Abstract
Buckwheat (Fagopyrum esculentum) is cultivated worldwide and its flour is used in a variety of food products. Although functional analyses of genes in buckwheat are highly desired, reliable methods to do it have yet to be developed. In this study we established a simple and efficient transient gene expression system using buckwheat protoplasts isolated from young hypocotyls using 96-well plates as a high-throughput platform. The transformation efficiency was comparable with that of similar systems, such as Arabidopsis mesophyll protoplasts. Stable results were obtained in a typical example of the experiment to examine transcription factor activity. This system shows potential for the large-scale analysis of gene function using protoplast isolated from fewer and younger plants than the conventional system and may provide novel information for efficient buckwheat breeding.
Keywords: buckwheat, effector–reporter analysis, hypocotyl protoplast, transient gene expression, 96-well plate
Introduction
Buckwheat (Fagopyrum esculentum) is an annual cross-pollinating crop widely cultivated all over the world, especially in Russia, Europe, and East Asia. Its flour is used in a variety of food products, such as noodles and cakes. Buckwheat contains an abundance of antioxidants, such as polyphenols and especially rutin, and is considered to be a functional food. However, problems with the cultivation of buckwheat include low yields, pre-harvest sprouting, and low resistance to excess water. In addition, buckwheat seeds contain allergenic proteins that cause immunoglobulin E-mediated allergenic reactions in humans (Park et al. 2000, Sano et al. 2014, Wieslander and Norbäck 2001). Hence, breeding to remove these inferior characters from cultivated buckwheat has long been desired. Recent advances in sequencing technology have stimulated buckwheat genomic research. Yasui et al. (2016) constructed the Buckwheat Genome Data Base (http://buckwheat.kazusa.or.jp/), which has been used to detect genes controlling important agronomical traits (Katsu et al. 2017, Matsui et al. 2016, 2018a, 2018b, Takeshima et al. 2019). However, molecular genetic tools and methods to validate gene function in buckwheat remain to be developed.
A stable transformation method to generate transgenic plants has been established in many plant species and has become powerful approach to assess gene function and transfer a beneficial agronomic trait to cultivated plants. In the case of buckwheat, successful generation of transgenic plants has been reported (Kim et al. 2010, Miljuš-Djukić et al. 1992). However, efficient production of transgenic buckwheat remains difficult. In addition, the transformation process and maintenance of established transgenic plants are time-consuming and therefore limit the utilization of this approach for large-scale study.
A transient gene expression system is an alternative convenient method to express exogenous genes in plant cells without stable transformation. A system using isolated protoplasts is among the most powerful tools to analyze gene function in diverse species (Cao et al. 2014, Tan et al. 2013, Yoo et al. 2007, Zhang et al. 2011). To date, it has been employed for analyses of the subcellular localization of proteins (Sheen et al. 1995), protein–protein interactions (Seidel et al. 2005, Walter et al. 2004), and signal transduction in plant cells (Sheen 2001). The protoplast system is suitable for use in large-scale experiments by employing 96/384-well microplates. In Arabidopsis, protein–protein and DNA–protein interactions in plant cells have been examined in a high-throughput manner (Fujikawa and Kato 2007, Wehner et al. 2011). In buckwheat, methods for protoplast isolation and transient gene expression have been reported previously, but have not been optimized for a high-throughput platform (Lei et al. 2017a, 2017b, Shen et al. 2002).
Here, we provide a method for simple protoplast isolation from common buckwheat grown in a growth chamber and its use for high-throughput transient gene expression. This method facilitates functional analysis of uncharacterized genes in buckwheat in large-scale experiments.
Materials and Methods
Plant materials
Seeds of Japanese common buckwheat ‘Sachiizumi’ (Matsui et al. 2013) were germinated on soil in a plant incubator (CHF 405, TOMY Inc.) having fluorescent lamps (FL40SEX-N-HG, NEC Lighting Inc.) under a diurnal cycle of 12-h light (60–80 μmol m–2 s–1) at 25°C/12-h dark at 16°C with 50% humidity.
Plasmid construction
To confirm gene transfection efficiency, the green fluorescent protein (GFP) driven by the Cauliflower mosaic virus 35S promoter was used as described previously (Fujiwara et al. 2014). For the analysis of transient transcriptional activation ability, cloned FeMYBF1 (Accession no. LC369592) in the vector pDONR207 was incorporated into pDEST430T1.2 (Ohta et al. 2000) by the LR reaction (ThermoFisher Inc.). Reporter and reference plasmids were used as described previously (Sakamoto et al. 2016). Plasmids used for reporter–effector analysis were prepared using primers listed in Table 1.
Table 1.
