Abstract
Bioengineered silver nanoparticles can emerge as a facile approach to combat plant pathogen, reducing the use of pesticides in an eco-friendly manner. The plants’ response during tripartite interaction of plant, pathogen, and nanoparticles remains largely unknown. This study demonstrated the use of bioengineered silver nanoparticles in combating black spot disease caused by necrotrophic fungus Alternaria brassicicola in Arabidopsis thaliana via foliar spray. The particles reduced disease severity by 70–80% at 5 μg/ml without showing phytotoxicity. It elicited plant immunity by a significant reduction in reactive oxygen species (ROS), decreases in stress enzymes by 0.6–19.8-fold, and emergence of autophagy. Comparative plant proteomics revealed 599 proteins expressed during the interaction, where 117 differential proteins were identified. Among different categories, proteins involved in bioenergy and metabolism were most abundant (44%), followed by proteins involved in plant defense (20%). Metabolic profiling by gas chromatography–mass spectroscopy yielded 39 metabolite derivatives in non-polar fraction and 25 in the polar fraction of plant extracts. It was observed that proteins involved in protein biogenesis and early plant defense were overexpressed to produce abundant antimicrobial metabolites and minimize ROS production. Bioengineered silver nanoparticles performed dual functions to combat pathogen attack by killing plant pathogen and eliciting immunity by altering plant defense proteome and metabolome.
Keywords: nanoparticles, pesticide, plant pathogen, proteomics, metabolomics
Graphical Abstract
GRAPHICAL ABSTRACT

Introduction
Plant diseases possess a potential threat toward the productivity and quality of food and fiber, putting agriculture on a higher pressure to feed the ever-increasing population. Among plants, the worldwide loss due to plant pathogens ranged from 50% in wheat to more than 80% in cotton (Oerke, 2006; Velásquez et al., 2018). Several management practices have been employed to overcome this problem, but the phenomenon of multidrug resistance acquired by pathogen and climate change has made the situation more difficult (Juroszek and Tiedemann, 2015; Kim et al., 2016).
Silver nanoparticles are potent antimicrobial agents, having proved their utility in medical applications (Mariana and De, 2019; Monir et al., 2019), however, their application in plant disease management is still in nascent stage (Ocsoy et al., 2013; Mishra et al., 2014). Stephano-Hornedo et al. (2020) and successfully controlled huanglongbing disease in Mexican limes (Citrus aurantifolia Swingle) by application of commercially available silver nanoparticles, found to be 3–60 times more efficient than conventional β-lactam antibiotics. Unlike chemical antimicrobial agents, silver nanoparticles exhibit multiple modes of actions of killing, which includes DNA unwinding, membrane disruption, changing permeability of membrane, and production of free radicals, making it very difficult for pathogens to acquire resistance against them (Verano-Braga et al., 2014; Kumari et al., 2017a, b).
Biogenic silver nanoparticles synthesized from biocontrol agent Trichoderma viride have proved their enhanced antimicrobial activity than their chemical counterparts by increased internalization inside cell in our previous studies (Kumari et al., 2017a). Recently, Mahawar et al. (2020) have also postulated a synergistic antifungal behavior of silver nanoparticles augmented with Calothrixelenkinii for controlling Alternaria blight disease by increase in polyphenol oxidase activity. Earlier, Ramalingam et al. (2016) had also observed higher ROS generation and increased changes in nano-mechanical properties of microbial cell surface after treatment with biogenic silver nanoparticles in comparison to chemically synthesized silver nanoparticles. Silver nanoparticles synthesized from chitin-induced exometabolites from a Trichoderma fusant were able to restrain the phytopathogen Sclerotium rolfsii at a minimum inhibitory concentration (MIC) of 20 μg/ml (Hirpara and Gajera, 2020) by modulating pathogenic gene expression. The effective role of silver nanoparticles in plant disease management in vitro and glass house conditions has been elucidated by some researchers, however, responses of plants toward the particles in fighting diseases remain unknown (Ocsoy et al., 2013; Ilaria et al., 2019).
An interaction between pathogen and plant follows a cascade of mechanism relying on the activation of the plant defense system and generation of reactive oxygen species (ROS), antioxidants, and stress enzymes (Gill and Tuteja, 2010). Several inducers and strategies have been employed to improve plant resistance toward pathogens (Ahuja et al., 2012; Hassan and Mathesius, 2012). Defense activities are elicited after attempted microbial infection including the deposition of the phenolics, ascorbic acid, and induction of antioxidant enzymes to overcome the stress (Singh et al., 2013).
A detailed investigation to decipher the changes occurring on the molecular level of the plant system during the tripartite interaction of plant, pathogen, and nanoparticles is needed. Development in the area of “proteomics” has opened new opportunities to explore large-scale data-rich biology of plants at protein levels. Mass spectrometry (MS)-based comparative proteomics can be a useful tool to determine the physiological status of plants (Verano-Braga et al., 2014). Metabolomics occupies a significant position among several omics approaches to know the physiological status of a cell, comprising the non-genetically encoded substrates, intermediates, and products of metabolic pathways, associated to a cell (Nielsen and Jewett, 2007; Chen et al., 2020).
In this study, the model pathosystem of Arabidopsis thaliana ecotype Columbia (col-0) and Alternaria brassicicola was investigated to assess the potential of biogenic silver nanoparticles in plant disease management. A. brassicicola causes black spot disease on virtually every important cultivated Brassica species. Proteomics, metabolomics, defense, and morphological response of plant during the tripartite interaction of plant, pathogen, and nanoparticles were also evaluated to get an insight into the mechanistic aspects of this interaction.
Materials and Methods
Nanoparticles
Spherical biogenic silver nanoparticles (2–5 nm) (SNP) were synthesized earlier from cell-free extract of T. viride (MTCC 5661) at specified conditions (Kumari et al., 2017b, 2019) and concentration was adjusted to 5 μg/ml before use. Particles were filtered through 0.22 μm syringe millipore filters and sonicated for 2 min before use for getting monodispersed population. Zeta size and zeta potential of nanoparticles were measured following the protocols of Kumari et al. (2017b) (Supplementary Figure S1).
Detached Leaf Assay
Detached leaf assay was performed with different concentrations of SNP (1, 2.5, 5, and 7.5 μg/ml). Briefly, 1-month-old A. thaliana leaves were washed with ultrapure water and surface sterilized by 0.4% sodium hypochlorite solution for 30 s, followed by rinsing with sterile water five times. Leaves were dipped in 5 ml of nanoparticles for 30 min and placed on 0.8% agar plates while leaves dipped in sterile water served as control. Spore suspension (105 spores/ml) of A. brassicicola was prepared in 1% gelatin and spotted on leaves (2 spots/leaf) with 10 μl of spore suspension. Reduction in disease severity was calculated by measuring the reduction in spore count as compared to control leaves.
Plant in vivo Experiments
Seeds of A. thaliana ecotype Columbia (Col-0) were surface sterilized by 0.4% sodium hypochlorite solution followed by repeated washing with distilled water. Surface-sterilized seeds were sown (10 seeds) on sterile soilrite and pots were transferred to temperature-controlled culture room (set at 22°C) in continuous light conditions after 3 days of cold treatment (Srivastava et al., 2012b). They were irrigated twice a week with the nutrient solution and the plants were allowed to grow for 1 month after germination. After 1 month, pots were divided into four groups with six replicates each:
-
1.
Control (Con);
-
2.
Plants treated with biogenic silver nanoparticles (SNP alone);
-
3.
Plants infected with A. brassicicola (AB alone);
-
4.
Plants with both silver nanoparticles and challenged with A. brassicicola (AB + SNP).
Control plants were sprayed with 1% gelatin, while SNP and AB + SNP treatments were sprayed with the biogenic nanoparticles of concentration 5 μg/ml until runoff. Two hours after treatment, AB and AB + SNP treatments were inoculated with spore suspension of A. brassicicola (105 spores/ml). Plants were bagged with a transparent plastic bag and kept for 48 h. After 48 h of infection, leaf samples were harvested immediately in liquid nitrogen and kept in -80°C till further analysis for biochemical and omics analysis. For staining, fresh leaves were collected and proceeded further with the mentioned protocols. Disease parameters and morphological data, viz., number of lesions/leaf, number of spores/leaf, chlorophyll content, and fresh and dry weight of leaves were also taken from fresh leaves 48 h post-infection.
Biochemical Analysis
Total chlorophyll content in plants was determined by the protocol given by Arnon (1949). Defense parameters of plants including proline, lipid peroxidation (LPX), hydrogen peroxide (H2O2), ascorbic acid, and total phenolics were measured following the protocols of Bates et al. (1973); Heath and Packer (1968), Sergiev et al. (1997); Mitusi and Ohata (1961), and Mallick and Singh (1980), respectively, with some modifications. Antioxidant enzymes, superoxide dismutase (SOD), catalase, GoPX, phenylalanine ammonia lyase (PAL), and polyphenol oxidase (PPO) were measured following the protocols of Beauchamp and Fridovich (1971); Aebi (1974), Hemeda and Klein (1990) and Lavania et al. (2006), respectively. All analyses were carried out on UV-vis spectroscopy (Perkin Elmer, MA, United States), keeping length of light path 1 cm.
Histochemical Staining
After 48 h of infection, leaves were preserved in acetic acid:ethanol (1:3) and stained with 0.1% trypan blue for fungal colonization and 0.1% lactophenol cotton blue for mycelia growth inhibition. For observation of the slides, light microscope (Leica Microsystems, GmbH, Germany) was used at CSIR-NBRI, Lucknow.
Scanning and Transmission Electron Microscopy
For evaluation of morphological differences between the treatments, transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were carried out with some modification of protocols (Tiwari et al., 2014) at the central instrument facility of CSIR–Indian Institute of Toxicology Research (IITR), Lucknow.
After 48 h of infection, fresh leaves of A. thaliana were cut and fixed in 4% formaldehyde and 2% glutaraldehyde in cacodylate buffer 0.1 M, pH 6.9. After 3 days, the samples were rinsed in 0.1 M cacodylate buffer and post-fixed in 1% osmium tetroxide in cacodylate buffer, pH 6.9, for 2 h at 4°C. The pieces were then rehydrated in propylene oxide, embedded in an araldite and dodecenyl succinic anhydride mixture, and baked for 48 h at 65°C. Ultrathin sections (5070 nm) were cut with a Leica EMUC7 ultramicrotome, stained with uranyl acetate and lead citrate, and examined under TEM (FEI, TechnaiG2 Spirit TWIN, United States).
For SEM studies, fixed and dehydrated samples were dried by critical point method. The samples were spur coated by gold metal by sputter coater and were analyzed in high vacuum mode by high-resolution field emission E.M., Quanta SEM field emission gun (FEG 450, Netherlands).
