Infections due to methicillin-resistant Staphylococcus aureus (MRSA) are present worldwide and represent a major public health concern. The capability of PCR followed by high-resolution melt (HRM) curve analysis for the detection of community-associated and livestock-associated MRSA strains and the identification of staphylococcal protein A (spa) locus was evaluated in 74 MRSA samples which were isolated from the environment, humans, and pigs on a single piggery. PCR-HRM curve analysis identified four spa types among MRSA samples and differentiated MRSA strains accordingly.
KEYWORDS: high-resolution melt curve analysis, MRSA, PCR
ABSTRACT
Infections due to methicillin-resistant Staphylococcus aureus (MRSA) are present worldwide and represent a major public health concern. The capability of PCR followed by high-resolution melt (HRM) curve analysis for the detection of community-associated and livestock-associated MRSA strains and the identification of staphylococcal protein A (spa) locus was evaluated in 74 MRSA samples which were isolated from the environment, humans, and pigs on a single piggery. PCR-HRM curve analysis identified four spa types among MRSA samples and differentiated MRSA strains accordingly. A nonsubjective differentiation model was developed according to genetic confidence percentage values produced by tested samples, which did not require visual interpretation of HRM curve results. The test was carried out at different settings, and result data were reanalyzed and confirmed with DNA sequencing. PCR-HRM curve analysis proved to be a robust and reliable test for spa typing and can be used as a tool in epidemiological studies.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) has been identified in health care, community, and animal production settings worldwide (1), and in general, different MRSA strains circulate within hospitals settings and community and livestock environments (2, 3). MRSA often causes skin infections; however, it also has the potential to cause bacteremia and other systemic infections such as pneumonia (4, 5).
MRSA can be categorized into three major types based on the setting they are found in and molecular characterization. The three types are hospital-associated (HA-MRSA), community-associated (CA-MRSA), and livestock-associated (LA-MRSA). While each type is predominantly found in the area that it is named after, the distinctions are blurred and the epidemiological features of each type can vary, with each type not only present in its dominant domain (community, hospital, or livestock) but also found in others. It is reported that variations in MRSA strains are mainly due to nucleotide mutation rather than extensive recombination, and this has been used for strain typing (6).
Molecular typing helps in the understanding of the epidemiology of MRSA, aiding in the study of the molecular evolution of MRSA. Typing also helps in defining and comparing local or regional epidemics and potential sources of MRSA spread. Researchers have used different typing methods, such as phage-open-reading typing (7–9), pulsed-field gel electrophoresis (PFGE) (10–13), multilocus sequence typing (MLST) (14–16), spa typing (http://www.seqnet.org), and high-resolution melt (HRM) curve analysis (17–19). These MRSA typing techniques have been used for outbreak investigations and population-based studies (6). Most MRSA typing methods rely on the detection of nucleotide variations in DNA sequences.
There are multiple reports comparing different typing methods in order to find the most applicable typing technique in terms of reliability, rapidity, reproducibility, and portability (20–23).
The polymorphic X region of the protein A gene (spa), which is present in the DNA sequence of all Staphylococcus aureus strains, has been successfully used for typing of MRSA isolates. The first report on spa typing was published in 1994 (24), and since then, this method has been frequently used for MRSA typing and in epidemiological studies (6, 25–28).
The spa typing method is a powerful technique preferentially used in epidemiological investigations due to its high discriminatory power and high interlaboratory reproducibility. The spa typing method is a single-locus sequence-based typing method that has been reported to be useful for evaluation of micro and macro variations, therefore facilitating MRSA typing in local and global epidemiological and outbreak investigations, and has been widely used for subtyping of Staphylococcus aureus in health care, community, and livestock settings (29).
HRM curve analysis is a post-PCR method used to detect nucleotide sequence variations without requiring DNA sequencing and has been used for differentiation of bacterial species and subspecies (30–32). The spa typing method based on PCR-HRM has also been reported to be rapid and cost-effective (17, 19, 33, 34). CA-MRSA contains different strains and, subsequently, several spa types. Rapid molecular characterization and spa typing of MRSA isolates could contribute to the control and prevention of bacterial spread in the area and communities.
The aim of this study was to evaluate the discriminatory power of PCR-HRM in nonsubjective differentiation of clonal structure and spa types on a diverse collection of MRSA isolates.
MATERIALS AND METHODS
Approval for the sample collection from pigs was granted by the Charles Sturt University Animal Care and Ethics Committee (protocol number 14/096), and all experiments were performed in accordance with the relevant guidelines and regulations. Approval for the recruitment of human participants into this study was obtained from the Charles Sturt University Human Research Ethics Committee (protocol number 2015/016). Informed consent was obtained from each participant prior to the experiments, and all methods were performed in accordance with the relevant guidelines and regulations.
Staphylococcus aureus field isolates.
Samples were collected from a commercial pig farm in Australia that experienced recurrent MRSA infections in piggery workers over a 3-year period, as described before (20). There were no clinical signs associated with infection in the workers or pigs. A total of 74 MRSA isolates consisting of isolates from the environment (n = 7), piggery workers (n = 33), and pigs (n = 34) and three methicillin-susceptible Staphylococcus aureus (MSSA) isolates from dairy cows were tested in this study (Table 1).
TABLE 1.