Primers used in this study
| Gene (Locus No. or GenBank No.) | Sequence (from 5′ to 3′) | Direction | Purpose |
|---|---|---|---|
| VAMP722 (AT2G33120) | ggggacaagtttgtacaaaaaagcagGCTCCATGGCGCAACAATCGTTGATCTACAGTT | Forward | Effector (control) |
| ggggaccactttgtacaagaaagctgGGTCTTTACCGCAGTTGAATCCCCCACAAATTG | Reverse | Effector (control) | |
| FeMYBF1 (LC369592) | ggggacaagtttgtacaaaaaagcagGCTCCATGGGAAGAGCACCTTGCTGTGA | Forward | Effector |
| ggggaccactttgtacaagaaagctgGGTCAGAAAGAAGCCAAGCAACCATTGC | Reverse | Effector | |
| FeFLS1 (HM357804) | ggggacaactttgtatagaaaagttgAATAATATAGTAGCAAGCTTTGA | Forward | Reporter |
| ggggactgcttttttgtacaaacttggTTCTTTGTGGTGTGTGAAGGAT | Reverse | Reporter | |
| FeFLS2 (HM357805) | ggggacaactttgtatagaaaagttgAAGAGCAGAACATGTAAATAAAG | Forward | Reporter |
| ggggactgcttttttgtacaaacttggCATTCTTTCTTTTGCTCTCCAG | Reverse | Reporter | |
| FeFLS (JF274261) | ggggacaactttgtatagaaaagttgCCTCCAGCATGCATCCTTCCTT | Forward | Reporter |
| ggggactgcttttttgtacaaacttggTACATGTACTCCTTGAGACTC | Reverse | Reporter | |
| FeDFR1a (LC216398) | ggggacaactttgtatagaaaagttgTTGTACATCTAGGGCGGCAAAG | Forward | Reporter |
| ggggactgcttttttgtacaaacttgggCATGGTGCGACGTCGTTTTGGT | Reverse | Reporter | |
| FeDFR2 (LC216399) | ggggacaactttgtatagaaaagttgCATTATTATATGAATGATACAC | Forward | Reporter |
| ggggactgcttttttgtacaaacttggCGTCGTCTTTCTTGCTTACA | Reverse | Reporter | |
| FeANS (HM149791) | ggggacaactttgtatagaaaagttgTACATTATAAACTAACTGATGT | Forward | Reporter |
| ggggactgcttttttgtacaaacttggCATTTTAATTCTCTTGGAATAT | Reverse | Reporter | |
| FeANR (LC107621) | ggggacaactttgtatagaaaagttgCTCATCTCACCAAATCACACTC | Forward | Reporter |
| ggggactgcttttttgtacaaacttggATTGAGGGATTAGTTCCAAATC | Reverse | Reporter |
Protoplast isolation
Protoplast isolation was performed by the slightly modified method described by Sakamoto et al. (2013) for hypocotyl cells and Wu et al. (2009) for cotyledon mesophyll cells. In brief, the hypocotyl of buckwheat seedlings grown for 7‒9 days was cut longitudinally and then cut into 1-cm segments with a razor blade. Sliced segments prepared from nine plants were transferred to 20 ml digestion buffer (pH 5.7) containing 1.0% (w/v) Cellulase Onozuka RS (Yakult Inc.), 0.25% (w/v) Macerozyme R10 (Yakult Inc.), 0.02% (w/v) Pectolyase Y23 (Kikkoman Inc.), 0.04% (w/v) BSA, 400 mM Mannitol, 20 mM KCl, 20 mM MES, 10 mM CaCl2, and 10 mM β-mercaptoethanol. After permeating the enzyme solution into the sliced hypocotyl by vacuum filtration, cell walls were digested at 25°C for 2 h with gentle shaking at 80 rpm in the dark. Released protoplasts were filtered through 70-μm nylon mesh (Cell-strainer, BD Bioscience Inc.) and collected in a 50-mL tube. The collected protoplasts were washed with W5 buffer (pH 5.7) containing 150 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES and collected by the centrifugation at 200 × g for 10 min. The washing process was repeated twice. After incubation at 4°C for 10 min, the protoplasts were collected by the centrifugation at 200 × g for 5 min and then resuspended in MMG (pH 5.7) buffer containing 400 mM mannitol, 15 mM MgCl2, and 4 mM MES, and the cell number was adjusted to 2.0‒3.0 × 105 cells/mL with MMG buffer.
Cotyledon mesophyll protoplasts were isolated by a slightly modified protocol of the tape-sandwich method (Wu et al. 2009). After peeling the abaxial epidermis, mesophyll cells were directly placed in digestion buffer and subjected to cell wall digestion and protoplast washing as described above.
