Abstract
The design of multitarget drugs (MTDs) has become an innovative approach for the search of effective treatments in complex diseases such as cancer. In this work, we communicate our efforts in the design of multi-targeting histone deacetylase (HDAC) and protein kinase CK2 inhibitors as a novel therapeutic strategy against cancer. Using tetrabromobenzotriazole (TBB) and 2-dimethylamino-4,5,6,7-tetrabromo-benzimidazole (DMAT) as scaffolds for CK2 inhibition, and a hydroxamate to coordinate the zinc atom present in the active site of HDAC (zinc binding group, ZBG), new multitarget inhibitors have been designed and synthesized. According to the in vitro assays, N-Hydroxy-6-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)hexanamide (11b) is the most interesting compound, with IC50 values of 0.66; 1.46 and 3.67 µM. for HDAC6; HDAC1 and CK2; respectively. Cellular assays on different cancer cell lines rendered promising results for N-Hydroxy-8-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)octanamide (11d). This inhibitor presented the highest cytotoxic activity, proapoptotic capability, and the best mitochondria-targeting and multidrug-circumventing properties, thus being the most promising drug candidate for further in vivo studies.
Keywords: HDAC, CK2, multi-target inhibitors, docking, molecular dynamics, CuAAC, cytotoxic activity
1. Introduction
The development of new efficient anticancer drugs still represents a challenge for medicinal chemists. Although significant progress has been made, single-target drugs still have significant drawbacks, such as induced toxicity and drug-resistance. Combination cancer therapy, a modality that combines two or more agents acting on different pharmacological targets, has expanded the treatment options. These therapies achieve the desirable results in a concerted manner through different mechanisms of action [1,2,3]. Nevertheless, anticancer drug cocktails have also some disadvantages, such as the possibility of drug–drug interactions, pharmacokinetic complexity, and low patient compliance [4,5]. As an alternative, mutitarget drugs (MTDs) have emerged as a novel strategy to overcome all these multicomponent cocktail limitations [6,7]. Moreover, multitarget drug discovery has economic advantages because it requires less clinical trials than multiple specific drugs. For all these reasons, the design of a single molecule, which can efficiently inhibit more than one biological target, constitutes an interesting and promising challenge in modern pharmacotherapy [8,9]. In fact, in the recent past years, several research groups have published successful dual inhibitors for the treatment of different disorders such as cancer, diabetes, psychiatric pathologies, inflammation and neurodegenerative diseases [10,11,12,13].
Protein kinase CK2 is a pleiotropic and ubiquitous serine/threonine protein kinase, which is composed of tetrameric complexes consisting of two catalytic subunits (CK2α, CK2α’) and a dimer of the regulatory β subunits [14,15]. CK2 phosphorylates a wide variety of substrates [16] and is essential for cell viability. It is localized in different cell compartments and is implicated in several essential functions, including signal transduction, cell growth and differentiation, gene expression and apoptosis [17,18]. CK2 is involved in several human pathologies, among them neurodegeneration and cancer. A wide variety of cancer cells show elevated levels of its enzymatic activity [19], and many cancers, including kidney, head and neck, mammary gland, lung, and prostate show overexpression of CK2 [20]. Anti-apoptotic and proliferative properties of this kinase create a favorable cellular environment for tumor maintenance and progression. For all these reasons, CK2 has emerged as a potential oncology target [21]. To date, several compounds, which potently inhibit CK2, have been described [22,23,24]. Among the ATP-competitive CK2 inhibitors reported, ellagic acid, coumarin derivatives, DMAT, TBB and Silmitasertib (CX-4945) (Figure 1) are distinguished by their high efficiency [25,26,27,28,29,30,31,32,33]. Benzonaphthyridine derivative CX-4945 is one of the most potent CK2 inhibitors and is currently in phase II clinical trials [34,35] (Figure 1).
Figure 1.
Representative CK2 inhibitors.
Histone deacetylases (HDAC) are a type of epigenetic enzymes that in recent years have aroused great interest in oncology. Its aberrant activity has been related to cell proliferation in a variety of human pathologies, especially solid tumors and myeloid neoplasia [36,37,38,39,40,41]. In humans, there are 18 HDAC isoforms that are divided into four different classes (I–IV). Class I includes HDAC1, 2, 3 and 8; class II includes 4, 5, 6, 7, 9 and 10; class III is made up of members of the sirtuin family and class IV is represented by HDAC11. Except class III, they are Zn-dependent enzymes. Class I HDACs are ubiquitously expressed and play a critical role in proliferation, while classes II and IV have tissue-specific roles [42]. These enzymes catalyze the deacetylation of the amino group of lysine residues in the N-terminal tails of histone and non-histone proteins and are involved in cell cycle progression, transcriptional regulation and apoptosis [43,44]. For a normal cell growth, a balance between acetylation by histone acetyl transferases (HATs) and deacetylation by HDACs is essential. HDAC inhibitors (HDACi), by inducing hyperacetylation of histones, alter gene transcription and exert antitumor effects through differentiation, growth arrest or/and apoptosis [45,46]. HDAC inhibition has become an important strategy in epigenetic drug discovery, and many HDAC inihibitors are currently under clinical investigation for cancer therapy [47,48]. Vorinosat (SAHA), Romidepsin (FK-228), Belinostat (PXD101) and Panobinostat (LBH589) have been approved by the FDA for the treatment of specific lymphoma and myeloma diseases [49] (Figure 2).
Figure 2.
FDA-approved histone deacetylase (HDAC) inhibitors [50].
Some examples of multitarget inhibitors that combine structural features of HDACi and other anticancer agents have been published in the recent years [51,52,53,54,55,56,57,58]. Combination of kinase inhibitors with the HDAC pharmacophore has been recently reported [59,60]. Two hybrid compounds are currently in clinical trials: an Erlotinib-based hybrid with activity in HDACs, EGFR and HER2 (IC50 = 4.4, 2.4 and 15.7 nM, respectively), and Fimepinostat (CUDC-907), which is an orally active pan PI3K/HDAC inhibitor [61].
Protein kinase CK2 constitutes an attractive option for a secondary target, owing not only to its role in the regulation of HDACs by post-translational modification [62,63], but also to its capacity to activate HDAC1 in tumors associated with hypoxia [64,65].
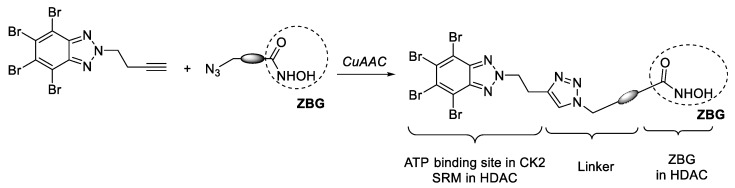
In this work, we communicate our efforts to develop new dual HDAC/CK2 inhibitors with utility as anticancer agents. With this aim, we selected TBB and DMAT (Figure 1), which are ATP-competitive inhibitors with a high affinity for the ATP binding site of CK2. These structures were functionalized with a terminal alkyne moiety and were connected to another subunit designed to interact with HDAC. This subunit is provided with an hydroxamic acid to act as a zinc-binding group (ZBG), and an azide for the Cu(I)-catalyzed alkyne azide cycloaddition reaction (CuAAC) (Scheme 1). On the other hand, the fragment of TBB or DMAT, besides occupying the ATP binding site in CK2, can interact with the residues in the rim of the active site in HDAC1, acting as a surface recognition moiety (SRM).
Scheme 1.
General approach for the synthesis of dual HDAC/CK2 based on TBB.
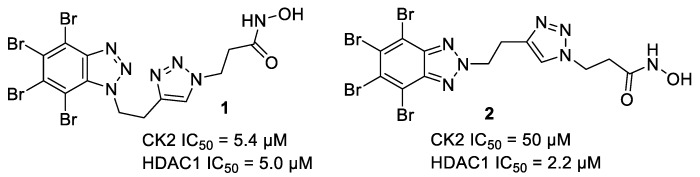
Previous results in our research group show that this strategy is valid. A series of compounds was synthesized, and promising inhibitors with an IC50 value in the order of 5 µM against both enzymes were reported [30]. Although the best inhibitory activity was obtained for compound 1, the corresponding N2-substituted isomer 2 (Figure 3) presented the best profile in cell-based assays, with cytotoxic activity in the low micromolar LC50 in two mammalian cell lines. Furthermore, this hybrid molecule induced apoptosis in leukemia cells in a concentration-dependent manner.
Figure 3.
Examples of previously described CK2/HDAC1 dual inhibitors [30].
2. Results and Discussion
2.1. Chemistry
The first modification that we introduced in the previously described inhibitors 1–2 (Figure 3) was the substitution of the TBB core by a 2-dimethylaminobenzoimidazole moiety, which is present in DMAT, a more potent and selective CK2 inhibitor (Figure 1).
