Abstract
Common buckwheat (Fagopyrum esculentum) is a heterostylous self-incompatible (SI) species with two different flower morphologies, pin and thrum. The SI trait is controlled by a single gene complex locus, S. Self-compatible (SC) lines were developed by crossing F. esculentum and F. homotropicum; these lines have an SC gene, Sh, which is dominant over the s allele and recessive to the S allele. S-ELF3 has been identified as a candidate gene in the S locus and is present in the S and Sh but not s alleles. A single-nucleotide deletion in the S-ELF3 gene of the Sh allele results in a frame shift. To develop co-dominant markers to distinguish between ShSh and Shs plants, we performed a next-generation sequencing analysis in combination with bulked-segregant analysis. We developed four co-dominant markers linked to the S locus. We investigated the polymorphism frequency between a self-compatible line and leading Japanese buckwheat cultivars. Linkage between a developed sequence-tagged-site marker and flower morphology was confirmed using more than 1000 segregating plants and showed no recombination. The developed markers would be useful for buckwheat breeding and also to produce lines for genetic analysis such as recombinant inbred lines.
Keywords: self-incompatibility, heterostyly, sequence-tagged sites (STS), bulked segregant analysis
Introduction
Common buckwheat (Fagopyrum esculentum) is a heteromorphic self-incompatible (SI) plant species; it needs insects for cross-pollination between flowers with different morphologies (pin and thrum flowers; Garber and Quisenberry 1927). The yields of buckwheat are influenced by the activity of insects, which is affected by weather. Production of new buckwheat cultivars takes a long time because it requires the loci for useful agricultural traits to be homozygous and loci that would cause inbreeding depression or undesirable traits to be heterozygous.
The self-incompatibility of buckwheat is controlled by a single gene complex locus, S, which is also called the S supergene: the short-style morph is a heterozygous thrum form (Ss) and the long-style morph is imparted by the recessive homozygous pin (ss) alleles (Sharma and Boyes 1961). Self-compatible (SC) lines have been developed by an interspecific cross between F. esculentum and F. homotropicum with embryo rescue (Aii et al. 1998, Campbell 1995, Matsui et al. 2003, Woo et al. 1999). Although the SC lines have a potential for high and stable yields, no such lines are practically available because of inbreeding depression and undesirable traits caused by the homozygous state of some loci. To overcome these problems, crosses between SC lines and leading cultivars would be needed.
The flower morphology of the SC lines, long homostyle (LH), is controlled by a single gene (Campbell 1995, Woo et al. 1999). The allele controlling the homomorphic flower type was designated Sh and the dominance relationship was found to be S > Sh > s (Woo et al. 1999). Sharma and Boyes (1961) postulated that the buckwheat S locus contains several genes and named it the S supergene, similar to that proposed for Primula (Dowrick 1956). The S supergene consists of at least five linked genes: Is and Ip govern the incompatibility reaction in the style and pollen, respectively, G controls style length, P controls pollen size, and A controls anther height (Dowrick 1956, Lewis and Jones 1992, Sharma and Boyes 1961). Each of these genes is di-allelic, and the five alleles are all recessive in pin plants and all dominant in thrum plants. These genes have not been identified but at least two loci are thought to exist (Matsui et al. 2003).
S LOCUS EARLY FLOWRING3 (S-ELF3), which may be a transcription factor, has been identified as a candidate gene for the Is or G gene; it is expressed only in the style of thrum plants and is linked to the S locus with no recombination in buckwheat (Yasui et al. 2012). Using genome assembly analysis with next-generation sequencing (NGS), we have developed a buckwheat genome database (BGDB) and also found that the region that includes this locus is absent in the genome of pin plants (Yasui et al. 2016). The Sh alleles have a deletion of a nucleotide in the S-ELF3 locus, which causes a frame shift.
