MALDI-TOF mass spectrometry (MS) identification of pathogenic filamentous fungi is often impaired by difficulties in harvesting hyphae embedded in the medium and long extraction protocols. The ID Fungi Plate (IDFP) is a novel culture method developed to address such difficulties and improve the identification of filamentous fungi by MALDI-TOF MS. We cultured 64 strains and 11 clinical samples on IDFP, Sabouraud agar-chloramphenicol (SAB), and ChromID Candida agar (CAN2). We then compared the three media for growth, ease of harvest, amount of material picked, and MALDI-TOF identification scores after either rapid direct transfer (DT) or a long ethanol-acetonitrile (EA) extraction protocol.
KEYWORDS: filamentous fungi, MALDI-TOF MS, identification, culture plates, culture media, filamentous fungi
ABSTRACT
MALDI-TOF mass spectrometry (MS) identification of pathogenic filamentous fungi is often impaired by difficulties in harvesting hyphae embedded in the medium and long extraction protocols. The ID Fungi Plate (IDFP) is a novel culture method developed to address such difficulties and improve the identification of filamentous fungi by MALDI-TOF MS. We cultured 64 strains and 11 clinical samples on IDFP, Sabouraud agar-chloramphenicol (SAB), and ChromID Candida agar (CAN2). We then compared the three media for growth, ease of harvest, amount of material picked, and MALDI-TOF identification scores after either rapid direct transfer (DT) or a long ethanol-acetonitrile (EA) extraction protocol. Antifungal susceptibility testing and microscopic morphology after subculture on SAB and IDFP were also compared for ten molds. Growth rates and morphological aspects were similar for the three media. With IDFP, harvesting of fungal material for the extraction procedure was rapid and easy in 92.4% of cases, whereas it was tedious on SAB or CAN2 in 65.2% and 80.3% of cases, respectively. The proportion of scores above 1.7 (defined as acceptable identification) were comparable for both extraction protocols using IDFP (P = 0.256). Moreover, rates of acceptable identification after DT performed on IDFP (93.9%) were significantly higher than those obtained after EA extraction with SAB (69.7%) or CAN2 (71.2%) (P = <0.001 and P = 0.001, respectively). Morphological aspects and antifungal susceptibility testing were similar between IDFP and SAB. IDFP is a culture plate that facilitates and improves the identification of filamentous fungi, allowing accurate routine identification of molds with MALDI-TOF-MS using a rapid-extraction protocol.
INTRODUCTION
From superficial infections to life-threatening invasive infections, filamentous fungi are involved in a large range of human diseases (1). Among them, invasive mold infections have been increasingly reported due to the expanded population of at-risk patients, i.e., those with immunosuppression, diabetes, or extensive burns (2, 3). Aspergillus fumigatus is still frequently involved, but because of the more intense and frequent immunosuppressive procedures and extensive use of antifungals, the epidemiology of medically important fungi is changing, and emerging species, including Fusarium spp., Scedosporium spp., and Mucorales are increasingly isolated from invasive infections (4–7). Moreover, there have been increasing reports of a large diversity of filamentous fungi encountered in respiratory samples of patients with cystic fibrosis (8, 9). In addition to Aspergillus spp. or Scedosporium/Lomentospora complex species, several unusual species, such as Rasamsonia argillacea, have recently emerged as clinically relevant molds (10).
In this context, rapid and reliable identification at the species level is crucial in terms of epidemiology, as well as for the optimal management of fungal infections (11). Indeed, the choice of antifungal therapy is driven by the identification of the fungus involved in the infection, as the susceptibility profile of filamentous fungi depends on the species (12, 13).
However, identification of filamentous fungi in clinical laboratories still frequently relies on morphological characteristics that, although they require specialized knowledge, are still subjective and time-consuming and are not always sufficiently discriminatory to enable correct identification at the species level (14). Indeed, cryptic species of the Aspergillus section Fumigati, e.g., Aspergillus fumigatus and Aspergillus lentulus, are not morphologically distinguishable, although they display distinct antifungal susceptibility profiles (12, 15, 16). Similarly, Rasamsonia strains are often misidentified as Paecilomyces or Penicillium (17, 18).
