV-1 and other primate lentiviruses. A single nucleotide polymorphism (SNP), rs45430, in an intron of the MX2 gene, was previously reported as a novel melanoma susceptibility locus in genome wide association studies. Functionally, however, it is still unclear if and how MX2 contributes to melanoma susceptibility and tumorigenesis. Here we show that MX2 is differentially expressed in melanoma tumors and cell lines, with most metastatic cell lines showing lower MX2 expression than primary melanoma cell lines and melanocytes. Furthermore, high expression of MX2 RNA in primary melanoma tumors is associated with better patient survival. Overexpression of MX2 reduces in vivo proliferation partially through inhibition of AKT activation, suggesting that it can act as a tumor-suppressor in melanoma. However, we have also identified a subset of melanoma cell lines with high endogenous MX2 expression where downregulation of MX2 leads to reduced proliferation. In these cells, MX2 downregulation interfered with DNA replication and cell cycle processes. Collectively our data for the first time show that MX2 is functionally involved in the regulation of melanoma proliferation but that its function is context-dependent.
Keywords: MX2, cell cycle, PI3K/AKT, SNP, melanoma specific survival
INTRODUCTION
Cutaneous melanoma makes up approximately 4% of skin cancers, yet it is responsible for more than 70% of skin cancer-related deaths (Sample & He, 2018). Somatic melanoma genetics are complex with tumors exhibiting high mutational load mostly attributed to UV induced DNA damage (Hodis et al., 2012). New germline genetic variants and genes contributing to melanoma susceptibility and progression are continually being discovered (Amos et al., 2011; Barrett et al., 2011; Bishop et al., 2009). Recently genome-wide association studies (GWAS) have linked rs45430 SNP, a major T to minor C allele change, intronic to MX2 (myxovirus resistance 2) gene with reduced risk to cutaneous melanoma and multiple primary tumors (Barrett et al., 2011; Gibbs et al., 2015). However, the functional role of this SNP or MX2 gene itself in the tumorigenesis has so far not been elucidated. MX2 protein is a dynamin-like GTPase2 identified as an interferon (IFN)-induced restriction factor for several primate lentiviruses including HIV-1 (Buffone, Schulte, Opp, & Diaz-Griffero, 2015; Goujon et al., 2013). Humans possess two MX genes, MX1 and MX2, with a high level of homology (Haller, Staeheli, Schwemmle, & Kochs, 2015). While MX1 protein is mainly induced after type I IFN (IFNα/β) stimulation during the antiviral response (Haller & Kochs, 2010; Kim, Shenoy, Kumar, Bradfield, & MacMicking, 2012), MX2 can be expressed at significant levels even in the absence of IFN (King, Raposo, & Lemmon, 2004). Unlike MX1, MX2 has an extended N-terminal domain, and exists as two isoforms. While the longer 78 kDa isoform displays anti-viral activity and is associated with the nuclear envelope, the shorter 76 kDa isoform is cytoplasmic without clearly defined cellular activity to date (Haller et al., 2015). MX2 found in association with nuclear pores contributes to the regulation of viral DNA nuclear import and/or integration into the host cell genome (Kane et al., 2013). One previous study suggested that MX2 could have additional, viral-independent cellular functions including regulation of cell-cycle progression (King et al., 2004).
Here we show for the first time, to best of our knowledge, that MX2 is functionally involved in cancer-related processes in melanoma. It is differentially expressed in melanoma tumors and cell lines, and it is a predictor of better patient survival. Interestingly, our data further show that MX2 function is complex, with both tumor suppressive and oncogenic features depending on the cellular context.
MATERIALS AND METHODS
Cell lines and culture conditions:
Primary human melanocytes (NHM9, NHM134, and NHM160) were isolated and cultured as previously described (Magnussen et al., 2012). Metastatic melanoma cell lines (MM) were established from melanoma patients treated at the Norwegian Radium hospital, Oslo University Hospital as described in (Flørenes et al., 2019). Melanoma cells were cultured in RPMI 1640 medium (Bio Whittaker, Verviers, Belgium) supplemented with 5% fetal bovine serum (FBS, Sigma), 2 mM/L L-glutamine (GibcoBRL, Paisley, UK) and maintained at 37°C in a humidified 5% CO2 atmosphere. Primary melanocytes were grown in 254CF melanocyte media purchased from Gibco Life Technologies (California, USA) supplemented with calcium chloride, HMGS-2 (human melanocytes growth supplement-2) and 10 ng/ml PMA. HEK293T cells (Clontech) were maintained in 4.5g/L glucose, 4mM L-glutamine Dulbecco’s Modified Eagle’s Medium (cat. No BE12–604F/U1; Lonza BioWhittaker, Verviers, Belgium) supplemented with 10% FBS and 25 mM HEPES (cat. No H0877, Sigma-Aldrich, UK).
siRNA knockdown:
Described in the Supporting information.
Double thymidine block:
Cells were synchronized at G1/S using a double thymidine block. At approximately 30% confluency MM382 cells were subjected to culturing media supplemented with 2mM thymidine for 16 hours (first block). Afterwards thymidine was washed off twice with PBS and cells were allowed to grow for 8 hours in normal conditions. Thymidine at final concentration of 2mM was added for additional 15 hours before final release. Cells were collected at 0, 2, 4, 6, 8, 10, and 12 hour time points after release.
Cell viability:
2×105 cells per well were seeded into 6 well-plates 24 h before treatment with siRNA. Cells were trypsinized, collected and the total number was counted after 72 h of treatment with siRNA. Viability values are presented as a mean percentage ±SE of three independent experiments normalized to the negative control siRNA.
RNA sequencing and analysis:
The RNA-seq files (fastq) prior to analysis were treated with Trimmomatic-0.38 (Bolger, Lohse, & Usadel, 2014) to remove sequence adapters. After trimming, the reads were (quasi)-mapped directly to the transcriptome using human (GRCh38, Ensemble version 94), Salmon software (Patro, Duggal, Love, Irizarry, & Kingsford, 2017). The DESeqDataSet were constructed by importing transcript abundance estimates from Salmon using the R txtimport package (Soneson, Love, & Robinson, 2015), differentially expressed genes detected by R DESeq2 package (Love, Huber, & Anders, 2014). For selection of differently expressed genes, a significance threshold based on adjusted p-value < 0.01 was applied. To further strengthen the selection, significant (p < 0.01) expressed genes from three groups; combined (knockout 1 and knockout 2) and individually were compared. From these a core of 520 genes was selected based on overlapping expression between the groups.
Data:
Sequence data are stored at Services for sensitive data (TSD) – University of Oslo. Access can be arranged by contacting the corresponding author (Ana S.) upon request.Graphical presentations: Heatmaps were constructed using aheapmap function in R package NMF (Gaujoux & Seoighe, 2010). The enrichGO function in the R-package Clusterprofiler (Yu, Wang, Han, & He, 2012) was used for the GO over-representation plots.