Proteomic Profiling
Protein Extraction
Proteins were isolated from leaves of A. thaliana from three replications to normalize the effect of biological variations. In brief, samples were ground in liquid N2, and the resulting powder was extracted with lysis buffer containing 50 mM Tris–HCl, pH 7.5, 60 mM KCl, 10 mM MgCl2, 0.5% Triton X, 2 mM PMSF, and 1 mM DTT. The extract was incubated overnight at 4°C on a rotary shaker at 180 rpm and further centrifuged at 15,000 rpm (25,200 g) for 15 min at 4°C. The extracted protein was precipitated with 0.1 M ammonium acetate (in 100% methanol) at -20°C and then centrifuged at 15,000 rpm for 15 min at 4°C. The pellet was washed thrice with chilled acetone. Further, it was dried and resuspended in a solubilization (rehydration) buffer consisting of 8 M urea, 2 M thiourea, 4% CHAPS, and 20 mM DTT and quantified by Bradford method (Agarwal et al., 2016).
Two-Dimensional Gel Electrophoresis, Gel Staining, and Protein Identification
Two-dimensional electrophoresis (2-DE) was carried out following the protocols of Agarwal et al. (2016) with some modifications [details provided in Supporting Information (SI)].
MS and MS/MS
A 4800 Proteomics Analyzer (Applied Biosystems, MA, United States) with TOF/TOF optics was used for all MALDI-MS and MS/MS applications. Samples were prepared by mixing 0.5 ml of sample with 0.5 ml of matrix solution (5 mg/ml a-Cyano-4-hydroxycinnamic acid in 50% ACN containing 0.1% TFA) and spotted on stainless steel 384-well target plate. They were air dried at room temperature and were then placed in the mass spectrometer and subjected to mass analysis. The mass spectrometer is externally calibrated with a mixture of angiotensin I, Glufibrino–peptide B, ACTH (1e17), and ACTH (18e39). For MS/MS experiments, the instrument was externally calibrated with a fragment of Glufibrino–peptide B. The monoisotopic peptide masses obtained from MALDI-TOF-TOF were analyzed by the 4000 Series Explorer software version 3.5 (ABI, MA, United States). Based on mass signals, protein identification was carried out with the Mascot software1 to search proteins against NCBInr databases. The parameters used for database searches were as follows: monoisotopic mass accuracy, <100 ppm; missed cleavages, 1; carbamidomethylation of cysteine as fixed modification and oxidation of methionine, and N-terminal acetylation (protein) as variable modifications. The only protein spots whose MOWSE score was above the significant threshold level determined by Mascot was considered to represent an identification. The protein functions were assigned using the Pfam2 and InterPro3 protein function databases. Self-organizing tree algorithm (SOTA) clustering was performed on the log-transformed fold induction expression values across different treatments using the Multi Experiment Viewer (MEV, 4.9) software. The clustering was done with the Pearson correlation as distance with 10 cycles and a maximum cell diversity of 0.8. Increase or decrease at least in one treatment was considered for differential spot identification. A criterion of p < 0.001 was used to determine the significant difference for analyzing the parallel spots between genotype with analysis of one-way variance (ANOVA).
Metabolic Profiling
Extraction of Metabolites
The lyophilized plant material (1.0 g) was extracted with hexane (35°C) in a ratio of 1:10 w/v. The solvent was separated by filtration and the procedure was repeated until the hexane layer became colorless. Thus, the separated solvent layer was concentrated under reduced pressure and the obtained sticky mass was stored at -20°C for further GC–MS analysis. The remaining solid plant material was further extracted thrice with fivefold excess (w/v) of 90% and then with 70% methanol–water. The extract obtained was reduced using rotavapor and defatted with the equal volume of hexane and further concentrated under reduced pressure. The samples were then lyophilized to dryness and the resulting solid was again stored at -80°C for further analyses (Srivastava et al., 2012a).
GC-MS Analysis
GC-MS analysis was performed following the protocols of Srivastava et al. (2012a) using Thermo Trace GC Ultra coupled with Thermo Fisher DSQ II mass spectrometers with electron impact ionization at 70 eV to generate mass spectra (details provided in Supplementary Material).
Quantification of Silver Ions in Plant Leaves
Quantification of silver ions present in leaves of the plant was done by the protocols of Srivastava et al. (2018) by inductively coupled plasma mass spectrometer (ICP-MS, Agilent 7500 cx). Briefly, 500 mg of dried Arabidopsis leaves were digested with 5:1 v/v of HNO3 and H2O2 at 120°C for 4 h. Total silver was quantified in the digested sample and analyzed by the ICP-MS. Silver was used as a standard and for calibration (300 ppb, Agilent).
Statistical Analysis
The results were expressed as the mean values ± SE (standard error of mean) of mg/g for non-polar compounds and μg/g for polar compound obtained in GC-MS analysis. The statistical significance for the four different treatments was determined by one-way ANOVA post hoc Bonferroni multiple comparison test (SPSS 11.5.0, United States) with the probability of P-value of ≤ 0.05.
Results and Discussion
Detached Leaf Assay
The spore count reduced to 32.66, 61.66, 85.86, and 86% in silver nanoparticle-treated A. brassicicola-infected plants (AB + SNP) when treated with 1, 2.5, 5, and 7.5 μg/ml of SNP, respectively, in comparison to A. brassicicola-infected plants (AB alone) (Supplementary Figure S2). Detached leaf assay is one of the easy methods that mimic the plants’ responses during stress conditions and in vitro conditions.
Based on the results obtained, 5 μg/ml of SNP was selected for further studies.
Evaluation of Biosynthesized Silver Nanoparticles on Pathogenesis of A. brassicicola on Model Plant A. thaliana
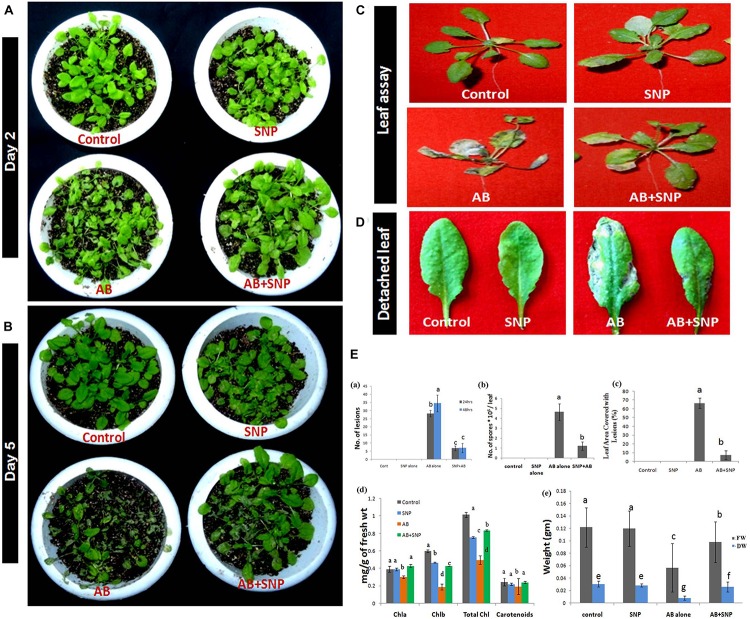
From visual observation, results similar to detached leaf assay were obtained indicating potency of SNPs as antifungal agents (Figure 1). After 48 h of infection, lesions and necrosis were observed in parts of the plant in AB alone (Su’udi et al., 2013), however, SNP pre-treated leaves were as healthy as control (Figure 1A). Leaf area covered with lesions in AB + SNP decreased significantly in comparison to AB alone after 48 h of infection (Figures 1A,E). After 5 days of infection, AB-alone plants showed severely damaged leaves, while AB + SNP plants controlled the visual symptoms of the disease (Figure 1B). There was a significant reduction of lesions (80%) and spores (74.13%) after 48 h of infection in AB + SNP as compared to AB alone (Figures 1C–E). Number of spores, lesions in leaf, and leaf area covered with lesions are important parameters to assess the disease severity in plants (Yan et al., 2002). This study corroborated well with the previous studies reporting biosynthesized silver nanoparticles as an efficient means to control plant diseases (Anandaradje et al., 2020; Shen et al., 2020). Chlorophyll content in AB only reduced by 51.2%, while the reduction in fresh and dry weight was 58.33 and 73.3%, respectively, as compared to control (Figures 1Ed,e). AB + SNP exhibited 68.68, 96, and 212.5% increase in chlorophyll content and fresh and dry weight, respectively, as compared to AB alone (Figures 1Ed,e). Loss of photosynthetic pigments is one of the major biochemical changes a plant undergoes during Alternaria infection (Meena et al., 2016). Increase in chlorophyll content in AB + SNP in comparison to AB alone showed the increased disease resistance of plant toward disease after treatment with SNPs. Further, no phytotoxicity of SNP was observed in terms of fresh and dry weight and chlorophyll content in plants (Figures 1Ed,e). Lactophenol cotton blue-stained leaves demonstrated a network of mycelia in AB only, which was not observed in AB + SNP (Supplementary Figures S3A,B). Lactophenol cotton blue is a stain used to distinguish fungal mycelia from plant cells, which stains fungal hyphae in blue color, leaving plant cell wall unstained (Marques et al., 2013). Similarly, trypan blue is another stain used to distinguish between dead and live tissues (Fernández-Bautista et al., 2016). Trypan blue-stained leaves showed higher viability in AB + SNP as compared to AB alone (Supplementary Figures S3C,D). The larger necrotic lesions formed after Alternaria infection in AB stained more profoundly in comparison to the smaller and lesser lesions in AB + SNP.
FIGURE 1.
Photograph showing effect of silver nanoparticles in reducing disease severity after (A) 48 h (day 2) post infection (B) Day 5 post infection (C) reduction in necrosis of leaves (D) reduction in number of lesions formed per leaf (E) Assessment of disease parameters in terms of (a) number of lesions (b) number of spores (c) leaf area covered with lesion (d) chlorophyll content (e) fresh and dry weight in silver nanoparticles pre-treated plants as compared to other treatments. Cont-Control, SNP-Biogenic silver nanoparticles lone, AB-A. brassicicola infected plants, AB + SNP-A. brassicicola infected, treated with SNP, FW-fresh weight, DW-Dry weight. Values are the means ± SD of three replicates. Means sharing different alphabets “a”, “b” differ significantly from each other at p ≤ 0.05.