Over the last few years, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has emerged as a powerful microorganism identification tool. This technology is accurate, rapid, and easy to use, and now sufficiently widespread to identify bacteria or yeast in clinical microbiology laboratories (29, 30). However, despite its democratization, the identification of pathogenic filamentous fungi by MALDI-TOF MS is still difficult. Indeed, molds may grow inside the solid medium and are difficult to properly scrape off. Harvesting them is tedious and the spectra are often contaminated by agar, leading to low identification scores. In addition, long extraction methods are frequently needed to obtain a clean and significant spectrum, making identification of molds by MALDI-TOF difficult and time-consuming for routine use in clinical mycology or microbiology laboratories.
ID Fungi Plate (Conidia, Quincieux, France) employs a new medium developed to facilitate the identification of filamentous fungi by MALDI–TOF MS platforms. This culture plate includes a transparent membrane deposited on the agar surface, allowing the growth of molds but impassable by fungal hyphae, thus facilitating a clean harvest. We evaluated these new plates in routine practice in a clinical mycology laboratory and compared them with two other media classically used in mycology laboratories.
MATERIALS AND METHODS
Filamentous fungi isolates and clinical samples.
Suspensions of 62 filamentous fungi from our cryopreserved bank of clinical isolates and two reference strains (Aspergillus brasiliensis ATCC 16404 and Aspergillus fumigatus IHEM 3007) were subcultured. The isolates from our clinical bank, previously identified by MALDI–TOF MS (Microflex LT instrument from Bruker Daltonics GmbH, Bremen, Germany) or based on morphological features, are presented in Table 1.
TABLE 1.
Clinical strains and species used in this study for the MALDI-TOF identification assay and comparison of morphology and antifungal susceptibility
| Species | No. of strains used for MALDI-TOF identification assay | No. of strains used for morphological comparison | No. of strains used for antifungal susceptibility comparison |
|---|---|---|---|
| Aspergillus | 28 | 4 | 8 |
| Aspergillus flavus/oryzae | 7 | 1 | |
| Aspergillus fumigatus | 10 | 2 | 4 |
| Aspergillus japonicus | 1 | 1 | |
| Aspergillus lentulus | 1 | ||
| Aspergillus nidulans | 2 | 1 | 1 |
| Aspergillus niger | 4 | 1 | 1 |
| Aspergillus terreus | 1 | ||
| Aspergillus ustus | 1 | ||
| Aspergillus versicolor | 1 | ||
| Fusarium | 9 | 1 | |
| Fusarium equisety | 1 | 1 | |
| Fusarium oxysporum | 1 | ||
| Fusarium petroliphilum | 1 | ||
| Fusarium proliferatum | 3 | ||
| Fusarium spp. | 3 | ||
| Mucorales | 4 | 1 | 1 |
| Lichtheimia spp. | 1 | ||
| Rhizomucor spp. | 1 | ||
| Rhizopus spp. | 2 | 1 | 1 |
| Penicillium | 2 | ||
| Penicillium spp. | 1 | ||
| Penicillium roqueforti | 1 | ||
| Dematiaceous | 3 | 1 | |
| Chaetomium sp. | 2 | 1 | |
| Fonsecaea nubica | 1 | ||
| Dermatophytes | 4 | 1 | |
| Microsporum canis | 1 | ||
| Nannizzia gypsea | 1 | ||
| Trichophyton mentagrophytes | 1 | 1 | |
| Trichophyton interdigitale | 1 | ||
| Others | 12 | 2 | 1 |
| Acremonium spp. | 1 | ||
| Purpureocillium lilacinum | 2 | 1 | |
| Rasamsonia argillacea | 2 | ||
| Scedosporium/Lomentospora spp. | 1 | ||
| Schizophyllum commune | 2 | ||
| Sporothrix schenckii | 1 | ||
| Trichoderma spp. | 1 | 1 | 1 |
| Unidentified | 2 |
The panel chosen was composed of fungi representative of a clinical microbiology laboratory, as well as much rarer and uncommon species. Moreover, 11 clinical samples (nine sputum and two nail samples) previously found to be positive for filamentous fungi, were selected, preserved at 4°C, and used retrospectively to inoculate the culture plates.
Fungal cultures.