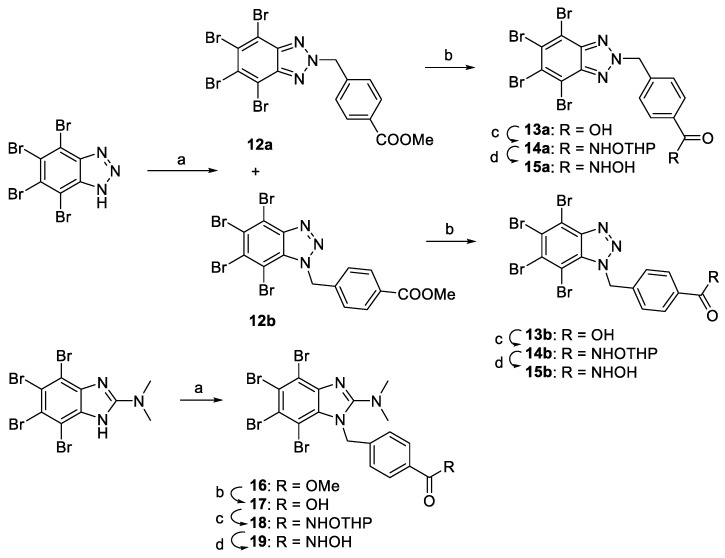
Connecting imidazole chains of different lengths were used in the design of hydroxamates 7. A convergent strategy was used for the synthesis of these compounds, involving a CuAAC [66] as the crucial step to connect HDAC and CK2 inhibition moieties (Scheme 2). Alkynes 4a–b were easily prepared by alkylation of polybrominated benzimidazole 3 [67] with the corresponding alkynyl bromide in the presence of K2CO3 [68]. Treatment of 4a–b with dimethylamine, yielded DMAT-derived alkynes 4c–d. Azides 5 were prepared according to our previous work [30]. CuAAC between azides 5a–e and the two above mentioned alkynes 4c–d using tris(benzyltriazolylmethyl)amine (TBTA) as co-catalyst, gave derivatives 6. Deprotection of the hydroxamate group gave compounds 7 with moderate to good yields and with a purity higher than 95% (HPLC) after column chromatography.
Scheme 2.
Synthetic route for compounds 7a–d. Reaction conditions: (a) K2CO3, acetone, MW, 110 °C, 60 min; (b) HNMe2, MeOH, 110 °C, 48 h; (c) CuSO4, sodium ascorbate, TBTA, RT, overnight; (d) AcCl, MeOH, 0 °C, 15 min.
To analyze if the presence of the triazole ring is mandatory for the inhibitory activity in one or both enzymes, DMAT-derivatives 11, containing hydrocarbon chain linkers of different sizes were synthesized. As depicted in Scheme 3, N1-alkylation of DMAT by reaction with the corresponding bromoester, followed by hydrolysis gave acids 9, which were coupled with THP O-protected hydroxylamine to give 10. Finally, deprotection of the hydroxamate group using in situ generated hydrochloric acid gave compounds 11.
Scheme 3.
Synthetic route for compounds 11a–d. Reactions conditions: (a) K2CO3, acetone, reflux, 72 h; (b) LiOH, THF/H2O, RT, 2h; (c) H2N-OTHP, EDCI, HOBt, NMM, DMF, RT, overnight; (d) AcCl, MeOH, 0 °C, 15 min.
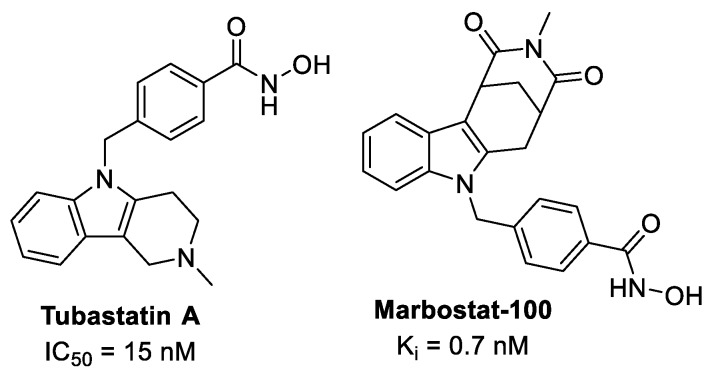
Many phenylhydroxamate-based HDAC inhibitors have been described, showing high potency and selectivity, especially for HDAC6 [69]. These structures are exemplified by Tubastatin A [70] and more recently by Marbostat-100 [71], a highly selective and potent inhibitor of HDAC6, being superior to the established HDACi Tubastatin A (Figure 4). HDAC6 has no in vivo activity against histones, but its targets are tubulin, HSP90 and other proteins involved in cancer development and progression [71]. Recently, a JAK2/HDAC6 dual inhibitor was identified to possess potent antiproliferative activity towards hematological cell lines and excellent in vivo antitumor efficacy in several acute myeloid leukemia models [72].
Figure 4.
Phenylhydroxamate-based HDAC-6 inhibitors.
Taking this into account, compounds 15a–b and 19 derived from TBB [73] and DMAT [74] respectively were synthetized (Scheme 4). N1-alkylation of the corresponding CK2i scaffold with methyl 4-(bromomethyl)benzoate was followed by hydrolysis, furnishing acids 13 and 17, which were coupled with THP O-protected hydroxylamine to obtain 14 and 18. Deprotection of the hydroxamate group gave final products 15a–b and 19.
Scheme 4.
Synthetic route for compounds 13–19. Reactions conditions: (a) methyl 4-(bromomethyl)benzoate, K2CO3, acetone, 150 °C, MW; (b) NaOH, THF, RT; (c) H2N-OTHP, EDCI, HOBt, NMM, DMF, RT, 24 h; (d) AcCl, MeOH, 0 °C, 15 min.
2.2. Enzymatic Inhibitory Evaluation
The inhibitory activities of the synthesized compounds were determined against CK2 and HDAC1 and the IC50 values are collected in Table 1. The activity towards human recombinant HDAC1 was tested using a fluorometric method. CK2 inhibitory activity was determined by a radiometric method, using a short peptide (RRRADDSDDDDD) as a substrate.
Table 1.
Inhibitory activity (IC50 (µM)) on HDAC1, HDAC6 and CK2.
| Compound | HDAC1 | HDAC6 | CK2 |
|---|---|---|---|
| 7a | 1.43 | - | 85.1 |
| 7b | 2.25 | 2.41 | 50.1 |
| 7c | 13.3 | 30.7 | 12.5 |
| 7d | 10.2 | - | 53.8 |
| 11a | 62.0 | - | 34.2 |
| 11b | 1.46 | 0.66 | 3.67 |
| 11c | 1.77 | 1.13 | 16.6 |
| 11d | 13.7 | 8.98 | 5.89 |
| 15a | 8.50 | 11.0 | 15.0 |
| 15b | 4.22 | 2.70 | 136 |
| 19 | 9.45 | 20.6 | 7.15 |
Compounds derived from DMAT were active in both enzymes. Compounds 7a and 7b, bearing a triazole ring in the connecting chain, presented an interesting activity in HDAC1 (1.43 and 2.25 µM), but they were 60-fold and 22-fold less active in CK2 respectively. A better result was obtained for compound 7c, which is a DMAT analogous of the best TBB-derivative previously described by our group [30]. In this case, the activity in both enzymes was very similar, which is the characteristic sought for a dual inhibitor. The introduction of another carbon atom between the imidazole ring and the hydroxamate (compound 7d) brought about a decrease in CK2 inhibitory activity.
In the series of compounds with alkyl connecting chains, the best result was obtained for 11b. The activity against HDAC1 (IC50 = 1.46 µM) and CK2 (IC50 = 3.67 µM) has improved our previous results on TBB derivatives 1 and 2. Longer chains of six and seven carbon atoms (11c and 11d) were also active, although 11c presented a 4.5-fold difference in the CK2 inhibitory activity compared to 11b, and 11d presented a ninefold difference with 11b in the HDAC1 inhibitory activity. Compound 11a, provided with a four-carbon atom chain, was less active in both enzymes. These results demonstrate that the presence of the triazole ring is not essential for inhibitory activity in this series of DMAT derivatives.
The introduction of an aromatic ring in the connecting chain of TBB derivative 15a and DMAT derivative 19 gave also interesting dual HDAC1/CK2 inhibitors, with IC50 in the low micromolar range. We had foreseen that these compounds, carrying an aryl hydroxamate such as the one present in Tubastatin A and Marbostat 100 (Figure 4), could be active in HDAC6. For this reason, we analyzed the activity of 15 and 19, together with other compounds from the alkyl and triazole series in HDAC6, tested using a fluorometric method (Table 1). Interestingly, we found that all of them were active in HDAC6, with the best result for 11b with IC50 of 0.66 µM. This compound presented also an interesting activity in HDAC1 and CK2 with IC50 values of 1.46 and 3.67 µM, respectively.
In summary, we have found interesting HDAC/CK2 multitarget compounds, which can inhibit three enzymes involved in proliferative events through different mechanisms.
2.3. Molecular Modelling
Computational modeling techniques were applied in order to rationalize the biological results shown in Table 1. For this purpose, docking techniques were used as a first approach, followed by Molecular Dynamics (MD) simulations with the aim of proposing a plausible binding mode for the complexes on the three enzymes, CK2, HDAC1 and HDAC6, and of evaluating the stability of the ligand-target complexes in an aqueous and dynamic environment.
CK2. The general binding mode of known selective inhibitors such as TBB, TBI and DMAT in CK2 has been established through X-ray studies [75,76,77]. Upon binding, these ligands occupy the adenine portion of the ATP through strong and selective interactions. Docking calculations performed in PDB code 5CQU as protein target with our set of compounds predicted an overall similar binding mode for the TBB and TBI moieties, that slightly differs from the reference TBB moiety of the bound ligand (JRJ) in the 5CQU structure [29]. The main differences arise from the orientation of the linker and the hydroxamic acid moiety. The general predicted binding mode of compounds 11a–d, 7a, 7c, 7d, 15a–b and 19, has the hydroxamic acid pointing towards the catalytic loop (Figure S1, SI), establishing different hydrogen bonds with the side chains of Lys158, Asn161, and Asp175. The minor differences among the predicted binding modes of these compounds come from the length and adjustment of the aliphatic or aromatic linkers inside the ATP binding site. Compounds 15a–b, (Figure S2, SI) could reach the catalytic loop through a slight rotation of the tetrabromo benzyl moiety, differently from compounds 11a–d, 7a, 7d and 19, that preserved the position of all bromine atoms when compared to JRJ from the reference crystal structure. Unlike the latter compounds, compound 7b is predicted to bind CK2 with the hydroxamic acid pointing towards the αD pocket and establishing hydrogen bond interactions with the side chains of Asn118 and Asp120, and the backbone of His160 (Figure S1, SI).