Transient gene transformation
Gene transfection in 96-well plates was performed by the slightly modified method described previously (Sakamoto et al. 2016, Yoshida et al. 2013). Briefly, 10 μL of plasmid mixture and 40 μL of protoplast suspension (ca. 1.0 × 104 cells) were dispensed into each well of a round bottom 96-well plate (Violamo Inc., catalog number 1-1601-03). Then, 50 μL of polyethylene-glycol (PEG)/Ca2+ solution containing 40% (w/v) PEG 4000 (Merck Inc. catalog number 81242), 200 mM mannitol, and 100 mM CaCl2 were added and immediately vortexed using a microplate shaker (NP-804, Nissin Rika Inc.) at 1300 rpm for 15 s. After kept at room temperature for 10 min, the transformed protoplast suspension was washed with W5 buffer three times and then incubated at 25°C for 18 h in the dark. Three plasmid concentrations (0.5, 2.0, and 10.0 μg/well) were tested for GFP fluorescence observation. For transient reporter–effector analysis, 400 ng effector (transcription factor) plasmid, 2000 ng reporter (driven by the promoter containing binding sites of the transcription factor) plasmid, and 40 ng reference plasmid (to monitor transfection efficiency) were mixed for each well.
Microscopic observation
Prepared protoplast cells were mounted on a hemocytometer to count cell numbers. The GFP fluorescence was visualized by a fluorescence microscope with emission at 525/50 nm and excitation at 470/40 nm after 24 h incubation at 25°C in the dark.
Determination of dual-luciferase activity
Dual-luciferase activity of transfected protoplasts was determined using the Pikkagene Dual Kit (TOYO B-Net Inc.). Transformed protoplasts were lysed in 20 μL cell lysis buffer provided in the Pikkagene Dual Kit with agitation using a platemixer (Shaking Incubator SI-300C, ASONE Inc.) at 900 rpm for 15 s. Next, 8 μL of cell lysate was transferred to each side wall of a flat-bottom 96-well plate and then was well mixed with 20 μL substrate solution for Firefly luciferase activity (the reporter). Luminescence of each well was measured using a microplate reader with an auto-dispensing device (Infinite F200, TECAN Inc.). Subsequently, the solution was further mixed with 20 μL substrate solution for Renilla luciferase activity and the luminescence was measured (internal control). The reporter activity value was normalized to the internal control activity and expressed as “relative luciferase activity”.
Results and Discussion
Establishment of simple protoplast isolation method from buckwheat hypocotyl
To establish a simple method to isolate protoplasts of buckwheat and transiently express genes in a 96-well plate, we first investigated the optimal tissue and sample amounts for protoplast isolation. Buckwheat plants grown for 7–9 days on soil were used for protoplast isolation (Fig. 1A, 1B). Cotyledon mesophyll cells were exposed by peeling the abaxial epidermis using the tape-sandwich method (Wu et al. 2009) (Fig. 1C, 1D) and the hypocotyl was cut longitudinally and then cut into 1-cm segments (Fig. 1F, 1G). Protoplast cells were isolated from both tissues by cell wall digestion in a 50-mL tube (Fig. 1E, 1H). Protoplast yield based on the fresh weight of cotyledons was two-times higher than that from the hypocotyl (Table 2). Approximately two to three cotyledons or 4–5 hypocotyls are required to isolate 1.0–1.5 × 106 protoplasts, which are sufficient for transient gene expression in a 96-well plate in general. The protoplasts isolated from cotyledons were smaller and showed less variation of cell type than those isolated from hypocotyls (Fig. 2). The buckwheat protoplasts were generally smaller than those isolated from Arabidopsis mesophyll cells (Yoo et al. 2007). Therefore, we slightly increased the volume of PEG-transformation solution (from 90 μL to 100 μL) and the speed of centrifugation (from 100 × g to 200 × g) from those of the protocol for Arabidopsis. Given that no surviving protoplast isolated from the cotyledon after overnight culture was observed with our protocol even if true leaves were taken from 2–3 weeks-old plants (data not shown), we adopted the protocol to isolate hypocotyl protoplasts hereafter.
Fig. 1.