To obtain self-fertilized F2 seeds in an F1 plant, the styles of pin plants (ss) need to be pollinated with pollen of SC plants (ShSh) because of the dominance relation, Sh > s, and to avoid production of seeds by self-pollination. In such a cross, the genotype of F1 plants would be Shs. In the F2 generation, flower morphology and SI/SC would segregate with a monogenic segregation ratio of 3 SC (long homostyle) to 1 SI (pin). Plants with the long homostyle would be either homozygous (ShSh) or heterozygous (Shs). A method to distinguish between the two genotypes is desirable to select SC homozygous plants in the F2 generation or to choose SC/SI segregating lines in each generation for recurrent selection. DNA markers to distinguish the ShSh and Shs genotypes would be powerful tools for this purpose.
Because the region of the S-ELF3 locus is missing in the genome of s plants (Yasui et al. 2016), we needed to detect the flanking regions of the Sh alleles that would be present in the s genome. In this study, we used NGS analysis in combination with bulked segregant analysis of segregating progeny of a cross between SI and SC plants. We developed co-dominant markers that can distinguish between homozygous (ShSh) and heterozygous (Shs) plants for marker-assisted selection.
Materials and Methods
Plant materials
To detect the s and Sh regions, the segregating line 16Aseg03 (Hara et al. 2019) was used; it consists of 142 F2 plants derived from a cross between a pin plant of the breeding line ‘Kyukei 29’ (KY29, ss) and an SC long-homostyle plant, ‘Kyukei SC7’ (KSC7, ShSh).
To develop co-dominant markers linked to the Sh allele, we used two different segregating lines, 16Aseg04 (Hara et al. 2019) and 16AsegA. Because buckwheat is a cross-pollinating species, each plant, even within the same cultivar, contains many heterozygous loci and has some non-fixed traits. The 16Aseg04 line was produced by crossing a pin plant of KY29 (ss) (different from the plant used to produce 16Aseg03) and KSC7 (ShSh). The 16AsegA line was developed by crossing a pin plant of a green stem mutant line (GSML, Matsui et al. 2008a, 2018) and an SC long-homostyle plant, 13AL130-4, which was developed using an SC line ‘Norin-PL1’ (Matsui et al. 2008b). The 16AsegA line was developed to confirm if a marker was valid for lines derived from a different SC line with the Sh allele. These lines were grown in a glasshouse at the NARO, Tsukuba, Japan.
To verify if a candidate region could be used to develop co-dominant markers, six 16AsegA plants were used for PCR amplification. To identify markers tightly linked to the S locus, we developed an F2 segregating line, 17KySeg01 (1009 plants), derived from a cross between a pin plant of ‘Sachiizumi’ (Matsui et al. 2013) and KSC7. The 17KySeg01 line was grown in a glasshouse at the Kyoto University, Kyoto, Japan.
To determine polymorphism rates between leading cultivars in Japan and KSC7, we used five cultivars, ‘Kitawasesoba’, ‘Hashikamiwase’, ‘Harunoibuki’ (Hara et al. 2012), ‘NARO-Fe1’, and ‘Sachiizumi’, and two breeding lines, KY28 and KY29. These plants were grown in a field at the Institute of Crop Science NARO, Tsukuba, Ibaraki, Japan.
DNA isolation and preparation of bulked DNA for NGS analysis
Total DNA was isolated from leaves of each plant from all segregating populations with a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Two sets of bulked DNA were made by mixing DNA of randomly selected 28 pin plants (PIN bulk) or 30 long-homostyle plants (LH bulk) of the 16ASeg03 line. The LH bulk was expected to contain two alleles (Sh and s) and the PIN bulk only the recessive allele (s), which would make it possible to detect the flanking regions by NGS analysis of the Sh allele. Genomic DNA was also extracted from the parental plants of the 16ASeg03 line and the five Japanese cultivars and two breeding lines mentioned above.