To better understand the stability of the different predicted binding modes and rationalize the in vitro results, all complexes were submitted to 20 ns Molecular Dynamics (MD) simulations and the ligand RMSD values were monitored along the simulation time (See Table S1, SI). The predicted binding mode of compounds 7a and 7d and 11a–d is preserved along the MD simulation, giving a mean RMSD value lower than 2.5 Å with only slight variations between them. These compounds establish a stable hydrogen bond between the hydroxamic acid and the side chain of Asp156 from the catalytic loop; whereas compound 7c was not able to maintain this hydrogen bond (Figure S3, SI). Compound 7b maintained the interaction with the αD pocket stabilized by the transient hydrogen bond with the side chain of Asn118, and a more stable hydrogen bond with the backbone of Thr119. (Figure S4, SI). In line with their smaller size, compounds 15a, 15b and 19 presented the lowest RMSD values (under 1.6 Å). However, only the latter located the four bromine atoms of the DMAT moiety in the same orientation as the reference structure JRJ, whereas compounds 15a and 15b needed to, either twist the TBB moiety, or relocate the bromine atoms in a different way (Figure S2, SI).
HDAC1 and HDAC6. We studied the general predicted binding mode of our set of compounds by docking techniques, and assessed the stability of their predicted binding mode by means of MD simulations. The overall behavior of these compounds in complex to HDAC1 and HDAC6 did not differ from those found in the literature [30]. All compounds established a bidentate chelation of the catalytic Zn2+, which was ion-, stabilized by hydrogen bonds with the side chains of His140/His610 and Tyr303/Tyr782 in HDAC1 and HDAC6, respectively (Figure S5, SI). The aliphatic linker interacted with the side chains of Phe205/Phe620 and Phe150/Phe680, both of which line the access tunnel to the metal site of both enzymes, and oriented the SRM to interact with amino acids in the surface of the protein. Thus, in the complexes with HDAC1, the aliphatic chain of compounds 11a–b and 11d oriented the DMAT moiety towards the side chains of Tyr204 and Phe205, allowing the establishment of π-π interactions; whereas for compound 11c the DMAT moiety was oriented to interact with the side chains of Leu271 and Phe150 (Figure S6, SI). The complexes of 11a–d with HDAC6 presented an overall similar predicted binding mode, which can be divided into two subgroups: compounds 11a and 11d interact with Phe679 and Met682, whereas compounds 11b and 11c interact with His500, Pro501 and Leu749 (Figure S7, SI). Compounds 7a–d, bearing the triazol linker, established a sandwiched π-π interaction with the side chains of Phe205/Phe620 and Phe150/Phe680, in HDAC 1 and 6, respectively. Their SRMs interact indistinctively with the amino acids of the perimeter of the entrance of the tunnel (Figures S6 and S7, SI). Despite their smaller and shorter structures, compounds 15a–b and 19 were able to reach the metal site, establishing strong π-π interactions with the side chains of Phe205/Phe620 and Phe150/Phe680, in HDAC 1 and 6, respectively. However, in this case, the TBB and DMAT moieties interacted, either with Phe205 and Leu271 on the surface of HDAC1, or with Pro501, Phe679, Met682 and Leu749 in HDAC6, all of them close to the tunnel entrance (Figures S6 and S7, SI).
During the MD simulations of the HDAC1 and HDAC6 complexes, the general behavior of the bound compounds was the exploration of the surface of the protein by the SRM, while the hydroxamate and the linker remained stably bound to the catalytic site of the HDACs. The largest RMSD values corresponded to compounds 7a–d, mainly due to the flexibility of the linker that exposed the SRM to the surface of the protein. This was translated into transient interactions with amino acids in the surface of the protein but with no clear destabilization of the predicted binding mode. Compounds 11a–d showed small rearrangements from the starting orientations but preserved the initial interactions described above along the simulation time. Compounds 15a–b and 19 presented small RMSD variations, in line with their small size, which lead to similar behavior to that of the CK2 complexes (Tables S2 and S3, SI).
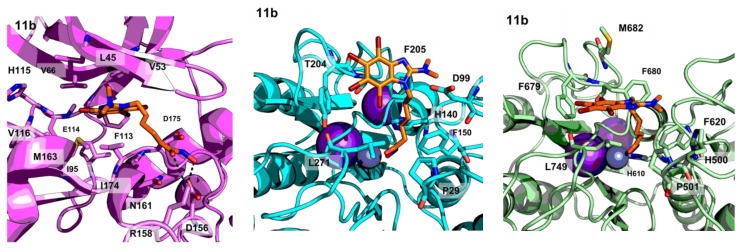
The in vitro activity of compound 11d is in agreement with the obtained models (Figure 5).
Figure 5.
PyMOL stick and cartoon representation of the proposed binding modes of compound 11b to CK2 (magenta), HDAC1 (cyan) and HDAC6 (light green). For the sake of clarity, only heavy atoms are shown.
The interaction of the SRM with CK2 does not vary from that in the reference crystal structure (PDB code 1ZOE), despite the presence of the linker and the hydroxamate group. The latter establishes additional hydrogen bonding interactions with the protein target. As for the binding to the HDACs, the hydroxamate establishes a bidentate chelation with the catalytic zinc ion such as the one found for SAHA (PDB code 1T69), while the linker allows the SRM to interact with the surface of the proteins in a way similar to that reported in the literature [30].
Additionally, the pan-assay interference compounds properties (PAINS) were examined by using the SWISS-ADME web server [78]. Interestingly, all compounds showed neither potential PAINS nor other promiscuity alerts, which makes them good drug candidates.
The best dual enzymatic inhibitors (11b–d), a representative of the triazole series (7c) and two aromatic linker containing compounds (15a and 19), were selected for in vitro studies.
2.4. In Vitro Evaluation
2.4.1. Proapoptotic Activity Against Tumor and Pseudonormal Cell Lines.
Cytotoxic activity of selected CK2-HDAC multitarget inhibitors was investigated in vitro towards human leukemia (Jurkat) and carcinoma (MCF-7, HCT-116) cell lines using trypan blue exclusion assay. For the evaluation of potential cancer selectivity of the selected compounds, their activity was also tested towards human embryonic kidney cells of HEK293 line under identical conditions (Figure 6). LC50 doses (concentration of drug killing 50% cells) of all compounds are summarized in Table 2.
Figure 6.
Comparison of cytotoxic activity of selected CK2-HDAC dual inhibitors (7c, 11b, 11c, 11d, 15a and 19) towards human leukemia (top left), carcinoma (top right and down left,) and pseudonormal (down right) cell lines. Trypan blue assay, 24 h incubation. Means and SD of at least three experiments in triplicate are shown.
Table 2.
LC50 values of studied CK2-HDAC dual inhibitors towards human tumor and pseudonormal cell lines in vitro.
| Comp | LC50 Values of Compounds for Cell Line, µM (M ± SD) | ||||||
|---|---|---|---|---|---|---|---|
| Jurkat | MCF-7 | HCT-116 | HEK293 | HL-60 | HL-60/adr | HL-60/vinc | |
| 11b | 10.15 ± 1.43 | 15.66 ± 2.54 | 4.22 ± 0.46 | 23.33 ± 4.38 | 7.67 ± 1.17 | 16.23 ± 1.68 | 16.79 ± 3.34 |
| 11c | 9.47 ± 1.10 | 9.97 ± 0.99 | 1.90 ± 0.24 | 13.61 ± 0.80 | 6.59 ± 0.96 | 10.63 ± 1.53 | 12.06 ± 3.27 |
| 11d | 5.30 ± 0.75 | 9.02 ± 0.90 | 3.10 ± 0.37 | 8.41 ± 1.26 | 4.69 ± 0.54 | 8.4 ± 1.21 | 2.32 ± 1.06 |
| 7c | 16.54 ± 2.33 | 27.75 ± 6.06 | 16.87 ± 3.90 | 42.71 ± 8.65 | 24.02 ± 8.66 | 18.58 ± 2.09 | 49.46 ± 9.47 |
| 15a | 43.58 ± 16.73 | 13.66 ± 2.67 | 8.67 ± 1.02 | 33.73 ± 7.52 | 34.42 ± 7.89 | 32.37 ± 3.44 | 36.55 ± 4.74 |
| 19 | 24.86 ± 3.80 | 13.55 ± 0.78 | 11.3 ± 1.01 | 33.36 ± 11.76 | 46.49 ± 17.31 | 32.31 ± 5.32 | 21.73 ± 3.39 |
DMAT-derived hydroxamates 11b–d showed twofold higher activity towards various types of tumor cells compared to derivatives containing aromatic linkers (15a and 19) (Figure 6, Table 2). Moreover, the length of the aliphatic chain in 11b–d inhibitors influenced their cytotoxic activity towards tumor cells. In particular, 11b with a five carbon aliphatic chain was the weakest compound (LC50 = 4.22–23.33 µM depending on cell line), while 11c (six-carbon chain) demonstrated higher activity (LC50 in 1.90–13.61 µM range), and 11d (seven-carbon chain) was the most active among all tested dual inhibitors (LC50 in 2.32–9.02 µM range). However, introduction of a triazole ring in the linker of DMAT derived hydroxamates (7c) significantly decreased the antiproliferative activity towards all tested cell lines, lowering it down to the level of CK2-HDAC inhibitors with aromatic linkers (15a and 19). HEK293 cell line was found to be two to three times more resistant to action the action of DMAT derived hydroxamates compared to leukemia and carcinoma cells, thus indicating of targeted action of synthesized inhibitors towards tumor cell lines.