Sample ID, MLST, and SCCmec typing, mean curve peak melting points ± SD, and HRM genotypes of tested samplesa
| Sample IDb | MLST | SCCmec | No. of times tested | Mean of the peak ± SD (°C) | Mean GCP when H029 (ST93) was used as reference genotype | Genotypes when H029 (ST93) was used as reference genotype with a cutoff value of ≥9 | Mean GCP when P030 (ST398) was used as reference genotype | Genotypes when P030 (ST398) was used as reference genotype with a cutoff value of ≥10 |
|---|---|---|---|---|---|---|---|---|
| E001 | ST93 | IV | 15 | 84.9 ± 0.5 | 70.2 ± 11.4 | ST93 | 0.0 ± 0.0 | Variation |
| E002 | ST93 | IV | 14 | 84.8 ± 0.5 | 77.6 ± 8.1 | ST93 | 0.0 ± 0.0 | Variation |
| E004 | ST398 | V | 15 | 82.9 ± 0.7 | 0.0 ± 0.0 | Variation | 51.5 ± 7.6 | ST398 |
| E008 | ST93 | IV | 15 | 84.7 ± 0.6 | 99.8 ± 0.1 | ST93 | 0.0 ± 0.0 | Variation |
| E017 | ST93 | IV | 16 | 84.8 ± 0.6 | 80.5 ± 9.0 | ST93 | 0.0 ± 0.0 | Variation |
| E021 | ST93 | IV | 12 | 84.7 ± 0.5 | 93.5 ± 2.6 | ST93 | 0.0 ± 0.0 | Variation |
| E026 | ST398 | V | 15 | 83.0 ± 0.6 | 0.0 ± 0.0 | Variation | 99.7 ± 6.9 | ST398 |
| H001 | ST93 | IV | 15 | 84.9 ± 0.6 | 71.5 ± 1.3 | ST93 | 0.0 ± 0.0 | Variation |
| H002 | ST93 | IV | 11 | 84.7 ± 0.5 | 94.2 ± 1.8 | ST93 | 0.0 ± 0.0 | Variation |
| H003 | ST93 | IV | 16 | 84.6 ± 0.5 | 86.5 ± 6.4 | ST93 | 0.0 ± 0.0 | Variation |
| H004 | ST93 | IV | 13 | 84.7 ± 0.5 | 89.4 ± 4.6 | ST93 | 0.0 ± 0.0 | Variation |
| H005 | ST93 | IV | 15 | 84.6 ± 0.6 | 82.8 ± 0.9 | ST93 | 0.0 ± 0.0 | Variation |
| H006 | ST93 | IV | 14 | 84.3 ± 0.5 | 40.8 ± 8.3 | ST93 | 0.0 ± 0.0 | Variation |
| H007 | ST93 | IV | 16 | 84.6 ± 0.4 | 87.3 ± 7.7 | ST93 | 0.0 ± 0.0 | Variation |
| H008 | ST93 | IV | 12 | 84.6 ± 0.5 | 91.8 ± 4.2 | ST93 | 0.0 ± 0.0 | Variation |
| H009 | ST93 | IV | 10 | 84.3 ± 0.5 | 34.9 ± 0.4 | ST93 | 0.0 ± 0.0 | Variation |
| H010 | ST398 | V | 14 | 82.7 ± 0.6 | 0.0 ± 0.0 | Variation | 14.6 ± 1.5 | ST398 |
| H011 | ST93 | IV | 12 | 84.6 ± 0.5 | 89.9 ± 1.9 | ST93 | 0.0 ± 0.0 | Variation |
| H012 | ST93 | IV | 13 | 84.7 ± 0.5 | 70.6 ± 4.8 | ST93 | 0.0 ± 0.0 | Variation |
| H013 | ST93 | IV | 15 | 84.8 ± 0.4 | 58.6 ± 4.7 | ST93 | 0.0 ± 0.0 | Variation |
| H014 | ST93 | IV | 16 | 84.8 ± 0.4 | 56.5 ± 6.3 | ST93 | 0.0 ± 0.0 | Variation |
| H015 | ST93 | IV | 12 | 84.7 ± 0.4 | 66.5 ± 3.3 | ST93 | 0.0 ± 0.0 | Variation |
| H016 | ST93 | IV | 11 | 84.7 ± 0.5 | 59.1 ± 8.1 | ST93 | 0.0 ± 0.0 | Variation |
| H017 | ST93 | IV | 10 | 84.7 ± 0.5 | 68.1 ± 3.7 | ST93 | 0.0 ± 0.0 | Variation |
| H018 | ST93 | IV | 13 | 84.7 ± 0.4 | 41.8 ± 5.8 | ST93 | 0.0 ± 0.0 | Variation |
| H019 | ST93 | IV | 17 | 84.9 ± 0.5 | 51.8 ± 3.3 | ST93 | 0.0 ± 0.0 | Variation |
| H020 | ST398 | V | 12 | 83.3 ± 0.9 | 0.0 ± 0.0 | Variation | 79.9 ± 9.8 | ST398 |
| H021 | ST398 | V | 11 | 83.0 ± 0.5 | 0.0 ± 0.0 | Variation | 66.3 ± 8.6 | ST398 |
| H022 | ST93 | IV | 15 | 84.7 ± 0.5 | 75.4 ± 5.4 | ST93 | 0.0 ± 0.0 | Variation |
| H023 | ST398 | V | 14 | 83.0 ± 0.5 | 0.0 ± 0.0 | Variation | 77.6 ± 1.4 | ST398 |
| H024 | ST93 | IV | 10 | 84.7 ± 0.3 | 48.5 ± 10.5 | ST93 | 0.0 ± 0.0 | Variation |
| H025 | ST93 | IV | 11 | 84.5 ± 0.5 | 32.9 ± 5.7 | ST93 | 0.0 ± 0.0 | Variation |
| H026 | ST93 | IV | 16 | 84.6 ± 0.5 | 57.4 ± 10.9 | ST93 | 0.0 ± 0.0 | Variation |
| H027 | ST93 | IV | 14 | 84.9 ± 0.4 | 27.6 ± 6.2 | ST93 | 0.0 ± 0.0 | Variation |
| H028 | ST93 | IV | 12 | 85.0 ± 0.3 | 19.2 ± 1.5 | ST93 | 0.0 ± 0.0 | Variation |
| H029 | ST93 | IV | 19 | 84.5 ± 0.5 | 99.7 ± 0.1 | ST93 | 0.0 ± 0.0 | Variation |
| H030 | ST93 | IV | 12 | 84.5 ± 0.6 | 54.6 ± 9.7 | ST93 | 0.0 ± 0.0 | Variation |
| H031 | ST93 | IV | 12 | 84.5 ± 0.5 | 42.6 ± 0.3 | ST93 | 0.0 ± 0.0 | Variation |
| H032 | ST398 | V | 15 | 82.8 ± 0.5 | 0.0 ± 0.0 | Variation | 14.6 ± 7.3 | ST398 |
| H033 | ST398 | V | 14 | 83.0 ± 0.5 | 0.0 ± 0.0 | Variation | 98.4 ± 0.2 | ST398 |
| P002 | ST398 | V | 14 | 82.6 ± 0.5 | 0.0 ± 0.0 | Variation | 61.7 ± 0.7 | ST398 |
| P004 | ST93 | IV | 14 | 83.6 ± 1.1 | 34.9 ± 6.7 | ST93 | 2.7 ± 0.3 | Variation |
| P009 | ST93 | IV | 16 | 84.8 ± 0.5 | 34.7 ± 3.8 | ST93 | 0.0 ± 0.0 | Variation |
| P015 | ST93 | IV | 13 | 84.8 ± 0.6 | 28.9 ± 7.1 | ST93 | 0.0 ± 0.0 | Variation |