Incucyte growth rate assessment:
Cells overexpressing MX2 and GFP as a control were seeded into 24 well-plate at a density of 25 000 cells per well. Cell proliferation was measured by a confluence assay using IncuCyte™ FLR(Essen Instruments, Ann Arbor, MI) live cell imaging system. Phase contrast images were generated every 3 hours over a period of 3 days (for melanoma cells) or 4 days (for melanocytes). Cell proliferation was determined analyzing cell confluence over time. The experiment was repeated three times in triplicate. Confluence values were normalized to an initial time point, data is presented as a mean value at a given time point ±SE.
Cytoplasmic and nuclear fractionation:
NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (cat. No. 78833; Thermo Fisher Scientific) was used to isolate cytoplasmic and nuclear proteins. Isolation was performed according to manufacturer’s instructions. Halt Protease Inhibitor Cocktail (cat. No. 87785; Thermo Fisher Scientific) was added to the CER I and NER extraction reagents before use.
Flow cytometric analysis:
For cell cycle analysis 2×105 cells per well were seeded into 6 well plates 24 hours before treatment with siRNA. 48 hours after transfection cells were harvested by trypsinization, washed twice in ice cold PBS and fixed resuspending cell pellets in 1 mL 70% ice-cold methanol. Fixated cells were stained with a ready to use DNA Labelling Solution (Cytognos, cat.no. CYT-PIR-25). Flow cytometric experiments were performed on BD FACSCalibur™ Flow cytometer (BD Biosciences). Data were analysed with FlowJo v.7.6.1 software (Treestar Inc. Ashland, OR, USA).
Quantitative real-time PCR:
Described in the Supporting information.
Rs45430 SNP genotyping:
qPCR reactions were performed in duplicate in 96-well plates. 5 ng of genomic DNA (gDNA) was mixed with TaqMan Genotyping Master Mix (cat. No 4371353; Applied Biosystems) and TaqMan SNP Genotyping Assay (cat. No 4351379, assay ID C_2564407_10; Applied Biosystems) specific for rs45430 polymorphism. PCR reactions were performed on a QuantStudio™5Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific) running the following program: (1) enzyme activation at 95°C for 10 minutes, (2) 40 cycles of PCR at 95°C for 15 second and 60°C for 1 minute. Genotypes of the samples were determined from the allelic discrimination and amplification plots.
Generation of MX2 and GFP expression constructs:
MX2 cDNA was purchased from Origene (Rockville, MD, USA), catalog no. SC127459. Entry vector encoding GFP – pENTRY-GFP – was a gift from William Hahn (Addgene plasmid #15301). A destination vector pLenti-CMV-Puro-DEST (w118–1) was a gift from Eric Campeau and Paul Kaufman; Addgene plasmid #17452. pCW57.1 construct was a gift from from David Root; Addgene plasmid #41393. Detailed procedures for plasmid construction are described in Supporting information.
Lentivirus production and generation of stable cell lines:
Described in the Supporting Information.
In vivo animal studies:
WM983b cells (2×106) stably expressing MX2 or GFP diluted in 200 μl serum free RPMI-1640 media were subcutaneously injected in the right flank of nude female mice (athymic nude foxn1 nu). Tumor sizes were measured once a week using a calliper, and the volume V was calculated as follows: V = W2 × L × 0.5 (where W and L are tumour width and length, respectively). The experimental protocol was evaluated and approved by the National Animal Research Authority and conducted in accordance with regulations of the European Laboratory Animals Science Association.
Immunoblotting:
Immunoblotting was performed as previously described (Magnussen et al., 2012) with few modifications. Cells were lysed with ice-cold NP-40 lysis buffer supplemented with phosphatase inhibitor (4906837001, Roche Diagnostics) and protease inhibitor (4693124001, Roche Diagnostics). Proteins were resolved on 4–20% or 10% gels (Biorad) by SDS-PAGE electrophoresis. List of antibodies used is presented in Supporting Table 1. Visualization was performed with Super Signal West Dura chemoluminescence kit (Pierce).
Clinical melanoma specimens for IHC:
Formalin-fixed, paraffin-embedded tissue from 42 benign nevi, 154 primary melanomas and 60 metastases was examined for expression of MX2 protein. Clinical follow-up was available for all patients. 72 male and 82 female, mean age 55.6 (range 19–97). The follow-up period ranged from 1 to 361 months (mean = 104.8 months, median = 126.5 months). The Regional Committee for Medical Research Ethics South of Norway (S-06151) and The Social and Health Directorate (06/2733) approved the current study protocol.
Immunohistochemical analysis:
Immunohistochemical staining procedure is described in the Supporting Information. Semiquantitative classification was used to describe staining intensity (absent = 0; weak = 1; moderate = 2; strong = 3) and percentage of positive tumor cell (absent = 0; 0–25% = 1; 25–50% = 2; 50–75% = 3; >75% = 4). By multiplying intensity score with percentage positive cell score, a total immunoreactivity score was calculated ranging from 0 to 12. Immunoscore >3 was considered as high in the statistical analyses.
Mitotic rate classification:
Mitotic rate was histologically assessed by count of mitoses per mm2, also described in (Poźniak et al., 2019).
Statistical analysis:
Statistical analysis was performed applying SPSS package Version 18, (SPSS inc., Chicago, IL) and STATA 14.2. Comparison between variables was performed using the χ2 test or Fisher exact test. Two-tailed paired Student’s t-test and Wilcoxon matched-pairs signed-rank test was used for evaluation of in vitro results. A P value of less than 0.05 was considered statistically significant. In the Leeds Melanoma Cohort (LMC) (Nsengimana et al., 2018), the relationship between MX2 expression and mean tumor thickness was evaluated using Mann-Whitney two sample test. Melanoma Specific Survival (MSS) analysis of MX2 gene expression was performed using univariate Cox proportional hazard model in the whole dataset, and in each of the Immune Subgroups (Low, Intermediate and High). The generation of the Immune Subgroups was defined in (Poźniak et al., 2019) in which immune cell infiltration was imputed using the expression of genes reported to be exclusively expressed by each immune cell. The Kaplan Meier curve was generated after dichotomizing MX2 expression by median (high and low). The difference of MX2 expression was tested among the 3 Immune Subgroups using Kruskal Wallis and Dunnett’s test. Kaplan-Meier survival estimates and log rank tests were used to evaluate the survival data.
Transcriptomic data:
Generation of gene expression data from 703 FFPE tumors of the LMC was as described elsewhere (Nsengimana et al., 2018). These data were deposited in the European Genome-Phenome Atlas (EGA), accession number EGAS00001002922. Gene expression from metastatic melanomas in The Cancer Genome Atlas (TCGA) were downloaded from c-bioportal (http://www.cbioportal.org/) and were classified into the 3 immune subgroups as reported previously (Poźniak et al., 2019).