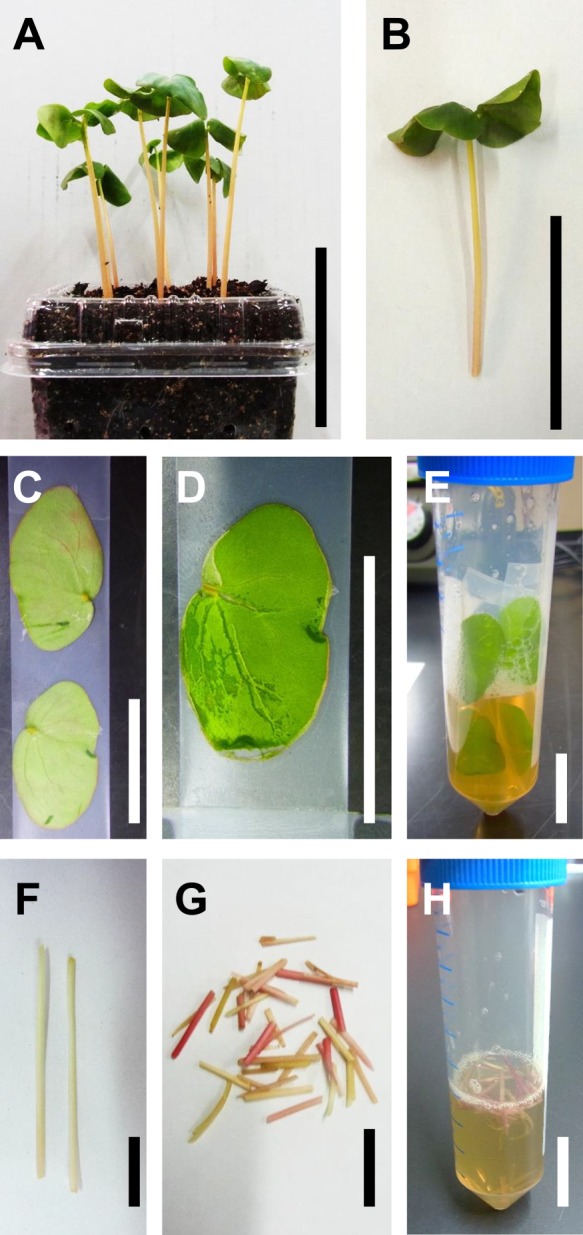
Protoplast isolation from buckwheat. A. Soil-grown buckwheat seedlings cultivated for 7 days after germination. B. A representative seedling used for protoplast isolation. C. Cotyledons placed on 3M adhering tape. D. A Cotyledon in which the adaxial epidermis was removed by peeling using the tape-sandwich method. E. The cotyledons attached to tape are incubated in a 50-ml tube to digest cell walls. F. Longitudinally cut hypocotyl. G. Segments of longitudinally cut hypocotyls for protoplast isolation. H. Hypocotyl segments digested with cellulose solution in a 50-ml tube. Scale bars = 5 cm (A and B) and 1 cm (C to H).
Table 2.
Yield and efficiency of protoplast isolation from buckwheat tissues
| Tissue | Cells/gFWa | Cells/Tissueb |
|---|---|---|
| Cotyledon | 1.7 ± 0.4 × 107 | 7.5 ± 2.8 × 105 |
| Hypocotyl | 2.6 ± 0.4 × 106 | 1.9 ± 0.2 × 105 |
a,b Score indicates mean value and standard deviation of four biological replicates.
Fig. 2.
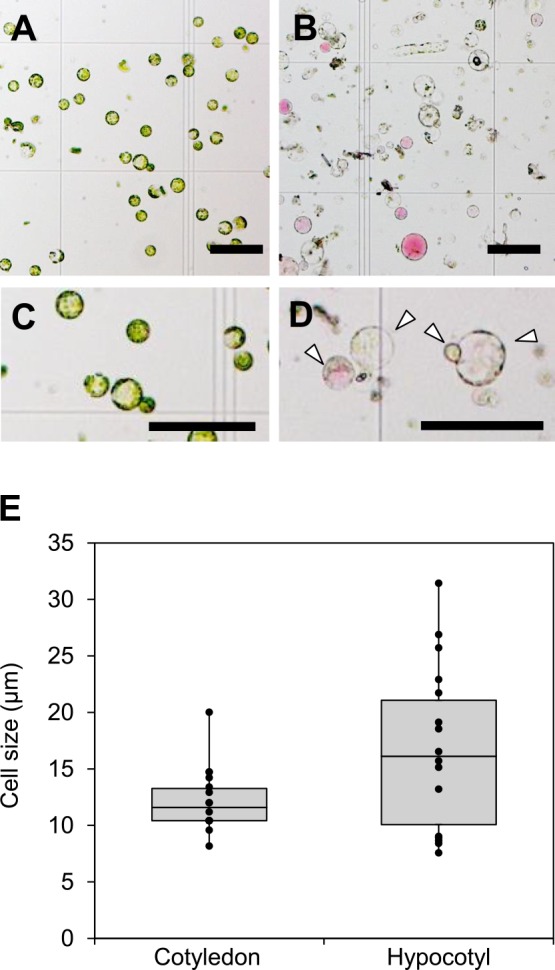
Size of isolated buckwheat protoplasts. A, B. Isolated protoplasts from cotyledon mesophyll cells (A) and hypocotyl (B). C, D. Magnified picture of representative protoplast of cotyledon (C) and hypocotyl (D). Arrow heads indicate the living protoplasts. Scale bars = 100 μm. F, Size of protoplasts isolated from the cotyledon and hypocotyl.