Scanning Electron Microscopy
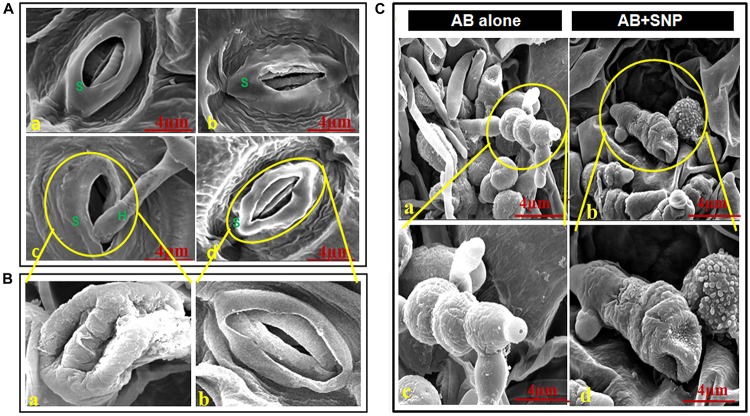
Adaxial surface of A. thaliana leaves with SNP and AB + SNP treatment showed no alteration in surface morphology (Figures 2A,b,d), however, AB-only plant showed healthy mycelia network over leaf surface (Figures 2C,a,c).
FIGURE 2.
Scanning electron micrographs of adaxial surface of A. thaliana leaf. (A) Stomata of (a) control, (b) SNP, (c) AB alone, and (d) AB + SNP. S represents stomata and H represents hyphae of fungal pathogen. (Ba) Enlarged image of damaged stomata in AB alone and (b) enlarged image of normal stomata in AB + SNP treatment. (Ca,c) Image showing healthy mycelia and spore network on plant surface in AB alone. (b,d) Absence of mycelia network and damaged spores in AB + SNP. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP.
The images also demonstrated the hyphae entering into the cell through stomata and consequent damage (Figures 2B,a). The AB + SNP treatment did not only inhibit the conidial germination, but also created structural aberrations in fungal conidia (Figure 2C). The spores of A. brassicicola in AB + SNP treatment were destroyed with severe deformation. Ocsoy et al. (2013) had observed similar deformations in tomato pathogen X. perforans by DNA-directed silver nanoparticles on graphene oxide. SNPs show multiple modes of actions including cell wall and cell membrane damage, denaturation of protein, and DNA unwinding to kill the pathogen (Ramalingam et al., 2016).
Determination of Antioxidant Activities of A. thaliana
Non-enzymatic Parameters
Phenolic compounds are the key of plant immune response, which participates in activating plant defense genes, production of phytoanticipins and phytoalexins, structural barriers, and modulation of pathogenicity (Shoresh and Harman, 2008). There was an increase of 47.82% in total phenolics in AB + SNP, as compared to AB-only plants (Supplementary Figure S4A). Krishnaraj et al. (2012) had demonstrated the elevated level of phenolics in the SNP-treated plants and speculated that mild stress might be beneficial to plants in protecting them against pathogen attacks. Content of ascorbic acid was highest in AB-only plants followed by AB + SNP and SNP treatment (Supplementary Figure S4B). Ascorbic acid is a well-known antioxidant and precursor of many defense molecules (Abdelrahmana et al., 2016). Higher content of ascorbic acid in pathogen-infected plants may be due to the higher requirement of antioxidants to combat biotic stress.
Content of thiobarbituric acid reactive substances (TBARS) and proline increased 55.3 and 368.3%, respectively, in AB-only plants, whereas the AB + SNP plants showed 28.47% reduction in TBARS content and 221.7% reduction in proline as compared to AB alone (Supplementary Figures S4C,D). TBARS content depicts the peroxidation of lipid membrane caused by ROS species (Singh et al., 2013). Reduction in TBARS content of AB + SNP indicated the healthier status of plants. Similarly, proline increases during the stress condition, which acts as a signaling molecule to modulate mitochondrial functions, influence cell proliferation or cell death, and trigger specific gene expression (Szabados and Savoure, 2009).
Further, to assess the ROS generation, H2O2 production was quantified. It was found that H2O2 level reduced to 28% in AB + SNP as compared to AB-only plants (Supplementary Figure S4E). There was slight increase in proline, TBARS, and H2O2 content in SNP alone, which might be an indicator of inducing plant immune response prior to pathogen infection.
Stress Enzyme Activity
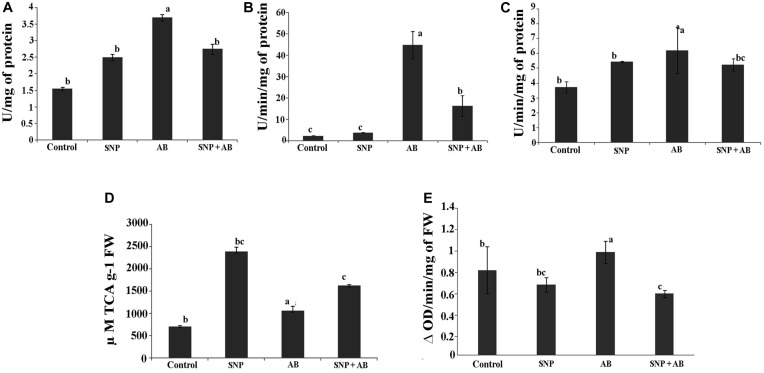
Among the four treatments, SOD, CAT, and GoPx were maximum in AB alone. SOD, CAT, and GoPx activities were enhanced by 1. 6-, 19. 8-, and 0.6-fold in AB alone while a decrease in stress enzymes was observed in AB + SNP plants (Figures 3A–C).
FIGURE 3.
Stress enzyme activities across all the treatments in terms of (A) SOD, (B) catalase, (C) GoPX, (D) PAL, and (E) PPO. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP. Values are the means ± SD of three replicates. Means sharing different letters “a” and “b” differ significantly from each other at p ≤ 0.05.
Infection of A. brassicicola induced the generation of ROS, thereby increasing the enzymatic activity to scavenge the ROS generated (Gill and Tuteja, 2010) in AB alone. The AB + SNP plants were able to combat the pathogen infection due to the antagonistic effects of SNP on A. brassicicola spores and activated systemic resistance of A. thaliana. Further, to study the effect of tripartite interaction of plant, pathogen, and nanoparticles on phenylpropanoid pathway, which stimulates disease resistance factors including phytoallexins and phenolics (Lavania et al., 2006), PAL and PPO activities were assessed. PAL activity was highest in the plants pre-treated with silver nanoparticles (SNP and AB + SNP) (Figures 3D,E), resulting in production of phenolic compounds (Supplementary Figure S4A), many of which are antimicrobial and can show a synergistic effect with silver nanoparticles to ameliorate the biotic stress. PPO activity was highest in the AB plants, followed by AB + SNP and SNP as compared to control (Figure 3E). The production of phenolic compounds before infection, due to the presence of SNP, might be a reason for strengthened immunity of plant to reduce the generation of ROS. Further, to gain an insight into the molecular responses of plants during the interaction, proteomics and metabolomics were carried out.
Transmission Electron Microscopy
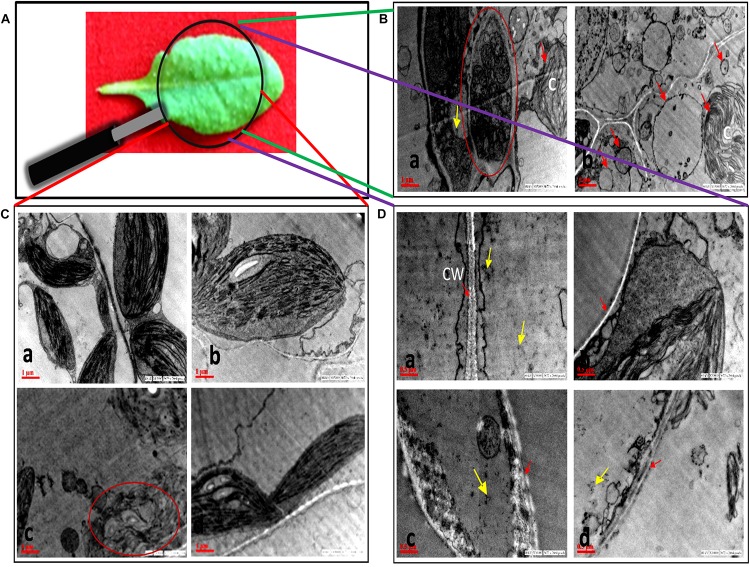
No morphological changes were observed in SNP-treated leaves as compared to control (Figure 4), while damage was observed in chloroplast and cell wall of AB alone (Figures 4B,Cc). The symptom reflected the infection of A. brassicicola and secretion of toxins (Otani et al., 1995). In AB alone, the internal lamellar system was retained but thylakoids were not as compactly arranged as in control. Damage in the cell wall of AB-alone plants was visible by scattered white deposition of cellulose, which was absent in AB + SNP treatment. The most interesting finding of this study was the observation of autophagic granules in AB + SNP treatment (Figure 4B). Autophagy has emerged as a powerful weapon of plants against necrotrophic pathogens (Kabbage et al., 2013). Single-membrane vesicles encapsulating several organelles (Figure 4Ba), depicting early lysosomal activities (Figure 4Bb), were observed near chlorophyll in AB + SNP treatment. In both figures, the activity was observed adjacent to chloroplast and demonstrates chlorophagy, which can be a mechanism to lower down ROS production and enhance plant immunity (Dong and Chen, 2013).
FIGURE 4.
Transmission electron micrographs of cell organelles of A. thaliana leaves across all treatments. (A) Sample image of A. thaliana leaf. (B) Autophagosomal structures in AB + SNP treatments. (a) Single membrane vesicles encapsulating several organelles (encircled with red) near chloroplast. (b) Membrane-bound autophagosomal structure near chloroplast. Arrow represents single-membrane autophagosomal structure, chloroplast is represented by (C). (C) Chloroplast of (a) control, (b) SNP, (c) AB alone, and (d) AB + SNP. Encircled image of AB represents damage in chloroplast. (D) Cell wall of (a) control, (b) SNP, (c) AB alone, and (d) AB + SNP. Arrow represents cell wall (CW). Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP. Yellow arrows represents SNPs.
Proteome Regulation by Biosynthesized Silver Nanoparticles
To study the role of proteins associated with resistance in Arabidopsis plant against A. brassicicola, comparative proteomics analysis was carried out from control, SNP alone, AB alone, and AB + SNP treatments.
For each of the treatments, three replicates gels were run and images were analyzed by the PDQuest software version 8.0.1. The mean value of the high-quality spots was used as the spot quantity on the master gel (Supplementary Figure S5 and Supplementary Table S1).