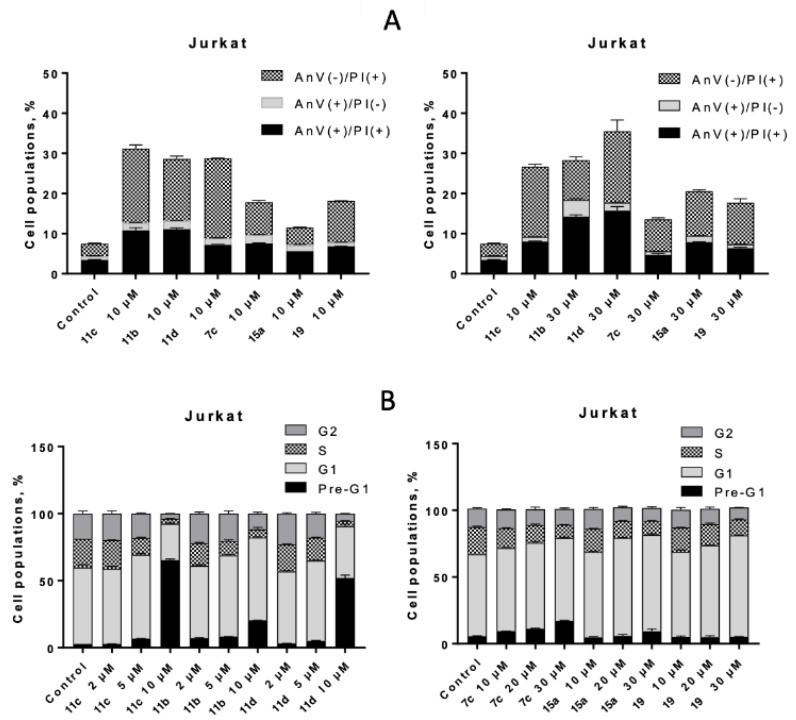
To reveal potential mechanisms of cell death induction, selected CK2-HDAC inhibitors were tested in a PI staining assay. All tested compounds demonstrated dose-dependent inhibitory effect on cell cycle progression in human leukemia cells, depleting cell population in S-phase in favor of G1 phase (Figure 7). However, this effect was masked for high doses of DMAT-hydroxamate derivatives (30 µM) due to their strong proapoptotic activity, leading to appearance of > 80% cells in pre-G1 phase (apoptotic). Thus, lower concentrations of 11b–d (2 µM and 5 µM) had to be addressed in order to observe their inhibitory effect on cell cycling. On the contrary, compounds with weaker cytotoxic potential (7c, 15a and 19) possessed mainly cytostatic activity, inhibiting growth of Jurkat T-leukemia cells at G1 phase of the cell cycle, while a number of apoptotic cells (pre-G1) increased insignificantly even at 30 µM dose (Figure 7). As G1/S cell arrest is considered a typical feature of HDAC inhibitors, including SAHA [79], the observed phenomenon is yet another confirmation of HDAC targeting by these novel dual inhibitors in cell culture.
Figure 7.
Cell cycle arrest and cell death induction by dual CK2-HDAC inhibitors. (A) the impact of the indicated compound doses on the distribution of Jurkat T-leukemia cells in the different phases of the cell cycle was determined by FACS of propidium iodide-stained cells after 24 h of continuous exposure. (B) evaluation of the impact of selected dual CK2-HDAC inhibitors (10 µM and 30 µM, 24 h) on phosphatidylserine externalization in Jurkat T-leukemia cells was determined by FACS, using FITC-labeled annexin V and PI staining. One of three experiments delivering comparable data is shown.
To confirm the pro-apoptotic activity of the studied inhibitors, Annexin V-PI double staining was addressed (Figure 7 down). Compounds with the highest cytotoxic activity (11b–d) also led to concentration-dependent twofold increase in number of annexin V-positive (apoptotic) cells compared to derivatives 15a and 19, possessing cytostatic activity. On contrary, introduction of the triazole motif in the linker (7c) led to a significant decrease in the amount of AnV(+) cells compared to 11d at 30 µM dose, indicating a negative impact of this group both on cytotoxic and pro-apoptotic activity of 7c.
2.4.2. Impact of ROS on Circumvention of Cancer Drug Resistance
Rapid development of cancer drug resistance both to traditional and targeted chemotherapies is considered one of the most crucial challenges in modern cancer treatment [80]. Usually, MDR is caused by overexpression of ABC-transporter proteins (namely, P-gp, MRP-1 and BCRP) in tumor cells and CK2 is directly involved in the regulation of their activity [81]. HDAC inhibitors were also shown to induce apoptosis in MDR cell lines [79], thus, combination of pharmacophore’s of CK2 and HDAC inhibitors in one molecule should gradually increase its MDR-circumventing properties.
Taking into consideration the high abundance of CK2 kinase in leukemia and lymphomas [82], the ability of selected CK2-HDAC dual inhibitors to circumvent MDR was investigated in human leukemia cells of HL-60 line and its doxorubicin-resistant (HL-60/adr, MRP-1 overexpression) and vincristine-resistant (HL-60/vinc, P-gp overexpression) sublines.
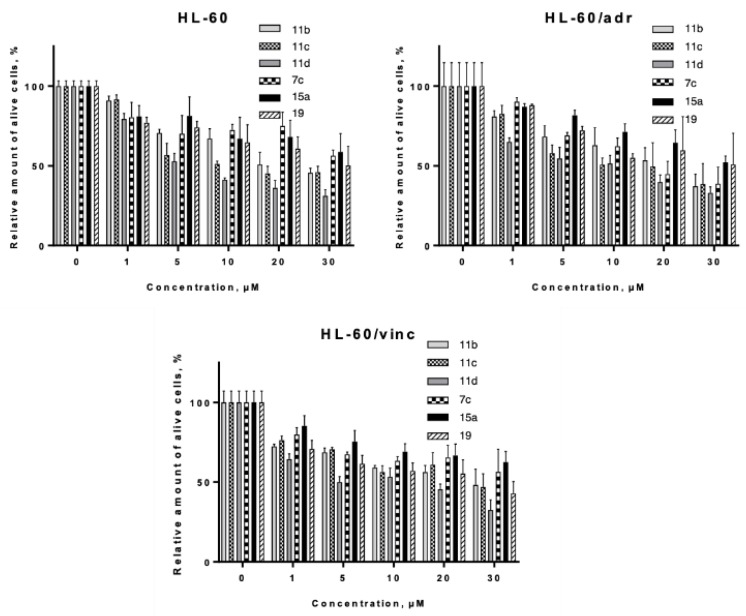
HL-60/wt cells were among the most sensitive to the action of DMAT derived hydroxamates 11b–d, as revealed by analysis of their LC50 values (Table 2, Figure 8). Incubation for 24 h with all the selected inhibitors led to effective killing of both MRP-1 and P-gp overexpressing cells, thus confirming our hypothesis. We observed only slight (~ twofold) decrease of activity of the tested compounds for both P-gp+ and MRP-1 overexpressing cells, except for compound 11d, which activity was even increased twofold towards the HL-60/vinc cell line. It should be stressed that the aforementioned MDR cell models demonstrated extreme resistance towards traditional anticancer drugs (e.g, 80-fold to doxorubicin for HL-60/adr line and 140-fold to doxorubicin for HL-60/vinc line) [83]. Thus the observed effects of dual CK2-HDAC inhibitors clearly put forward to their potent activity to circumvent MDR in vitro.
Figure 8.
Evaluation of cytotoxic activity of selected CK2-HDAC dual inhibitors towards human leukemia cells of the HL-60 line and its drug-resistant sublines HL-60/adr (MRP-1 overexpression) and HL-60/vinc (P-glycoprotein overexpression). Trypan blue assay, 24 h incubation. Means and SD of at least three experiments in triplicate are shown.
To reveal potential mechanisms underlying this phenomenon, we studied the impact of the selected compounds on ROS production in both sensitive and MDR cell lines (Figure S8). It was found that all compounds, except 7c, had no statistically significant impact on hydrogen peroxide production in all the tested cell lines, as revealed by a DCFDA assay (Figure S8, SI). On the contrary, all the dual inhibitors led to a moderate (30%–50%) increase in superoxide production after 24 h of incubation (revealed by DHE assay), with the exception of compound 7c on HL-60/vinc cells. As mitochondria are considered the main source of cellular superoxides, the next aim of our studies was to check whether CK2-HDAC inhibitors induce oxidative stress in tumor cells via depolarization of mitochondria, using the JC-1 assay. However, it was technically impossible to measure JC-1 accumulation on HL-60/adr and HL-60/vinc cells, because JC-1 is a substrate for these ABC-transporter proteins and is actively pumped out into extracellular medium.
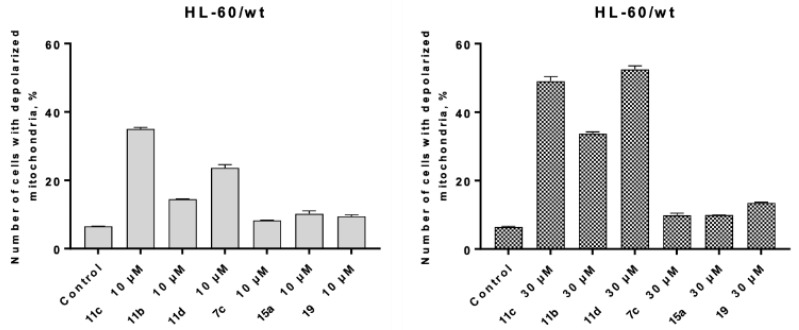
Compounds showing the highest cytotoxic and pro-apoptotic activities in the previous studies, also led to major depolarization of mitochondria in a dose-dependent manner (Figure 9). Notably, 11b (five methylene groups) led to 14.55% of depolarization of mitochondria at 10 µM, and 34.06% at 30 µM. Compounds with longer aliphatic chains led to even more profound effects (34.73% of depolarized mitochondria at 10 µM of 11c, and 49.98% at 30 µM). The highest level of depolarized mitochondria was observed for 11d (53.23%), while in the case of the three other compounds it remained at very low level (9%–10%). This indicates that mitochondria are not involved in apoptosis induction by 7c, 15a and 19.