Transfection efficiency of buckwheat protoplast
We next investigated the transfection efficiency of hypocotyl protoplasts in 96-well-plate-based gene transformation. The transformation efficiency was increased dependent on the plasmid concentration, i.e., 13% ± 3% (0.5 μg), 33% ± 5% (2.0 μg), and 58% ± 5% (10.0 μg), whereas no fluorescence was observed in untransformed protoplasts (Fig. 3). Considering the transformation efficiency of Arabidopsis mesophyll cells in a 96-well plate is 54% ± 7% (Fujikawa and Kato 2007), the present transient gene expression system using buckwheat hypocotyl protoplasts is sufficiently efficient for large-scale experiments.
Fig. 3.
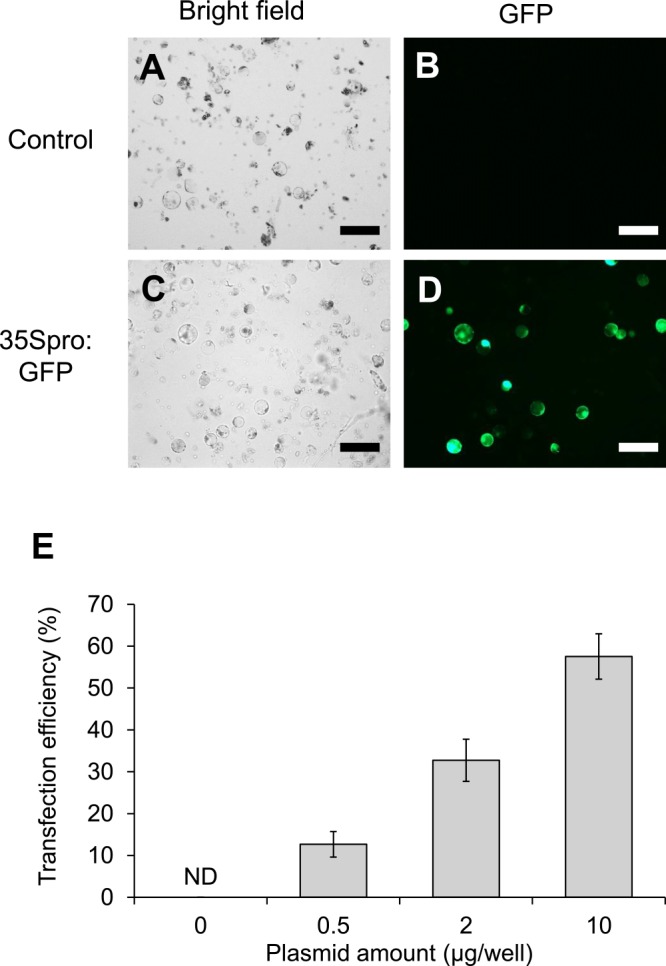
Gene transfection efficiency of buckwheat hypocotyl protoplasts. Green fluorescent protein (GFP) was transiently expressed in buckwheat protoplasts isolated from 7-day-old plants using a PEG/Ca2+ method in a 96-well plate. Transformed protoplasts were incubated at 22°C for 24 h and visualized under a fluorescent microscope. A, B. Non-transformed hypocotyl protoplasts observed by brightfield (A) and fluorescence (B) microscopy. C, D. Hypocotyl protoplasts expressing GFP were observed by brightfield (C) and fluorescence (D) microscopy. E, Transformation efficiency was calculated as the number of living cells and GFP-fluorescent cells. Mean values and standard deviations were calculated from four biological replicates. ND indicates not detected. Scale bars = 100 μm.
Availability of buckwheat protoplast for the effector–reporter assay in 96-well plate
We examined the feasibility of the buckwheat hypocotyl protoplast system for a 96-well-plate-based dual-luciferase assay using transient effector–reporter analysis. The Gal4 DNA-binding domain fused with VP16 (Gal4DB-VP16) and Gal4DB-SRDX effectors, which are an artificial transcriptional activator and repressor, respectively, significantly changed the reporter activity driven by the promoter containing Gal4 binding sites from the control (Gal4DB alone) (Fig. 4). In addition, Gal4DB-FeMYBF1, which is a R2R3 MYB family transcription factor suggested to be a possible transcriptional activator for flavonol biosynthesis in buckwheat (Matsui et al. 2018a), also significantly activated the same reporter activity (n = 4). The relative SD calculated by SD/mean in each experiment was 0.20 (control), 0.11 (Gal4DB-VP16), 0.37 (Gal4DB-SRDX), and 0.22 (Gal4DB-FeMYBF1), which is acceptable for a transient effector–reporter experiment.