RESULTS
MX2 is constitutively and differentially expressed in melanoma tumors and cell lines
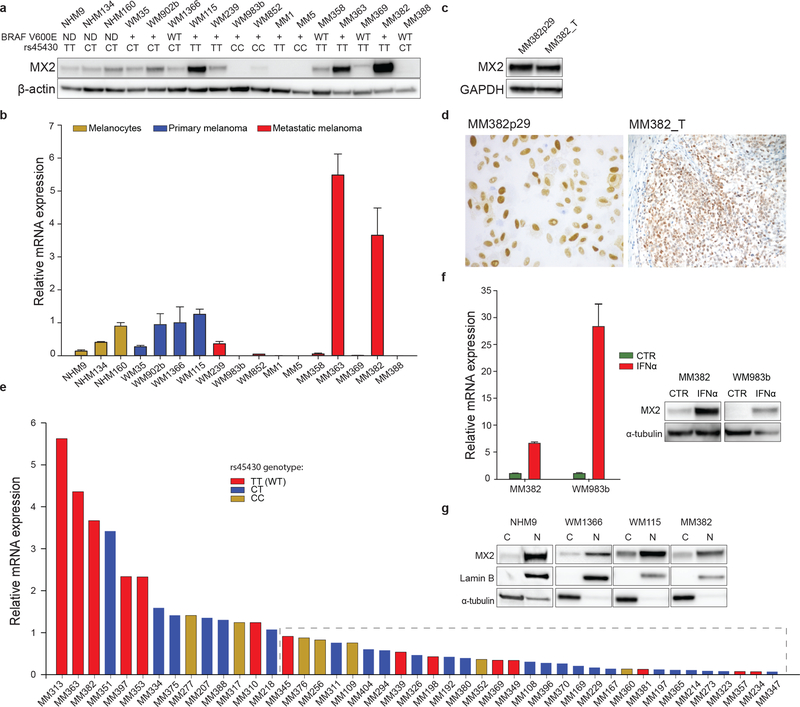
To investigate the potential role of MX2 in melanoma, we first examined its RNA and protein expression in a panel of human melanocytes, established primary and metastatic melanoma cell lines. Immunoblot analysis revealed constitutive, yet differential MX2 protein expression that correlated with RNA levels (Figure 1a and b). Most metastatic lines expressed lower levels of MX2 compared to the primary melanoma and cultured melanocyte lines. Furthermore, an apparent reduction of MX2 protein level was seen in metastatic WM239 line compared to primary WM115 line, both derived from the same patient, suggesting that MX2 is downregulated during disease progression.
Figure 1. Characterization of MX2 expression.
a) Analysis of MX2 protein expression by immunoblotting (β actin used as a loading control). BRAF V600E and rs45430 status specified under the cell names: (ND) – not determined, (WT) – wild type, (+) – mutation is present. b) MX2 mRNA expression in normal human melanocytes (NHM), primary and metastatic melanoma lines (mRNA expression is presented as a mean value ±SE of three independent experiments). MX2 mRNA expression is normalized to primary melanoma WM1366 cell line. c) Comparison of MX2 protein expression in established melanoma WM382 line and original tumor sample by immunoblotting and d) immunohistochemistry. e) MX2 mRNA expression in metastatic melanoma tumor samples. Tumors expressing lower MX2 mRNA levels compared to primary WM1366 are inside the dashed rectangle. Columns are colored according to rs45430 genotype. f) Increase of MX2 mRNA and protein expression after treatment with IFNα 1000 IU/ml for 24 h (mRNA expression is presented as a mean value ±SE of three independent experiments). g) Cytoplasmic and nuclear expression of MX2 in normal human melanocytes, primary and metastatic melanoma cell lines examined by immunoblotting. Each MX2 blot was visualized separately.
Interestingly, the highest MX2 protein expression was seen in the recently established early passage metastatic MM382 line. To rule out that this could be an in vitro culturing artifact we also examined MX2 expression in the original tumor sample that was histologically dissected and evaluated to contain more than 80% of tumor cells, and found it to be comparable to the cell line (Figure 1c, d). Variable expression of MX2 RNA was also observed in 45 fresh metastatic melanoma tumor samples, derived from lymph nodes. The majority (31/45) of samples had lower relative RNA levels when normalized to primary WM1366 cell line (Figure 1e). Furthermore, there was no statistically significant difference in MX2 expression between BRAFV600E mutant and wild type samples (Supporting Table 2).
We also investigated if rs45430 SNP is associated with MX2 expression both in cell lines and metastatic melanoma samples (Figure 1a and e). While we observed a tendency for TT genotype to be associated with a higher MX2 expression, it was not statistically significant.
In other cell types, MX2 expression is shown to be induced by IFN signaling. To examine if this is valid in melanoma, we incubated the low and high MX2 expressing cell lines WM983b and MM382 with IFNα or IFNγ for 24h. IFNα/γ treatment resulted in upregulation of both RNA and protein MX2 level (Figure 1f and Supporting Figure 1a), confirming that MX2 is an IFN response gene in melanoma, though it can also be constitutively expressed independently of INF stimulation.
We also examined possible association between MX2 expression and related MX1, in the same cell lines and tumor samples. We observed no correlation between MX2 and MX1 protein (Supporting Figure 1b) and mRNA expression (Supporting Figure 1c and d) in the cell lines. However, in tumor samples MX2 and MX1 mRNA expression significantly correlated, possibly due to contribution of microenvironment derived IFN (Supporting Figure 1e and f).
Antiviral functions of MX2 have been associated with its localization to the nuclear envelope; however cytoplasmic localization has also been reported (Dicks et al., 2018; Melén et al., 1996). Cytoplasmic and nuclear fractionation of melanoma cell lines showed that MX2 protein is mainly found in the nuclear fraction, but a weak cytoplasmic localization was also detected (Figure 1g).
MX2 expression is associated with longer melanoma specific survival
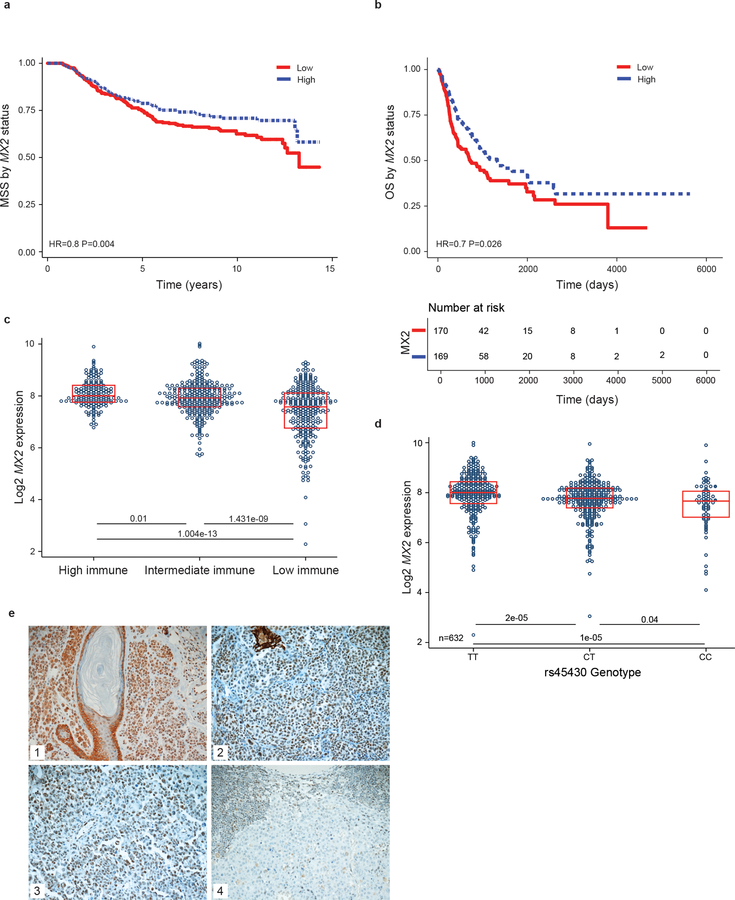
Using previously described whole transcriptome data derived from 703 primary melanomas from the Leeds Melanoma Cohort (LMC) (Nsengimana et al., 2018) we investigated the expression and association of MX2 mRNA level with melanoma specific survival (MSS). A Kaplan Meier curve was generated after dichotomization of MX2 expression to high and low groups with respect to the median showing that higher MX2 expression was associated with longer melanoma specific survival (HR=0.8, P=0.004) (Figure 2a). Similar results were observed in the TCGA melanoma metastases – application of Cox proportional hazards model after median-based dichotomization revealed that high MX2 expression was associated with better overall patient survival (N=339, HR=0.7, P=0.026) (Figure 2b).