Figure 9.
Depolarization of mitochondria in HL-60/wt cells under the action of selected CK2-HDAC dual inhibitors (10 µM and 30 µM, 24 h) was determined by FACS using the Mitochondrial Membrane Potential Assay JC-1. One of three experiments delivering comparable data is shown.
Specific hydrogen peroxide burst, observed under the action of 7c, may be explained by the presence of a triazole in its structure. The 3-amino-1,2,4-triazole has been shown to be a specific catalase inhibitor and to lead to major increase in cellular H2O2 levels [84].
Altogether, circumvention of cancer drug resistance by DMAT derived hydroxamates is accompanied by oxidative stress in tumor cells due to depolarization of mitochondria.
3. Materials and Methods
3.1. Chemistry
Unless stated otherwise, purchased starting materials used were high-grade commercial products. NMR spectra were recorded at 400 MHz (1H) and 100 MHz (13C) on a Bruker 400-AC magnetic resonance spectrometer at RT in CDCl3 [calibrated at 7.26 ppm (1H) and 77.0 ppm (13C)] and in DMSO-d6 [calibrated at 2.50 ppm (1H) and 39.5 ppm (13C)]. Data are presented as follows (1H): chemical shift (ppm), multiplicity (s singlet, bs broad singlet, d doublet, t triplet, q quartet, m multiplet), coupling constant J (Hz) and integration. Data for 13C-NMR are reported in terms of chemical shifts (ppm). Mass spectra (MS) were determined on a Bruker Esquire 3000 spectrometer (ionizing voltage = 70 eV). Analytical purity of the tested compounds was determined by using an Agilent 1260 Infinity II HPLC system. Melting points (uncorrected) were taken in open-end capillary tubes and were determined on a Stuart Scientific SMP3 apparatus. Thin-layer chromatography (TLC) was run on Merck silica gel 60 F254 plates. Column chromatography was performed using silica gel Merk-60 (230–400 mesh).
2,4,5,6,7-Pentabromo-1-(prop-2-yn-1-yl)-1H-benzo[d]imidazole (4a)
To a solution of 3 [67] (1.95 mmol, 1 g) in acetone (20 mL) in a 20 mL ΜW vessel, K2CO3 (12.29 μmol, 1.7 g) and 3-bromoprop-1-yne (3.9 mmol, 0.43 mL) were added. The reaction mixture was heated at 110 °C by MW irradiation for 60 minutes. After allowing the reaction to cool to RT, the crude was filtered, removing solid and being the resulting solution concentrated under vacuum and recrystallized in EtOH to afford compound 4a (906 mg, 84%) as a pale yellow solid (m.p. 200–202 °C). 1H-NMR (400 MHz, DMSO): δ 5.37 (d, J = 2.4 Hz, 2H), 3.63 (t, J = 2.4 Hz, 1H). 13C-NMR (100 MHz, DMSO): δ 142.6, 135.6, 132.2, 123.8, 121.9, 115.6, 106.42, 77.6, 77.5, 37.7.
2,4,5,6,7-Pentabromo-1-(but-3-yn-1-yl)-1H-benzo[d]imidazole (4b)
Following the same procedure as before, starting from 3 [67] (1.95 mmol, 1 g), K2CO3 (12.29 mmol, 1.7 g) and 4-bromobut-1-yne (3.9 mmol, 0.41 mL), and after recrystallization in EtOH, compound 4b (437 mg, 40%) was obtained as a yellow solid (m.p. 190–192 °C). 1H-NMR (400 MHz, DMSO): δ 4.68 (t, J = 7.2 Hz, 2H), 2.96 (t, J = 2.6 Hz, 1H), 2.75 (td, J = 7.2, 2.6 Hz, 2H). 13C-NMR (100 MHz, DMSO): δ 143.1, 136.7, 132.8, 124.0, 122.0, 115.9, 106.9, 79.8, 74.8, 45.5, 20.7.
4,5,6,7-tetrabromo-N,N-dimethyl-1-(prop-2-yn-1-yl)-1H-benzo[d]imidazol-2-amine (4c)
To a stirred solution of 4a (1.19 mmol, 655 mg) in metanol (5 mL), dimethylamine (40% in water, 4.5 mL) was added. The reaction mixture was placed in a sealed tube and refluxed at 110 °C for 48 h. After removing the solvent under reduced pressure, the crude material was purified by column chromatography on silica gel (hexane/EtOAc 8:2) to afford compound 4c (432 mg, 71%) as white solid (m.p. 194-195 °C). 1H-NMR (400 MHz, DMSO): δ 4.95 (d, J = 2.4 Hz, 2H), 3.53 (t, J = 2.4 Hz, 1H), 3.07 (s, 6H). 13C-NMR (100 MHz, DMSO): δ 161.9, 142.9, 133.4, 120.2, 118.9, 112.5, 105.7, 79.2, 76.9, 41.4, 36.9.
4,5,6,7-Tetrabromo-1-(but-3-yn-1-yl)-N,N-dimethyl-1H-benzo[d]imidazol-2-amine (4d)
Following the same procedure as before, starting from 4b (1.24 mmol, 700 mg) and dimethylamine (40% in water, 4.5 mL), and after purification by column chromatography on silica gel (hexane/EtOAc 8:2), compound 4d (574 mg, 88%) was obtained as white solid (m.p. 190–192 °C). 1H-NMR (400 MHz, DMSO): δ 4.41 (t, J = 6.7 Hz, 2H), 2.99 (s, 6H), 2.78 (t, J = 2.6 Hz, 1H), 2.49–2.45 (m, 2H). 13C-NMR (100 MHz, DMSO): δ 163.1, 143.5, 132.8, 120.2, 119.0, 112.9, 105.9, 79.7, 73.3, 44.5, 41.7, 19.2.
4-(4-((4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)butanamide (6a)
To a solution of alkyne 4c (0.153 mmol, 35 mg) and azide 5b [30] (0.168 mmol, 86.6 mg) in DMF (2.5 mL), sodium ascorbate (0.092 mmol, 18 mg), CuSO4 × 5 H2O (0.05 mmol, 13 mg) and TBTA (0.015 mmol, 1 mg) were added. The resulting mixture was stirred at RT overnight. Then, water was added (10 mL) and the precipitate formed was filtered and purified by column chromatography on silica gel (DCM/MeOH 97.5:2.5) affording compound 6a (77.7 mg, 70%) as a white solid. 1H-NMR (400 MHz, CDCl3): δ 10.93 (bs, 1H), 7.97 (s, 1H), 5.55 (s, 2H), 4.78 (s, 1H), 4.29 (t, J = 6.0 Hz, 2H), 3.96–3.82 (m, 1H), 3.55–3.43 (m, 1H), 2.98 (s, 6H), 2.00-1.89 (m, 4H), 1.70-1.43 (m, 6H). 13C-NMR (100 MHz, DMSO): δ 168.9, 162.7, 144.6, 143.7, 133.2, 122.3, 121.8, 120.7, 114.4, 105.8, 102.4, 62.6, 49.1, 42.8, 42.4, 29.3, 28.0, 26.0, 25.1, 18.5.
5-(4-((4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)pentanamide (6b)
Following the same procedure as before, starting from alkyne 4c (0.291 mmol, 150 mg), azide 5c [30] (0.265 mmol, 64.2 mg), sodium ascorbate (0.175 mmol, 32 mg), CuSO4 × 5 H2O (0.087 mmol, 20 mg) and TBTA (0.03 mmol, 1.8 mg), and after purification by column chromatography on silica gel (DCM/MeOH 97.5:2.5), compound 6b (98.16 mg, 49%) was obtained as a white solid (m.p. 151–153 °C). 1H- NMR (400 MHz, DMSO): δ 10.89 (bs, 1H), 7.94 (s, 1H), 5.54 (s, 2H), 4.77 (s, 1H), 4.28 (t, J = 7.0 Hz, 2H), 3.94–3.79 (m, 1H), 3.53-3.40 (m, 1H), 2.97 (s, 6H), 1.97 (t, J = 7.2 Hz, 2H), 1.80–1.17 (m, 10H). 13C-NMR (100 MHz, DMSO): 165.9, 162.5, 143.4, 142.2, 132.8, 122.9, 120.0, 119.0, 112.9, 105.8, 100.9, 61.3, 45.8, 45.3, 41.6, 32.8, 27.7, 25.9, 24.6, 18.2.
3-(4-(2-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)ethyl)-1H-1,2,3-triazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)propanamide (6c)
Following the same procedure as before, starting from alkyne 4d (0.284 mmol, 160 mg), azide 5a [30] (0.258 mmol, 55 mg), sodium ascorbate (0.135 mmol, 30 mg), CuSO4 · 5 H2O (0.077 mmol, 19 mg) and TBTA (0.0258 mmol, 1.5 mg), and after purification by column chromatography on silica gel (DCM/MeOH 97.5:2.5), compound 6c (121.4 mg, 63%) was obtained as a white solid (m.p. 140–142 °C). 1H- NMR (400 MHz, DMSO: δ 11.08 (bs, 1H), 7.71 (s, 1H), 4.76 (s, 1H), 4.59-4.42 (m, 4H), 3.96–3.80 (m, 1H), 3.54–3.40 (m, 1H), 2.92 (s, 6H), 2.87 (t, J = 8 Hz, 2H), 2.57 (t, J = 6.7 Hz, 2H), 1.73-1.46 (m, 6H). 13C- NMR (100 MHz, DMSO): δ 165.9, 162.6, 143.4, 142.2, 132.8, 122.9, 120.0, 119.0, 112.9, 105.8, 100.9, 61.3, 45.8, 45.3, 41.7, 32.8, 27.7, 25.9, 24.6, 18.2.
