Leprosy is caused by Mycobacterium leprae, and it remains underdiagnosed in Burkina Faso. We investigated the use of fluorescent in situ hybridization (FISH) for detecting M. leprae in 27 skin samples (skin biopsy samples, slit skin samples, and skin lesion swabs) collected from 21 patients from Burkina Faso and three from Côte d’Ivoire who were suspected of having cutaneous leprosy. In all seven Ziehl-Neelsen-positive skin samples (four skin biopsy samples and three skin swabs collected from the same patient), FISH specifically identified M. leprae, including one FISH-positive skin biopsy sample that remained negative after testing with PCR targeting the rpoB gene and with the GenoType LepraeDR assay.
KEYWORDS: fluorescent hybridization, Mycobacterium, Mycobacterium leprae, leprosy, skin
ABSTRACT
Leprosy is caused by Mycobacterium leprae, and it remains underdiagnosed in Burkina Faso. We investigated the use of fluorescent in situ hybridization (FISH) for detecting M. leprae in 27 skin samples (skin biopsy samples, slit skin samples, and skin lesion swabs) collected from 21 patients from Burkina Faso and three from Côte d’Ivoire who were suspected of having cutaneous leprosy. In all seven Ziehl-Neelsen-positive skin samples (four skin biopsy samples and three skin swabs collected from the same patient), FISH specifically identified M. leprae, including one FISH-positive skin biopsy sample that remained negative after testing with PCR targeting the rpoB gene and with the GenoType LepraeDR assay. Twenty other skin samples and three negative controls all remained negative for Ziehl-Neelsen staining, FISH, and rpoB PCR. These data indicate the usefulness of a microscopic examination of skin samples after FISH for first-line diagnosis of cutaneous leprosy. Accordingly, FISH represents a potentially useful point-of-care test for the diagnosis of cutaneous leprosy.
INTRODUCTION
Leprosy caused by Mycobacterium leprae is endemic in some developing countries, including Burkina Faso (192 new cases in 2017), where 31% of newly diagnosed patients present with grade 2 disabilities. Burkina Faso has the highest prevalence in this region of Africa (1), partially due to difficulties in early diagnosis, as routine laboratory diagnosis is limited to the microscopic observation of acid-fast bacilli in skin biopsy and slit-skin samples, which has an acknowledged poor sensitivity of 104 bacilli/g of tissue (2). Fluorescence microscopy is known to offer benefits in terms of sensitivity compared to optical microscopy and Ziehl-Neelsen staining (3, 4).
In a previous study, we established that rpoBMTC probe-based fluorescent in situ hybridization (FISH) alone or FISH combined with Ziehl-Neelsen staining specifically “FISHes out” M. tuberculosis complex mycobacteria in sputum samples (5). Here, we investigated FISH as a confirmatory technique for detecting M. leprae in skin biopsy specimens to diagnose leprosy.
MATERIALS AND METHODS
Clinical samples.
This prospective, cross-sectional study was authorized by the Hauts Bassins Regional Health Authority (no. 0014, 2017) and the Ethics Committee of the Burkina Faso National Institute of Public Health (no. 23, 2019). After the merits of the study were explained to patients with cutaneous lesions suggestive of leprosy, informed consent was collected before skin sampling was performed at the Hospital University Centre Souro Sanou of Bobo-Dioulasso, Burkina Faso, by a dermatologist as part of the routine medical investigation of patients. No biopsy specimens and slit skin smears were collated specifically for the present study. Written authorization for the use of photos for scientific or academic purposes was obtained from the patients. Samples were collected in two time periods: the first from January to March 2018 and the second in August 2019. Fifteen males and nine females (age range, 15 to 77 years; mean, 40 ± 18 years) were enrolled. Twenty-one patients, including two with the multibacillary (MB) form of clinical leprosy (more than five lesions), were from Burkina Faso, and three patients, including one with the MB leprosy form, were from Côte d’Ivoire (Table 1). Skin biopsy specimens, slit-skin samples, and skin lesion swabs were affixed to slides that had previously been disinfected with 70% alcohol, as previously described (6). A first Ziehl-Neelsen heat stain performed at the Hospital University Centre Souro Sanou mycobacteriology laboratory for diagnostic purposes was repeated at the Institut Hospitalier Universitaire Méditerranée Infection, Marseille, France, using the Quick-TB kit (cold staining) (RAL Diagnostics, Martillac, France).