Figure 2. Expression of MX2 is associated with a better melanoma specific survival.
a) Kaplan-Meier melanoma specific survival analysis of 703 primary melanomas and b) Kaplan-Meier overall survival analysis of 339 TCGA metastatic melanomas stratified by median MX2 RNA expression where low is defined as bellow median. Analysis performed applying univariate Cox proportional hazard model. c) MX2 RNA expression in High, Intermediate and Low Immune subgroups. d) Expression quantitative trait loci (eQTL) analysis of MX2 gene single nucleotide polymorphism rs45430 in 703 primary melanomas e) Representative immunohistochemistry staining of MX2 in 1) nevi, 2) primary and 3) and 4) metastatic melanoma.
Furthermore, weak yet significant negative correlation was observed between MX2 expresion and Breslow thickness (R=−0.2, P=5.4×10−8) as well as MX2 and the mitotic rate (R=−0.13, P=0.002) in the LMC (Supporting Figure 2a and b).
Since interferon signaling, which might induce MX2, is involved in immune cell infiltration in tumors, we analyzed MX2 expression in the LMC stratified by strength of immune signal resulting in 3 immune subgroups. The generation of the Immune Subgroups was defined in (Poźniak et al., 2019). MX2 expression was significantly lowest in the low immune and highest in the high immune subgroup (Figure 2c). Comparable results were seen in the TCGA melanoma metastases cohort (Supporting Figure 2c).
Analysis of LMC also showed associations between MX2 expression and histologically detected tumor-infiltrating lymphocytes (TILs) (Supporting Figure 2d). MX2 expression was significantly higher in tumors with TILs in comparison to tumors that had no TILs. We then compared the association between MX2 expression and MSS in the LMC primary tumors stratified by strength of immune signal. MX2 expression was borderline protective in the Low Immune Subgroup. The results for the Intermediate and high Immune Subgroups were not significant but show similar estimates of the hazard ratio so their lack of significance may simply reflect relatively small sample size (Table 1). There was no association between MX2 expression and MSS in the TCGA stratified by immune status (data not shown).
Table 1.
Association between MX2 expression and MSS in the LMC primary tumors stratified by the strength of immune signal.
| Subgroup stratified by the strength of immune signal | HR | P | 95% CI |
|---|---|---|---|
| Whole dataset (N=703) | 0.80 | 0.004 | 0.69–0.93 |
| Low immune (N=268) | 0.83 | 0.064 | 0.69–1.01 |
| Intermediate immune (N=256) | 0.92 | 0.626 | 0.65–1.29 |
| High immune (N=144) | 1.11 | 0.765 | 0.56–2.19 |
We also tested whether rs45430 SNP is associated with MX2 expression in the primary melanomas. The SNP data were generated as previously described (Law et al., 2015). Expression of MX2 was significantly lower in participants homozygous for the C allele in comparison to the CT or TT genotype (Figure 2d).
Protein expression of MX2 was also analyzed by immunohistochemistry in a second melanoma data set consisting of 42 paraffin-embedded nevi, 154 primary and 60 metastatic melanomas. As shown in Figure 2e cytoplasmic and/or nuclear expression was observed. Note that a variable MX2 staining was also observed in infiltrating immune cells.
Comparably high MX2 expression (immunoscore >3) was seen in nevi (21.4%), primaries (26.6%) and metastases (24%). However, complete lack of immunoreactivity was observed in 2.4% nevi, 7.8% primary and 15% metastatic tumors respectively, suggesting that MX2 is downregulated during disease progression in a proportion of tumors. Analysis of disease-specific and progression-free survival in this cohort showed no significant correlation with MX2 expression. There was a significant positive correlation observed between MX2 expression and extent of tumor infiltrating immune cells in the tumors (R= 0.23 P=0.008).
Overexpression of MX2 reduces melanoma proliferation by reducing activation of the PI3K/AKT pathway
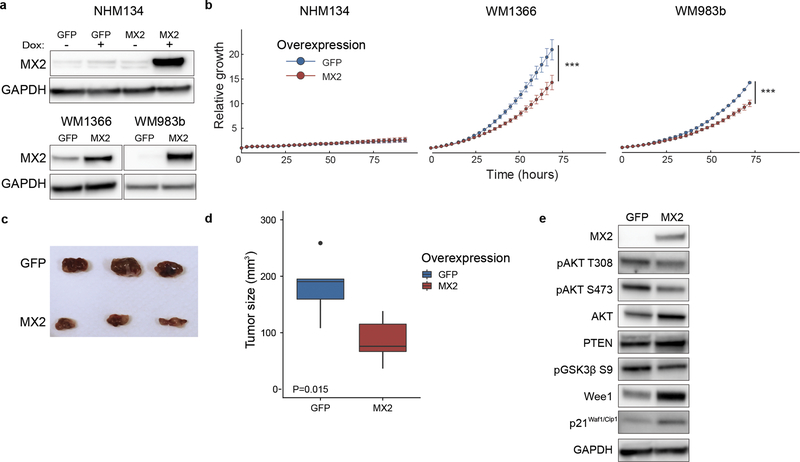
To further investigate the functional role of MX2 in melanoma, we stably overexpressed MX2 in normal human melanocytes NHM134, NRAS mutant WM1366 and BRAF mutant metastatic WM983b cell line. To develop a stable melanocyte cell line expressing MX2 or GFP we used the Tet-On doxycycline-inducible system. As shown in Figure 3a, a clear increase in MX2 protein levels was observed in all cell lines after selection or induction when compared to GFP expressing control vector transfected cells. In vitro effects of increased MX2 protein levels on proliferation were assessed using IncuCyte™ analyzing the area occupied by the cells (% confluence). The relative confluence of cells growing under normal conditions for 72 h was significantly reduced in MX2 overexpressing WM1366 and WM983b cells compared to their respective GFP controls (Figure 3b). This inhibitory growth effect was not observed in normal melanocytes. No visibly detectable phenotypical changes were observed in the engineered melanocytes overexpressing GFP and MX2 (Supporting Figure 3a). Furthermore, we found no effects on expression of melanocyte differentiation markers Melan-A and MITF (Supporting Figure 3b).