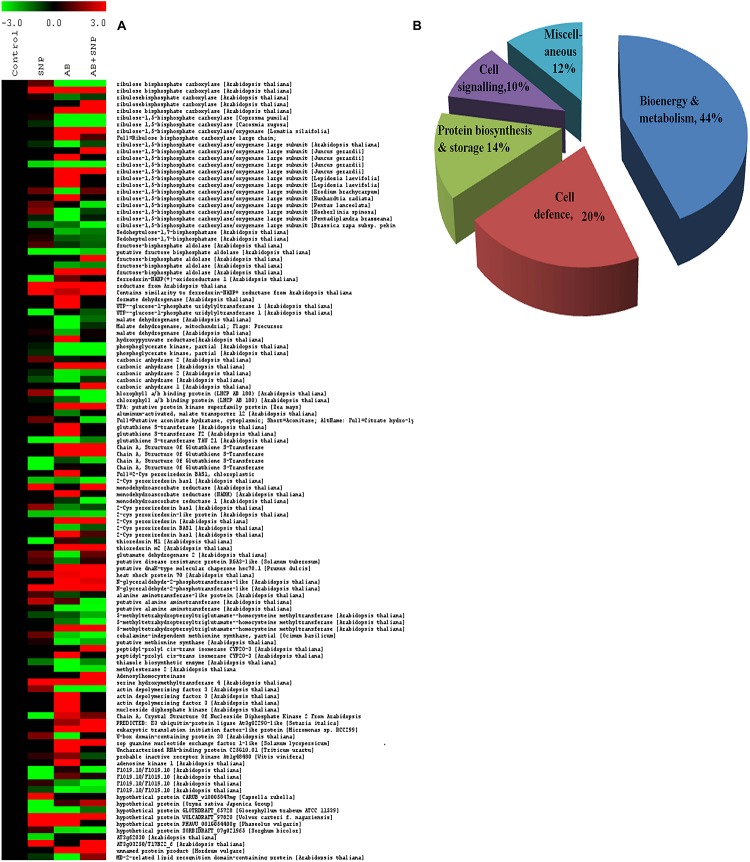
A total of 599 high-quality spots were detected in master gel image in which 144 spots were found to be differentially expressed by more than 2.5-fold. The differential spots obtained were further subjected to MALDI MS/MS in which 117 proteins were identified with the significant score (Supplementary Table S2). Out of 117 identified, 50 proteins were upregulated, 33 were downregulated, and 34 demonstrated a mixed pattern of differential protein expression among four treatments (Figure 5A). A total of 63 proteins were unique in identification, while the rest were the result of post-translation or members of multigene families (Agrawal et al., 2013).
FIGURE 5.
(A) Clustering of differentially expressed proteins. Each protein is represented by a single row of colored boxes. Induction to suppression ranges from red to green. (B) Functional cataloging of differentially expressed proteins. The identified proteins were assigned a putative function using protein function databases and functionally grouped in five different categories as represented in the pie chart. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola- infected plants; AB + SNP, A. brassicicola-infected, treated with SNP.
The details of the MS/MS analyses, including protein identification, the score of the identified protein, threshold score, number of matched peptides, and % sequence coverage, are listed in Supplementary Table S3. Result analysis showed that most of the protein spots quantitatively differ in different treatment along with qualitative difference. Supplementary Figure S6 represents some typical gel regions showing protein spots in the enlarged dotted box of Supplementary Figure S5 with quantitatively and qualitatively altered expression.
Differentially Expressed Proteins Based on Functional Classes
To get an insight into the differentially expressed proteins, identified proteins were further divided into five functional classes based on their putative function, viz., bioenergy and metabolism (BEM) with highest number of protein (44%), plant defense (20%), cell signaling (14%), storage and biogenesis (10%), and miscellaneous (12%) (Figure 5B). Detailed information regarding differentially expressed proteins has been provided in Supplementary Material.
Hierarchical Clustering of Differentially Expressed Proteins
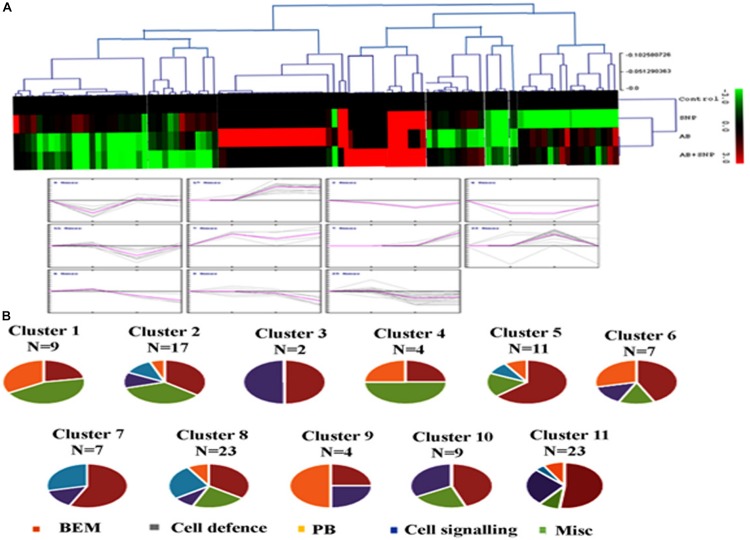
To know the expression pattern of different proteins, and group them accordingly, unbiased clustering was performed using the SOTA algorithm. Proteins were grouped according to the pattern of their expression, using the Pearson correlation coefficient as the distance function. The data used in the study was fold change in expression of proteins with respect to the control protein expression value. The analysis yielded 11 different clusters of proteins, representing a similar expression pattern for each cluster (Figures 6A,B), however, only clusters having n ≥ 5 were taken into consideration for further analysis.
FIGURE 6.
Clusterogram of differentially expressed proteins showing 11 clusters based on their expression profiles. (A) The SOTA cluster trees and (B) expression profiles of 11 different clusters. Pie chart represents respective contribution of proteins of different functional groups. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP.
Cluster 1 was represented by nine proteins, dominated by the proteins related to plant defense and stress. As observed earlier, the maximum expression was found in pathogen-infected plants while proteins were downregulated in the treatment of silver nanoparticles. Cluster 2 represented 17 proteins in which most of them were involved in BEM and plant defense and rescue. This cluster also showed upregulation in infected plants, while the expression pattern of SNP was similar to control. Clusters 5, 6, 10, and 11 were also dominated by proteins involved in BEM, downregulated in infected plants, and upregulated in SNP and AB + SNP treatment as compared to AB alone. Cluster 7 had proteins involved in cell signaling, which were highly upregulated in AB + SNP treatment. Apart from proteins involved in BEM, clusters 9 and 10 also showed proteins of protein biogenesis, showing upregulation in SNP as observed earlier in this study. Altogether, the hierarchical clustering suggests the different pattern of expression of all proteins, which, along with function classification, can help in elucidation of the mechanism involved behind nanoparticle-mediated plant protection against pathogenic fungi.
Domain Analysis
Further domain analysis was also carried out to determine the putative functions of unknown proteins (Supplementary Table S4). AtC 2024, 3015, 1030, 8431, and 8529 were found to have ribulose bisphosphate carboxylase small-chain domain, while AtC 4229 contained myosin S1 fragment, N-terminal domain. AtC 6230 was found to have S-adenosyl-L-methionine-dependent methyltransferase-like domain while AtC6619 had nucleotide-diphospho-sugar transferases domain. Interestingly, AtC 7415, 8411, and 4236 had ABC-transporter extracellular N-terminal domain, zinc finger CCHC-type domain, and cyclophilin-like domain, respectively. AtC 7415 and 8411 were upregulated in the plants with SNP treatment.
ABC transporters are one of the largest protein families involved in transport across cell membrane, performing a wide range of functions (Kang et al., 2011). Recently, their role in conferring plant resistance against pathogens and tolerance to abscisic acid has been elucidated (Ji et al., 2014). Similarly, cyclophilins are plant proteins involved in a variety of functions including development and stress responsiveness (Romano et al., 2004). Zinc finger CCHC-type domain-containing proteins comprise a large conserved protein family across the eukaryotic system and play a critical role in mRNA metabolism (Bogamuwa and Jang, 2014). Many of the zinc finger proteins are potential transcription factors that regulate plant responses toward biotic and abiotic stress (Nakai et al., 2013; Yang et al., 2014). Differential expression of proteins containing domains described above needs a detailed investigation to decipher the nanoparticle-mediated changes in plant proteome during plant–pathogen–nanoparticles interaction.
Metabolic Profiling
GC-MS Analysis
A total of 39 metabolite derivatives in non-polar (CHCl3) fraction (Table 1) and 25 in polar (water) fraction (Table 2) of plant extracts with four treatments (Control, SNP, AB, and AB + SNP) were detected by GC-MS analysis. The polar metabolites included diverse groups of organic acids, sugars, and intermediates of Krebs cycle, while non-polar metabolites mainly consisted of organic acids, fatty acids, and sterols. GC-MS analysis allows the metabolites to separate on the basis of their mass/charge ratio, which together can serve as a tag for that metabolite (Srivastava et al., 2012a).
TABLE 1.
Quantitative variability in non-polar metabolites of A. thaliana leaves during plant–pathogen–nanoparticles interaction.