| P022 | ST398 | V | 13 | 82.7 ± 0.5 | 0.0 ± 0.0 | Variation | 13.3 ± 3.2 | ST398 |
| P028 | ST93 | IV | 12 | 84.6 ± 0.5 | 51.8 ± 7.7 | ST93 | 0.0 ± 0.0 | Variation |
| P030 | ST398 | V | 20 | 83.0 ± 0.5 | 0.0 ± 0.0 | Variation | 99.9 ± 0.0 | ST398 |
| P037 | ST93 | IV | 12 | 84.6 ± 0.6 | 33.7 ± 3.9 | ST93 | 0.0 ± 0.0 | Variation |
| P042 | ST93 | V | 15 | 84.8 ± 0.6 | 12.1 ± 0.2 | ST93 | 0.0 ± 0.0 | Variation |
| P061 | ST93 | IV | 14 | 84.8 ± 0.4 | 14.8 ± 0.4 | ST93 | 0.0 ± 0.0 | Variation |
| P071 | ST30 | V | 16 | 83.9 ± 0.6 | 8.4 ± 1.5 | Variation | 0.0 ± 0.0 | Variation |
| P082 | ST398 | V | 10 | 83.2 ± 0.8 | 0.0 ± 0.0 | Variation | 55.1 ± 0.4 | ST398 |
| P087 | ST398 | V | 11 | 83.1 ± 0.7 | 0.0 ± 0.0 | Variation | 88.1 ± 0.4 | ST398 |
| P115 | ST398 | V | 12 | 83.1 ± 0.6 | 0.0 ± 0.0 | Variation | 98.4 ± 1.2 | ST398 |
| P116 | ST93 | IV | 13 | 84.7 ± 0.4 | 9.7 ± 3.6 | ST93 | 0.0 ± 0.0 | Variation |
| P129 | ST398 | V | 14 | 83.1 ± 0.7 | 0.0 ± 0.0 | Variation | 88.5 ± 0.5 | ST398 |
| P141 | ST93 | IV | 11 | 84.6 ± 0.4 | 10.2 ± 1.2 | ST93 | 0.0 ± 0.0 | Variation |
| P151 | ST93 | IV | 12 | 84.2 ± 0.4 | 53.1 ± 2.2 | ST93 | 0.0 ± 0.0 | Variation |
| P156 | ST93 | IV | 16 | 84.6 ± 0.4 | 14.1 ± 3.5 | ST93 | 0.0 ± 0.0 | Variation |
| P183 | ST93 | IV | 12 | 84.6 ± 0.4 | 14.7 ± 0.5 | ST93 | 0.0 ± 0.0 | Variation |
| P186 | ST398 | V | 15 | 83.4 ± 0.4 | 2.9 ± 0.5 | Variation | 94.4 ± 0.3 | ST398 |
| P216 | ST398 | V | 14 | 83.5 ± 0.4 | 5.7 ± 0.8 | Variation | 84.5 ± 3.3 | ST398 |
| P221 | ST398 | V | 12 | 83.3 ± 0.3 | 3.8 ± 0.9 | Variation | 94.2 ± 3.9 | ST398 |
| P223 | ST398 | V | 14 | 83.4 ± 0.4 | 4.1 ± 0.2 | Variation | 92.9 ± 1.4 | ST398 |
| P225 | ST93 | IV | 15 | 85.1 ± 0.4 | 95.8 ± 0.5 | ST93 | 2.8 ± 0.4 | Variation |
| P236 | ST398 | V | 15 | 83.2 ± 0.3 | 3.0 ± 0.3 | Variation | 96.3 ± 0.9 | ST398 |
| P244 | ST398 | V | 11 | 83.2 ± 0.3 | 3.2 ± 1.1 | Variation | 96.2 ± 3.8 | ST398 |
| P257 | ST398 | V | 12 | 83.2 ± 0.3 | 3.1 ± 0.7 | Variation | 97.3 ± 2.1 | ST398 |
| P269 | ST93 | IV | 14 | 85.1 ± 0.4 | 99.7 ± 0.4 | ST93 | 2.6 ± 0.4 | Variation |
| P270 | ST93 | IV | 16 | 85.1 ± 0.4 | 99.4 ± 0.6 | ST93 | 2.8 ± 0.4 | Variation |
| P271 | ST93 | IV | 12 | 84.9 ± 0.3 | 99.6 ± 0.4 | St93 | 2.1 ± 0.5 | Variation |
| P306 | ST93 | IV | 14 | 85.0 ± 0.3 | 99.9 ± 0.1 | ST93 | 2.3 ± 0.1 | Variation |
| P325 | ST93 | IV | 12 | 84.9 ± 0.4 | 99.8 ± 0.2 | ST93 | 2.2 ± 0.1 | Variation |
| P330 | ST398 | V | 15 | 83.3 ± 0.3 | 2.3 ± 0.1 | Variation | 99.7 ± 1.3 | ST398 |
| 20-L | NA | NA | 18 | 81.6 ± 0.6 | 0.0 ± 0.0 | Variation | 0.0 ± 0.0 | Variation |
| 117-L | NA | NA | 18 | 81.5 ± 0.6 | 0.0 ± 0.0 | Variation | 0.0 ± 0.0 | Variation |
NA, not applicable.
ID, identification. E, H, and P indicate collected samples from environment, human and pig, respectively.
DNA isolation.
Total genomic DNA was extracted from all samples using the Wizard Genomic DNA purification kit (Promega, Australia) according to the manufacturer’s instructions. Each DNA was quantified using NanoDrop 2000 (Thermo Scientific, Australia), and the DNA concentration was adjusted to 5 ng/μl and used in the PCR immediately or stored at –20°C for future use.
PCR.
PCR amplifications were performed in 25-μl reaction volume on a Rotor-Gene 6000 thermal cycler (Qiagen, Australia). For each reaction, the assay contained 2 μl extracted genomic DNA, 25 μM of each primer, 1.5 mM Mgcl2, 1,250 μm of each dNTP, 5 μM SYTO 9 green fluorescent nucleic acid stain (Invitrogen, Australia), 5× GoTaq Green Flexi reaction buffer, and 1 U GoTaq DNA polymerase (Promega, Australia). PCR conditions consisted of one cycle of 95°C for 4 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final cycle of 72°C for 2 min. The primers used to amplify and sequence the polymorphic X region of protein A were Spa1113F (5′-TAAAGACGATCCTTCGGTGAGC-3′) and sps-1514R (5′-TAAAGACGATCCTTCGGTGAGC-3′), which were published previously (35) with slight modification as mentioned in the Ridom SpaServer database (http://www.ridom.de/staphtype/spa_sequencing.shtml). The spa types were confirmed by sequencing and through the Ridom SpaServer database (http://www.spaserver.ridom.de).