Figure 3. MX2 overexpression inhibits melanoma cell growth both in vitro and in vivo.
a) Immunoblot analysis of MX2 protein expression in normal human melanocytes and melanoma cells after lentiviral transduction. Tet-On system was used to achieve doxycycline inducible expression of MX2 and GFP in normal human melanocytes. Increased MX2 protein expression seen in melanocytes after administration of 500 ng/ml doxycycline for 48 hours and stable expression in melanoma lines. b) Growth curves of cells overexpressing GFP or MX2 were obtained using Incucyte Zoom live-cell imaging system. Curves represent fold increase of cell growth versus time at 3-hour intervals. Results are expressed as mean ±SE of three independent experiments. Wilcoxon matched-pairs signed-rank test was used for comparison between the groups. Statistically significant results are marked with asterisk, *** p<0.001. c) Image of harvested tumors at day 50 post subcutaneous injection of 2×106 WM983b cells stably expressing GFP or MX2. d) Tumor volume at day 50 after injection. t-test was applied to assess significance, * p<0.05. e) Assessment of AKT pathway activity by immunoblotting in lysates of the xenograft tumors overexpressing MX2 or GFP.
To study whether the effect of MX2 overexpression was relevant for tumor formation in vivo, WM983b and WM1366 cells overexpressing MX2 or control protein GFP were subcutaneously injected into the right flank of athymic nude mice. As shown in Figure 3 panels c and d, overexpression of MX2 significantly suppressed tumor growth compared to GFP control group of WM983b cells. WM1366 MX2 and GFP expressing cells displayed poor in vivo growth properties and no tumor growth was detected for up to 50 days.
In attempt to explain the observed MX2 growth inhibitory effects, we analyzed known survival and proliferation signaling pathways including the MAPK and AKT pathways in extracted tumor xenograft lysates. We found that overexpression of MX2 led to reduced phosphorylation of AKT regulatory residues Thr308 and Ser473, decreased levels of AKT downstream phosphoprotein GSK3β and a minor increase in PTEN protein levels, suggesting that activity of the pathway is reduced. Furthermore, we observed elevated expression levels of Wee1 and the tumor suppressor p21Cip1 (Figure 3e) suggesting abrogation of the cell cycle. We did not observe significant changes to the MAPK pathway. The same results were obtained in vitro for both WM1366 and WM983b cells (Supporting Figure 3c) suggesting that MX2 contributes to the regulation of the cell cycle and proliferation, displaying tumor suppressor features.
MX2 function in melanoma is cell line dependent
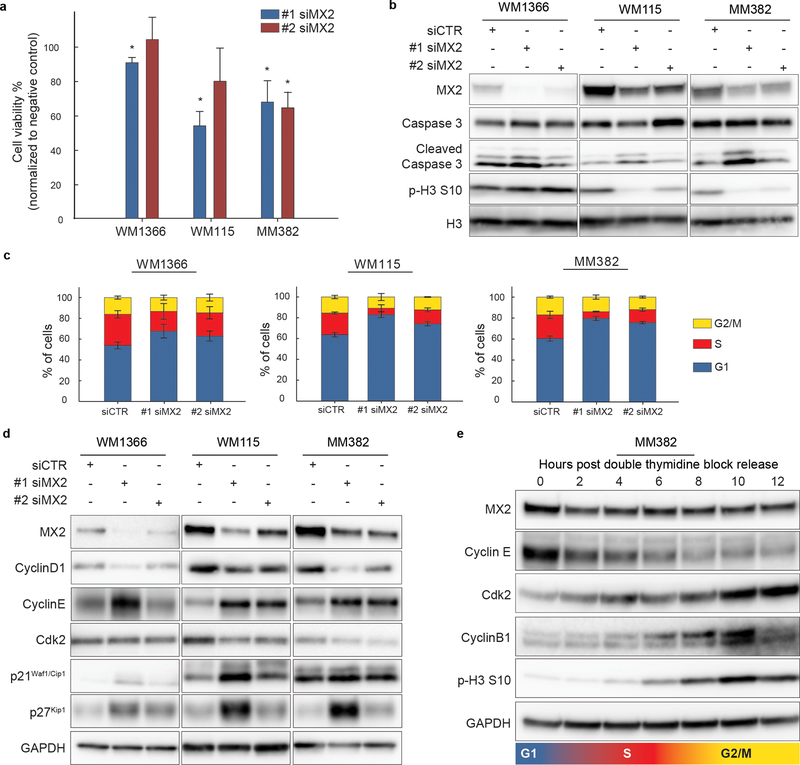
Since we observed that a subset of melanoma cell lines displays high constitutive MX2 expression, it is possible that these cells have adapted to circumvent its growth inhibitory effects or that MX2 has a different functional role in these cells. To investigate these possibilities, we down-regulated MX2 using two different MX2 targeting siRNA oligos. A clear reduction in MX2 mRNA levels was seen after siRNA transfection without effecting MX1 (Supporting Figure 4a and b). Interestingly, 72 h post transfection a significant viability decrease was seen in high MX2 expressing WM115 and MM382 cells while subtle or no effects were seen in low MX2 expressing WM1366 cells (Figure 4a). To identify if the observed decrease in viability was due to decreased proliferation or apoptosis, we examined the expression the of mitosis marker phospho-Histone H3 (pHH3) and the apoptotic marker cleaved Caspase 3. As seen in Figure 4b, no notable increase in Caspase 3 cleavage was observed in any of the cell lines following MX2 downregulation while a clear reduction in phosphorylation of HH3 at serine 10 was evident in WM115 and MM382 suggesting cell cycle related effect in these lines. Activation of the AKT and MAPK signaling pathways was also examined, but surprisingly no significant changes were observed (Supporting Figure 5).
Figure 4. MX2 downregulation perturbs cell cycle and reduces proliferation in a subset of melanoma lines.
a) Trypan Blue dye exclusion viability test 72 hours post transfection with two independent siRNAs for MX2 (#1 siMX2 and #2 siMX2) and negative control (siCTR). Viability counts are normalized to negative control. Results are expressed as mean ±SE of three independent experiments. Two-tailed paired t-test was used to test statistical significance. * p<0.05. b) Immunoblot analysis of affected apoptosis and proliferation associated proteins upon MX2 knockdown. GAPDH was used as a loading control c) Evaluation of cell cycle distribution by flow cytometry using propidium iodide staining. Cells were transfected with #1 siMX2, #2 siMX2 and siCTR 48 hours prior to flow cytometry analysis. Bar graphs represent percentages of cells in different cell cycle phases (average from three independent experiments ±SE). d) Immunoblot analysis of proteins involved in G1/S transition of the cell cycle. e) Oscillation of MX2 protein level during cell cycle was examined by releasing MM382 cells synchronized in G1/S phase from double thymidine block.