Next-generation sequencing analysis
Paired-end reads of 100 bp from the two bulks and the KSC7 and KY29 plants were obtained on an Illumina HiSeq 2000 System at Macrogen Japan (Kyoto, Japan). The raw reads are available from the DDBJ/EMBL/NCBI under the accession numbers DRX178921-DRX178924. Low-quality reads and adaptors (CACGACGCTCTTCCGATCT and ACCGCTCTTCCGATCTGTAA) were trimmed using Trimmomatic-0.32 (Bolger et al. 2014) with the following settings: HEADCROP, 2; SLIDINGWINDOW, 4:25; LEADING, 25; TRAILING, 25; MINLEN, 50. Trimmed reads were mapped to the reference sequences using BWA 0.7.15 (Li and Durbin 2009) with the ‘bwa aln’ option with -l 32 -k 2 -n 5 and the ‘bwa sampe’ option with default settings. Only genome sequences (Yasui et al. 2016) of ≥1 kb were selected as reference sequences. Mapping results were processed with SAMtools 0.1.18 (Li et al. 2009). SNPs were detected using UnifiedGenotyper in GATK 3.7 (DePristo et al. 2011) with the –glm BOTH option.
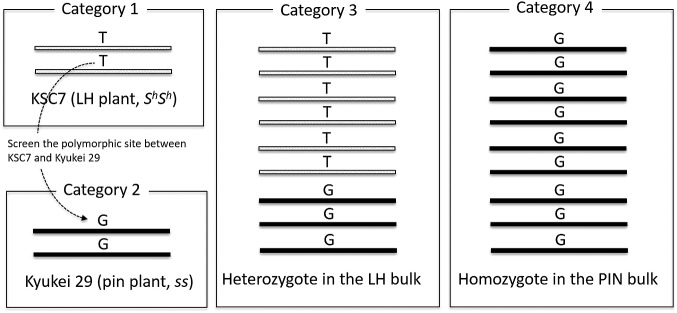
The Sh-linked SNPs were detected using the following criteria: 1) homozygous in KSC7, the nucleotide differs from that in the reference sequence because the reference sequence in BGDB was developed using a thrum type of plant; 2) homozygous in KY29, the nucleotide is the same as in the reference sequence; 3) heterozygous in the LH bulk; 4) homozygous in the PIN bulk, the nucleotide is the same as in the reference sequence (Fig. 1). The reference sequences in BGDB were short (N50 = 25.1 kb; Yasui et al. 2016), and it is difficult to obtain a graphical change of the number of Sh-linked SNP sites through scaffolds. Hence, we counted the Sh-linked SNPs in all reference sequences and finally obtained the ratio of the number of Sh-linked SNPs to the number of all SNPs, named Sh-linked SNP index, for each scaffold.
Fig. 1.
Scheme of screening for Sh allele–linked SNPs. Hatched and solid bars indicate genome sequences harboring the Sh and s allele, respectively. Sequences were screened for SNPs between KSC7 and the reference sequence (category 1, Yasui et al. 2016) and between KSC7 and KY29 (category 2). Heterozygous sites in the LH bulk (category 3) and homozygous sites in the PIN bulk (category 4) were treated as Sh-linked SNPs.
Development of sequence-tagged-site markers from NGS data
Sequences of ca. 1000 bp were randomly selected in each of the 50 candidate regions (Supplemental Table 1) and primers were designed with Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) to satisfy the following conditions: product length, ca. 600–650 bp; primer length, 22–26 bp; Tm, 55–64°C. Amplification with genomic DNA as a template was performed with the designed specific primers as follows: 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s. Amplification was confirmed by agarose gel electrophoresis, and the DNA fragments were digested with three randomly chosen enzymes with four recognition sites (AluI, HaeIII, and MspI).
Co-dominant markers were developed using six plants from the 16ASegA segregating population to find highly versatile markers.
Confirmation of linkage relations
To check whether the markers are linked to the S locus and are useful for different populations, we checked the linkage relation in the 16ASeg03 and 16ASeg04 populations. Using 50 plants from each population, the amplification with the corresponding primers and subsequent digestion of the PCR products with the corresponding restriction enzymes were performed. For a marker that seemed to be useful in linkage analysis, additional linkage analysis with 17KySeg01 which is F2 segregating lines (1009 plants) with the corresponding specific primers and enzymes was performed. The genotype of the S locus was determined based on the flower morphology. Electrophoresis was performed in a capillary electrophoresis system (LabChip GX, PerkinElmer). A DNA5K/RNA/CZE chip was used with a HT DNA5K Reagent Kit (PerkinElmer).