Differentiation of clonal structures using HRM curve analysis.
On completion of each PCR, HRM curve analysis was continued on the same machine (Rotor-Gene 6000 thermal cycler; Qiagen, Australia). In order to determine the optimal melting conditions for differentiation of Staphylococcus aureus isolates, the PCR products were subjected to 0.5°C/s ramping between 75°C and 95°C. All samples were tested in triplicate, their melt profiles were analyzed using Rotor Gene 1.7.27 software, and the HRM algorithm was provided. Normalization regions of 77 to 78°C and 85 to 86°C were applied for analysis. Samples H029 and P030 were selected as representative of sequence type 93 (ST93) and ST398 clones, respectively, and set as “genotype,” and the average genotype confidence percentage (GCP; the value related to each isolate as being compared to the genotype, with a value of 100% indicating an exact match) for the replicates was predicted by the software.
Sample H029 contained additional virulence factors, such as Panton-Valentine leucocidin (PVL), and human immune evasion genes (IEG), such as staphylokinase (SAK), staphylococcal complement inhibitor (SCIN), and chemotaxis inhibitory protein (CHIP), which are typical characteristics of a CA- MRSA such as ST93. Sample P030 was a typical LA-MRSA ST398 and was resistant to non-beta-lactam antimicrobial classes regularly used in pigs, such as macrolides, lincosamides, and tetracycline. In addition, this isolate was lacking human evasion genes. Therefore, these two isolates were selected as reference genotypes, and all other isolates were compared with these two selected genotypes.
The GCPs for all ST93 and ST398 clones were averaged individually, and the standard deviation (SD) was calculated and used to establish the GCP range for each type cutoff point. Each cutoff point was applied in HRM analysis to evaluate the differentiation power of the test to genotype field isolates.
The discrimination power or sensitivity of the test for MRSA type differentiation was also determined using receiver operating characteristic (ROC) analysis at the calculated cutoff points for the assay.
Results from HRM typing were compared with results from MLST and staphylococcal cassette chromosome mec (SCCmec) typing (20), which includes the major element of methicillin resistance.
Sequencing and nucleotide sequence analysis of amplicons.
The PCR amplicons of selected samples were purified using the Wizard SV gel and PCR clean-up system (catalog no. A9281; Promega, Australia) following the manufacturer’s instructions. Purified amplicons were subjected to automated sequencing (Australian Genome Research Facility Ltd., Brisbane, Australia) in both directions, using the same primers used for PCR. The nucleotide sequences were analyzed using CLUSTAL W (36) and BioEdit Sequence Alignment Editor version 6.0.9.0. Multiple sequence alignment was generated using a gap open penalty of 10 and a gap extension penalty of 1. The phylogenetic tree was generated using the maximum likelihood (ML) method.
Confirmation of HRM curve profiles.
The melt curve patterns produced with PCR-HRM were validated using a Web-based melt curve prediction method (uMelt; Wittwer Laboratory, University of Utah; https://www.dna-utah.org/umelt/umelt.html). The unified thermodynamic parameters were used for prediction of melt curves using individual sequences of different clones (ST93, ST398, and ST30) of MRSA isolates (37).
Differentiation of spa types using HRM curve analysis.
To identify the spa types of Staphylococcus aureus isolates, open source software (DNAGear; http://w3.ualg.pt/~hshah/DNAGear/) was used (38). DNA sequences of amplicons were submitted, and the spa types were identified based on spa type nomenclature used by Ridom at Ridom SpaServer (http://spaserver.ridom.de/).
The capability of PCR-HRM in the detection and differentiation of spa types was investigated using isolates H029 (spa type t202) and P030 (spa type t034) as reference genotypes, and all samples were tested.
To further assess the capability of the PCR-HRM in the differentiation of spa types among tested samples, four samples, representative of each spa type (E001 as t202, E004 as t011, E026 as t034, and P071 as t318), were used individually as a reference genotype, HRM data were reanalyzed, and the rest of the isolates were retyped using a cutoff value of 10 to 100. Three MSSA strains/isolates were also tested.
Finally, randomly selected samples from all seven spa types (t011, t318, t034, t202, t6367, t529, and t021) were used individually and all together as reference genotypes in one run, and all HRM curve data were reanalyzed to evaluate the consistency of the results.
RESULTS
Discrimination of ST93 and ST398 clones.
Following PCR, amplicons were subjected to HRM curve analysis and were visually assessed. Results showed distinct conventional and normalized melt curves for the three MLST types.
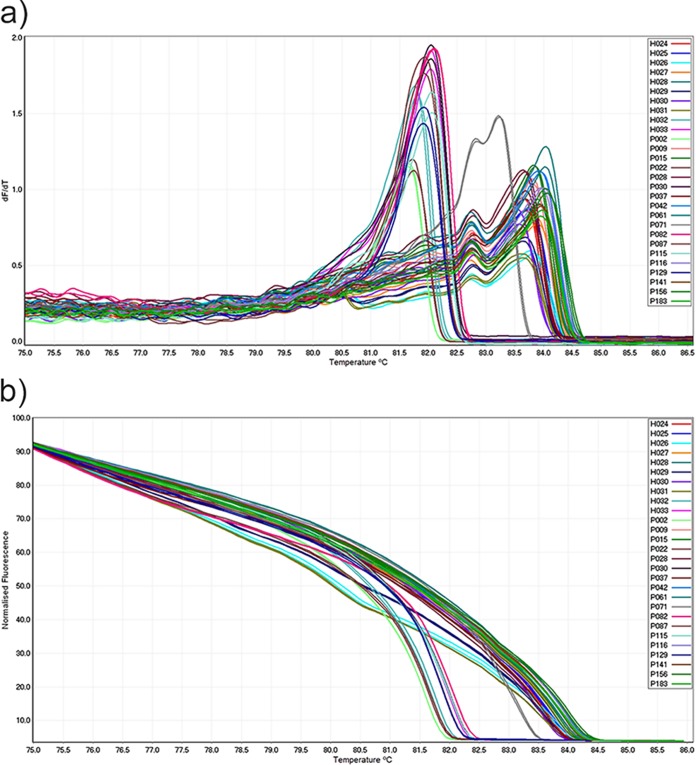
All CA-MRSA ST93 clones produced a melt curve with a peak of 82.7°C to 84.6°C (mean, 83.8 ± 0.4°C) and a shoulder peak at lower temperatures, while all LA-MRSA ST398 clones generated only one peak of 81.6°C to 83.1°C (mean, 82.3 ± 0.5°C). All curves produced with ST93 were distinct from curves produced with ST398. Sample P071 produced two peaks, at 82.8 ± 0.03°C and 83.2 ± 0.03°C, which were unique and different from ST93 and ST398 (Fig. 1). This sample (P071) was previously typed as ST30 using the MLST method (20).