Effects of MX2 downregulation on the cell cycle distribution were assessed by flow cytometry 48 h post siRNA transfections. Analysis revealed that MX2 knockdown increased the proportion of cells in G1 phase, including weak effect in WM1366 suggesting induction of the cell cycle arrest (Figure 4c). As expected, G1 arrest was accompanied with decreased levels of Cyclin D1 and kinase Cdk2 expression and increased levels of Cdk inhibitory proteins p27Kip1 and p21Cip1 (Figure 4d). Again, observed effects were more prominent in WM115 and MM382 cell lines than in WM1366.
Due to these notable effects on the cell cycle, we investigated if MX2 expression itself could be oscillating during the cycle. Synchronization of the cells at G1/S boundary by double thymidine block showed expected oscillation of the cyclins and mitotic phosphohistone H3 after release. MX2 levels, however, did not change during the cell cycle progression suggesting that it’s expression is cell cycle phase independent (Figure 4f).
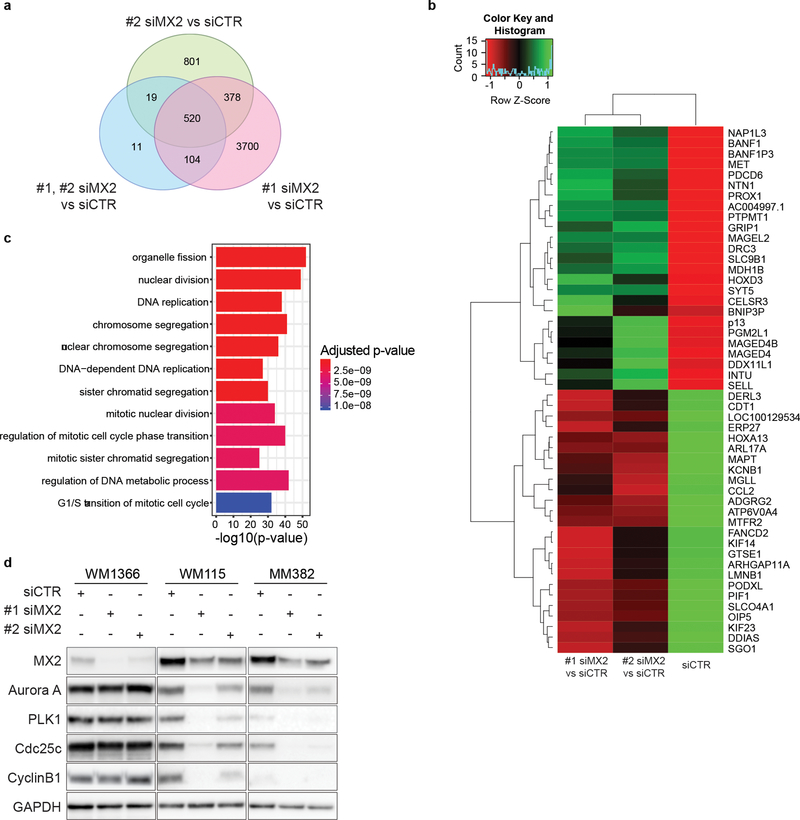
To further investigate which cellular processes are influenced by MX2 downregulation we also performed RNAseq of MM382 cells 48 h after siRNA transfection. A core of 520 differentially expressed genes (Supporting Table 3) overlapping between two siRNA oligos was selected for further GO enrichment analysis (Figure 5a) while fifty most up and downregulated genes are presented in Figure 5b. Analysis showed that highly over-represented GO terms included processes involved in cell cycle regulation and progression (Figure 5c). Among significantly downregulated genes after MX2 downregulation was a major mitotic protein kinase Aurora A. Validation of RNAseq data by western blot indeed confirmed that in WM115 and MM382 lines Aurora A and its downstream target PLK1, which control centrosome maturation and spindle assembly at G2/M transition are downregulated following MX2 siRNA transfection (Figure 5d). As a result, protein levels of downstream members of Aurora A – PLK1 axis, including cdc25c and cyclin B1, were also reduced leading to impeded progression through G2/M. Jointly our data suggest that even if MX2 is downregulated in metastatic samples and displays tumor-suppressive function in the majority of melanoma lines, in a subset of melanomas, it displays proto-oncogenic features and is an important factor necessary for cell cycle regulation and proliferation of these cells.
Figure 5. Gene expression analysis of MM382 melanoma cells after MX2 siRNA transfection.
a) Venn diagram of differentially expressed genes (DEGs) between #1 siMX2, #2 siMX2 and siCTR. Pink circle represents the number of genes with different expression levels between #1 siMX2 vs. siCTR. Green circle represents the number of genes with different expression levels between #2 siMX2 vs. siCTR. Blue circle represents the number of genes with different expression levels between #1 siMX2 and #2 siMX2 vs. siCTR. b) Heatmap of 25 most upregulated (green) and downregulated (red) genes. c) GO enrichment analysis of biological processes for the 520 differentialy expressed genes overlapping between #1 siMX2 and #2 siMX2. d) Validation of RNA-seq transcriptome analysis by immunoblotting.
DISCUSSION
In recent years several GWAS have identified novel melanoma susceptibility SNPs, including in the intron of MX2 gene that have no previously defined functional roles in cancer-related processes. Thus, the overall objective of our study was to investigate if and how MX2 function can influence melanoma tumorigenesis. So far MX2 has been mainly defined by its antiviral functions, highlighting its induction by type I IFN and ability to interfere with the replication of different types of negative-stranded RNA viruses. Our expression data from melanocytes, primary and metastatic melanoma show that MX2 can be constitutively expressed independently of IFN induction, which is in agreement with two previous studies in HeLa and T98G cells (King et al., 2004; Melén et al., 1996).
While we detected MX2 expression in all melanocyte and primary melanoma cell lines, 8 out of 10 metastatic cell lines showed lower or lack of expression. There was also an apparent reduction of expression in a metastatic vs. primary cell line derived from the same patient suggesting that MX2 is downregulated during disease progression. Furthermore, an increasing percentage of MX2 IHC negative samples was observed in metastatic lesions. The exact mechanism of this downregulation needs further elucidation, but inactivation of the IFN pathway and suppression of its target genes during disease development has been reported in melanoma as well as in other cancers (Katlinskaya et al., 2016; Katlinski et al., 2017; Walter et al., 2017). Interestingly, a study of breast cancer by Han et al. found that transcription factor and chromatin organizer SATB1 reprograms gene expression profile of cancer cells to promote tumor growth and that MX2 is among the repressed genes (Han, Russo, Kohwi, & Kohwi-Shigematsu, 2008).