Results
Identification of flanking regions of the Sh allele and development of co-dominant markers
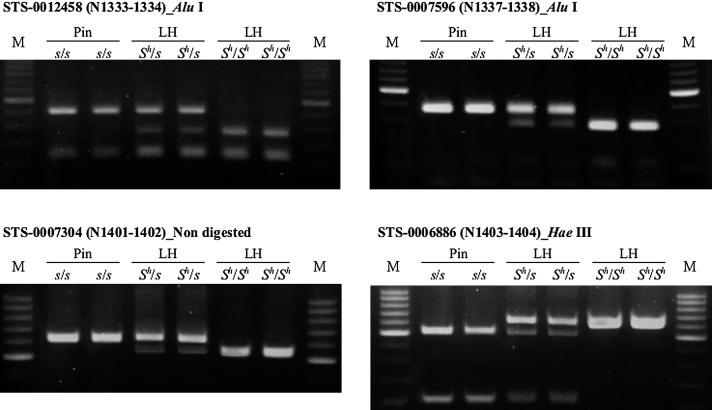
By comparing the sequences between the LH and PIN DNA bulks, we obtained 263 scaffolds with high values of the Sh-linked SNP index (>0.5; Supplemental Table 1). Because it takes time to design markers at the detected SNPs, and actual buckwheat breeding does not require many markers for the selection of a trait, we chose the top 50 scaffolds and made primers to amplify each region (Supplemental Table 1). Amplification in six 16ASegA plants showed that 16 primer sets resulted in a weak band, more than two bands, or no amplification in some plants (Supplemental Table 1). The 34 amplicons that showed good amplification were digested with AluI, HaeIII, and MspI. Four primer sets showed polymorphisms consistent with flower morphology (Fig. 2, Supplemental Table 1), and the primer sets with the restriction enzymes were named as S linked sequence-tagged-site (STS) markers (STS-0012458, STS-007596, STS-0007304, and STS-0006886; Fig. 2, Supplemental Table 1). Polymorphism was detected by AluI digestion in STS-0012458 and STS-007596, and by HaeIII digestion in STS-0006886. Sizes of the undigested fragments amplified in STS-0007304 differed between pin and LH plants, but the band intensity and stability were low (Supplemental Table 1).
Fig. 2.
PCR products from F2 pin and long-homostyle (LH) plants (16ASeg3). PCR products were digested with AluI or HaeIII. Undigested amplification products of STS-0007304 differed between pin and LH. The genotypes of S locus, Shs and ShSh were shown based on the genotype of each marker.
Confirmation of linkage using several segregating lines
To check if these markers are really linked to the S locus and are useful for different segregating populations, we performed linkage analysis with randomly selected 50 plants from each of the 16ASeg3 and 16ASeg4 populations. Segregation of the four markers fit flower morphology with no recombination (Table 1). STS-0012458 was polymorphic in all populations that we used and was further investigated using the segregating line, 17KySeg01 (1009 plants). Flower morphology segregated as 745 LH and 264 pin, fitting a segregating ratio of 3:1 (Table 2; χ2 = 0.7298, 0.40 < P < 0.50); STS-0012458 segregated as AA:AB:BB = 240:505:264, fitting a segregating ratio of 1:2:1 (χ2 = 1.1427, 0.40 < P < 0.50).
Table 1.
Linkage relation between flower morphology and developed markers evaluated with 50 randomly selected plants from each segregating line
| Segregating line | Long homostyle | Pin | χ2 value, P | |||||
|---|---|---|---|---|---|---|---|---|
| Aa | H | B | A | H | B | Flower morphology (3:1) | Marker (1:2:1) | |
| Seg3b | 15 | 23 | 0 | 0 | 0 | 12 | 0.027, 0.80 < P < 0.90 | 0.680, 0.70 < P < 0.80 |
| Seg4 | 14 | 26 | 0 | 0 | 0 | 10 | 0.667, 0.40 < P < 0.50 | 0.720, 0.60 < P < 0.70 |
a A, H and B are the genotypes of STS markers. All STS markers (0012458, 0007596, 0007304, and 0006886) showed the same segregation pattern in each segregating line.
b Seg3, 16ASeg3 line; Seg4, 16ASeg4 line.