FIG 1.
Conventional and normalized melt curve analysis of MRSA isolates. (a) Conventional and (b) normalized melt curve analysis of PCR amplicons from CA-MRSA (ST93) and LA-MRSA (ST398) isolates. All LA-MRSA isolates produced a single peak (mean 82.3°C), while CA-MRSA isolates produced one peak at higher temperature (mean 83.8°C) and a shoulder peak at lower temperature. The P071 (ST30) isolate was distinct from the ST93 and ST398 isolates.
MSSA samples produced curves distinct from the curves of MRSA samples. The two MSSA samples (20-L and 117-L) produced similar curves with a mean melting point at 81.5 ± 0.6°C, which was lower than the mean melting points of the ST93 and ST398 strains (Fig. 2).
FIG 2.
Differentiation of MRSA and MSSA isolates using conventional and normalized melt curve analysis. (a) Conventional and (b) normalized HRM curve analysis of PCR amplicons for CA-MRSA (ST93) (red), LA-MRSA (ST398) (blue), and MSSA (green) isolates.
High-resolution melt curve analysis showed that there were some differences between the ST93 and ST398 HRM curve patterns. Compared to ST398, ST93 melt curves were relatively congested and difficult for visual differentiation of intraspecies isolates, while the curve profiles of isolates from the ST398 clone showed more distinctive differences between the isolates based on their melting point temperatures or normalized curves (Fig. 1). Isolates from ST398 consisted of t011 and t034 spa types, while all samples identified as ST93 were typed as t202.
Nonsubjective differentiation of ST93 and ST398 strains based on genotype confidence percentage values.
The genotype confidence percentage (GCP) values obtained from all ST93 and ST398 samples from repeated runs of PCR-HRM were averaged separately, and two cutoff points were calculated to be used as a mathematical model to analyze the relationship of the isolates and to differentiate between the two MRSA clones without visual assessment of the HRM curves by the operator (nonobjective). The mean GCP for all ST93 isolates was 64.6 with a mean standard deviation (SD) of 27.8. To calculate the cutoff point value for ST93 isolates, the 2SD value (2 × 27.8 = 55.6) was subtracted from the mean GCP (64.6 − 55.6 = 9), and a cutoff value of 9 was calculated. Therefore, the GCP range for the ST93 isolates was determined to be 9 to 100. This cutoff range was used for genotyping of all isolates.
When one of the ST93 isolates, such as H029, was used as a reference genotype and a cutoff value (9 to 100) was applied, all ST93 isolates produced a GCP of 10.2 to 99.9 and, therefore, were genotyped as ST93, and all ST398 isolates produced a GCP of 0.0 to 5.7 and automatically were genotyped as “variation.” Isolate P071 (ST30) produced a GCP of 8.4 and was also genotyped as variation (Table 1).
Similarly, the mean GCP ± SD for all ST398 isolates was found to be 74.8 ± 32.5, and a cutoff value of GCP subtracted by 2SD was calculated (10 to 100). When one of the ST398 isolates (P030) was set as a reference genotype and the cutoff point 10 to 100 was applied, all ST398 isolates produced a GCP of 13.3 to 99.9 and were genotyped as ST398, while all ST93 isolates and P071 (ST30) produced a GCP of ≤2.8 and were automatically genotyped as variation by the software (Table 1).
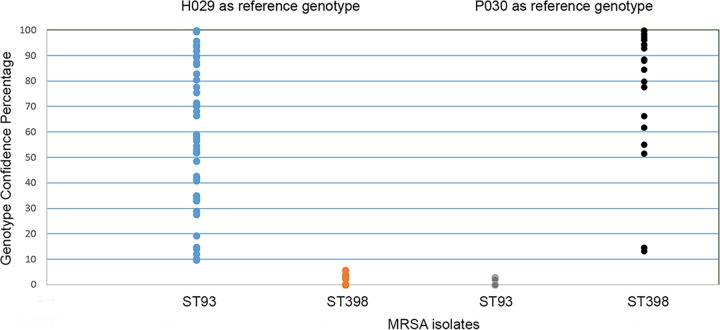
In PCR-HRM curve analysis, when isolate H029 (ST93) was set as a genotype and a cutoff value of 9 to 100 was applied, all ST93 isolates were correctly genotyped as ST93, and the rest of samples were genotyped as variation. The lowest GCP of the ST93 isolates was 10.2, and the highest GCP for ST398 and ST30 isolates was 5.7; therefore, the gap between these GCPs was as low as 5.0. However, when ST393 isolate P030 was used as a reference genotype with a cutoff value of 10 to 100, all ST398 isolates were genotyped as ST398, and all other samples (ST93 and ST30) were genotyped as variation. The lowest GCP of ST398 and the highest GCP of other genotypes were 13.3 and 2.8, respectively. The GCP gap between these two genotypes was 10.5. To show the differentiation power of PCR-HRM in discriminating between ST93 and ST398 clones, the mean GCP value of each MRSA isolate was plotted in a dot plot, which shows slightly better discrimination when an ST398 isolate (P030) is used as a reference genotype (Fig. 3). The PCR-HRM had a higher discrimination power in differentiation between MRSA and MSSA isolates. All MSSA isolates had a GCP of 99.7 to 46.8 and were genotyped as MSSA, all MRSA isolates had a GCP of 0.0 to 0.36, and the gap between the highest MRSA and lowest MSSA isolates was approximately 46 GCP (Fig. 4).
FIG 3.
Comparison of the distribution of GCPs from CA-MRSA (ST93) and LA-MRSA (ST398) isolates by individual value plot when H029 and P030, respectively, were used as reference genotypes.