Here we also showed that reintroduction of MX2 expression in endogenously low expressing cell lines leads to downregulation of AKT activity and inhibition of tumor growth in vitro and in vivo. These effects were profound in a metastatic WM983b suggesting that downregulation of MX2 is important during disease progression. Since it is demonstrated that type I and II IFNs used in melanoma treatment due to their antiproliferative effects can regulate AKT activity in a complex manner (Kaur, Sassano, Dolniak, et al., 2008; Kaur, Sassano, Joseph, et al., 2008) one can speculate if some of these effects can partially be mediated by MX2.
Analysis of transcriptomic data from the Leeds Melanoma Cohort of 703 tumors and TCGA metastases showed that high expression of MX2 mRNA was associated with better prognosis. We did not observe similar effects of protein expression in our second validation cohort, which might be due to much smaller sample size. It is difficult to exclude the possibility that a significant component of the MX2 gene expression signal in the Leeds Melanoma Cohort is derived from TILs which them self are a favorable prognostic marker in melanoma (Fu et al., 2019). Expression of MX2 in tumor cells did, however, correlate with the amount of immune cell infiltration implying that IFN secretion by TILs leads to induction of MX2 and other IFN dependent genes like MX1. Indeed, we did observe a correlation between MX2 and MX1 expression in tumor samples. However, we did not see such correlation in our panel of cell lines, suggesting that even though IFNs are major regulatory factors of MX2, there are also other mechanisms involved. For instance, a recent study by Punia et al. found that Engrailed-2 (EN2) transcription factor secreted by prostate tumors can induce MX2 expression in stromal cells (Punia, Primon, Simpson, Pandha, & Morgan, 2019), and MX2 was a single gene showing a dose-response relationship to recombinant EN2 treatment.
Interestingly, the observation that MX2 expression is borderline protective even in the Low Immune subgroup combined with fact that some tumors display high MX2 immunoreactivity while lacking TILs, argues for its immune independent functions. Jointly these results support the hypothesis that MX2 has tumor-suppressive features in melanoma.
Inheritance of the minor C allele rs45430 SNP in the intron of MX2 was reported to be protective for melanoma and multiple primaries in the GWAS studies (Barrett et al., 2011; Gibbs et al., 2015). Here we found that the homozygous C allele is associated with lower expression of MX2 in primary melanoma tumors, and a similar trend was seen in metastatic samples. Since we also report that higher expression levels of MX2 are seen in thinner primaries with a lower mitotic rate and better survival, these data seem somewhat difficult to explain. However, it is known that expression quantitative trait loci (eQTL) can display opposite directional effects in a tissue-specific manner (Mizuno & Okada, 2019). Indeed, minor C allele is associated with lower MX2 expression in whole blood while the opposite is seen for sun-exposed skin (Supporting Figure 6) (TheGTExConsortium, 2015). Currently it is unclear what functional role MX2 plays in different immune cell types or how this relates to melanoma risk, therefore further studies are warranted.
Interestingly, we have observed similar discrepancies previously for an inherited SNP in the PARP1 gene. The SNP was associated with higher PARP1 levels, increased risk of melanoma, and related to PARP1 induced cell proliferation mediated through MITF (Choi et al., 2017). Yet, the same SNP was found to be associated with a lower risk of death from melanoma (Davies et al., 2014).
Remarkably, despite its growth inhibitory effects and downregulation in metastatic cell lines, a subset of melanomalines in our panel exhibited high endogenous MX2 expression. Knockdown of MX2 in these lines decreased proliferation and lead to perturbation of the cell cycle, which is inconsistent with observations from our overexpression experiments. However, MX2 belongs to dynamin-like GTPase family proteins, which are also known to be involved in the regulation of cell cycle progression and it is likely that MX2 function is complex and cell type and context-dependent. In support of these observations, one previous study has reported that depletion of endogenous MX2 in cancer cells results in delayed progression through G1/S phase of the cell cycle (King et al., 2004). We have observed similar G1 arrest accompanied with Cyclin D1 degradation, Cyclin E upregulation in p21 dependent manner as reported (Sandor et al., 2000). In addition, our RNAseq analysis revealed that MX2 is also involved in DNA replication and mitosis processes partially by regulation of Aurora A and PKL1. A study by Kane et al. investigating MX2 potency to inhibit HIV-1 showed that arresting the cell cycle in osteosarcoma and myelogenous leukemia cells increases MX2 viral inhibitory activity (Kane et al., 2013). We can speculate that antiviral MX2 potency in non-dividing cells increases when it does not engage in other cellular processes, including DNA replication and/or mitosis as suggested by our study. These results further support the hypothesis of a cellular type and setting dependent MX2 function.
In summary, we have demonstrated that widely accepted antiviral MX2 gene has tumor-suppressive features in melanoma, where it regulates the growth of tumor cells partially through negative modification of AKT activity, and it is downregulated during disease progression. However, its role seems to be complex and cell context-dependent since we found that in a subset of melanoma cell lines it is highly expressed and necessary for cell cycle progression. Further elucidation of this dual mechanism of action is needed to understand its complex roles in tumorigenesis.
Supplementary Material
Significance.
The study provides the first evidence that antiviral MX2 gene is associated with the tumorigenesis process in melanoma. It has an IFN independent role in the regulation of cell cycle and the PI3K/AKT pathway. However, MX2 function is clearly cell type and context-dependent. Our findings are adding a functional explanation to previous genome-wide association studies that reported an association between MX2 gene and reduced risk for melanoma.
Acknowledgements
M. Juraleviciute and J.Pozniak were funded by Horizon2020 Research and Innovation Programme under grant agreement No.641458 (MELGEN). The collection of samples in the Leeds Melanoma Cohort Study was funded by Cancer Research UK (project grant C8216/A6129, Programme Awards C588/A4994 and C588/A10589, and by the NIH (R01 CA83115). The project also received support from the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and from the National Cancer Institute (NCI) of the US National Institutes of Health (NIH) (CA83115).