Table 2.
Linkage relation between flower morphology and a developed marker, STS-0012458
| Long homostyle | Pin | χ2 value, P | |||||
|---|---|---|---|---|---|---|---|
| Aa | H | B | A | H | B | Flower morphology (3:1) | Marker (1:2:1) |
| 240 | 505 | 0 | 0 | 0 | 264 | 0.7298, 0.40 < P < 0.50 | 1.1427, 0.40 < P < 0.50 |
a A, H and B are the genotypes of STS markers.
Polymorphism between leading cultivars in Japan and KSC7
It is important to show high rates of polymorphism between parental lines. Among the four primer sets, STS-0012458, STS-0007304, and STS-0006886 showed high frequency of polymorphisms between KSC7 and pin plants of leading Japanese cultivars and breeding lines (Table 3), indicating that these three markers would be useful for introducing the Sh into many varieties.
Table 3.
Polymorphism frequency between a self-compatible line, KSC7, and Japanese cultivars and breeding lines
| Marker | Enzyme | KSC7 | KTW | HKW | HAR | NF1 | SAC | K28 | K29 | Total by the band patterna | Total number of plants | Percentage of polymorphism relative to KSC7 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | H | – | + | H | – | + | H | – | + | H | – | + | H | – | + | H | – | + | H | – | + | H | – | |||||
| STS-0012458 | AluI | + | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 4 | 0 | 0 | 38 | 38 | 100.0 |
| STS-0007596 | MspI | – | 0 | 0 | 6 | 0 | 0 | 6 | 6 | 0 | 0 | 3 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 4 | 14 | 0 | 23 | 37 | 37.8 |
| STS-0007304 | 0 | 0 | 6 | 0 | 0 | 4 | 0 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 5 | 0 | 0 | 4 | 0 | 0 | 30 | 30 | 100.0 | ||
| STS-0006886 | HaeIII | – | 6 | 0 | 0 | 6 | 0 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 35 | 0 | 0 | 35 | 100.0 |
a Only pin plants of each variety were used. KTW, Kitawasesoba; HKW, Hashikamiwase; HAR, Harunoibuki; NRF, NARO-Fe1; SAC, Sachiizumi; K28, Kyukei 28; K29, Kyukei 29. +, digested; –, not digested; H, heterozygous.
Discussion
Marker-assisted selection can improve the efficiency and accuracy of conventional plant breeding in many crops. Co-dominant markers are particularly powerful in selecting the desirable traits without missing heterozygous plants because they can distinguish between homozygous and heterozygous plants. Co-dominant DNA markers can usually be developed on the basis of the sequence of a gene controlling targeted traits or tightly linked regions. Because the region of the S locus of buckwheat is hemizygous (S-ELF3 in the S locus is present only on a chromosome carrying the S or Sh allele; Yasui et al. 2012, 2016), the S-ELF3 sequence cannot be used for developing co-dominant markers. Furthermore, the exact length of the deletion in the s allele are still unknown because the scaffold that includes S-ELF3 is short.
Bulked segregant analysis is a powerful tool to identify the flanking regions of a target gene. Several markers linked to genes controlling agricultural traits in buckwheat have been developed with this technique (Aii et al. 1998, Matsui et al. 2004). Aii et al. (1998) have developed a co-dominant marker linked to the Sh allele on the basis of a random amplified polymorphic DNA (RAPD) marker. However, because of a large distance between this marker and the S locus (ca. 6 cM), the genotype of some plants determined with the marker did not match their SC/SI morphology. The ability of RAPD or similar markers such as amplified fragment length polymorphism (AFLP) markers to reveal polymorphism depends on the primer sequences.