FIG 4.

Comparison of the distribution of GCPs from MRSA and MSSA isolates by individual value plot when the MSSA isolate (20-L) was used as the reference genotype.
Identification of spa types using HRM curve analysis and DNA sequencing.
The DNA sequences of isolates were submitted to GenBank. All isolates were correctly typed when H029 and P030 were used as the reference genotype at the calculated cutoff values in HRM curve analysis (Table 2).
TABLE 2.
spa types and repeats of S. aureus isolates using sequencing (Ridom) and HRM curve analysis
| Sample ID | spa types (Ridom) | spa repeats | spa types (HRM) | MRSA/MSSA | GenBank accession no. |
|---|---|---|---|---|---|
| E001 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431211 |
| E002 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431212 |
| E004 | [t011] | 08-16-02-25-34-24-25 | MRSA | MG431236 | |
| E008 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431213 |
| E021 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431214 |
| E026 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431237 |
| H005 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431215 |
| H006 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431216 |
| H007 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431217 |
| H008 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431218 |
| H009 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431219 |
| H010 | [t011] | 08-16-02-25-34-24-25 | t011 | MRSA | MG431238 |
| H018 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431220 |
| H020 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431239 |
| H028 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431221 |
| H029 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431222 |
| H030 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431223 |
| H031 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431224 |
| H032 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431240 |
| H033 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431241 |
| P002 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431242 |
| P004 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431225 |
| P009 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431226 |
| P028 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431227 |
| P030 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431243 |
| P061 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431228 |
| P071 | [t318] | 15-12-16-16-02-16-02-25-17-24 | t318 | MRSA | MG431246 |
| P151 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431229 |
| P186 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431244 |
| P216 | [t034] | 08-16-02-25-02-25-34-24-25 | t034 | MRSA | MG431245 |
| P225 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431230 |
| P269 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431231 |
| P270 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431232 |
| P271 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431233 |
| P306 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431234 |
| P325 | [t202] | 11-17-23-17-17-16-16-25 | t202 | MRSA | MG431235 |
| ATCC 25923 | [t021] | 15-12-16-02-16-02-25-17-24 | t021 | MSSA | MG431247 |
| 20-L | [t6367] | 07-23-12-21-17 | t6367 | MSSA | MG431248 |
| 117-L | [t529] | 04-34 | t529 | MSSA | MG431249 |
For genotypic comparisons, when one of each spa type (E001 as t202, E004 as t011, E026 as t034, and P071 as t318) was used individually as a reference genotype with a cutoff value of 10 to 100 and HRM data were reanalyzed, the rest of the samples were genotyped correctly (data not shown).
Finally, when all spa types were assigned as reference genotypes in one run, all samples were typed accordingly. In all three above-mentioned different settings of HRM, results were in agreement with spa types using DNA sequencing (Ridom) (Table 2).
In the phylogenetic analysis, all similar spa types were grouped together (Fig. 5). The MRSA and MSSA isolates (20-L and 117-L) were placed in two different clades. The t202 and t034 spa types were genetically closer to each other than to the t011 spa type. P071 (t318) was different from the rest of the MRSA spa types and was positioned in a different clade, genetically closer to the t202 spa type.
FIG 5.
Phylogenetic relationship of spa types and MRSA/MSSA isolates based on the nucleotide sequence of the spa gene.
DISCUSSION
Staphylococcus aureus is an important cause of bacterial infections in animals and humans. Control of infections caused by MRSA isolates of Staphylococcus aureus is particularly challenging due to limited treatment options. Isolation of MRSA from pigs and workers in a piggery shows that the bacteria are colonized on the skin or nose without producing active infection or clinical signs. This illustrates that MRSA can be considered part of the body microbial flora. However, colonization of MRSA in different parts of the body may increase the risk of infection. Underlying diseases, extensive antibiotic use, trauma, and wounds can facilitate the introduction of colonized bacteria into the body and lead to MRSA infection (39). To study the spread of community-, health care-, and livestock-associated MRSA isolates, different molecular techniques have been used. In this study, 74 field isolates were differentiated and typed based on HRM curve analysis. Results from HRM were in agreement with MLST and staphylococcal cassette chromosome mec (SCCmec).
Slight changes in the melting points of HRM curves were observed in different runs of PCR-HRM and on different days; however, the shape of conventional melt curves and normalized graphs were unchanged. The quality and quantity of genomic DNA are the two important factors in PCR-HRM curve analysis (31, 40–42). Therefore, to achieve comparable and reproducible HRM curves, the DNA concentrations of samples were adjusted.
The capacity of HRM curve analysis to identify spa genotypes was further investigated using each of the spa types as a reference genotype individually or all together in one run. This consisted of five reevaluations of the test, and results from these analyses were similar.
The sensitivity and specificity of the PCR-HRM in the differentiation of MRSA clones and spa typing was 100% based on ROC analysis at the calculated cutoff points. The sensitivity of PCR-HRM in the detection and differentiation of DNA sequence variations among samples varied based on factors such as the quality and quantity of DNA, PCR thermal conditions, the type of fluorescence dye, and HRM ramping. For example, HRM is sensitive to the salt concentration in the reaction mixture, and variable DNA extraction methods can affect the melting profiles. In HRM curve analysis, removing negative samples and those that did not amplify can positively affect the melting profile of other samples (43).
The resolution of HRM curves in the differentiation of samples with a small number of nucleotide changes within the target gene was increased by reducing of the ramp time. This will result in higher resolution on differences in HRM profiles. In this study, a ramping of 0.5°C was found to be optimal to generate distinctive curves and to discriminate samples.
PCR-HRM has been used for MRSA spa typing; however, these studies were based on visual differentiation of spa within either ST93 (19) or ST398 (18) of MRSA clones. In the current study, MRSA clones ST93, ST398, and ST30 were tested, differentiated, and correctly identified. In addition, unlike previous studies, all 74 samples were typed based on spa gene variations using a nonsubjective differentiation model according to GCP values, which did not require visual interpretation of HRM curve results. Testing isolates from different sources, hosts, or geographical regions with different spa types may require calculating a new cutoff value. Hence, it is recommended to determine the cutoff point based on generated GCP and SD values as described above.