TABLE 1.
Characteristics of the 24 patients suspected of having cutaneous leprosya
| Patient code | Sex | Age (yrs) | Location of the patient | Antileprosy treatment before sampling | Clinical form (WHO criteria) |
|---|---|---|---|---|---|
| P1 | M | 64 | Bobo-Dioulasso (BF) | No | PB |
| P2 | M | 26 | Dogoma (BF) | No | MB |
| P3 | F | 57 | Bobo-Dioulasso (BF) | No | PB |
| P4 | M | 60 | Vavoua (CI) | No | MB |
| P5 | M | 46 | Koundougou (BF) | Yes | PB |
| P6 | F | 65 | Bobo-Dioulasso (BF) | No | PB |
| P7 | F | 20 | Karangasso (BF) | No | PB |
| P8 | F | 60 | Deguelin (BF) | No | PB |
| P9 | F | 57 | Bobo-Dioulasso (BF) | No | PB |
| P10 | M | 30 | Dano (BF) | No | PB |
| P11 | M | 21 | Bobo-Dioulasso (BF) | No | PB |
| P12 | F | 29 | Bobo-Dioulasso (BF) | No | PB |
| P13 | M | 45 | Bobo-Dioulasso (BF) | No | PB |
| P14 | M | 64 | Bobo-Dioulasso (BF) | No | PB |
| P15 | M | 24 | Bobo-Dioulasso (BF) | No | PB |
| P16 | M | 21 | Bobo-Dioulasso (BF) | No | PB |
| P17 | M | 77 | Vavoua (CI) | No | PB |
| P18 | M | 15 | Boundiali (CI) | No | PB |
| P19 | F | 29 | Baré (BF) | Yes | PB |
| P20 | F | 41 | Bobo-Dioulasso (BF) | No | PB |
| P21 | M | 20 | Bama (BF) | No | MB |
| P22 | F | 35 | Bobo-Dioulasso (BF) | No | PB |
| P23 | M | 28 | Bobo-Dioulasso (BF) | No | PB |
| P24 | M | 33 | Bobo-Dioulasso (BF) | No | PB |
F, female; M, male; BF, Burkina Faso; CI, Côte d’Ivoire; MB, multibacillary; PB, paucibacillary.
Molecular analyses.
DNA was extracted through a combination of chemical lysis (Qiagen, Hilden, Germany), treatment with glass powder, heating at 56°C for 2 h, sonication for 30 min, and automatic elution with an EZ1 DNA tissue kit (Qiagen, Hilden, Germany). The extracted DNA was used for partial PCR amplification and sequencing of the rpoB gene, as previously described (7). In these experiments, sterile water was used as the negative control, and the beta-actin gene was amplified to assess the quality of DNA extraction. The GenoType LepraeDR test (Hain Lifescience GmbH, Nehren, Germany) was performed in the Laboratory of Bacteriology of Hospital Arnaud de Villeneuve in Montpellier, France, for the molecular detection of M. leprae resistant to antileprosy drugs (8).
FISH analysis.