In this study, we sequenced the whole genome using bulked DNA. By setting screening criteria (see Materials and Methods), we can determine the area linked to the hemizygous region. We developed tightly linked markers, and the linkage distance of one of them was less than 0.1% because any recombination was not recognized in the segregating line, 17KySeg01 (1009 plants). It would be very useful for buckwheat breeding. Among multiple candidate regions, we used the top 50 only. If we knew the candidate regions tightly linked to the S locus, we could develop more co-dominant markers more efficiently. Recently, the NGS-based target re-sequencing AmpliSeq technology (Thermo Fisher Scientific, Waltham, MA, USA) has been used to sequence plant DNA (Ogiso-Tanaka et al. 2019, Stevanato et al. 2017). Such methods would also increase the efficiency of co-dominant marker development. Identification of the region deleted in the s allele would allow us to accurately develop co-dominant markers by designing primers to cover the deleted region. Unfortunately, the tightly linked marker STS-0012458 we developed is not on the same scaffold as S-ELF3. The BGDB consists of more than 300,000 scaffolds, so the missing region of the s allele remains unidentified. Upgrade and improvement of BGDB would help to solve the issue.
All markers except STS-0007596 showed a high rate of polymorphism between the SC line KSC7 and cultivars and breeding lines in Japan, probably because KSC7 contains genomic regions derived from F. homotropicum. KSC7 was developed using ‘Norin-PL1’ (Matsui et al. 2008b), which was generated by a cross between F. esculentum and F. homotropicum (Matsui et al. 2003). A low rate of polymorphism of the STS-0007596 may be because it was developed on a genomic region that was derived from the F. esculentum, or on a conserved area between F. esculentum and F. homotropicum. Both SC lines, ‘Norin-PL1’ and KSC7, have inferior traits that make them unsuitable for cultivation by farmers. Identification of the genomic region of F. homotropicum in ‘Norin-PL1’ may reveal regions related to these inferior traits such as preharvest sprouting (Hara et al. 2019).
SC lines are beneficial for buckwheat cultivation and breeding, because they do not need pollinators, have stable yield, and their useful agronomical traits can be fixed easily. Because some traits including beneficial traits are hidden by heterozygosity in normal cultivated and indigenous natural populations, SC lines can be used to reveal these traits easily by homozygous. Furthermore, SC lines can be easily used to produce segregating and analytical lines such as F2, recombinant inbred lines, near isogenic lines, and mutated populations. With the development of sequencing technology, it is now possible to obtain varieties with new desirable traits such as non-allergens and glutinous starches from artificially mutated or unique natural populations based on DNA sequences.
To breed a high-yielding variety adapted to a particular cultivation area, a line with desirable traits would need to be crossed with a leading local cultivar. Co-dominant markers such as those developed in this study would be useful for distinguishing between SI and SC lines to avoid inbreeding depression often found in the latter.
Author Contribution Statement
KM and YY conceived of the study and designed the experiments. YY, MN and UM performed the NGS analysis. KM developed co-dominant markers. KM, YY, MN, UM and TR performed linkage analysis. KM and TR investigated the frequency of polymorphisms. KM and YY wrote the manuscript. MN, UM and TR edited and revised the manuscript.
Supplementary Material
Acknowledgments
We thank Mr. Yoshioka, Mr. Yokotsuka and Mr. Nakamura in NARO for growing buckwheat plants. This work was supported by the NARO, Japan.