Cryopreserved clinical and reference strains were subcultured on Sabouraud agar with chloramphenicol (SAB) (Becton, Dickinson GmbH, Heidelberg, Germany). A suspension of each strain was prepared in 3 ml pure water with 0.05% Tween 20. Three drops of suspension were then inoculated in parallel on ID Fungi Plates (IDFP), SAB, and ChromID Candida agar (CAN2) (bioMérieux, Marcy-l’Etoile, France). Clinical samples were used to inoculate the same media, using a dry swab for nail samples and a 10-μl inoculation loop for sputum. Plates were incubated at 27°C for one to eight days and checked daily until sufficient mycelium growth was obtained.
Culture and harvest parameters.
Growth intensity, ease of harvest, and the amount of material obtained were qualitatively evaluated by the same operator for each medium and reported on a form as follows. Growth intensity was characterized as rich, mild, or poor by a trained operator, according to a predefined abacus; harvest practicability was defined as easy when harvesting took <10 s, moderate when it took 10 to 60 s, or tedious when it took >60 s; and pellet size (i.e., the amount of material recovered from each medium) was characterized as invisible, small, or large by the same trained operator according to a predefined abacus.
MALDI-TOF MS sample preparation and MS acquisition.
MALDI-TOF MS was performed using a Microflex LT instrument (Bruker Daltonics GmbH, Bremen, Germany). Colonies were gently scraped using a sterile plastic tip and two preparation protocols were performed and analyzed in duplicate for each of the following methods: (i) the direct transfer to the target using formic acid method (DT), as previously described (19); and (ii) the ethanol/formic acid extraction (EA) (20). Briefly, the DT method consisted of directly smearing the colony onto the target, overlaid with 1 μl formic acid (70% vol/vol). The EA method consisted of first resuspending the fungal material, scraped from the plate with a calibrated 10-μl plastic disposable loop, in 300 μl pure water until the solution was cloudy (visual estimation). Sample were then mixed with 900 μl ethanol, centrifuged, and an equal volume of 70% formic acid (added to completely cover the pellet, with a maximum of 50 μl as recommended) and 100% acetonitrile was added to the pellet. One microliter of the supernatant was spotted onto the target and overlaid with 1 μl of matrix (α-cyano-4-hydroxy-cinnamic acid solution in 50% acetonitrile and 2.5% trifluoroacetic acid [Bruker Daltonics GmbH, Bremen, Germany]). After scraping the fungal colony off the plate, the DT process was approximately 2 min long for one strain versus 15 min for the EA process.
Spectra were submitted to two fungal libraries in a single run: (i) the Bruker Filamentous Fungi Library (Version 2.0) (364 MSP) and (ii) the NIH fungal library database (365 MSP) (21). A score between 0.00 and 3.00 was obtained, depending on the degree of similarity to a given spectra in one of the reference databases. The higher score of each duplicate was used. Scores were categorized as follows: score >2, highly reliable identification at the species level; score between 1.7 and 2, acceptable identification at the species level based on the recommendations of previous studies (22–24); and score <1.7, unreliable. Culture scraping failure resulted in spectra without peaks.
Morphological identification and antifungal susceptibility testing.
Macroscopic and microscopic identification and antifungal susceptibility were tested on 10 randomly chosen isolates cultured in parallel on SAB and IDFP to ensure that the entire mold diagnosis workflow was possible when subculturing on IDFP.
Voriconazole (VO), anidulafungin (ANI), amphotericin B (AB), and posaconazole (POS) MICs were evaluated using Etest strips (bioMérieux, Marcy I’Etoile, France). Itraconazole (IT) and isavuconazole (ISA) MICs were evaluated using other strips (Liofilchem, Roseto Degli Abruzzi, Italy).
Statistical analysis.
Statistical analyses were performed using XLSTAT statistical and data analysis solution v 2019.1 (Addinsoft, NY, USA, https://www.xlstat.com). Fisher’s exact test was used to compare the distribution of culture characteristics. Chi-square tests were used to compare the proportion of scores at >2 and >1.7. The Kruskal-Wallis test and multiple pairwise comparisons using Dunn’s procedure were used to compare identification scores. Student’s t test on matched samples was used to compare susceptibility results.