| Rt (min) | Metabolites | Fragmentation patterns | Control (mg/g) | SNP (mg/g) | AB (mg/g) | SNP + AB (mg/g) |
| 6.79 | Ethylene glycol | m/z 206,193,191,147,103,73 | 0.450 ± 0.047a | 0.290 ± 0.024c | 0.390 ± 0.041ab | 0.350 ± 0.036bc |
| 9.33 | Propanoic acid | m/z 234,220,219,147,117,73,66,55 | NDa | 0.190 ± 0.020b | NDa | 0.380 ± 0.040b |
| 9.55 | Caproic acid | m/z 188,174,131,75,73,69 | NDb | NDb | NDb | 0.040 ± 0.004a |
| 16.01 | Glycerol | m/z 308,293,219,205,148,117,75,73,55 | NDb | NDb | NDb | 0.960 ± 0.100a |
| 18.09 | Non-anoic acid | m/z 230,216,171,155,117,85,73,55 | NDb | 0.200 ± 0.021a | NDb | NDb |
| 18.84 | Cyclotetradecane | m/z 196,168,126,97,83,69,55,51 | NDb | 0.150 ± 0.016a | NDb | NDb |
| 19.06 | Tetradecane | m/z 198,127,113,99,71,57 | NDb | 3.270 ± 0.340a | 0.110 ± 0.011b | NDb |
| 21.59 | Pentadecane | m/z 212,127,113,99,71,55,54 | NDc | 4.310 ± 0.449a | 0.060 ± 0.006c | 0.680 ± 0.071b |
| 23.98 | Hexadecane | m/z 226,127,113,99,85,71,57 | NDc | 7.810 ± 0.813a | NDc | 2.600 ± 0.271b |
| 25.24 | Lauric acid | m/z 272,259,229,185,117,83,73,55 | NDc | 0.690 ± 0.072a | 0.060 ± 0.006c | 0.430 ± 0.045b |
| 26.22 | Heptadecane | m/z 240,169,127,113,99,85,71,55,53 | NDc | 4.590 ± 0.478a | NDc | 1.890 ± 0.197b |
| 26.35 | Pristane | m/z 268,249,141,113,85,71,57,56 | NDb | 1.340 ± 0.139a | NDb | NDb |
| 28.37 | Octadecane | m/z 254,169,141,113,99,85,71,57 | NDc | 7.390 ± 0.769a | NDc | 3.730 ± 0.388b |
| 28.55 | Phytane | m/z 282,249,183,155,127,85,71,55 | NDb | 0.770 ± 0.080a | NDb | NDb |
| 29.19 | Neophytadiene | m/z 278,263,195,138,95,68,57,55,51 | 9.800 ± 1.020a | NDd | 3.970 ± 0.413b | 1.790 ± 0.186c |
| 29.42 | Myristic acid | m/z 300,287,201,148,132,117,73,55 | NDd | 0.700 ± 0.073c | 0.990 ± 0.103b | 1.160 ± 0.121a |
| 30.39 | Non-adecane | m/z 268,197,155,127,113,99,85,71,57 | NDc | 4.140 ± 0.431a | NDc | 2.160 ± 0.225b |
| 31.39 | n-Pentadecanoic acid | m/z 314,300,255,145,117,85,75,73,55 | NDb | NDb | NDb | 0.390 ± 0.041a |
| 32.34 | Eicosane | m/z 282,211,141,113,99,85,71,57 | NDc | 5.760 ± 0.600a | NDc | 3.360 ± 0.350b |
| 32.77 | cis-5,8,11-Eicosatrienoic acid | m/z 378,348,232,129,117,75,73,55 | 0.390 ± 0.041c | 0.580 ± 0.060c | 1.290 ± 0.134b | 1.640 ± 0.171a |
| 34.19 | Heneicosane | m/z 296,225,141,127,99,85,71,57 | NDc | 3.630 ± 0.378a | NDc | 1.030 ± 0.107b |
| 35.65 | Phytol | m/z 368,353,213,157,143,123,75,73,57 | 0.870 ± 0.091d | 1.490 ± 0.155c | 1.990 ± 0.207b | 2.350 ± 0.245a |
| 35.97 | Docosane | m/z 310,211,183,127,99,71,57 | NDc | 4.160 ± 0.433a | NDc | 2.540 ± 0.264b |
| 36.35 | Linolenic acid | m/z 350,336,321,260,117,108,75,73 | 2.660 ± 0.277b | 2.940 ± 0.306b | 2.500 ± 0.260b | 14.370 ± 1.496a |
| 36.77 | Stearic acid | m/z 356,341,146,117,83,73,54 | 0.980 ± 0.102c | 1.890 ± 0.197b | 0.890 ± 0.093c | 2.700 ± 0.281a |
| 37.67 | Tricosane | m/z 324,239,169,127,99,85,71,57 | NDc | 2.540 ± 0.264a | NDc | 1.590 ± 0.165b |
| 39.31 | Tetracosane | m/z 338,309,239,155,127,113,71,57 | NDc | 3.020 ± 0.314a | NDc | 1.850 ± 0.193b |
| 40.89 | Pentacosane | m/z 352,295,197,155,127,85,71,57 | NDc | 1.740 ± 0.181a | NDc | 1.150 ± 0.120b |
| 42.41 | Hexacosane | m/z 366,253,155,99,85,71,57 | NDc | 1.650 ± 0.172a | NDc | 0.930 ± 0.097b |
| 43.04 | Docosanoic acid | m/z 412,398,243,149,129,117,73,55 | NDb | 0.210 ± 0.022a | NDb | 0.230 ± 0.024a |
| 43.87 | Heptacosane | m/z 380,323,281,127,99,85,71,57 | 0.620 ± 0.065b | 1.170 ± 0.122a | 0.350 ± 0.036c | 0.690 ± 0.072b |
| 44.91 | 2-Monolinolein | m/z 498,482,408,234,191,129,73,55 | 2.010 ± 0.209a | NDc | 0.750 ± 0.078b | NDc |
| 45.29 | Octacosane | m/z 394,337,183,127,113,99,85,71,57 | NDc | 1.060 ± 0.110a | NDc | 0.680 ± 0.071b |
| 45.86 | Tetracosanoic acid | m/z 440,426,381,257,149,129,117,75,73 | 0.270 ± 0.028b | 0.180 ± 0.019c | 0.310 ± 0.032b | 0.460 ± 0.048a |
| 50.02 | α-Tocopherol | m/z 502,487,354,237,123,95,73 | NDb | 0.510 ± 0.053a | NDb | NDb |
| 50.17 | Cholesterol | m/z 458,416,329,247,129,95,73,57 | 1.320 ± 0.137a | 0.820 ± 0.085b | 0.880 ± 0.092b | 0.550 ± 0.057c |
| 51.51 | Campesterol | m/z 472,457,343,289,247,145,73,55 | 4.750 ± 0.494b | 1.410 ± 0.147c | 11.200 ± 1.166a | 2.150 ± 0.224c |
| 51.93 | Stigmasterol | m/z 484,355,213,161,129,83,69,73,53 | 0.940 ± 0.098c | 1.290 ± 0.134c | 11.480 ± 1.195a | 3.890 ± 0.405b |
| 52.76 | β-Sitosterol | m/z 486,398,275,213,159,129,73,57 | 34.070 ± 3.546a | 9.230 ± 0.961c | 24.850 ± 2.586b | 11.550 ± 1.202c |
Mean values ± SD of mg/g of the dry weight of samples. ND, not detected; a, b, c, and d denote statistical significance P < 0.05. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP.
TABLE 2.
Quantitative variability in polar metabolites of A. thaliana leaves during plant pathogen nanoparticles interaction.
| Rt (Min) | Metabolites | Fragmentation patterns | Control (μg/g) | SNP (μg/g) | AB (μg/g) | SNP + AB (μg/g) |
| 6.79 | Ethylene glycol | m/z 206,193,191,147,103,73 | 0.710 ± 0.074b | 0.270 ± 0.028d | 0.500 ± 0.052c | 1.370 ± 0.143a |
| 16.07 | Glycerol | m/z 308,293,219,205,148,117,75,73,55 | 1.360 ± 0.142d | 2.590 ± 0.270c | 5.990 ± 0.623b | 9.470 ± 0.986 a |
| 16.72 | Maleic acid | m/z 260,247,245,147,75,73,66 | NDb | 0.290 ± 0.030a | NDb | NDb |
| 16.95 | Succinic acid | m/z 262,249,247,172,148,129,75,73,55 | 1.460 ± 0.152a | 1.300 ± 0.135a | NDb | NDb |
| 17.63 | Glyceric acid | m/z 332,308,292,205,189,147,103,73 | 0.900 ± 0.094b | 2.140 ± 0.223a | NDc | NDc |
| 17.89 | Fumaric acid | m/z 260,248,217,147,133,75,73 | NDb | 11.350 ± 1.18a | NDb | NDb |
| 21.77 | Malic acid | m/z 350,335,307,245,148,133,73,55 | NDb | 7.680 ± 0.799a | NDb | NDb |
| 22.39 | Erythritol | m/z 410,348,307,217,205,189,147,117,103,73 | 0.570 ± 0.059a | 0.251 ± 0.026c | NDc | 0.240 ± 0.02b |
| 23.26 | Erythronic acid | m/z 424,409,379,294,220,149,117,73 | 0.280 ± 0.029a | 0.320 ± 0.03a | NDb | NDb |
| 23.68 | L-Threonic acid | m/z 424,409,379,294,220,189,147,117,73 | 1.060 ± 0.110b | 3.120 ± 0.325a | NDc | NDc |
| 26.12 | D-Xylose | m/z 467,421,362,307,147,103,73,59 | 1.520 ± 0.158a | 0.940 ± 0.098c | 1.220 ± 0.127b | 0.970 ± 0.10c |
| 26.48 | D-ribose | m/z 467,467,362,218,189,103,73 | 4.350 ± 0.453a | 0.410 ± 0.043bc | 0.660 ± 0.069b | NDc |
| 27.38 | Xylitol | m/z 512,395,307,217,147,103,73 | 1.030 ± 0.107 b | 0.300 ± 0.031c | 3.050 ± 0.317a | 1.240 ± 0.129b |
| 28.36 | Ribonic acid | m/z 526,511,421,333,292,217,147,103,73 | 1.680 ± 0.175b | 2.220 ± 0.231a | NDc | NDc |
| 28.56 | Galactonic acid | m/z 628,511,393,292,217,189,147,103,73 | NDb | 1.860 ± 0.194a | NDb | NDb |
| 30.65 | D-Fructose (MeOX1) | m/z 569,408,307,263,217,173,147,103,73 | 2.100 ± 0.219c | 8.160 ± 0.849b | 16.840 ± 1.753a | NDd |
| 30.86 | D-Fructose (MeOX2) | m/z 569,408,307,263,217,173,147,103,73 | NDc | 5.840 ± 0.608b | 11.220 ± 1.168a | NDc |
| 31.13 | D-Glucose (MeOX2) | m/z 569,466,390,319,217,160,147,73 | 2.480 ± 0.258c | 14.130 ± 1.47b | 17.170 ± 1.787a | 1.160 ± 0.121c |
| 31.49 | D-Galactose | m/z 569,376,217,160,147,103,73 | NDb | 4.200 ± 0.437 a | 4.680 ± 0.487 a | NDb |
| 31.76 | D-Glucitol | m/z 614,422,346,320,217,157,103,73 | NDc | 0.430 ± 0.045 c | 12.370 ± 1.288a | 4.290 ± 0.447b |
| 32.03 | Ribitol | m/z 512,379,218,157,103,73,59 | NDb | NDb | 1.810 ± 0.188a | NDb |
| 32.18 | Gulonic acid | m/z 466,361,218,147,129,75,73 | 1.710 ± 0.178b | 0.150 ± 0.016c | 2.400 ± 0.250a | 1.080 ± 0.112b |
| 33.16 | Gluconic acid (Di TMS) (6TMS) | m/z 628,437,334,292,277,147,73 | NDb | 5.220 ± 0.543 a | 0.420 ± 0.044b | NDb |
| 34.71 | Inositol | m/z 612,508,362,337,217,191,147,73 | 11.550 ± 1.202b | 3.160 ± 0.329c | 1.460 ± 0.360d | 29.910 ± 3.113a |
| 38.75 | α-D Galactopyranoside (6TMS) | m/z 686,362,217,204,191,147,73 | 2.320 ± 0.241b | 2.480 ± 0.258b | 3.200 ± 0.333 b | 18.350 ± 1.910a |
Mean values ± SD of μg/g of the dry weight of samples. ND, not detected; a, b, c, and d denote statistical significance P < 0.05. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP.