4-(4-(2-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)ethyl)-1H-1,2,3-triazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)butanamide (6d)
Following the same procedure as before, starting from alkyne 4d (0.283 mmol, 160 mg), azide 5b [30] (0.258 mmol, 59 mg), sodium ascorbate (0.135 mmol, 30 mg), CuSO4 × 5 H2O (0.077 mmol, 19 mg) and TBTA (0.0258 mmol, 1.5 mg), and after purification by column chromatography on silica gel (DCM/MeOH 97.5:2.5) compound 6d (83.4 mg, 43%) was obtained as a white solid (m.p. 125–127 °C). 1H-NMR (400 MHz, DMSO): δ 10.95 (bs, 1H), 7.73 (s, 1H), 4.81 (s, 1H), 4.53 (t, J = 7.2 Hz, 2H), 4.25 (t, J = 6.5 Hz, 2H), 3.95-3.83 (m, 1H), 3.54–3.40 (m, 1H), 2.91 (s, 6H), 2.88 (t, J = 7.4 Hz, 2H), 2.52 (t, J = 1.9 Hz, 2H), 1.96 (bs, 2H), 1.721.43 (m, 6H). 13C- NMR (100 MHz, DMSO): δ 168.8, 163.7, 143.7, 132.7, 121.8, 120.3, 113.3, 106.4, 102.0, 62.2, 49.2, 46.8, 41.8, 29.7, 28.0, 26.2, 26.0, 25.0, 18.4.
N-Hydroxy-4-(4-((4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)butanamide (7a)
To a solution of 6a (0.1 mmol, 72 mg) in MeOH (5 mL), a pre-cooled solution of AcCl (1 mmol, 0.07 mL) in MeOH (1 mL) was slowly added over 30 minutes at 0 °C. The reaction mixture was stirred at RT for 15 minutes. Then, NaHCO3 (saturated solution) was added until neutral pH. After extraction with EtOAc (3 × 20 mL), the combined organic layers were dried over MgSO4. The solvent was evaporated under reduced pressure, and the resulting crude material was purified by column chromatography (DCM/MeOH 98:2) to give 7a (45 mg, 70%) as a white solid (mp 191–193 °C). 1H-NMR (400 MHz, DMSO): δ 10.37 (bs, 1H), 8.71 (bs, 1H), 7.99 (s, 1H), 5.55 (s, 2H), 4.28 (t, J = 6.7 Hz, 2H), 2.97 (s, 6H), 1.97–1.89 (m, 4H). MS (ESI) m/z 682 [M+ + 23]. HPLC purity 99%.
N-Hydroxy-5-(4-((4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)pentanamide (7b)
Following the same procedure as before, starting from 6b (0.21 mmol, 158 mg) and AcCl (2.1 mmol, 0.15 mL), and after purification by column chromatography (DCM/MeOH 98:2), 7b (14.8 mg, 11%) was obtained as a white solid (mp 203–205 °C). 1H- NMR (400 MHz, DMSO): δ 10.33 (bs, 1H), 8.66 (bs, 1H), 7.95 (s, 1H), 5.55 (s, 2H), 4.27 (t, J = 7.0 Hz, 2H), 2.98 (s, 6H), 1.93 (t, J = 7.3 Hz, 2H), 1.74–1.67 (m, 2H), 1.39–1.32 (m, 2H). MS (ESI) m/z 696 [M+ + 23], 674 [M+ + 1]. HPLC purity 95%.
N-Hydroxy-3-(4-(2-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)ethyl)-1H-1,2,3-triazol-1-yl)propanamide (7c)
Following the same procedure as before, starting from 6c (0.16 mmol, 121.4 mg) and AcCl (1.6 mmol, 0.11 mL) and after purification by column chromatography (DCM/MeOH 98:2), 7c (18.9 mg, 18%) was obtained as a white solid (mp 168-170 °C). 1H- NMR (400 MHz, DMSO): δ 10.49 (bs, 1H), 8.83 (bs, 1H), 7.73 (s, 1H), 4.53–4.45 (m, 4H), 2.92 (s, 6H), 2.87 (t, J = 7.3 Hz, 2H), 2.56-2.52 (m, 2H). MS (ESI) m/z 682 [M+ + 23], 660 [M+ + 1]. HPLC purity 95%.
N-Hydroxy-4-(4-(2-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)ethyl)-1H-1,2,3-triazol-1-yl)butanamide (7d)
Following the same procedure as before, starting from 6d (0.1 mmol, 79 mg) and AcCl (1 mmol, 0.07 mL), and after purification by column chromatography (DCM/MeOH 98:2), 7d (31 mg, 47%) was obtained as a white solid (159–161 °C). 1H- NMR (400 MHz, DMSO): δ 10.39 (bs, 1H), 8.73 (bs, 1H), 7.74 (s, 1H), 4.53 (“t”, 2H), 4.23–4.27 (m, 2H), 2.91 (s, 6H), 2.89 (“t”, 2H), 2.60 (m, under DMSO, 4H). MS (ESI) m/z 696 [M+ + 23], 674 [M+ + 1]. HPLC purity 98%.
Methyl 5-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)pentanoate (8a).
DMAT [74] (0.5 mmol, 238 mg), K2CO3 (3.5 mmol, 0.48 g), methyl 5-bromopentanoate (2 mmol, 0.39 g) and acetone (10 mL) were placed in a sealed tube and the mixture was heated at 80 °C for 72 h. After removing the solvent under reduced pressure, the crude was purified by column chromatography on silica gel (hexane/EtOAc 8:2) affording compound 8a (183 mg, 62%) as a colorless oil. 1H- NMR (400 MHz, CDCl3): δ 4.27 (t, J = 7.2 Hz, 2H), 3.64 (s, 3H), 3.06 (s, 6H), 2.28 (t, J = 7.3 Hz, 2H), 1.63–1.60 (m, 2H), 1.42–1.49 (m, 2H). 13C-NMR (100 MHz, CDCl3): δ 173.4, 162.9, 143.5, 132.7, 121.2, 120.5, 114.1, 105.9, 51.6, 46.1, 42.4, 33.3, 29.7, 21.4.
Ethyl 6-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)hexanoate (8b)
Following the same procedure as before, and starting from DMAT [74] (0.5 mmol, 238 mg), K2CO3 (3.5 mmol, 0.48 g) and ethyl 6-bromohexanoate (2 mmol, 0.35 mL), 8b (222 mg, 72%) was obtained as a colorless oil. 1H- NMR (400 MHz, CDCl3): δ 4.26–4.22 (m, 2H), 4.10 (q, J = 7.1 Hz, 2H), 3.02 (s, 6H), 2.23 (t, J = 7.4 Hz, 2H), 1.62–1.54 (m, 4H), 1.23 (t, J = 7.1 Hz, 3H), 1.18–1.13 (m, 2H). 13C-NMR (100 MHz, CDCl3): δ 173.3, 162.9, 143.6, 132.8, 121.0, 120.3, 114.1, 105.9, 60.3, 46.3, 42.3, 34.0, 29.9, 25.6, 24.3, 14.2.
Ethyl 7-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)heptanoate (8c)
Following the same procedure as before, and starting from DMAT [74] (0.5 mmol, 238 mg), K2CO3 (3.5 mmol, 0.48 g) and ethyl 7-bromoheptanoate (2 mmol, 0.39 mL), 8c (246 mg, 78%) was obtained as a colorless oil. 1H- NMR (400 MHz, CDCl3): δ 4.24 (t, J = 7.4 Hz, 2H), 4.11 (q, J = 7.1 Hz, 2H), 3.03 (s, 6H), 2.24 (t, J = 7.4 Hz, 2H), 1.60–1.52 (m, 4H), 1.32–1.22 (m, 5H), 1.18–1.13 (m, 2H). 13C-NMR (100 MHz, CDCl3): δ 173.5, 163.0, 143.6, 132.8, 120.9, 120.3, 114.1, 105.9, 60.2, 46.5, 42.3, 34.1, 30.1, 28.5, 25.8, 24.7, 14.2.
Methyl 8-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)octanoate (8d)
Following the same procedure as before, and starting from DMAT [74] (0.42 mmol, 201 mg), K2CO3 (3 mmol, 0.41 g) and methyl 8-bromooctanoate (2 mmol, 0.39 g), 8d (177 mg, 67%) was obtained as a colorless oil. 1H- NMR (400 MHz, CDCl3): δ 4.23 (t, J = 7.2 Hz, 2H), 3.66 (s, 3H), 3.04 (s, 6H), 2.27 (t, J = 7.5 Hz, 2H), 1.59–1.53 (m, 4H), 1.28–1.24 (m, 4H), 1.15–1.11 (m, 2H).
5-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)pentanoic acid (9a)
To a solution of 8a (0.27 mmol, 160 mg) in THF/H2O (2.5 mL/1 mL), LiOH (2 mL, 2.5N in water) was added and the reaction mixture was stirred at RT for 2 h. After removing solvents under reduced pressure, H2O (20 mL) was added and EtOAc (20 mL) to wash. The aqueous phase was acidified with HCl (1 M) and extracted with EtOAc (2 × 20 mL). Combined organic layers were washed with brine, dried over MgSO4, filtered and the solvent was evaporated under reduced pressure to afford compound 9a (116 mg, 75%) as a white oil. 1H-NMR (400 MHz, DMSO): δ 12.01 (bs, 1H), 4.26 (t, J = 7.1 Hz, 2H), 2.98 (s, 6H), 2.17 (t, J = 7.3 Hz, 2H), 1.57–1.50 (m, 2H), 1.28–1.23 (m, 2H). 13C-NMR (100 MHz, DMSO): δ 174.1, 162.6, 143.3, 132.9, 120.0, 119.1, 112.9, 106.0, 45.9, 41.8, 32.9, 29.1, 21.0.