RESULTS
Among the 64 collection isolates, two were not identified by MALDI-TOF MS after subculture on the three media. Microscopy also failed to identify these two strains.
Among the 11 clinical samples directly inoculated onto the plates, seven (63.6%) could not be analyzed. Three were negative after culture on the three media of the study and four were positive for various fungi (yeasts and/or molds), resulting in the need for a subculture to proceed correctly. Finally, the results for 66 isolates could be correctly evaluated (60 clinical isolates from our cryopreserved bank, 2 reference strains, and 4 clinical samples).
Culture characteristics.
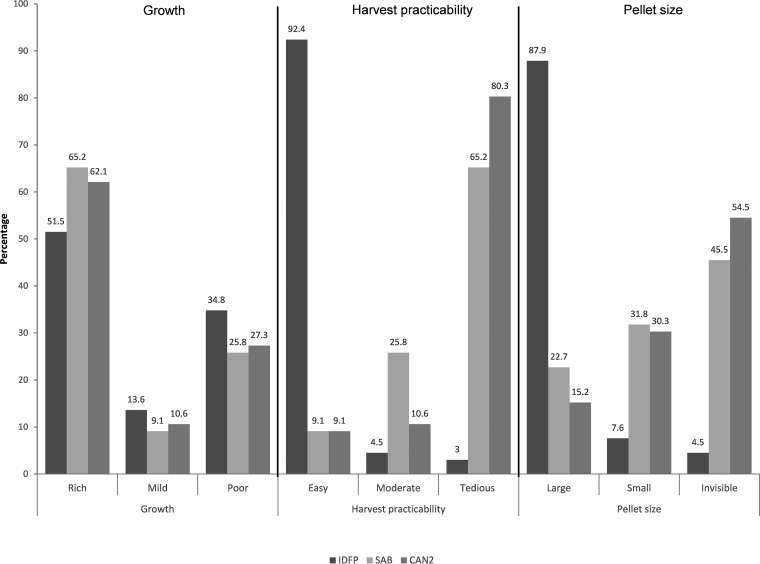
Growth intensity was not significantly different between the three plates (P = 0.592). IDFP enabled easy harvesting of 92.4% of the isolates versus 9.1% for SAB or CAN2. Moreover, the size of the pellet was large for 87.9% of the strains cultured on IDFP versus 22.7 and 15.2% for those cultured on SAB and CAN2, respectively (P < 0.0001) (Fig. 1).
FIG 1.
Distribution of cultural characteristics after culture on IDFP, SAB, or CAN2. IDFP, ID fungi plates (Conidia, Lyon, France); SAB, Sabouraud agar with chloramphenicol (Becton, Dickinson GmbH, Heidelberg, Germany); CAN2, ChromID Candida agar (bioMérieux, Marcy-l’Etoile, France).
Identification.
A comparison of the extraction protocols showed the proportion of scores >2 to be not significantly different after EA or DT following culture on IDFP or CAN2 (P = 0.256 and 0.292, respectively). For culture on SAB, more scores >2 were obtained after EA (P = 0.032). However, a comparable proportion of scores above 1.7 (i.e., acceptable identification) was obtained after EA or DT for cells cultured on the three media (P = 1.000 on IDFP and SAB, and P = 0.088 on CAN2) (Table 2).
TABLE 2.
Distribution of MALDI Biotyper scores depending on the culture plate and extraction protocola
| Score | IDFP DT |
IDFP EA |
SAB DT |
SAB EA |
CAN2 DT |
CAN2 EA |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Score ≥2 | 42 | 63.6 | 49 | 74.2 | 23 | 34.8 | 36 | 54.5 | 34 | 51.5 | 27 | 40.9 |
| Score between 1.7 and 2 | 20 | 30.3 | 14 | 21.2 | 23 | 34.8 | 10 | 15.2 | 22 | 33.3 | 20 | 30.3 |
| Unreliable score | 3 | 4.5 | 3 | 4.5 | 20 | 30.3 | 15 | 22.7 | 8 | 12.1 | 8 | 12.1 |
| Spectra without peaks | 1 | 1.5 | 0 | 0.0 | 0 | 0.0 | 5 | 7.6 | 2 | 3.0 | 11 | 16.7 |
IDFP, ID fungi plates (Conidia, Lyon, France); SAB, Sabouraud agar with chloramphenicol (Becton, Dickinson GmbH, Heidelberg, Germany); CAN2, ChromID Candida agar (bioMérieux, Marcy-l’Etoile, France); DT, direct transfer-formic acid method; EA, formic acid-acetonitrile extraction method.