The concentration of sterols including β-sitosterol, stigmasterol, and compesterol increased significantly in AB alone as compared to control. Sterols are known to accumulate during pathogen infection, thereby promoting plant disease susceptibility (Berger et al., 2004; Griebel and Zeier, 2010). A significant decrease in β-sitosterol, stigmasterol, and compesterol in AB + SNP as compared to AB treated leaves was observed. Similarly, concentrations of sugars, D-xylose, D-ribose, D-fructose, D-glucose, D-galactose, and sugar alcohols such as ribitol, glucitol, and xylitol were found to increase significantly in infection of AB. Sugar accumulation in plants is a sign of plant’s strategy to fight infection, which can be manipulated and utilized by fungal pathogen to acquire food and nutrition for their survival (Moghaddam and Ende, 2012). Decrease in sugar level in SNP-treated plants is also an evidence of the successful inhibition of plant pathogen by these tiny particles (Table 1).
Hydrocarbons including cyclotetradecane, tetradecane, pentadecane, hexadecane, heptadecane, and non-adecane are known for their antimicrobial activity and have been extracted from many medicinal plants (Ozdemir et al., 2004; Lin et al., 2012). Similarly, eicosane; heneicosane; tri-, tetra-, penta-, hexa-, hepta-, octa-, and docosane; and tetracosanoic acid compounds demonstrate antagonistic activities toward pathogens (Kalegari et al., 2012; Zhang et al., 2014). The levels of these compounds were significantly enhanced in SNP-treated plants. The enhanced concentration of the compounds in SNP treatment demonstrates their positive effect on plant immune response, which may be an important factor for plant survival after fungal infection. The AB + SNP treatment showed increased concentration of these metabolites, indicating preparation of plants to combat biotic stress.
Organic acids of antimicrobial potential (Kabara et al., 1972; Schnürer and Magnusson, 2005) including propanoic acid, caproic acid, lauric acid, docasanoic acid, erythronic acid, L-threonic acid, non-anoic acid, galactonic acid, gluconic acid, and ribonic acid were not detected in control and pathogen-infected plants while silver nanoparticle-treated plants emerged as significant producers of these organic compounds. Similarly, antimicrobial stearic acid (Da Silva et al., 2002) was significantly enhanced with treatment of silver nanoparticles. Compounds having antimicrobial properties not only helped plants to combat the attack of A. brassicicola but also acted as vaccines to plants, which keeps them ready to fight any further biotic stress and activated defense mechanism.
TCA cycle intermediates such as maleic acid, succinic acid, glyceric acid, fumaric acid, and malic acid concentration increased in leaves of SNP treatment. The concentrations of pristane and phytane, saturated terpenoids that are known for their antifungal potential (Yin et al., 2011), were also enhanced in leaves treated with silver nanoparticles, whereas they were completely absent in control and pathogen-infected leaves.
Metabolites showing antioxidant potential as phytol and α-tocopherol (Kontush et al., 1996; Santos et al., 2013) were significantly increased in SNP-treated leaves. The concentration of α-D galactopyranoside and inositol was much higher in AB + SNP as compared to other treatments. Inositol acts as an osmolyte, which balances the osmotic pressure during infection (Khan et al., 2012).
Inductively Coupled Plasma-Mass Spectroscopy (ICP-MS) Analysis
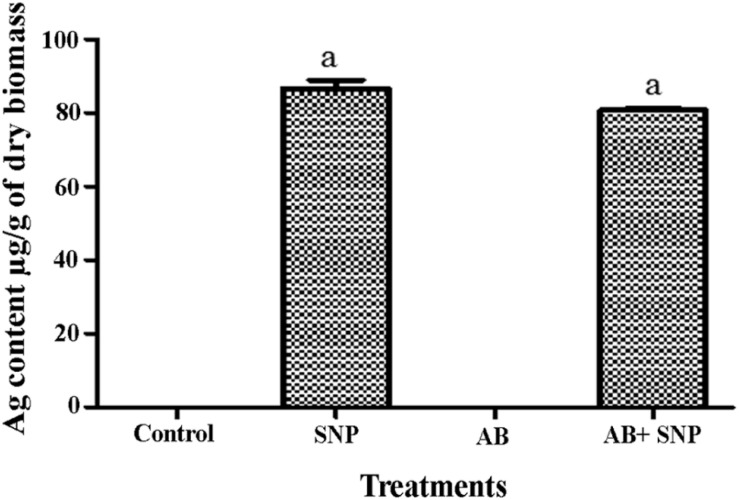
To study the internalization of silver in leaf tissues, ICP-MS studies were carried out. In both SNP and AB + SNP treatments, the concentration of silver inside the leaves was 87.6 and 80.7 μg/g of dry biomass, respectively (Figure 7). Larue et al. (2014) have also found an accumulation of silver in the leaves of Lactuca sativa after foliar exposure without observing any phytotoxicity. Our further studies on the interaction of studied plant pathosystem–silver nanoparticles with soil show that negligible effects were found on native soil microflora (Kumari et al., 2017c). Das et al. (2016) have found iron oxide nanomaterials as a sustainable and eco-friendly source of slow iron release without showing phytotoxicity and effect on seed germination. Agglomeration and oxidation of nanoparticles inside plant cells and in soil might be some of the reasons that nullify the toxic effects of silver nanoparticles on plants. Silver nanoparticle phytotoxicity has been observed by several studies on higher concentration, exclusively after root exposure (Larue et al., 2014); this study using 5 μg/ml SNP by foliar exposure can be considered as safe, though further studies are required to understand the fate of nanoparticles inside cells.
FIGURE 7.
Total Ag content in leaves of A. thaliana. Cont, control; SNP, biogenic silver nanoparticles alone; AB, A. brassicicola-infected plants; AB + SNP, A. brassicicola-infected, treated with SNP. Values are the means ± SD of three replicates. Means sharing different letters “a” and “b” differ significantly from each other at p ≤ 0.05.
Mechanism Involved in Silver Nanoparticle-Mediated Plant Protection Against Pathogens
Biosynthesized silver nanoparticles not only were able to kill plant pathogen A. brassicicola after their foliar spray but also enhanced plants’ immunity employing a variety of mechanisms (Scheme 1).
SCHEME 1.
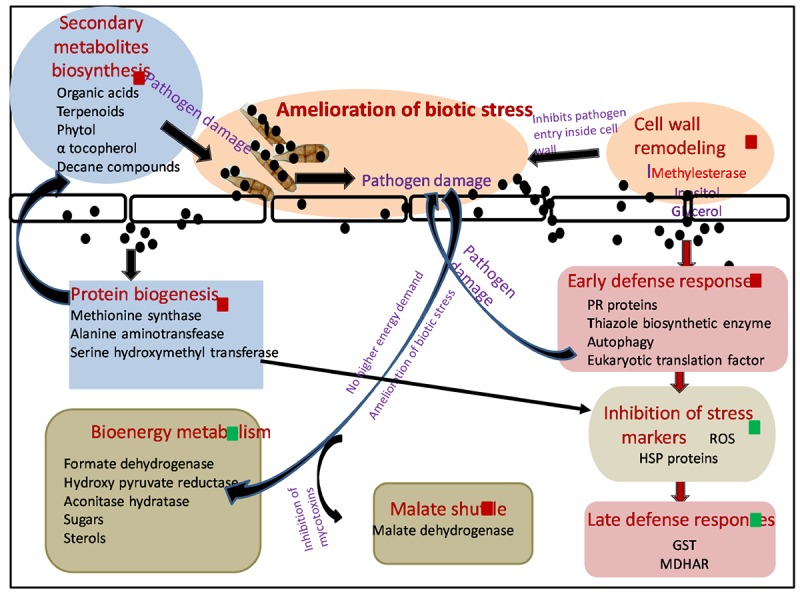
Molecular mechanism involved in amelioration of biotic stress by SNP during tripartite interaction of plant–pathogen–nanoparticles. Black spheres represent SNP, red squares represent proteins and metabolites upregulated, and green squares represent proteins and metabolites downregulated. Red arrows shows the differentially expressed proteins involved in different pathways. Black arrows demonstrate hypothesized mechanism linking differentially expressed proteome and metabolome. ROS, reactive oxygen species; HSP, heat shock proteins; GST, glutathione S transferase; MDHAR, monodehydroascorbatereductase.
In this study, A. brassicicola infection (AB only) caused upregulation of several proteins involved in BEM and thus increase in sugar and sterol content was observed in GC-MS. Successful establishment of necrotrophic infection starts with damaging plant cells, thereby increasing the energy constraint over the metabolic pathways of the host, which is further utilized by the fungus itself for its nourishment and growth, leading toward the death of host plant (Divon and Fluhr, 2007). Application of SNP resulted in amelioration of biotic stress by damaging fungal pathogen and thus resulting in a downregulation in the expression of proteins involved in sugar metabolism and sugar metabolites as compared to AB-only plants. SNPs were also able to protect the host plant by the mycotoxins released by A. brassicicola affecting malate shuttle as observed by an upregulation in the expression of protein malate dehydrogenase after treatment of SNP in comparison with AB alone.
Interaction of SNP with host plant resulted in the elevation of several proteins involved in early defense response of the plants, thus minimizing the production of ROS and collateral damage induced by them. Induction of disease resistance protein, thiazole biosynthetic enzyme, and serine hydroxymethyl transferase resulted in decreased production of ROS and several stress markers such as proline, TBARS, and dnaK-type molecular chaperone hsc70.1 as observed by biochemical and proteomics study. As the production of ROS was minimized by the application of SNP, expression of ROS scavenging enzymes such as GST was also not elevated, while in AB alone, induced biotic stress multifold expression of many GST proteins was observed.
Autophagy also emerged as a probable mechanism that could contribute toward enhanced early defense response of plants against the pathogen after application of SNP. Autophagy plays a very important role in inducing cell death including necrotrophic fungi, depriving them of energy. It also regulates salicylic acid and jasmonic acid, thus influencing plant basal resistance against necrotrophic pathogens (Zhou et al., 2012). Increased level of U-box domain-containing protein 38 and E3 ubiquitin ligase was observed in AB + SNP, indicating the upregulation of proteins involved in autophagy. This result was strongly supported by the TEM images where autophagy and chlorophagy were observed in AB + SNP treatment.
Significant enhancements in proteins involved in protein biogenesis were observed in SNP and SNP + AB treatments. Synthesis of a diverse pool of amino acids and related proteins, especially methionine, can be further utilized in synthesis of antimicrobial proteins, secondary metabolite, and organic acids, which was evident from the results of GC-MS. Higher expression of the protein involved in the amino acid synthesis and methionine biosynthetic pathway in SNP and AB + SNP was able to produce a range of antimicrobial compounds that can fight the pathogen before establishment of infection, thus providing a higher level of immunity to plants. The higher expression level of methyl esterase was also observed in AB + SNP as compared to AB, which might be responsible for enhanced resistance of plant cell wall by maintaining its osmoregulation by increasing the production of osmolytes such as inositol.