Fig. 4.
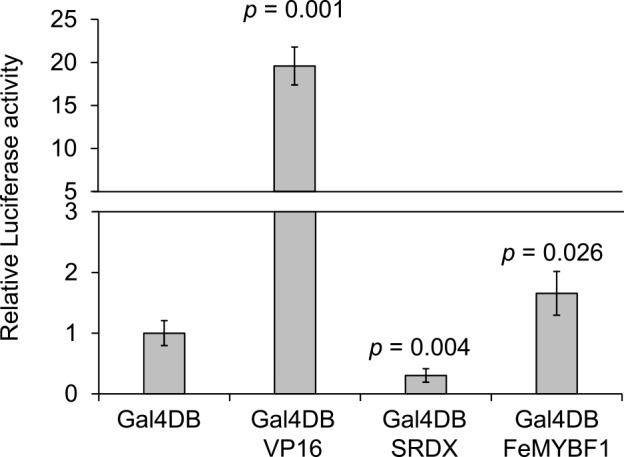
Transcriptional activation/repression ability assay using Gal4 system. Dual-luciferase activity was determined for co-transformed buckwheat protoplasts by effector (Gal4DB-VP16, Gal4DB-SRDX, or Gal4DB-FeMYBF1), reporter, and reference plasmids after incubation for 16 h at 22°C in the dark. Mean values and standard deviations were calculated from the results of four biological replicates. P-values were calculated using Welch’s test (two-sided) with Bonferroni–Holm correction when Gal4DB was considered as a negative control.
We further examined the suitability of the system for functional analysis of buckwheat transcription factors by employing FeMYBF1 and several putative target genes as an example. FeMYBF1 activated reporter gene expression driven by the FeCHS, FeFLS, FeFLS1, FeFLS2, and FeANS promoters to varying extents (Fig. 5), which is similar to the result obtained in a previous study (Matsui et al. 2018a). Interestingly, while FeDFR1a promoter activity in buckwheat protoplasts was induced only 1.58-fold and less than FeFLSs promoters by FeMYBF1, that in Arabidopsis mesophyll protoplasts was induced more than 10-fold and higher than the activation of FeFLSs reporter activity by FeMYBF1 (Matsui et al. 2018a). Agius et al. (2005) reported that a heterologous promoter does not necessarily function in different plants due to differences in the regulatory mechanism. The FeDFR1a promoter might be irregularly activated by FeMYBF1 in Arabidopsis mesophyll protoplasts owing to the difference in cell identity and/or plant species.
Fig. 5.
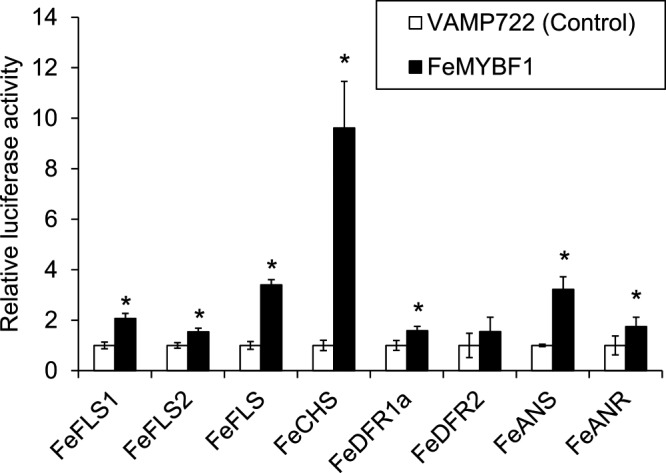
Effector–reporter analysis of FeMYBF1 for flavonol-related gene promoters. FeMYBF1 effector was co-expressed with a reporter construct including Firefly luciferase driven by a flavonol-related gene promoter and a reference construct harboring Renilla luciferase. Relative luciferase activity was determined when the value of VAMP722 activity, which is a negative control localized to the vacuole, was set to 1. Mean values and standard deviations were calculated from four biological replicates. An asterisk indicates that the P-value calculated by Welch’s test (two-sided) was lower than 0.05.
Conclusion
Given the absence of a stable and simple protocol for genetic transformation of buckwheat, establishment of an alternative efficient method to examine gene function in buckwheat is important. This study presents an efficient transient gene expression system using buckwheat protoplasts, which is applicable to a large-scale experiment. Although most functional analyses of buckwheat transcription factors have been carried out to prepare transgenic Arabidopsis (Matsui et al. 2018a, Yao et al. 2017), at least two months are required to perform the analysis. Furthermore, heterologous expression between Arabidopsis and buckwheat may cause difficulties. The present system provides rapid and more reliable information for gene function studies in buckwheat. We consider the present buckwheat system is superior to the previous protoplast system established by Shen et al. (2002) in buckwheat and conventional Arabidopsis mesophyll cell system (Yoo et al. 2007) in terms of shorter period of plant growth and fewer plant number to isolate enough protoplast cells. We propose that the present system could be a general basic method for many studies on plant species including buckwheat and Arabidopsis.