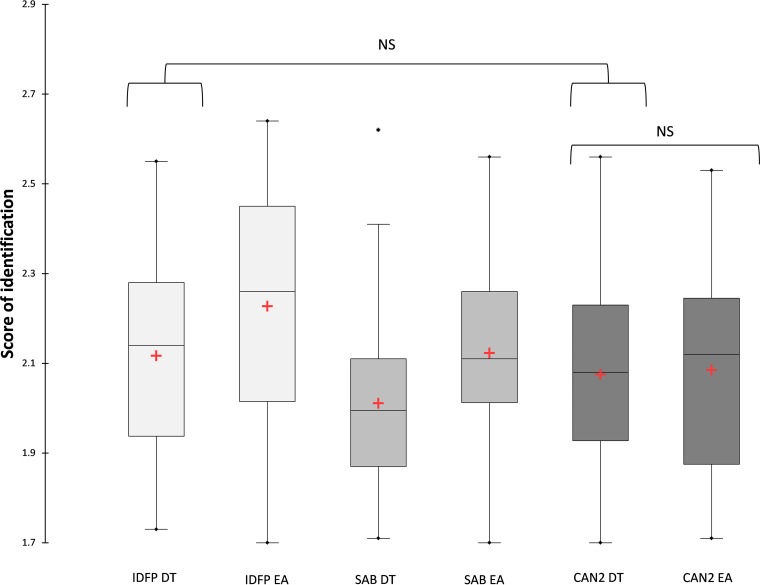
After DT, the proportion of scores >2 was significantly higher for cultures grown on IDFP than on SAB (63.7% versus 34.8%, P = 0.001). The trend was similar when we compared IDFP to CAN2, but the difference did not reach statistical significance, possibly due to the small number of samples (63.7% versus 51.5%, P = 0.214). Similarly, the proportion of scores >1.7 for cultures grown on IDFP was significantly higher than those grown on SAB (93.9% versus 69.7%, P < 0.001), and the trend was similar when comparing IDFP and CAN2, but the difference did not reach statistical significance (84.8%, P = 0.153). Considering exclusively scores >1.7, the scores were significantly higher for cultures grown on IDFP than SAB after DT (P = 0.008). Values were not significantly different between cultures grown on IDFP or CAN2 after DT (P = 0.287) (Fig. 2).
FIG 2.
Distribution of scores above 1.7 by culture plate and extraction protocol. Red plus signs correspond to means. Lines through boxes correspond to medians. Scores were significantly higher after EA than after DT on IDFP and SAB (P = 0.021 and 0.016, respectively). Scores after DT were significantly higher on IDFP than on SAB (P = 0.008). Scores after EA were significantly higher on IDFP than on SAB or CAN2 (P = 0.033 and 0.005, respectively). NS, nonsignificant differences.
After EA, the proportion of scores >2 was significantly higher for cultures grown on IDFP than SAB or CAN2 (P = 0.026 and P < 0.001, respectively). We observed the same trend for the proportion of scores >1.7 (P < 0.001 with CAN2 and SAB). The average of scores above 1.7 was significantly higher for cultures grown on IDFP than SAB (P = 0.033) or CAN2 (P = 0.005) (Fig. 2).
In addition, the proportion of acceptable identifications was significantly higher after DT for molds grown on IDFP than after EA for those grown on SAB (P < 0.001) or CAN2 (P = 0.001).
Morphological identification and susceptibility testing.
Comparative macroscopic and microscopic observation of 10 fungi cultured on either SAB or IDFP showed the duplicate cultures to have the same color, structure, and fructification on both media after the same period of culture.
We tested the MICs of 10 fungi to ANI, VO, POS, AB, ISA, and IT after culture on IDFP or SAB. The results were not significantly different depending on the culture plate (P = 0.22).