Conclusion
In this study, antifungal activity of biosynthesized silver nanomaterial was tested in the model pathosystem of A. thaliana and A. brassicicola. SNP demonstrated excellent antimicrobial activity and helped host plant to combat the disease as evident by disease parameters, enzymatic and non-enzymatic assays, and light and electron microscopy. It enhanced the phenolic content; ameliorated biotic stress as observed by a decrease in H2O2, LPX, and proline content; and subsequently decreased the concerned defense enzyme without showing any phytotoxicity.
Proteomics and metabolomics study of A. thaliana leaves during tripartite interaction of plant, pathogen, and nanoparticles revealed that many proteins and metabolites differentially expressed during the interaction. Proteins related to bioenergy, metabolism, and plant defense were most abundant to be differentially expressed, resulting in differential regulation of sugar, steroid, and secondary metabolite synthesis during tripartite interaction. This study provides an insight into the role of SNP in plant disease management. The nanomaterials were not only capable of killing the pathogen; they also enhanced antimicrobial property of plant and induced primary and secondary immune responses by modulating proteome and metabolome.
Supporting Information
This contains further information regarding characterization of SNP, detached leaf assay, lactophenol staining, stress and antioxidative parameters, 2-DE gel analysis, zoomed-in gel images, reproducibility of 2-DE gels, MS/MS details of identified differential proteins, and putative domain analysis of identified unknown proteins, as well as detailed result and discussion of differentially expressed proteins and detailed methodology of MS/MS and GC-MS studies.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
The idea was conceived by CN and AM. The work was done by MK, SP, SM, and VG. The data were interpreted by MK, SP, LA, and AP. The manuscript was written by MK, SP, and VG. The manuscript was critically analyzed by LA, SD, AM, and CN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank CSIR-Indian Institute of Toxicology Research for providing TEM and SEM facility. MK and SP thank CSIR for awarding Fellowship (JRF and SRF). We would also like to thank Dr. Samir Sawant, Dr. O. P. Sidhu, and Dr. Vivek Pandey for kindly providing seeds of A. thaliana ecotype Columbia (Col-0) and help during metabolomics and proteomics work, respectively. We are thankful to Geetgovind Sinam for ICP-MS analysis at Central Instrumentation Facility, CSIR-NBRI. Thanks are due to Director, CSIR–National Botanical Research Institute, Lucknow, for the necessary support.
Funding. This study was partially funded by a network project of the Council of Scientific and Industrial Research (CSIR) “Root SF BSC0204.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00242/full#supplementary-material
References
- Abdelrahmana M., Abdel-Motaal F., El-Sayed M., Jogaiah S., Shigyo M., Ito S., et al. (2016). Dissection of Trichoderma longibrachiatum-induced defence in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Sci. 246 128–138. 10.1016/j.plantsci.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Aebi H. (1974). “Catalase,” in Methods in Enzymatic Analysis, ed. Bergmeyer H. V. (New York, NY: Academic press; ). [Google Scholar]
- Agarwal L., Gupta S., Mishra S. K., Pandey G., Kumar S., Chauhan P. S., et al. (2016). Elucidation of complex nature of PEG induced drought-stress response in rice root using comparative proteomics approach. Frontiers in Plant Science. 7:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal L., Narula K., Basu S., Shekhar S., Ghosh S., Datta A., et al. (2013). Comparative proteomics reveals a role for seed storage protein ama1 in cellular growth, development, and nutrient accumulation. J. Proteome Res. 12 4904–4930. 10.1021/pr4007987 [DOI] [PubMed] [Google Scholar]
- Ahuja I., Kissen R., Bones A. M. (2012). Phytoalexins in defence against pathogens. Trends Plant Sci. 17 73–90. 10.1016/j.tplants.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Anandaradje A., Meyappan V., Kumar I., Sakthivel N. (2020). “Microbial synthesis of silver nanoparticles and their biological potential,” in Nanoparticles in Medicine, ed. Shukla A. (Singapore: Springer; ). [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39 205–207. 10.1016/j.dental.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp C., Fridovich I. (1971). Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. Rev. 44 276–287. 10.1016/0003-2697(71)90370-8 [DOI] [PubMed] [Google Scholar]
- Berger S., Papadopoulos M., Schreiber U., Kaiser W., Roitsch T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant. 122 419–428. 10.1111/j.1399-3054.2004.00433.x [DOI] [Google Scholar]
- Bogamuwa S. P., Jang J. C. (2014). Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 55 1367–1375. 10.1093/pcp/pcu074 [DOI] [PubMed] [Google Scholar]
- Chen C., Kai N., Chen J., Wan C. (2020). UHPLC-Q-TOF/MS-based metabolomics approach reveals the antifungal potential of Pinocembroside against citrus green moldphytopathogen. Plants 9:17. 10.3390/plants9010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva L. L., Nascimento M., Siqueira S. D. H., Furlan M., daSilva B. V. (2002). Antibacterial activity of a stearic acid derivative from Stemodiafoliosa. Planta Med. 68 1137–1139. 10.1055/s-2002-36346 [DOI] [PubMed] [Google Scholar]
- Das P., Sarmah K., Hussain N., Pratihar S., Das S., Bhattacharyy P., et al. (2016). Novel synthesis of an iron oxalate capped iron oxide nanomaterial: a unique soil conditioner and slow release eco-friendly source of iron sustenance in plants. RSC Adv. 6:103012 10.1039/c6ra18840k [DOI] [Google Scholar]
- Divon H. H., Fluhr R. (2007). Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiol. Lett. 266 65–74. 10.1111/j.1574-6968.2006.00504.x [DOI] [PubMed] [Google Scholar]
- Dong J., Chen W. (2013). The role of autophagy in chloroplast degradation and chlorophagy in immune defences during pst dc3000 (avrrps4) infection. PLoS ONE 8:e73091. 10.1371/journal.pone.0073091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Bautista N., Domínguez-Núñez J. A., Moreno M. C., Berrocal-Lobo M. (2016). Plant tissue trypan blue staining during phytopathogen infection. Bio-Protocol. 6:e2078. [Google Scholar]
- Gill S. S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Griebel T., Zeier J. (2010). A role for b-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J. 63 254–268. 10.1111/j.1365-313x.2010.04235.x [DOI] [PubMed] [Google Scholar]
- Hassan S., Mathesius U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63 3429–3444. 10.1093/jxb/err430 [DOI] [PubMed] [Google Scholar]
- Heath R., Packer L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid per- oxidation. Arch. Biochem. Biophys. 196 385–395. [DOI] [PubMed] [Google Scholar]
- Hemeda H. M., Klein B. P. (1990). Effects of naturally occurring antioxidants on peroxidase activity of vegetable extracts. J. Food Sci. 55 184–185. 10.1111/j.1365-2621.1990.tb06048.x [DOI] [Google Scholar]
- Hirpara D. G., Gajera H. P. (2020). Green synthesis and antifungal mechanism of silver nanoparticles derived from chitin- induced exometabolites of Trichoderma interfusant. Appl. Organ. Chem. 34:e5407. [Google Scholar]
- Ilaria S., Antonietta L., Alfredo A. (2019). Nanotechnology in plant science: to make a long story short. Front. Bioeng. Biotechnol. 7:120. 10.3389/fbioe.2019.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Peng Y., Meckes N., Allen S., Stewart N., Traw M. B. (2014). ABC transporter AtABCG16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 114:248153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juroszek P., Tiedemann A. (2015). Linking plant disease models to climate change: scenarios to project future risks of crop diseases: a review. J. Plant Dis. Protect. 122 3–15. 10.1007/bf03356525 [DOI] [Google Scholar]
- Kabara J. J., Swieczkowski D. M., Conley A. J., Truant J. P. (1972). Fatty acids and derivatives as antimicrobial agents. Antimicrob. Agents Chemother. 2 23–28. 10.1128/aac.2.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M., Williams B., Dickman M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotiniasclerotiorum. PLoS Pathogen. 9:e1003287. 10.1371/journal.ppat.1003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalegari M., Miguel M. D., Philippsen A. F., Dias J. F. G., Zanin S. A. W., de Lima C. P., et al. (2012). Antibacterial, allelopathic and antioxidant activity of extracts and compounds from Roureainduta planch. (Connaraceae). J. Appl. Pharmaceut. Sci. 2 061–066. [Google Scholar]
- Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., et al. (2011). Plant ABC transporters. Arabidopsis Book. 9:e0153. 10.1199/tab.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Mishra A., Nautiyal C. S. (2012). Paenibacilluslentimorbus B-30488r controls early blight disease in tomato by inducing host resistance associated gene expression and inhibiting Alternariasolani. Biol. Control. 62 65–74. 10.1016/j.biocontrol.2012.03.010 [DOI] [Google Scholar]
- Kim S. H., Qi D., Ashfield T., Helm M., Innes R. W. (2016). Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351 684–687. 10.1126/science.aad3436 [DOI] [PubMed] [Google Scholar]
- Kontush A., Finckh B., Karten B., Kohlschütter A., Beisiegel U. (1996). Antioxidant and prooxidant activity of alpha-tocopherol in human plasma and low density lipoprotein. J. Lipid Res. 37 1436–1448. [PubMed] [Google Scholar]
- Krishnaraj C., Jagan E. G., Ramachandran R., Abirami S. M., Mohan N., Kalaichelvan P. T. (2012). Effect of biologically synthesized silver nanoparticles on Bacopamonnieri (Linn.). Process. Biochem. 47 651–658. 10.1016/j.procbio.2012.01.006 [DOI] [Google Scholar]
- Kumari M., Giri V. P., Pandey S., Kumar M., Katiyar R., Nautiyal C. S., et al. (2019). An insight into the mechanism of antifungal activity of biogenic nanoparticles than their chemical counterparts. Pest. Physiol. Biochem. 157 45–52. 10.1016/j.pestbp.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Kumari M., Shukla S., Pandey S., Giri V. P., Tripathi T., Bhatia A., et al. (2017a). Enhanced cellular internalization: A bactericidal mechanism more relative to biogenic nanoparticles than chemical counterparts. ACS-Appl. Mater. Interf. 9 4519–4533. 10.1021/acsami.6b15473 [DOI] [PubMed] [Google Scholar]