References
- Amos CI, Jing K, Wang L-E, Wei Q, Vattathil S, Fang S, … Mann GJ (2011). Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Human Molecular Genetics, 20(24), 5012–5023. doi: 10.1093/hmg/ddr415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, … Bishop DT (2011). Genome-wide association study identifies three new melanoma susceptibility loci. Nature Genetics, 43, 1108–1113. doi: 10.1038/ng.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, … Newton Bishop JA (2009). Genome-wide association study identifies three loci associated with melanoma risk. Nature Genetics, 41, 920–925. doi: 10.1038/ng.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, & Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffone C, Schulte B, Opp S, & Diaz-Griffero F (2015). Contribution of MxB Oligomerization to HIV-1 Capsid Binding and Restriction. Journal of Virology, 89(6), 3285–3294. doi: 10.1128/JVI.03730-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Xu M, Makowski MM, Zhang T, Law MH, Kovacs MA, … Brown KM (2017). A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nature Genetics, 49, 1326–1335. doi: 10.1038/ng.3927 [DOI] [PubMed] [Google Scholar]
- Davies JR, Jewell R, Affleck P, Anic GM, Randerson-Moor J, Ozola A, … Newton-Bishop J (2014). Inherited variation in the PARP1 gene and survival from melanoma. International Journal of Cancer, 135(7), 1625–1633. doi: 10.1002/ijc.28796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks MDJ, Betancor G, Jimenez-Guardeño JM, Pessel-Vivares L, Apolonia L, Goujon C, & Malim MH (2018). Multiple components of the nuclear pore complex interact with the amino-terminus of MX2 to facilitate HIV-1 restriction. PLOS Pathogens, 14(11), e1007408. doi: 10.1371/journal.ppat.1007408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flørenes VA, Flem-Karlsen K, McFadden E, Bergheim IR, Nygaard V, Nygård V, … Mælandsmo GM (2019). A Three-dimensional Ex Vivo Viability Assay Reveals a Strong Correlation Between Response to Targeted Inhibitors and Mutation Status in Melanoma Lymph Node Metastases. Translational Oncology, 12(7), 951–958. doi: 10.1016/j.tranon.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Chen N, Ge C, Li R, Li Z, Zeng B, … Li G (2019). Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. Oncoimmunology, 8(7), 1593806–1593806. doi: 10.1080/2162402X.2019.1593806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, & Seoighe C (2010). A flexible R package for nonnegative matrix factorization. BMC Bioinformatics, 11(1), 367. doi: 10.1186/1471-2105-11-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DC, Orlow I, Kanetsky PA, Luo L, Kricker A, Armstrong BK, … Thomas NE (2015). Inherited Genetic Variants Associated with Occurrence of Multiple Primary Melanoma. Cancer Epidemiology Biomarkers & Prevention, 24(6), 992–997. doi: 10.1158/1055-9965.EPI-14-1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, … Malim MH (2013). Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature, 502, 559–562. doi: 10.1038/nature12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, & Kochs G (2010). Human MxA Protein: An Interferon-Induced Dynamin-Like GTPase with Broad Antiviral Activity. Journal of Interferon & Cytokine Research, 31(1), 79–87. doi: 10.1089/jir.2010.0076 [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, & Kochs G (2015). Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends in Microbiology, 23(3), 154–163. doi: 10.1016/j.tim.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Han H-J, Russo J, Kohwi Y, & Kohwi-Shigematsu T (2008). SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature, 452, 187–193. doi: 10.1038/nature06781 [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson Ian R., Kryukov Gregory V., Arold Stefan T., Imielinski M, Theurillat J-P, … Chin L (2012). A Landscape of Driver Mutations in Melanoma. Cell, 150(2), 251–263. doi: 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, … Bieniasz PD (2013). MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature, 502, 563–566. doi: 10.1038/nature12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlinskaya YV, Katlinski KV, Yu Q, Ortiz A, Beiting DP, Brice A, … Fuchs SY (2016). Suppression of Type I Interferon Signaling Overcomes Oncogene-Induced Senescence and Mediates Melanoma Development and Progression. Cell Reports, 15(1), 171–180. doi: 10.1016/j.celrep.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlinski KV, Gui J, Katlinskaya YV, Ortiz A, Chakraborty R, Bhattacharya S, … Fuchs SY (2017). Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell, 31(2), 194–207. doi: 10.1016/j.ccell.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, … Platanias LC (2008). Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proceedings of the National Academy of Sciences, 105(12), 4808–4813. doi: 10.1073/pnas.0710907105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, … Platanias LC (2008). Dual Regulatory Roles of Phosphatidylinositol 3-Kinase in IFN Signaling. The Journal of Immunology, 181(10), 7316–7323. doi: 10.4049/jimmunol.181.10.7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-H, Shenoy Avinash R., Kumar P, Bradfield Clinton J., & MacMicking John D. (2012). IFN-Inducible GTPases in Host Cell Defense. Cell Host & Microbe, 12(4), 432–444. doi: 10.1016/j.chom.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Raposo G, & Lemmon MA (2004). Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proceedings of the National Academy of Sciences, 101(24), 8957–8962. doi: 10.1073/pnas.0403167101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MH, Bishop DT, Lee JE, Brossard M, Martin NG, Moses EK, … Iles MM (2015). Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nature Genetics, 47, 987–995. doi: 10.1038/ng.3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnussen GI, Holm R, Emilsen E, Rosnes AKR, Slipicevic A, & Flørenes VA (2012). High Expression of Wee1 Is Associated with Poor Disease-Free Survival in Malignant Melanoma: Potential for Targeted Therapy. PLOS ONE, 7(6), e38254. doi: 10.1371/journal.pone.0038254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melén K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, & Julkunen I (1996). Human MxB Protein, an Interferon-α-inducible GTPase, Contains a Nuclear Targeting Signal and Is Localized in the Heterochromatin Region beneath the Nuclear Envelope. Journal of Biological Chemistry, 271(38), 23478–23486. [DOI] [PubMed] [Google Scholar]
- Mizuno A, & Okada Y (2019). Biological characterization of expression quantitative trait loci (eQTLs) showing tissue-specific opposite directional effects. European Journal of Human Genetics. doi: 10.1038/s41431-019-0468-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsengimana J, Laye J, Filia A, O’Shea S, Muralidhar S, Poźniak J, … Newton-Bishop J (2018). β-Catenin–mediated immune evasion pathway frequently operates in primary cutaneous melanomas. The Journal of Clinical Investigation, 128(5), 2048–2063. doi: 10.1172/JCI95351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, & Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods, 14, 417–419. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poźniak J, Nsengimana J, Laye JP, O’Shea SJ, Diaz JMS, Droop AP, … Newton-Bishop J (2019). Genetic and Environmental Determinants of Immune Response to Cutaneous Melanoma. Cancer Research, 79(10), 2684–2696. doi: 10.1158/0008-5472.CAN-18-2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punia N, Primon M, Simpson GR, Pandha HS, & Morgan R (2019). Membrane insertion and secretion of the Engrailed-2 (EN2) transcription factor by prostate cancer cells may induce antiviral activity in the stroma. Scientific Reports, 9(1), 5138. doi: 10.1038/s41598-019-41678-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample A, & He Y-Y (2018). Mechanisms and prevention of UV-induced melanoma. Photodermatology, photoimmunology & photomedicine, 34(1), 13–24. doi: 10.1111/phpp.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor V, Senderowicz A, Mertins S, Sackett D, Sausville E, Blagosklonny MV, & Bates SE (2000). P21-dependent G1arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. British Journal of Cancer, 83(6), 817–825. doi: 10.1054/bjoc.2000.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, & Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research, 4, 1521. doi: 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TheGTExConsortium. (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science, 348(6235), 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter KR, Goodman ML, Singhal H, Hall JA, Li T, Holloran SM, … Hagan CR (2017). Interferon-Stimulated Genes Are Transcriptionally Repressed by PR in Breast Cancer. Molecular Cancer Research, 15(10), 1331–1340. doi: 10.1158/1541-7786.MCR-17-0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, & He Q-Y (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A Journal of Integrative Biology, 16(5), 284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.