In addition, a transgene-free genome editing technology using a protoplast system provides a powerful tool for plant breeding (Lin et al. 2018, Woo et al. 2015). Lin et al. (2018) reported that mutagenesis efficiency by transiently expressed CRISPR-Cas9 together with guide RNA ranged from 1.1% to 75.2% when transformation efficiency of the transgene ranged from 43% to 63%. Considering the maximum transformation efficiency shown in Fig. 2, the present method is suitable for transgene-free genome editing. An additional task yet to be established is a stable method for plant regeneration from protoplasts. A previous study reported that plant regeneration from hypocotyl protoplasts of common buckwheat was successful, although the efficiency was only approximately 1% (Adachi et al. 1989). Combination of the transient gene expression system established in the present study and a more efficient system for plant regeneration from protoplasts would be a useful tool for buckwheat to elucidate gene function and facilitate efficient buckwheat breeding.
In this study, we established a method for simple protoplast isolation and an efficient transient gene expression system employing 96-well plates. The present system will enable functional analysis of buckwheat genes in native cells and accelerate buckwheat breeding in the future.
Author Contribution Statement
SS performed all experiments. KM and YO performed protoplast isolation. SS, KM, and NM wrote the manuscript. All authors contributed to the design of the experiment and revised the manuscript.
Acknowledgments
We thank Ms Miyoko Yamada (AIST) and Yoshimi Sugimoto (AIST) for their skillful technical support. We thank Robert McKenzie, PhD, from Edanz Group (www.edanzediting.com/ac), for editing a draft of this manuscript.
Literature Cited
- Adachi T., Yamaguchi A., Miike Y. and Hoffmann F. (1989) Plant regeneration from protoplasts of common buckwheat (Fagopyrum esculentum). Plant Cell Rep. 8: 247‒250. [DOI] [PubMed] [Google Scholar]
- Agius F., Amaya I., Botella M.A. and Valpuesta V. (2005) Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. J. Exp. Bot. 56: 37‒46. [DOI] [PubMed] [Google Scholar]
- Cao J.M., Yao D.M., Lin F. and Jiang M.Y. (2014) PEG-mediated transient gene expression and silencing system in maize mesophyll protoplasts: a valuable tool for signal transduction study in maize. Acta Physiol. Plant. 36: 1271–1281. [Google Scholar]
- Fujikawa Y. and Kato N. (2007) Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52: 185‒195. [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Sakamoto S., Kigoshi K., Suzuki K. and Ohme-Takagi M. (2014) VP16 fusion induces the multiple-knockout phenotype of redundant transcriptional repressors partly by Med25-independent mechanisms in Arabidopsis. FEBS Lett. 588: 3665‒3672. [DOI] [PubMed] [Google Scholar]
- Katsu K., Suzuki R., Tsuchiya W., Inagaki N., Yamazaki T., Hisano T., Yasui Y., Komori T., Koshio M., Kubota S. et al. (2017) A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 17: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Xu H., Park W.T., Park N.I., Lee S.Y. and Park S.U. (2010) Genetic transformaiton of buckwheat (Fagopyrum esculentum M.) with Agrobacterium rhizogenes and production of ruin in transformed root cultures. Aust. J. Crop Sci. 4: 485–490. [Google Scholar]
- Lei G.J., Yokosho K., Yamaji N., Fujii-kashino M. and Ma J.F. (2017a) Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 215: 1080‒1089. [DOI] [PubMed] [Google Scholar]
- Lei G.J., Yokosho K., Yamaji N. and Ma J.F. (2017b) Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat. Plant Cell Physiol. 58: 2179‒2189. [DOI] [PubMed] [Google Scholar]
- Lin C.S., Hsu C.T., Yang L.H., Lee L.Y., Fu J.Y., Cheng Q.W., Wu F.H., Hsiao H.C., Zhang Y., Zhang R. et al. (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16: 1295‒1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Hara T., Tetsuka T. and Morishita T. (2013) New common buckwheat cultivar “Sachi-izumi” for warm areas in Japan. Bull. Natl. Agric. Res. Cent. Kyushu Okinawa 59: 23‒37. [Google Scholar]
- Matsui K., Hisano T., Yasui Y., Mori M., Walker A.R., Morishita T. and Katsu K. (2016) Isolation and characterization of genes encoding leucoanthocyanidin reductase (FeLAR) and anthocyanidin reductase (FeANR) in buckwheat (Fagopyrum esculentum). J. Plant Physiol. 205: 41‒47. [DOI] [PubMed] [Google Scholar]