- Kumari M., Pandey S., Giri V. P., Bhattacharya A., Shukla R., Nautiyal C. S., et al. (2017b). Tailoring shape and size of biogenic silver nanoparticles to enhance antimicrobial efficacy against MDR bacteria. Microb. Pathog. 105 346–355. 10.1016/j.micpath.2016.11.012 [DOI] [PubMed] [Google Scholar]
- Kumari M., Pandey S., Mishra S. K., Nautiyal C. S., Mishra A. (2017c). Effect of biosynthesized silver nanoparticles on native soil microflora via plant transport during plant–pathogen–nanoparticles interaction. 3 Biotech. 7:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue C., Castillo-Michel H., Sobansk S., Cécillon L., Bureau S., Barthès V., et al. (2014). Foliar exposure of the crop Lactucasativa to silver nanoparticles: evidence for internalization and changes in Ag speciation. J. Hazardous Mater. 264 98–106. 10.1016/j.jhazmat.2013.10.053 [DOI] [PubMed] [Google Scholar]
- Lavania M., Chauhan P. S., Chauhan S. V. S., Singh H. B., Nautiyal C. S. (2006). Induction of plant defence enzymes and phenolics by treatment with plant growth-promoting rhizobacteria Serratiamarcescens NBRI1213. Curr. Microbiol. 52 363–368. 10.1007/s00284-005-5578-2 [DOI] [PubMed] [Google Scholar]
- Lin J., Dou J., Xu J., Aisa H. K. (2012). Chemical composition, antimicrobial and antitumor activities of the essential oils and crude extracts of Euphorbia macrorrhiza. Molecules 17 5030–5039. 10.3390/molecules17055030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawar H., Prasanna R., Gogoi R., Bala Singh S., Chawla G., Kumar A., et al. (2020). Synergistic effects of silver nanoparticles augmented Calothrix elenkinii for enhanced biocontrol efficacy against Alternaria blight challenged tomato plants. 3 Biotech 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick C. P., Singh M. B. (1980). Plant Enzymology and Histoenzymology. New Delhi: Kalyani publishers. [Google Scholar]
- Mariana G., De L. R. (2019). Synthesis of silver nanoparticles mediated by fungi: a review. Front. Bioeng. Biotechnol. 7:287. 10.3389/fbioe.2019.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques J. P. R., Soares M. K. M., Gloria B. A. (2013). New staining technique for fungal-infected plant tissues. Turkish J. Bot. 37 1204–1209. [Google Scholar]
- Meena M., Zehra A., Dubey M. K., Aamir M., Gupta V. K., Upadhyay R. S. (2016). Comparative evaluation of biochemical changes in tomato (Lycopersicon esculentum Mill.) infected by alternaria alternata and its toxic metabolites (TeA, AOH, and AME). Front. Plant Sci. 7:1408. 10.3389/fpls.2016.01408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Kumari M., Pandey S., Chaudhry V., Gupta K. C., Nautiyal C. S. (2014). Biocatalytic and antimicrobial activities of gold nanoparticles synthesized by Trichoderma sp. Bioresour. Technol. 166 235–242. 10.1016/j.biortech.2014.04.085 [DOI] [PubMed] [Google Scholar]
- Mitusi A., Ohata T. (1961). Photooxidative consumption and photoreductive formation of ascorbic acid in green leaves. Plant Cell Physiol. 2 31–44. 10.1093/oxfordjournals.pcp.a077661 [DOI] [Google Scholar]
- Moghaddam M. R. B., Ende E. (2012). Sugars and plant innate immunity. J. Exp. Bot. 63 3989–3998. 10.1093/jxb/ers129 [DOI] [PubMed] [Google Scholar]
- Monir H. M., Ahmed P. S., Masato T., Datta S. R., Tanushree S., Zinia I., et al. (2019). Investigation of the antibacterial activity and in vivo cytotoxicity of biogenic silver nanoparticles as potent therapeutics. Front. Bioeng. Biotechnol. 7:239. 10.3389/fbioe.2019.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y., Nakahira Y., Sumida H., Takebayashi K., Nagasawa Y., Yamasaki K., et al. (2013). Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J. 73 761–775. 10.1111/tpj.12069 [DOI] [PubMed] [Google Scholar]
- Nielsen J., Jewett M. C. (2007). The role of metabolomics in systems biology. Topics Curr. Genet. 18 1–10. 10.1007/4735_2007_0228 [DOI] [Google Scholar]
- Ocsoy I., Paret M. L., Ocsoy M. A., Kunwar S., Chen T., You M., et al. (2013). Nanotechnology in plant disease management: DNA-directed silver nanoparticles on graphene oxide as an antibacterial against Xanthomonasperforans. ACS Nano 7 8972–8980. 10.1021/nn4034794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke E. C. (2006). Crop losses to pests. J. Agric. Sci. 144 31–43. 10.1017/s0021859605005708 [DOI] [Google Scholar]
- Otani H., Kohmoto K., Kodama M. (1995). Alternaria toxins and their effects on host plants. Can. J. Bot. 73 453–458. 10.1139/b95-282 [DOI] [Google Scholar]
- Ozdemir M., Miller G., Song L., Kim J., Sodek A., Koussevitzky S., et al. (2004). Thiamine confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 151 421–432. 10.1104/pp.109.140046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam B., Parandhaman T., Das S. K. (2016). Antibacterial effects of biosynthesized silver nanoparticles on surface ultra structure and nanomechanical properties of gram negative bacteria viz. Escherichia coli and Pseudomonas aeruginosa. ACS Appl. Mater. Interf. 8 4963–4976. 10.1021/acsami.6b00161 [DOI] [PubMed] [Google Scholar]
- Romano P. G. N., Horton P., Gray J. E. (2004). The Arabidopsis cyclophilin gene family. Plant Physiol. 134 1268–1282. 10.1104/pp.103.022160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C., Claure P., Salvadori M. S., Mota V. G., Costa L. M., de Almeida A. A. C., et al. (2013). Antinociceptive and antioxidant activities of phytol in vivo and in vitro models. Neurosci. J. 9:949452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnürer J., Magnusson J. (2005). Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16 70–78. 10.1016/j.tifs.2004.02.014 [DOI] [Google Scholar]
- Sergiev I., Alxieva V., Karanov E. (1997). Effect of spermone, atrazine and combination between them on some endogenous protective systems and stress markers in plants. ComptesRendus De L AcademieBulgare Des Sci. 51 121–124. [Google Scholar]
- Shen T., Wang Q., Li C., Zhou B., Li Y., Liu Y. (2020). Transcriptome sequencing analysis reveals silver nanoparticles antifungal molecular mechanism of the soil fungi Fusarium solani species complex. J. Hazard. Mater. 388:122063. 10.1016/j.jhazmat.2020.122063 [DOI] [PubMed] [Google Scholar]
- Shoresh M., Harman G. E. (2008). The molecular basis of shoot responses of maize seedlings to Trichoderma harzianum T22 inoculation of the root: a proteomic approach. Plant Physiol. 147 2147–2163. 10.1104/pp.108.123810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Sarma B. K., Upadhyay R. S., Singh H. B. (2013). Compatible rhizosphere microbes mediated alleviation of biotic stress in chickpea through enhanced antioxidant and phenylpropanoid activities. Microbiol. Res. 168 33–40. 10.1016/j.micres.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Bisht H., Sidhu O. P., Srivastava A., Singh P. C., Pandey R. M., et al. (2012a). Changes in the metabolome and histopathology of Amaranthushypochondriacus L. in response to Ageratum enation virus infection. Phytochemistry 80 8–16. 10.1016/j.phytochem.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Chaudhry V., Mishra A., Chauhan P. S., Rehman A., Yadav A., et al. (2012b). Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 7 235–245. 10.4161/psb.18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Srivastava S., Bist V., Awasthi S., Chauhan R., Chaudhry V., et al. (2018). Chlorella vulgaris and Pseudomonas putida interaction modulates phosphate trafficking for reduced arsenic uptake in rice (Oryza sativa L.). J. Hazard. Mater. 351 177–187. 10.1016/j.jhazmat.2018.02.039 [DOI] [PubMed] [Google Scholar]
- Stephano-Hornedo J. L., Torres-Gutiérrez O., Toledano-Magaña Y., Gradilla-Martínez I., Pestryakov A. (2020). ArgovitTM silver nanoparticles to fight huanglongbing disease in Mexican limes (Citrus aurantifolia Swingle). RSC Adv. 10 6146–6155. 10.1039/c9ra09018e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su’udi M., Park J., Park S., Hwang D., Bae S., Kim S., et al. (2013). Quantification of Alternariabrassicicola infection in the Arabidopsis thaliana and Brassica rapa subsp. pekinensis. Microbiology 159 1946–1955. 10.1099/mic.0.068205-0 [DOI] [PubMed] [Google Scholar]
- Szabados L., Savoure A. (2009). Proline: a multifunctional amino acid. Trends Plant Sci. 15 89–97. 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Tiwari S. K., Agarwal S., Seth B., Yadav A., Nair S., Bhatnagar P., et al. (2014). Curcumin-loaded nanoparticles potently induce adult neurogenesis and reverse cognitive deficits in alzheimer’s disease model via canonical wnt/β-catenin pathway. ACS Nano 8 76–103. 10.1021/nn405077y [DOI] [PubMed] [Google Scholar]
- Velásquez A. C., Castroverde C., He S. Y. (2018). Plant-pathogen warfare under changing climate conditions. Cur. Biol. 28 R619–R634. 10.1016/j.cub.2018.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verano-Braga T., Miethling-Graff R., Wojdyla K., Rogowska-Wrzesinska A., Brewer J. R., Erdmann H., et al. (2014). Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics. ACS Nano 8 2161–2175. 10.1021/nn4050744 [DOI] [PubMed] [Google Scholar]
- Yan Z., Reddy M. S., Ryu C., McInroy J. A., Wilson M., Kloepper J. W. (2002). Induced systemic protection against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 92:1329. 10.1094/PHYTO.2002.92.12.1329 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ma C., Xu Y., Wei Q., Imtiaz M., Lan H., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26 2038–2054. 10.1105/tpc.114.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Bi Y., Chen S., Li Y., Wang Y., Ge Y., et al. (2011). Chemical composition and antifungal activity of cuticular wax isolated from Asian pear fruit (cv. Pingguoli). Sci. Horticult. 229 577–582. 10.1016/j.scienta.2011.04.028 [DOI] [Google Scholar]
- Zhang Z., Han X., Wei J., Xue J., Yang Y., Liang L., et al. (2014). Compositions and antifungal activities of essential oils from agarwood of Aquilariasinensis(Lour.) gilg induced by Lasiodiplodiatheobromae (Pat.). J. Braz. Chem. Soc. 25 20–26. [Google Scholar]
- Zhou Y., Kong Y., Kundu S., Cirillo J. D., Liang H. (2012). Antibacterial activities of gold and silver nanoparticles against Escherichia coli and bacillus Calmette-Guérin. J. Nanobiotechnol. 10:19. 10.1186/1477-3155-10-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.