SVARAP software (9) was used for the in silico design of a M. leprae rpoB gene probe after alignment of 100 rpoB sequences from different mycobacteria available in GenBank with MEGA-X (www.megasoftware.net/dload_win_gui). A highly conserved region in M. leprae was used to design a probe with Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). We verified that the nucleotide sequence of the designed probe did not target regions harboring missense mutations for rifampin resistance, as previously described (10). The specificity of the probe was ensured in silico by conducting a BLAST search of the probe sequence against the GenBank database and aligning it with 12 bacterial genomes (Table 2). Then, the specificity of this Alexa Fluor 555-labeled fluorescent oligonucleotide probe (5′-CAACTCCTCAGGCAAGTTGA-3′) was experimentally tested in a series of microorganisms known to be associated with chronic cutaneous lesions in West African patients, including Mycobacterium ulcerans, Mycobacterium marinum, Mycobacterium tuberculosis complex, Staphylococcus aureus, Streptococcus pyogenes, and Klebsiella pneumoniae. The FISH detection of M. leprae in the skin samples was performed using previously described methods (5). Briefly, smears were fixed with 4% paraformaldehyde and treated with 10 mg/ml lysozyme and 10 μg/ml proteinase K. After an overnight incubation with a 10-μl suspension containing the specific probe, smears were serially washed and mounted with ProLong Diamond Antifade containing 4,6-diamidino-2-phenylindole (DAPI) (Fisher Scientific, Illkirch, France). Microscopic observations using the 100× lens objective of a Leica DMI3000 microscope (Leica Microsystèmes, Nanterre, France) were confirmed using a Zeiss LSM 800 confocal microscope (with a 63×, 1.4-numerical-aperture [NA] oil immersion objective and a 568-nm excitation laser) (Zeiss, Marly-le-Roi, France). Images were acquired using ZEN software (Zeiss). Three skin biopsy specimens devoid of any evidence of mycobacteria were included as negative controls in Ziehl-Neelsen staining and FISH experiments.
TABLE 2.
In silico and experimental tests of the specificity of the Alexa Fluor 555-labeled fluorescent oligonucleotide probea
| Test and bacterium | GenBank accession no. or source | In silico hybridization (%) or exptl hybridization resultb |
|---|---|---|
| In silico | ||
| Mycobacterium leprae | CP029543.1 | 100 |
| Mycobacterium lepromatosis | EU203594.2 | 85 |
| Mycobacterium haemophilum | CP011883.2 | 65 |
| Mycobacterium tuberculosis | CP023640.1 | 65 |
| Mycobacterium bovis BCG | CP033311.1 | 65 |
| Mycobacterium ulcerans | LR135168.1 | 60 |
| Mycobacterium marinum | CP024190.1 | 60 |
| Mycobacterium abscessus | CP022234.1 | 60 |
| Mycobacterium fortuitum | CP014258.1 | 65 |
| Staphylococcus aureus | LR134267.1 | 65 |
| Streptococcus pyogenes | AY583221.1 | 65 |
| Pseudomonas aeruginosa | LR130534.1 | 60 |
| Total | 12 | |
| Experimental specificity | ||
| Mycobacterium ulcerans | Our laboratory | ND |
| Mycobacterium marinum | Our laboratory | ND |
| Mycobacterium tuberculosis complex | Our laboratory | ND |
| Staphylococcus aureus | Our laboratory | ND |
| Streptococcus pyogenes | Our laboratory | ND |
| Pseudomonas aeruginosa | Our laboratory | ND |
| Klebsiella pneumoniae | Our laboratory | ND |
| Total | 7 |
5′-CAACTCCTCAGGCAAGTTGA-3′.
ND, hybridization was not detected by fluorescence microscopy.
RESULTS
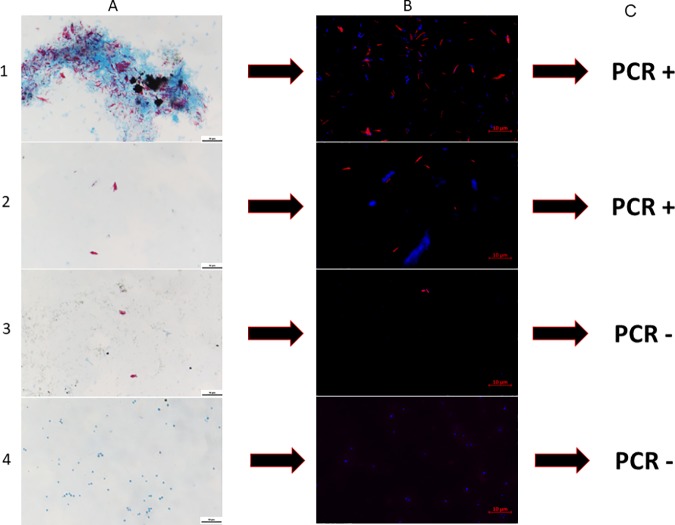
Four skin biopsy specimens and three skin lesion swabs (collected from the same patient, P21) exhibited acid-fast bacilli, as confirmed by two independent rounds of Ziehl-Neelsen staining, in the presence of three negative controls that remained free of acid-fast bacilli. After we ensured the in silico and experimental specificity of the rpoB gene sequence-based FISH probe, FISH highlighted Ziehl-Neelsen-positive mycobacteria as red-fluorescent bacilli (Fig. 1) in these four skin biopsy specimens and the three skin lesion swab smears by both fluorescence microscopy and confocal microscopy, whereas no FISH-positive mycobacteria were