The PCR-HRM successfully differentiated ST93 and ST398 isolates within each clone; however, the discrimination power of PCR-HRM in differentiating MSSA and MRSA isolates proved to be higher than differentiation of ST93 and ST398 isolates. This was in agreement with results obtained from phylogenetic analysis. The two MSSA isolates were included as controls to evaluate their curve profiles. The two MSSA isolates showed curve profiles distinct from those of MRSA isolates, with high discrimination power. However, to be able to use the assay for differentiation of MRSA and MSSA isolates, more MSSA samples need to be tested.
All isolates typed as ST93 were identified as one spa type (t202). The low variability of the spa types among ST93 could be due to the close genetic relatedness of isolates in the pig farm. It is anticipated that more spa types could be detected if samples were collected from different geographical locations or sources.
In conclusion, spa typing using PCR-HRM curve analysis is superior to the other typing methods, such as MLST and PFGE, due to the flexibility of the assay, automatic and easy interpretation of results, reproducibility, speed, and low cost. This method can also be used to differentiate ST93 and ST398, as well as MRSA and MSSA isolates at the same time.
ACKNOWLEDGMENTS
We thank the Australian pig producers who participated in this study by providing samples.
This work was supported by the Graham Centre for Agricultural Innovation (grant no. 40825) and the School of Animal and Veterinary Sciences, Charles Sturt University (grant no. 40702).
S.A.G. designed the study and performed the DNA sequencing and analysis. S.S. and Q.Z. performed the experiment. S.A.G. wrote the manuscript, and J.H. reviewed the manuscript. All authors reviewed and approved the final manuscript.
We declare that we have no competing interests.
REFERENCES
- 1.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, Moore CE, Day NP. 2003. How clonal is Staphylococcus aureus? J Bacteriol 185:3307–3316. doi: 10.1128/jb.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeremiah CJ, Kandiah JP, Spelman DW, Giffard PM, Coombs GW, Jenney AW, Tong SY. 2016. Differing epidemiology of two major healthcare-associated meticillin-resistant Staphylococcus aureus clones. J Hosp Infect 92:183–190. doi: 10.1016/j.jhin.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Coombs GW, Nimmo GR, Pearson JC, Collignon PJ, Bell JM, McLaws ML, Christiansen KJ, Turnidge JD, Australian Group on Antimicrobial Resistance. 2013. Australian Group on Antimicrobial Resistance Hospital-onset Staphylococcus aureus Surveillance Programme annual report, 2011. Commun Dis Intell Q Rep 37:E210–E218. [DOI] [PubMed] [Google Scholar]
- 5.Dantes R, Emerging Infections Program–Active Bacterial Core Surveillance MRSA Surveillance Investigators, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S. 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol 42:792–799. doi: 10.1128/jcm.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi H, Seki M, Yamamoto N, Hamaguchi S, Ojima M, Hirose T, Yoshiya K, Toyokawa M, Nishi I, Ogura H, Shimazu T, Tomono K. 2015. Validation of a phage-open reading frame typing kit for rapid identification of methicillin-resistant Staphylococcus aureus (MRSA) transmission in a tertiary hospital. Infect Drug Resist 8:107–111. doi: 10.2147/IDR.S83509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osawa K, Shigemura K, Jikimoto T, Shirakawa T, Fujisawa M, Arakawa S. 2014. Comparison between phage-open-reading frame typing and automated repetitive-sequence-based PCR for typing MRSA isolates. J Antibiot (Tokyo) 67:565–569. doi: 10.1038/ja.2014.41. [DOI] [PubMed] [Google Scholar]
- 9.Kawamura H, Tokuda K, Imuta N, Kubota T, Koriyama T, Miyanohara H, Hashiguchi T, Kawano Y, Nishi J. 2016. Epidemiological analysis of nosocomial MRSA outbreaks using phage open-reading frame typing in a tertiary-care hospital. Jpn J Infect Dis 69:523–524. doi: 10.7883/yoken.JJID.2015.320. [DOI] [PubMed] [Google Scholar]
- 10.Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. 2006. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev 19:512–530. doi: 10.1128/CMR.00025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore G, Cookson B, Gordon NC, Jackson R, Kearns A, Singleton J, Smyth D, Wilson AP. 2015. Whole-genome sequencing in hierarchy with pulsed-field gel electrophoresis: the utility of this approach to establish possible sources of MRSA cross-transmission. J Hosp Infect 90:38–45. doi: 10.1016/j.jhin.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Crnich CJ, Duster M, Warrack S, Maki D, Safdar N. 2014. Comparison of pulsed-gel electrophoresis and a commercial repetitive-element PCR method for assessment of methicillin-resistant Staphylococcus aureus clustering in different health care facilities. J Clin Microbiol 52:2027–2032. doi: 10.1128/JCM.03466-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohadian Moghadam S, Pourmand MR, Douraghi M, Sabzi S, Ghaffari P. 2017. Utilization of PFGE as a powerful discriminative tool for the investigation of genetic diversity among MRSA strains. Iran J Public Health 46:351–356. [PMC free article] [PubMed] [Google Scholar]
- 14.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machuca MA, Sosa LM, Gonzalez CI. 2013. Molecular typing and virulence characteristic of methicillin-resistant Staphylococcus aureus isolates from pediatric patients in Bucaramanga, Colombia. PLoS One 8:e73434. doi: 10.1371/journal.pone.0073434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemeghaire S, Argudin MA, Haesebrouck F, Butaye P. 2014. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet Res 10:153. doi: 10.1186/1746-6148-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JH, Cheng VC, Chan JF, She KK, Yan MK, Yau MC, Kwan GS, Yam WC, Yuen KY. 2013. The use of high-resolution melting analysis for rapid spa typing on methicillin-resistant Staphylococcus aureus clinical isolates. J Microbiol Methods 92:99–102. doi: 10.1016/j.mimet.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Mayerhofer B, Stoger A, Pietzka AT, Fernandez HL, Prewein B, Sorschag S, Kunert R, Allerberger F, Ruppitsch W. 2015. Improved protocol for rapid identification of certain spa types using high resolution melting curve analysis. PLoS One 10:e0116713. doi: 10.1371/journal.pone.0116713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong SY, Lilliebridge RA, Holt DC, McDonald MI, Currie BJ, Giffard PM. 2009. High-resolution melting analysis of the spa locus reveals significant diversity within sequence type 93 methicillin-resistant Staphylococcus aureus from northern Australia. Clin Microbiol Infect 15:1126–1131. doi: 10.1111/j.1469-0691.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- 20.Sahibzada S, Abraham S, Coombs GW, Pang S, Hernandez-Jover M, Jordan D, Heller J. 2017. Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA ST398 between humans and pigs in Australia. Sci Rep 7:1–11. doi: 10.1038/s41598-017-04789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Li G, Xia X, Yang B, Xi M, Meng J. 2014. Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog Dis 11:281–286. doi: 10.1089/fpd.2013.1643. [DOI] [PubMed] [Google Scholar]