DISCUSSION
The integration of MALDI-TOF MS within routine clinical laboratories has revolutionized medical microbiology, but its use to identify filamentous fungi is still jeopardized by practical difficulties. Here, we demonstrate that the use of the ID Fungi Plate (Conidia), a culture medium designed specifically for this purpose, facilitates and improves the identification of filamentous fungi by MALDI-TOF MS.
Indeed, we found that the proportion of acceptable identifications and identification score values were higher after culture on IDFP than on SAB. The difference in terms of identification score was less obvious when IDFP was compared to CAN2, but CAN2 is not suitable for subculturing filamentous fungi, as this medium was not initially designed to culture molds and their morphological aspect can be altered on this plate. Furthermore, IDFP provided a clear-cut benefit for fungal harvesting relative to both SAB and CAN2.
Sample preparation protocols leading to a generous amount of clean material and improved databases are two key factors for performing reliable fungal identification by MALDI-TOF. Although a larger pellet is not necessary to obtain good-quality spectra and identifications, we observed that the spectra obtained with IDFP were of higher quality (not shown) when comparing spectra profiles of the different culture conditions and provided more specific peaks, which probably led to the better identification scores.
Using their homemade reference spectra library and an ethanol-acetonitrile extraction protocol, Ranque et al. demonstrated the improvement of identification of clinical strains of filamentous fungi (mostly for non-Aspergillus species) by MALDI-TOF MS over conventional methods (14). Also, Riat et al. demonstrated that identification is possible directly from solid isolation media, but they emphasized that this approach requires fungal material free of culture medium (25). However, due to the ability of molds to grow into the medium, this is particularly tedious and consequently time-consuming to achieve using the usual culture plates. This specific aspect of culturing molds triggered Bruker’s recommendations to use an overnight subculture in liquid media before identification by MALDI-TOF MS when DT and EA are unsuccessful. However, this supplemental step delays the diagnosis and is not adapted to routine and rapid mold identification. IDFP is the first plate-based medium that proposes a solution to facilitate the harvest of fungal material with the same time course as other subculture media, such as SAB plates.
Interestingly, we did not find any added value of using the long extraction (EA) versus short DT protocol with IDFP. A similar proportion of acceptable identification at the species level was obtained after DT or EA (93.9 and 95.5%, respectively). However, Zvezdanova et al. obtained significantly better results using classical culture media following mechanical lysis of the molds and subsequent protein extraction than by directly smearing the sample on the MALDI-TOF MS target (26). Moreover, a preparation protocol similar to EA or involving a bead-beating step is generally recommended by authors and providers of filamentous fungi libraries (21, 27). With IDFP, the DT protocol was sufficient to reliably identify filamentous fungi by MALDI-TOF MS, leading to significant time savings.
In this study, spectra were compared to those in two different libraries in a single experiment: the NIH library was used as a complement to the Bruker database to enhance the rate of acceptable identifications. Our aim was to evaluate the media (and not the libraries) in the setting of our routine clinical laboratory practice. Thus, we choose to compare only spectra obtained after DT or EA, which are the recommended protocols in the first line when using Bruker databases. We did not use the mechanical method recommended by the provider of the NIH library, which could have led to more “best-match” identifications from this library. It was important to verify that the new plate allowed the identification of molds based on macroscopic and microscopic features, as microscopic examination is still commonly used in medical mycology. We observed similar morphological aspects and susceptibility profiles for fungi cultured on either SAB or IDFP. These results suggest that IDFP could be used as a single subculture plate, replacing the usual SAB media, for example.
One remaining question was whether IDFP is suitable in the first line for clinical samples, but the high failure rate with clinical samples directly inoculated onto the identification media in this study did not allow us to clearly address this point. This issue has to be addressed in a study with more clinical samples prospectively inoculated on IDFP to evaluate this plate as a primary culture medium. Indeed, the use of such a medium as a first-line culture plate may have particular time savings. However, most clinical samples transmitted to clinical mycology or microbiology laboratories are negative for filamentous fungi and thus IDFP would be useless for such samples. In addition, colony isolation and subcultures are frequently needed for polymorphic cultures, which are frequent when dealing with respiratory samples of patients with cystic fibrosis (9, 28), and the use of IDFP in the first line in this context would also be of limited value. Furthermore, IDFP is almost 12 times more expensive than SAB, and five times more expensive than CAN2. Given these considerations and for maximum cost efficiency, IDFP is, in our opinion, more suitable for second-step subculture and mold reisolation, when needed, than for direct use on clinical samples.