6-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)hexanoic acid (9b)
Following the same procedure as before, and starting from 8b (0.28 mmol, 172 mg) and LiOH (2 mL, 2.5N in water), 9b (129 mg, 79%) was obtained as a white solid. 1H-NMR (400 MHz, DMSO): δ 4.24 (t, J = 7.3 Hz, 2H), 2.98 (s, 6H), 2.12 (t, J = 7.3 Hz, 2H), 1.53–1.42 (m, 4H), 1.11–1.07 (m, 2H). 13C- NMR (100 MHz, DMSO): δ 174.3, 162.8, 143.3, 132.9, 119.9, 119.0, 112.9, 105.8, 46.0, 41.8, 33.6, 29.4, 25.0, 24.0.
7-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)heptanoic acid (9c)
Following the same procedure as before, and starting from 8c (0.38 mmol, 243 mg) and LiOH (2 mL, 2.5N in water), 9c (201 mg, 88%) was obtained as a white solid. 1H- NMR (400 MHz, DMSO): δ 4.24 (t, J = 7.2 Hz, 2H), 2.98 (s, 6H), 2.15–2.11 (m, 2H), 1.53–1.45 (m, 2H), 1.44–1.36 (m, 2H), 1.25–1.17 (m, 2H), 1.09–1.02 (m, 2H). 13C- NMR (100 MHz, DMSO): δ 174.4, 163.0, 143.4, 133.0, 120.0, 119.0, 112.9, 105.9, 46.1, 41.8, 33.5, 29.5, 27.9, 25.2, 24.3.
8-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)octanoic acid (9d)
Following the same procedure as before, and starting from 8d (0.49 mmol, 312 mg) and LiOH (4.6 mL, 2.5N in water), 9d (183 mg, 60%) was obtained as a colorless oil. 1H- NMR (400 MHz, DMSO): δ 11.96 (bs, 1H), 4.24 (t, J = 7.2 Hz, 2H), 2.98 (s, 6H), 2.15 (t, J = 7.3 Hz, 2H), 1.52–1.48 (m, 2H), 1.43–1.40 (m, 2H), 1.20–1.15 (m, 4H), 1.05–1.02 (m, 2H).
5-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)pentanamide (10a)
To a solution of 9a (0.42 mmol, 242 mg) in DMF (10 mL) were successively added HOBt (0.5 mmol, 78 mg), NMM (1.26 mmol, 0.14 mL), NH2OTHP (0.84 mmol, 98 mg) and EDCI (0.59 mmol, 0.1 mL). The resulting mixture was stirred at RT overnight. H2O (20 mL) and EtOAc (3 × 20 mL) were added. After extractions, combined organic layers were washed with brine, dried over MgSO4, filtered and evaporated under reduced pressure. The crude obtained was purified by column chromatography on silica gel (EtOAc/hexane 8:2) affording compound 10a (136 mg, 50%) as a white solid. 1H-NMR (400 MHz, DMSO): δ 10.86 (bs, 1H), 4.71 (s, 1H), 4.24 (m, 2H), 3.84 (t, J = 10.0 Hz, 1H), 3.40 (m, 1H), 2.98 (s, 6H), 1.93 (t, J = 7.1 Hz, 2H), 1.64–1.47 (m, 8H), 1.30–1.26 (m, 2H). 13C- NMR (100 MHz, DMSO): δ 168.6, 162.9, 143.4, 132.9, 120.0, 119.0, 112.9, 106.0, 100.8, 61.3, 46.0, 41.8, 31.6, 29.2, 27.7, 24.7, 21.6, 18.3.
6-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)hexanamide (10b)
Following the same procedure as before, and starting from 9b (0.32 mmol, 193 mg), HOBt (0.38 mmol, 58 mg), NMM (0.96 mmol, 0.11 mL), NH2OTHP (0.64 mmol, 75 mg) and EDCI (0.44 mmol, 0.08 mL), 10b (61 mg, 28%) was obtained as a white solid. 1H- NMR (400 MHz, CDCl3): δ 8.25 (bs, 1H), 4.91 (s, 1H), 4.23 (t, J = 7.2 Hz, 2H), 3.93–3.89 (m, 1H), 3.63–3.60 (m, 1H), 3.02 (s, 6H), 2.09–1.98 (m, 2H), 1.81–1.76 (m, 2H), 1.67–1.55 (m, 8H), 1.19–1.14 (m, 2H). 13C- NMR (100 MHz, CDCl3): δ 170.0, 163.0, 143.6, 132.8, 121.0, 120.3, 114.0, 105.9, 102.5, 62.5, 46.3, 42.3, 32.9, 29.9, 27.9, 25.6, 24.9, 24.6, 18.5.
7-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)heptanamide (10c)
Following the same procedure as before, and starting from 9c (0.51 mmol, 313 mg), HOBt (0.61 mmol, 91 mg), NMM (1.53 mmol, 0.16 mL), NH2OTHP (1.02 mmol, 117 mg) and EDCI (0.71 mmol, 0.12 mL), 10c was obtained (182 mg, 51%) as a white solid. 1H- NMR (400 MHz, CDCl3): δ 8.19 (bs, 1H), 4.92 (s, 1H), 4.23 (t, J = 7.2 Hz, 2H), 3.95–3.89 (m, 1H), 3.63–3.60 (m, 1H), 3.03 (s, 6H), 2.05 (m, 2H), 1.80–1.77 (m, 2H), 1.62–1.52 (m, 8H), 1.30–1.25 (m, 2H), 1.17–1.12 (m, 2H). 13C- NMR (100 MHz, CDCl3): δ 170.0, 163.0, 143.6, 132.9, 121.0, 120.3, 114.0, 105.9, 102.5, 62.6, 46.5, 42.4, 42.3, 33.1, 30.1, 28.5, 28.0, 25.9, 25.0, 18.5.
8-(4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)-N-((tetrahydro-2H-pyran-2-yl)oxy)octanamide (10d)
Following the same procedure as before, and starting from 9d (0.29 mmol, 183 mg), HOBt (0.34 mmol, 53 mg), NMM (0.87 mmol, 0.09 mL), NH2OTHP (0.58 mmol, 89 mg) and EDCI (0.39 mmol, 0.07 mL), 10d (32.4 mg, 16%) was obtained as a white solid. 1H- NMR (400 MHz, CDCl3): δ 4.92 (s, 1H), 4.22–4.17 (m, 2H), 3.92–3.87 (m, 1H), 3.56–3.54 (m, 1H), 3.03 (s, 6H), 2.09–2.06 (m, 2H), 1.79–1.69 (m, 2H), 1.57–1.51 (m, 10H), 1.18–1.14 (m, 2H), 1.08–1.13 (m, 2H).
N-Hydroxy-5-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)pentanamide (11a)
Following the same procedure that we used for the synthesis of 7a, starting from 10a (0.21 mmol, 136 mg) and AcCl (2.1 mmol, 0.15 mL), and after purification by column chromatography (EtOAc/Hexane 4:1), 11a (36 mg, 30%) was obtained as a white solid (m.p.168 - 170 °C). 1H- NMR (400 MHz, DMSO): δ 10.30 (bs, 1H), 8.67 (bs, 1H), 4.25 (t, J = 7.0 Hz, 2H), 2.98 (s, 6H), 1.89 (t, J = 7.3 Hz, 2H), 1.50–1.46 (m, 2H), 1.29–1.25 (m, 2H). MS (ESI) m/z 615 [M+ + 23], 593 [M+ + 1]. HPLC purity 98%.
N-Hydroxy-6-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)hexanamide (11b)
Following the same procedure that we used for the synthesis of 7a, starting from 10b (0.09 mmol, 61 mg), and AcCl (0.9 mmol, 0.07 mL) and after purification by column chromatography (EtOAc/Hexane 4:1), 11b (38 mg, 70%) was obtained as a white solid (m.p. 167–169 °C). 1H- NMR (400 MHz, DMSO): δ 10.29 (bs, 1H), 8.64 (bs, 1H), 4.23 (t, J = 7.1 Hz, 2H), 2.99 (s, 6H), 1.87 (t, J = 7.3 Hz, 2H), 1.50–1.44 (m, 4H), 1.08–1.06 (m, 2H). MS (ESI) m/z 629 [M+ + 23], 607 [M+ + 1]. HPLC purity 97%.
N-Hydroxy-7-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)heptanamide (11c)
Following the same procedure that we used for the synthesis of 7a, starting from 10c (0.25 mmol, 180 mg) and AcCl (2.5 mmol, 0.18 mL), and after purification by column chromatography (DCM/MeOH 98:2), 11c (50 mg, 33%) was obtained as a white solid (m.p. 120-122 °C). 1H- NMR (400 MHz, DMSO): δ 10.3 (bs, 1H), 8.62 (bs, 1H), 4.23 (t, J = 7.3 Hz, 2H), 2.99 (s, 6H), 1.89 (t, J = 7.2 Hz, 2H), 1.51–1.48 (m, 2H), 1.42–1.38 (m, 2H), 1.21–1.17 (m, 2H), 1.08–1.04 (m, 2H). MS (ESI) m/z 643 [M+ + 23], 621 [M++ 1]. HPLC purity 98%.
N-Hydroxy-8-(4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)octanamide (11d)
Following the same procedure that we used for the synthesis of 7a, starting from 10d (0.12 mmol, 89.9 mg) and AcCl (1.2 mmol, 0.08 mL), and after purification by column chromatography (DCM/MeOH 98:2), 11d (19 mg, 25%) was obtained as a white solid (m.p. 118 °C). 1H- NMR (400 MHz, DMSO): δ 10.3 (bs, 1H), 8.64 (bs, 1H), 4.24 (t, J = 7.3 Hz, 2H), 2.99 (s, 6H), 1.89 (t, J = 7.3 Hz, 2H), 1.51–1.48 (m, 2H), 1.43–1.40 (m, 2H), 1.19–1.13 (m, 4H), 1.06–1.02 (m, 2H). MS (ESI) m/z 657 [M+ + 23], 635 [M++ 1]. HPLC purity 95%.