detected in the remaining samples. Two of the four FISH-positive skin biopsy specimens were further confirmed to contain M. leprae based on the positive results of the GenoType LepraeDR assay, which indicated the absence of detectable antileprosy drug resistance. Three of four FISH-positive skin biopsy specimens were also positive for the rpoB gene in the PCR assay, and sequencing indicated 99% gene sequence similarity with the reference M. leprae sequence (GenBank accession no. CP029543.1). The fourth (P5) skin biopsy specimen remained negative for M. leprae, despite positivity for the beta-actin gene (in all skin samples) and the positivity of Ziehl-Neelsen staining and FISH. In these experiments, the three negative controls remained negative for Ziehl-Neelsen staining, FISH, and rpoB-PCR. The three FISH-positive skin lesion swabs were also positive for the rpoB gene in the PCR assay, and sequencing confirmed the presence of M. leprae (Table 3).
FIG 1.
Fluorescent in situ hybridization (FISH) of Mycobacterium leprae in skin biopsy specimens collected from patients suspected of having cutaneous leprosy in Burkina Faso. Rows 1, 2, and 3 show positive skin biopsy specimens from patients P2, P4, and P5, respectively (Table 1); row 4 shows a negative-control skin biopsy specimen from patient 8111270308 (Table 1). (A) Ziehl-Neelsen staining; (B) FISH-DAPI staining; (C) results of the GenoType LepraeDR test and PCR amplification of the rpoB gene. +, positive; −, negative. Bar, 10 μm.
TABLE 3.
Results of the laboratory investigation of skin samplesa
| Patient code | Sample | Result of: |
GenoType LepraeDR result |
|||||
|---|---|---|---|---|---|---|---|---|
| Ziehl-Neelsen microscopy (BI) | Fluorescent microscopy | PCR for rpoB | Detection of M. leprae | Rifampin resistance | Dapsone resistance | Ofloxacin resistance | ||
| P1 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P2 | Skin biopsy | Pos (4+) | Pos | ML | ML | Sensitive | Sensitive | Sensitive |
| P3 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P4 | Skin biopsy | Pos (3+) | Pos | ML | ML | Sensitive | Sensitive | Sensitive |
| P5 | Skin biopsy | Pos (1+) | Pos | Neg | Neg | NA | NA | NA |
| P6 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P7 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P8 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P9 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P10 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P11 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P12 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P13 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P14 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| P15 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P16 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P17 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P18 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P19 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P20 | Slit skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P21 | Skin biopsy | Pos (5+) | Pos | ML | NT | NT | NT | NT |
| Skin swab (3) | Pos (3+) | Pos | ML | NT | NT | NT | NT | |
| P22 | Swab of ulcerative skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P23 | Swab of ulcerative skin | Neg | Neg | Neg | NA | NA | NA | NA |
| P24 | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| 8111270308* | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| 8121367852* | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
| 9011404509* | Skin biopsy | Neg | Neg | Neg | NA | NA | NA | NA |
NA, not applicable; Neg, negative; Pos, positive; *, negative-control sample collected from patient admitted to the Institut Hospitalier Universitaire Méditerranée Infection, Marseille, France, and presenting skin lesions devoid of any evidence of mycobacteria; NT, not tested; 3, three swabs collected from three types of skin lesions (skin wound, scaly skin evoking cutaneous leishmaniasis, and skin papules with raised extremities) in patient P21; BI, bacterial index determined using Ridley’s logarithmic scale (22).