Literature Cited
- Aii J., Nagano M., Penner G., Campbell G.C. and Adachi T. (1998) Identification of RAPD markers linked to the Homostylar (Ho) gene in buckwheat. Breed. Sci. 48: 59–62. [Google Scholar]
- Bolger A.M., Lohse M. and Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, C. (1995) Inter-specific hybridization in the genus Fagopyrum. In: Matano, T. and A. Ujihara (eds.) Proceedings of the 6th International Symposium on Buckwheat, Shinshu University Press, Ina, pp. 255–263. [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowrick V.P.J. (1956) Heterostyly and homostyly in Primula obconica. Heredity (Edinb) 10: 219–236. [Google Scholar]
- Garber R.J. and Quisenberry K.S. (1927) The inheritance of length of style in buckwheat. J. Agric. Res. 34: 181–183. [Google Scholar]
- Hara T., Tetsuka T. and Matsui K. (2012) New buckwheat cultivar “Harunoibuki”. Bull. Natl. Agric. Res. Cent. Kyushu Okinawa Reg. 58: 37–48. [Google Scholar]
- Hara T., Takeshima R. and Matsui K. (2019) Genes with different modes of inheritance regulate seed germination in preharvest-sprouting-tolerant lines of buckwheat (Fagopyrum esculentum). Jpn. Agric. Res. Q. (in press) [Google Scholar]
- Lewis, D. and D.A. Jones (1992) The genetics of heterostyly. In: Barrett, S.C.H. (ed.) Evolution and Function of Heterostyly. Springer-Verlag, Berlin Heidelberg, pp. 129–150. [Google Scholar]
- Li H. and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and Genome Project Data P. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Tetsuka T., Nishio T. and Hara T. (2003) Heteromorphic incompatibility retained in self-compatible plants produced by a cross between common and wild buckwheat. New Phytol. 159: 701–708. [DOI] [PubMed] [Google Scholar]
- Matsui K., Kiryu Y., Komatsuda T., Kurauchi N., Ohtani T. and Tetsuka T. (2004) Identification of AFLP makers linked to non-seed shattering locus (sht1) in buckwheat and conversion to STS markers for marker-assisted selection. Genome 47: 469–474. [DOI] [PubMed] [Google Scholar]
- Matsui K., Eguchi K. and Tetsuka T. (2008a) A novel gene that diverts the anthocyanin biosynthetic pathway towards the production of proanthocyanidins in common buckwheat (Fagopyrum esculentum). Breed. Sci. 58: 143–148. [Google Scholar]
- Matsui K., Tetsuka T., Hara T. and Morishita T. (2008b) Breeding and characterization of a new self-compatible common buckwheat parental line, “Buckwheat Norin-PL1”. Bull. Natl. Agric. Res. Cent. Kyushu Okinawa Reg. 49: 1–17. [Google Scholar]
- Matsui K., Hara T., Tetsuka T. and Morishita T. (2013) New common buckwheat cultivar “Sachi-izumi” for warm areas in Japan. Bull. Natl. Agric. Res. Cent. Kyushu Okinawa Reg. 59: 23‒37. [Google Scholar]
- Matsui K., Tomatsu T., Kinouchi S., Suzuki T. and Sato T. (2018) Identification of a gene encoding glutathione S-transferase that is related to anthocyanin accumulation in buckwheat (Fagopyrum esculentum). J. Plant Physiol. 231: 291–296. [DOI] [PubMed] [Google Scholar]
- Ogiso-Tanaka E., Shimizu T., Hajika M., Kaga A. and Ishimoto M. (2019) Highly multiplexed AmpliSeq technology identifies novel variation of flowering time-related genes in soybean (Glycine max). DNA Res. 26: 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K.D. and Boyes J.W. (1961) Modified incompatibility of buckwheat following irradiation. Can. J. Bot. 39: 1241–1246. [Google Scholar]
- Stevanato P., Broccanello C., Pajola L., Biscarini F., Richards C., Panella L., Hassani M., Formentin E., Chiodi C., Concheri G. et al. (2017) Targeted next-generation equencing identification of mutations in disease resistance gene analogs (RGAs) in wild and cultivated beets. Genes (Basel) 8: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S.H., Adachi T., Jong S.K. and Campbell C.G. (1999) Inheritance of self-compatibility and flower morphology in an inter-specific buckwheat hybrid. Can. J. Plant Sci. 79: 483–490. [Google Scholar]
- Yasui Y., Mori M., Aii J., Abe T., Matsumoto D., Sato S., Hayashi Y., Ohnishi O. and Ota T. (2012) S-LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-incompatibility. PLoS ONE 7: e31264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Hirakawa H., Ueno M., Matsui K., Katsube-Tanaka T., Yang S.J., Aii J., Sato S. and Mori M. (2016) Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 23: 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.