The advantages of identification by MALDI-TOF MS are well established for yeast, bacteria, or molds, for which technical issues have been an obstacle for routine laboratory use of this technology. Here, we found that the use of the new ID Fungi Plate as a single-subculture plate both facilitates and improves the identification of filamentous fungi. This simplification and optimization may allow routine use and rapid identification of molds by MALDI-TOF MS in clinical laboratories, which is of increasing importance for appropriate patient management.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Bitar D, Lortholary O, Le Strat Y, Nicolau J, Coignard B, Tattevin P, Che D, Dromer F. 2014. Population-based analysis of invasive fungal infections, France, 2001–2010. Emerg Infect Dis 20:1149–1155. doi: 10.3201/eid2007.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaal JV, Leclerc T, Soler C, Donat N, Cirrode A, Jault P, Bargues L. 2015. Epidemiology of filamentous fungal infections in burned patients: a French retrospective study. Burns 41:853–863. doi: 10.1016/j.burns.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Slavin M, Australia and New Zealand Mycoses Interest Group, van Hal S, Sorrell TC, Lee A, Marriott DJ, Daveson K, Kennedy K, Hajkowicz K, Halliday C, Athan E, Bak N, Cheong E, Heath CH, Orla Morrissey C, Kidd S, Beresford R, Blyth C, Korman TM, Owen Robinson J, Meyer W, Chen S-A. 2015. Invasive infections due to filamentous fungi other than Aspergillus: epidemiology and determinants of mortality. Clin Microbiol Infect 21:490.e1-490–e10. doi: 10.1016/j.cmi.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 5.Shoham S. 2013. Emerging fungal infections in solid organ transplant recipients. Infect Dis Clin North Am 27:305–316. doi: 10.1016/j.idc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park BJ, Pappas PG, Wannemuehler KA, Alexander BD, Anaissie EJ, Andes DR, Baddley JW, Brown JM, Brumble LM, Freifeld AG, Hadley S, Herwaldt L, Ito JI, Kauffman CA, Lyon GM, Marr KA, Morrison VA, Papanicolaou G, Patterson TF, Perl TM, Schuster MG, Walker R, Wingard JR, Walsh TJ, Kontoyiannis DP. 2011. Invasive non-Aspergillus mold infections in transplant recipients, United States, 2001–2006. Emerg Infect Dis 17:1855–1864. doi: 10.3201/eid1710.110087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas AP, Chen S-A, Slavin MA. 2016. Emerging infections caused by non-Aspergillus filamentous fungi. Clin Microbiol Infect 22:670–680. doi: 10.1016/j.cmi.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Engel TGP, Dutch Cystic Fibrosis Fungal Collection Consortium, Slabbers L, de Jong C, Melchers WJG, Hagen F, Verweij PE, Merkus P, Meis JF. 2019. Prevalence and diversity of filamentous fungi in the airways of cystic fibrosis patients—a Dutch, multicentre study. J Cyst Fibros 18:221–226. doi: 10.1016/j.jcf.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Pihet M, Carrere J, Cimon B, Chabasse D, Delhaes L, Symoens F, Bouchara J-P. 2009. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—a review. Med Mycol 47:387–397. doi: 10.1080/13693780802609604. [DOI] [PubMed] [Google Scholar]
- 10.Giraud S, Favennec L, Bougnoux M-E, Bouchara J-P. 2013. Rasamsonia argillacea species complex: taxonomy, pathogenesis and clinical relevance. Future Microbiol 8:967–978. doi: 10.2217/fmb.13.63. [DOI] [PubMed] [Google Scholar]
- 11.Tissot F, Agrawal S, Pagano L, Petrikkos G, Groll AH, Skiada A, Lass-Flörl C, Calandra T, Viscoli C, Herbrecht R. 2017. ECIL-6 guidelines for the treatment of invasive candidiasis, aspergillosis and mucormycosis in leukemia and hematopoietic stem cell transplant patients. Haematologica 102:433–444. doi: 10.3324/haematol.2016.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2008. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based Identification. Antimicrob Agents Chemother 52:1244–1251. doi: 10.1128/AAC.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackner M, de Hoog GS, Verweij PE, Najafzadeh MJ, Curfs-Breuker I, Klaassen CH, Meis JF. 2012. Species-specific antifungal susceptibility patterns of Scedosporium and Pseudallescheria species. Antimicrob Agents Chemother 56:2635–2642. doi: 10.1128/AAC.05910-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranque S, Normand A-C, Cassagne C, Murat J-B, Bourgeois N, Dalle F, Gari‐Toussaint M, Fourquet P, Hendrickx M, Piarroux R. 2014. MALDI‐TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses 57:135–140. doi: 10.1111/myc.12115. [DOI] [PubMed] [Google Scholar]