DISCUSSION
We report the utility of FISH for determining the diagnosis of cutaneous leprosy when applied to smears prepared from skin samples collected from patients suspected of having cutaneous leprosy. Indeed, the observations reported here were authenticated by the negativity of negative controls analyzed in the experiments, the agreement of observations obtained using different techniques, and the reproducibility of Ziehl-Neelsen staining. The report confirms the usefulness of microscopic examination of skin sample smears after FISH for the diagnosis of cutaneous leprosy, as only two similar reports have been published previously (11, 12). FISH is increasing the specificity of microscopy in a context where other cutaneous mycobacterioses prevail, such as in Burkina Faso (13). In addition, we observed one FISH-positive sample with a clinical paucibacillary leprosy form that remained negative for the detection of M. leprae DNA using two different molecular assays. This discrepancy might be explained by several factors, including the small size of the tissue section extracted, which contained a low number of bacilli (14). DNA amplification inhibitors from human surgical tissue also may explain the false-negative PCR result (15). Another potential explanation is the low sensitivity of the PCR techniques targeting a single copy of a gene in the genome (16–18) in a paucibacillary specimen (19).
We propose that FISH is an efficient method for the first-line diagnosis of cutaneous leprosy. FISH provides greater specificity than Ziehl-Neelsen staining, and a single observation is sufficient for the precise diagnosis of leprosy. Based on a previous estimate of a cost of $5 (U.S.) per FISH test (20), we anticipate the implementation of FISH as a new diagnosis tool for point-of-care testing in one of the Burkina Faso health care centers that is already using fluorescence microscopy, including the possibility of a remote interpretation of smartphone photos (21).
ACKNOWLEDGMENTS
This study was supported by the French Government under the Investissements d’Avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (National Agency for Research) (reference: Méditerranée Infection 10-IAHU-03). This study was also supported by Région Le Sud (Provence Alpes Côte d’Azur) and a European funding agency (FEDER PA 0000320 PRIMI). Anselme Millogo is supported by a Ph.D. grant from IHU Méditerranée Infection, Marseille, France, and Université de Montpellier, Montpellier, France.
We acknowledge medical practitioners at the Souro Sanou University Hospital dermatology service for receiving and collecting samples and treating diagnosed patients. We acknowledge Arthur Diakourga Djibougou and Clément Ziemle Méda for their advice and contributions in the process of submitting the protocol for this study to the ethics committee.
We have no conflicts of interest to declare.
REFERENCES
- 1.Ouédraogo N, Ouédraogo M, Tapsoba G, Traore FF, Bassole A, Zeba/Lompo S, Tioye YL, Ilboudo L, Kabore N, Zigani E, Drabo F, Serme M, Nassa C, Kafando C, Korsaga/Some NN, Barro/Traore F, Niamba P, Traore A. 2018. Dépistage de la lèpre en stratégie avancée au Burkina Faso. Bull Assoc Lepr Langue Fr 33:4–7. https://www.leprosy-information.org/media/838/download. [Google Scholar]
- 2.Shepard CC, McRae DH. 1968. A method for counting acid-fast bacteria. Int J Lepr Other Mycobact Dis 36:78–82. [PubMed] [Google Scholar]
- 3.Marais BJ, Brittle W, Painczyk K, Hesseling AC, Beyers N, Wasserman E, van Soolingen D, Warren RM. 2008. Use of light-emitting diode fluorescence microscopy to detect acid-fast bacilli in sputum. Clin Infect Dis 47:203–207. doi: 10.1086/589248. [DOI] [PubMed] [Google Scholar]
- 4.Elston DM, Liranzo MO, Scollard DM. 2017. Comparing the sensitivity of auramine-rhodamine fluorescence to polymerase chain reaction in the detection of Mycobacterium leprae in Fite-negative tissue sections. J Am Acad Dermatol 76:992–993. doi: 10.1016/j.jaad.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 5.Loukil A, Kirtania P, Bedotto M, Drancourt M. 2018. FISHing Mycobacterium tuberculosis complex by use of a rpoB DNA probe bait. J Clin Microbiol 56:e00568-18. doi: 10.1128/JCM.00568-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darriet F, Bernioles P, Loukil A, Saidani N, Eldin C, Drancourt M. 2018. Fluorescence in situ hybridization microscopic detection of Bacilli Calmette Guérin mycobacteria in aortic lesions: a case report. Medicine (Baltimore)) 97:e11321. doi: 10.1097/MD.0000000000011321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adékambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 41:5699–5708. doi: 10.1128/jcm.