- Matsui K., Oshima Y., Mitsuda N., Sakamoto S., Nishiba Y., Walker A.R., Ohme-Takagi M., Robinson S.P., Yasui Y., Mori M. et al. (2018a) Buckwheat R2R3 MYB transcription factor FeMYBF1 regulates flavonol biosynthesis. Plant Sci. 274: 466‒475. [DOI] [PubMed] [Google Scholar]
- Matsui K., Tomatsu T., Kinouchi S., Suzuki T. and Sato T. (2018b) Identification of a gene encoding glutathione S-transferase that is related to anthocyanin accumulation in buckwheat (Fagopyrum esculentum). J. Plant Physiol. 231: 291‒296. [DOI] [PubMed] [Google Scholar]
- Miljuš-Djukić J., Nešković M., Ninković S. and Crkvenjokov R. (1992) Agrobacterium-mediated transformation and plant regeneration of buckwheat (Fagopyrum esculentum Moench.). Plant Cell Tissue Organ Cult. 29: 101‒108. [Google Scholar]
- Ohta M., Ohme-Takagi M. and Shinshi H. (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22: 29‒38. [DOI] [PubMed] [Google Scholar]
- Park J.W., Kang D.B., Kim C.W., Koh S.H., Yum H.Y., Kim K.E., Hong C.S. and Lee K.Y. (2000) Identification and characterization of the major allergens of buckwheat. Allergy 55: 1035‒1041. [DOI] [PubMed] [Google Scholar]
- Sakamoto S., Fujikawa Y. and Esaka M. (2013) Analysis of ascorbic acid biosynthesis using a simple transient gene expression system in tomato fruit protoplasts. Biosci. Biotechnol. Biochem. 77: 673‒675. [DOI] [PubMed] [Google Scholar]
- Sakamoto S., Takata N., Oshima Y., Yoshida K., Taniguchi T. and Mitsuda N. (2016) Wood reinforcement of poplar by rice NAC transcription factor. Sci. Rep. 6: 19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M., Nakagawa M., Oishi A., Yasui Y. and Katsube-Tanaka T. (2014) Diversification of 13S globulins, allergenic seed storage proteins, of common buckwheat. Food Chem. 155: 192‒198. [DOI] [PubMed] [Google Scholar]
- Seidel T., Golldack D. and Dietz K.J. (2005) Mapping of C-termini of V-ATPase subunits by in vivo-FRET measurements. FEBS Lett. 579: 4374‒4382. [DOI] [PubMed] [Google Scholar]
- Sheen J., Hwang S., Niwa Y., Kobayashi H. and Galbraith D.W. (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8: 777‒784. [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466‒1475. [PMC free article] [PubMed] [Google Scholar]
- Shen R., Ma J.F., Kyo M. and Iwashita T. (2002) Compartmentation of aluminum in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 215: 394‒398. [DOI] [PubMed] [Google Scholar]
- Takeshima R., Nishio T., Komatsu S., Kurauchi N. and Matsui K. (2019) Identification of a gene encoding polygalacturonase expressed specifically in short styles in distylous common buckwheat (Fagopyrum esculentum). Heredity 123: 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B., Xu M., Chen Y. and Huang M. (2013) Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 114: 11–18. [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke S., Blazevic D., Grefen C., Schumacher K., Oecking C. et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428‒438. [DOI] [PubMed] [Google Scholar]
- Wehner N., Hartmann L., Ehlert A., Böttner S., Oñate-Sánchez L. and Dröge-Laser W. (2011) High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 68: 560‒569. [DOI] [PubMed] [Google Scholar]
- Wieslander G. and Norbäck D. (2001) Buckwheat allergy. Allergy 56: 703‒704. [DOI] [PubMed] [Google Scholar]
- Woo J.W., Kim J., Kwon S.I., Corvalán C., Cho S.W., Kim H., Kim S.G., Kim S.T., Choe S. and Kim J.S. (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33: 1162‒1164. [DOI] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T. and Lin C.S. (2009) Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P.F., Li C.L., Zhao X.R., Li M.F., Zhao H.X., Guo J.Y., Cai Y., Chen H. and Wu Q. (2017) Overexpression of a tartary buckwheat gene, FtbHLH3, enhances drought/oxidative stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 8: 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Hirakawa H., Ueno M., Matsui K., Katsube-Tanaka T., Yang S.J., Aii J., Sato S. and Mori M. (2016) Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 23: 215‒224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H. and Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565‒1572. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sakamoto S., Kawai T., Kobayashi Y., Sato K., Ichinose Y., Yaoi K., Akiyoshi-Endo M., Sato H., Takamizo T. et al. (2013) Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D. et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]