- 22.Buyukcangaz E, Velasco V, Sherwood JS, Stepan RM, Koslofsky RJ, Logue CM. 2013. Molecular typing of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) isolated from animals and retail meat in North Dakota, United States. Foodborne Pathog Dis 10:608–617. doi: 10.1089/fpd.2012.1427. [DOI] [PubMed] [Google Scholar]
- 23.Aguadero V, Gonzalez Velasco C, Vindel A, Gonzalez Velasco M, Moreno JJ. 2015. Evaluation of rep-PCR/DiversiLab versus PFGE and spa typing in genotyping methicillin-resistant Staphylococcus aureus (MRSA). Br J Biomed Sci 72:120–127. doi: 10.1080/09674845.2015.11666808. [DOI] [PubMed] [Google Scholar]
- 24.Frenay HM, Theelen JP, Schouls LM, Vandenbroucke-Grauls CM, Verhoef J, van Leeuwen WJ, Mooi FR. 1994. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J Clin Microbiol 32:846–847. doi: 10.1128/JCM.32.3.846-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenner L, Widmer AF, Dangel M, Frei R. 2008. Distribution of spa types among meticillin-resistant Staphylococcus aureus isolates during a 6 year period at a low-prevalence university hospital. J Med Microbiol 57:612–616. doi: 10.1099/jmm.0.47757-0. [DOI] [PubMed] [Google Scholar]
- 26.Church DL, Chow BL, Lloyd T, Gregson DB. 2011. Comparison of automated repetitive-sequence-based polymerase chain reaction and spa typing versus pulsed-field gel electrophoresis for molecular typing of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis 69:30–37. doi: 10.1016/j.diagmicrobio.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Harmsen D, Claus H, Vogel U. 2005. DNA sequence-based tandem repeat analysis of the clfB gene is less discriminatory than spa typing for methicillin-resistant Staphylococcus aureus. Int J Med Microbiol 294:525–528. doi: 10.1016/j.ijmm.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Narukawa M, Yasuoka A, Note R, Funada H. 2009. Sequence-based spa typing as a rapid screening method for the areal and nosocomial outbreaks of MRSA. Tohoku J Exp Med 218:207–213. doi: 10.1620/tjem.218.207. [DOI] [PubMed] [Google Scholar]
- 29.Hallin M, Friedrich AW, Struelens MJ. 2009. spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol Biol 551:189–202. doi: 10.1007/978-1-60327-999-4_15. [DOI] [PubMed] [Google Scholar]
- 30.Banowary B, Dang VT, Sarker S, Connolly JH, Chenu J, Groves P, Raidal S, Ghorashi SA. 2018. Evaluation of two multiplex PCR-high-resolution melt curve analysis methods for differentiation of Campylobacter jejuni and Campylobacter coli intraspecies. Avian Dis 62:86–93. doi: 10.1637/11739-080417-Reg.1. [DOI] [PubMed] [Google Scholar]
- 31.Ghorashi SA, Noormohammadi AH, Markham PF. 2010. Differentiation of Mycoplasma gallisepticum strains using PCR and high-resolution melting curve analysis. Microbiology 156:1019–1029. doi: 10.1099/mic.0.031351-0. [DOI] [PubMed] [Google Scholar]
- 32.Saeidabadi MS, Nili H, Dadras H, Sharifiyazdi H, Connolly J, Valcanis M, Raidal S, Ghorashi SA. 2017. Evaluation of PCR and high-resolution melt curve analysis for differentiation of Salmonella isolates. Avian Pathol 46:319–331. doi: 10.1080/03079457.2016.1268676. [DOI] [PubMed] [Google Scholar]
- 33.Mazi W, Sangal V, Sandstrom G, Saeed A, Yu J. 2015. Evaluation of spa-typing of methicillin-resistant Staphylococcus aureus using high-resolution melting analysis. Int J Infect Dis 38:125–128. doi: 10.1016/j.ijid.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Stephens AJ, Inman-Bamber J, Giffard PM, Huygens F. 2008. High-resolution melting analysis of the spa repeat region of Staphylococcus aureus. Clin Chem 54:432–4366. doi: 10.1373/clinchem.2007.093658. [DOI] [PubMed] [Google Scholar]
- 35.Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/jcm.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SantaLucia J., Jr 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A 95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Tam F, Brunel A-S, Bouzinbi N, Corne P, Bañuls A-L, Shahbazkia HR. 2012. DNAGear: a free software for spa type identification in Staphylococcus aureus. BMC Res Notes 5:642. doi: 10.1186/1756-0500-5-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradley SF. 2015. MRSA colonisation (eradicating colonisation in people without active invasive infection). BMJ Clin Evid 2015:0923. [PMC free article] [PubMed] [Google Scholar]
- 40.Ghorashi SA, Kanci A, Noormohammadi AH. 2015. Evaluation of the capacity of PCR and high-resolution melt curve analysis for identification of mixed infection with Mycoplasma gallisepticum strains. PLoS One 10:e0126824. doi: 10.1371/journal.pone.0126824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banowary B, Dang VT, Sarker S, Connolly JH, Chenu J, Groves P, Ayton M, Raidal S, Devi A, Vanniasinkam T, Ghorashi SA. 2015. Differentiation of Campylobacter jejuni and Campylobacter coli using multiplex-PCR and high resolution melt curve analysis. PLoS One 10:e0138808. doi: 10.1371/journal.pone.0138808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S, Sarker S, Ghorashi SA, Forwood JK, Raidal SR. 2016. A comparison of PCR assays for beak and feather disease virus and high resolution melt (HRM) curve analysis of replicase associated protein and capsid genes. J Virol Methods 237:47–57. doi: 10.1016/j.jviromet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 43.Daniels R, Hamilton EJ, Durfee K, Ndiaye D, Wirth DF, Hartl DL, Volkman SK. 2015. Methods to increase the sensitivity of high resolution melting single nucleotide polymorphism genotyping in malaria. J Vis Exp :e52839. [DOI] [PMC free article] [PubMed] [Google Scholar]