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cambau E, Chauffour-Nevejans A, Tejmar-Kolar L, Matsuoka M, Jarlier V. 2012. Detection of antibiotic resistance in leprosy using GenoType LepraeDR, a novel ready-to-use molecular test. PLoS Negl Trop Dis 6:e1739. doi: 10.1371/journal.pntd.0001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson P, Tamalet C, Raoult D. 2006. SVARAP and aSVARAP: simple tools for quantitative analysis of nucleotide and amino acid variability and primer selection for clinical microbiology. BMC Microbiol 6:21. doi: 10.1186/1471-2180-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honore N, Cole ST. 1993. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother 37:414–418. doi: 10.1128/aac.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musso D, Rovery C, Loukil A, Vialette V, Nguyen NL. 2019. Leprosy in French Polynesia. New Microbes New Infect 29:100514. doi: 10.1016/j.nmni.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefmann M, Schweickert B, Buchholz P, Gobel UB, Ulrichs T, Seiler P, Theegarten D, Moter A. 2006. Evaluation of peptide nucleic acid-fluorescence in situ hybridization for identification of clinically relevant mycobacteria in clinical specimens and tissue sections. J Clin Microbiol 44:3760–3767. doi: 10.1128/JCM.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andonaba JB, Barro-Traoré F, Yaméogo T, Diallo B, Korsaga-Somé N, Traoré A. 2013. La tuberculose cutanée: observation de six cas confirmés au CHU Souro SANOU (CHUSS) de Bobo-Dioulasso (Burkina Faso.). Pan Afr Med J 16:50. doi: 10.11604/pamj.2013.16.50.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez AN, Talhari C, Moraes MO, Talhari S. 2014. PCR-based techniques for leprosy diagnosis: from the laboratory to the clinic. PLoS Negl Trop Dis 8:e2655. doi: 10.1371/journal.pntd.0002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. 2012. Methods of mycobacterial DNA isolation from different biological material: a review. Veterinarni Medicina 51:180–192. doi: 10.17221/5538-VETMED. [DOI] [Google Scholar]
- 16.Sharma R, Singh P, McCoy RC, Lenz SM, Donovan K, Ochoa MT, Estrada-Garcia I, Silva-Miranda M, Jurado-Santa Cruz F, Balagon MF, Stryjewska B, Scollard DM, Pena MT, Lahiri R, Williams DL, Truman RW, Adams LB. 16 November 2019. Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish M. leprae and M. lepromatosis. Clin Infect Dis doi: 10.1093/cid/ciz1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Xing Y, He J, Tan F, You Y, Wen Y. 2019. Develop and field evolution of single tube nested PCR, SYBRGreen PCR methods, for the diagnosis of leprosy in paraffin-embedded formalin fixed tissues in Yunnan province, a hyper endemic area of leprosy in China. PLoS Negl Trop Dis 13:e0007731. doi: 10.1371/journal.pntd.0007731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathak VK, Singh I, Turankar RP, Lavania M, Ahuja M, Singh V, Sengupta U. 2019. Utility of multiplex PCR for early diagnosis and household contact surveillance for leprosy. Diagn Microbiol Infect Dis 95:114855. doi: 10.1016/j.diagmicrobio.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Gurung P, Gomes CM, Vernal S, Leeflang M. 2019. Diagnostic accuracy of tests for leprosy: a systematic review and meta-analysis. Clin Microbiol Infect 25:1315–1327. doi: 10.1016/j.cmi.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Baliga S, Murphy C, Sharon L, Shenoy S, Biranthabail D, Weltman H, Miller S, Ramasamy R, Shah J. 2018. Rapid method for detecting and differentiating Mycobacterium tuberculosis complex and non-tuberculous mycobacteria in sputum by fluorescence in situ hybridization with DNA probes. Int J Infect Dis 75:1–7. doi: 10.1016/j.ijid.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Drancourt M, Michel-Lepage A, Boyer S, Raoult D. 2016. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev 29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridley DS, Hilson GR. 1967. A logarithmic index of bacilli in biopsies. I. Method. Int J Lepr Other Mycobact Dis 35:184–186. [PubMed] [Google Scholar]