- 15.Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot Cell 4:625–632. doi: 10.1128/EC.4.3.625-632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balajee SA, Gribskov J, Brandt M, Ito J, Fothergill A, Marr KA. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J Clin Microbiol 43:5996–5999. doi: 10.1128/JCM.43.12.5996-5999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud S, Pihet M, Razafimandimby B, Carrere J, Degand N, Mely L, Favennec L, Dannaoui E, Bouchara J-P, Calenda A. 2010. Geosmithia argillacea: an emerging pathogen in patients with cystic fibrosis. J Clin Microbiol 48:2381–2386. doi: 10.1128/JCM.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong G, White M, Lechtzin N, West NE, Avery R, Miller H, Lee R, Lovari RJ, Massire C, Blyn LB, Liang X, Sutton DA, Fu J, Wickes BL, Wiederhold NP, Zhang SX. 2017. Fatal disseminated Rasamsonia infection in cystic fibrosis post-lung transplantation. J Cyst Fibros 16:e3–e7. doi: 10.1016/j.jcf.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Theel ES, Schmitt BH, Hall L, Cunningham SA, Walchak RC, Patel R, Wengenack NL. 2012. Formic acid-based direct, on-plate testing of yeast and Corynebacterium species by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:3093–3095. doi: 10.1128/JCM.01045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl H-G. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol 47:2912–2917. doi: 10.1128/JCM.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. 2013. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:828–834. doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulthess B, Ledermann R, Mouttet F, Zbinden A, Bloemberg GV, Böttger EC, Hombach M. 2014. Use of the Bruker MALDI Biotyper for identification of molds in the clinical mycology laboratory. J Clin Microbiol 52:2797–2803. doi: 10.1128/JCM.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L'Ollivier C, Cassagne C, Normand A-C, Bouchara J-P, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. 2013. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol 51:713–720. doi: 10.3109/13693786.2013.781691. [DOI] [PubMed] [Google Scholar]
- 24.Normand A-C, Cassagne C, Gautier M, Becker P, Ranque S, Hendrickx M, Piarroux R. 2017. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol 17:25. doi: 10.1186/s12866-017-0937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riat A, Hinrikson H, Barras V, Fernandez J, Schrenzel J. 2015. Confident identification of filamentous fungi by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry without subculture-based sample preparation. Int J Infect Dis 35:43–45. doi: 10.1016/j.ijid.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Zvezdanova ME, Escribano P, Ruiz A, Martínez-Jiménez MC, Peláez T, Collazos A, Guinea J, Bouza E, Rodríguez-Sánchez B. 2019. Increased species-assignment of filamentous fungi using MALDI-TOF MS coupled with a simplified sample processing and an in-house library. Med Mycol 57:63–70. doi: 10.1093/mmy/myx154. [DOI] [PubMed] [Google Scholar]
- 27.Cassagne C, Ranque S, Normand A-C, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6:e28425. doi: 10.1371/journal.pone.0028425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paugam A, Baixench M-T, Demazes-Dufeu N, Burgel P-R, Sauter E, Kanaan R, Dusser D, Dupouy-Camet J, Hubert D. 2010. Characteristics and consequences of airway colonization by filamentous fungi in 201 adult patients with cystic fibrosis in France. Med Mycol 48:S32–S36. doi: 10.3109/13693786.2010.503665. [DOI] [PubMed] [Google Scholar]
- 29.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel R. 2015. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem 61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]