Methyl 4-((tetrabromo-2H-benzo[d][1,2,3]triazol-2-yl)methyl)benzoate (12a) and methyl 4-((tetrabromo-1H-benzo[d][1,2,3]triazol-1-yl)methyl)benzoate (12b)
A solution of TBB [73] (200 mg, 0.45 mmol), K2CO3 (9 eq) and methyl 4-(bromomethyl)benzoate (105 mg, 0.65 mmol) in acetone (5 mL) was irradiated under MW conditions at 150 °C for one minute. The solvent was removed under vacuum and the crude was solved in EtOAc. The organic phase was washed with brine and H2O, dried over MgSO4, filtered and evaporated. The crude material was purified by column chromatography on silica gel (hexane/DCM 35:65), giving 12a (215 mg, 80%) and 12b (50 mg, 18%). For 12a: 1H-NMR (400 MHz, DMSO) δ 7.98 (d, J = 8.3 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H), 6.19 (s, 2H), 3.84 (s, 3H). For 12b: 1H-NMR (400 MHz, DMSO) δ 7.84 (d, J = 8.4 Hz, 2H), 7.23 (d, J = 8.3 Hz, 2H), 6.30 (s, 2H), 3.83 (s, 3H).
4-((Tetrabromo-2H-benzo[d][1,2,3]triazol-2-yl)methyl)benzoic acid (13a)
A solution of 12a (50 mg, 0.09 mmol) in 1:1 THF:NaOH 1M (40 eq) was stirred at RT till completion of the reaction (TLC). The crude was then washed with EtOAc and the aqueous phase was acidified with HCl 3M. The precipitate obtained was filtered and used in the next reaction without further purification. 1H-NMR (400 MHz, DMSO) δ 13.06 (bs, 1H), 7.95 (d, J = 8.2 Hz, 2H), 7.49 (d, J = 8.2 Hz, 2H), 6.17 (s, 2H). 13C NMR (100 MHz, DMSO) δ 166.8, 142.9, 138.9, 131.0, 129.8, 128.4, 125.9, 113.7, 59.8.
4-((Tetrabromo-1H-benzo[d][1,2,3]triazol-1-yl)methyl)benzoic acid (13b)
Following the same procedure as before and starting from 12b (70 mg, 0.11 mmol), 13b (60 mg, 98%) was obtained as a white solid. 1H-NMR (400 MHz, DMSO) δ 7.89 (d, J = 8.4 Hz, 2H), 7.21 (d, J = 8.4 Hz, 2H), 6.29 (s, 2H).
4-((Tetrabromo-2H-benzo[d][1,2,3]triazol-2-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (14a)
Following the same procedure that was used for the synthesis of 10a and starting from 13a (0.21 mmol, 188 mg), HOBt (0.25 mmol, 39 mg), NMM (0.63 mmol, 0.07 mL), NH2OTHP (0.42 mmol, 49 mg) and EDCI (0.29 mmol, 0.05 mL), 14a (60 mg, 98%) was obtained, after column chromatography on silica gel (EtOAc/hexane 3:7), as a white solid. 1H-NMR (400 MHz, DMSO) δ 8.71 (bs, 1H), 7.76 (d, J = 8.3 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H), 5.92 (s, 2H), 5.06 (bs, 1H), 4.03–3.89 (m, 1H), 3.69–3.57 (m, 1H), 1.93–1.57 (m, 6H).
4-((Tetrabromo-1H-benzo[d][1,2,3]triazol-1-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (14b)
Following the same procedure that was used for the synthesis of 10a and starting from 13b (0.11 mmol, 60 mg), HOBt (0.12 mmol, 20 mg), NMM (0.31 mmol, 0.04 mL), NH2OTHP (0.21 mmol, 25 mg) and EDCI (0.15 mmol, 0.03 mL), 14b (60 mg, 98%) was obtained, after column chromatography on silica gel (EtOAc/hexane 2:1 to 1:1), as a white solid. 1H-NMR (400 MHz, DMSO) δ 9.03 (bs, 1H), 7.71 (d, J = 8.2 Hz, 2H), 7.15 (d, J = 8.0 Hz, 2H), 6.18 (s, 2H), 5.04 (bs, 1H), 3.98–3.90 (m, 1H), 3.72–3.51 (m, 1H), 1.91–1.47 (m, 6H).
N-Hydroxy-4-((tetrabromo-2H-benzo[d][1,2,3]triazol-2-yl)methyl)benzamide (15a)
Following the same procedure that we used for the synthesis of 7a, starting from 14a (0.12 mmol, 80 mg) and AcCl (1.2 mmol, 0.09 mL), crude reaction was obtained. A precipitate formed after addition of Et2O was filtered and solved in DCM/MeOH (5%) and precipitated again with hexane. The solid was filtered, giving 15a (70 mg, quantitative conversion) as a white solid (m.p. 218–220 °C). 1H-NMR (400 MHz, DMSO) δ 11.23 (bs, 1H), 9.07 (bs, 1H), 7.75 (d, J = 8.3 Hz, 2H), 7.46 (d, J = 8.3 Hz, 2H), 6.12 (s, 2H). MS (ESI) m/z 584.6 [M++1]. HPLC purity 94%.
N-Hydroxy-4-((tetrabromo-1H-benzo[d][1,2,3]triazol-1-yl)methyl)benzamide (15b)
Following the same procedure as before starting from 14b (0.067 mmol, 50 mg,), and AcCl (0.67 mmol, 0.05 mL), 15b (30 mg, 77%) was obtained as a white solid (m.p. 139-142 °C). 1H-NMR (400 MHz, DMSO) δ 11.22 (bs, 1H), 7.70 (d, J = 8.4 Hz, 2H), 7.18 (d, J = 8.4 Hz, 2H), 6.25 (s, 2H). MS (ESI) m/z 584.6 [M++1]. HPLC purity 97%.
Methyl 4-((4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)benzoate (16)
A mixture of DMAT [74] (150 mg, 0.314 mmol), methyl 4-(bromomethyl)benzoate (72.22 mg, 0.314 mmol) and K2CO3 (130 mg, 0.942 mmol) in acetone (2.5 mL) was refluxed for 48 h. The reaction mixture was evaporated to dryness, diluted in EtOAc (10 mL) and washed with brine (2×10 mL). The crude material was dried with MgSO4, filtered, evaporated to dryness and purified by column chromatography (hexane/EtOAc 9:1 to 1:1) to give 16 (182 mg, 93%) as a white solid (m.p. 106–108 °C). 1H-NMR (400 MHz, CDCl3) δ 7.98 (d, J = 8.4 Hz, 2H), 7.07 (d, J = 8.4 Hz, 2H), 5.55 (s, 2H), 3.89 (s, 3H), 2.97 (s, 6H).
4-((4,5,6,7-Tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (18)
Following the same procedure that we used for the synthesis of 14a, and starting from 16 (170 mg, 0.27 mmol), 17 (145 mg, 88%) was obtained as an oil which was used in the next step without further purification. 17 (140 mg, 0.23 mmol) was treated as described for 15a, and after purification by column chromatography on silica gel (hexane/EtOAc 8:2 to 1:1) gave 18 (100 mg, 61%) as a white solid. 1H-NMR (400 MHz, CDCl3) δ 8.84 (s, 1H), 7.69 (d, J = 8.2 Hz, 2H), 7.06 (d, J = 8.2 Hz, 2H), 5.53 (s, 2H), 5.05 (s, 1H), 3.99–3.82 (m, 1H), 3.70–3.52 (m, 1H), 2.96 (s, 6H), 1.92–1.56 (m, 6H).
N-Hydroxy-4-((4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzo[d]imidazol-1-yl)methyl)benzamide (19)
Following the same procedure that we used for the synthesis of 7a, and starting from 18 (90 mg, 0.126 mmol), 19 (60 mg, 79%) was obtained as a white solid (m.p. 210–213 °C). 1H-NMR (400 MHz, DMSO) δ 7.67 (d, J = 8.3 Hz, 2H), 7.07 (d, J = 8.3 Hz, 2H), 5.57 (s, 2H), 2.94 (s, 6H). MS (ESI) m/z 648.7 [M++23]. HPLC purity 95%.
3.2. Molecular Modelling
To date, 158 crystal structures of the CK2 alpha catalytic subunit have been deposited in the PDB database (UniProt Entry NX_P68400 Human CK2 alpha). Four of them (3RPS, 4kWP, 5CQW, 5CQU) are bound to TBB derivatives, including compound JRJ that is the most similar to our set of molecules. Interestingly, JRJ is crystallized in two different conformations: in 5CQU the flexible chain points to the ATP phosphate-binding region, whereas in 5CQW it points to the α-D helix. Of these two 3D structures, we selected 5CQU as the protein shows a more open G-loop compared to 5CQW. For HDAC1, despite its biological interest, only two 3D-structures have been deposited in the PDB: 4BKX and 5ICN (UniProt Entry Q13547 Human Histone deacetylase 1). Of those, PDB code 5ICN was selected due to the presence of a hydroxamic derivative bound to the holo structure [85]. For HDAC6 there is only one available crystal structure, PDB code 5EDU (UniProt entry Q9UBN7 human histone deacetylase 6). This structure presents both domains (histone deacetylase I & II) in the holo conformation bound to the inhibitor Trichostatin A [86]. The hydroxamic-zinc interaction in metalloproteinases has been intensively studied and it has been established that upon binding, the pKa of the hydroxamic acid decreases, resulting in the spontaneous deprotonation of the terminal hydroxyl group, transferring the proton to the histidine (His 140 in HDAC1 and His610 in HDAC6) within the active site [87]. All receptors were processed with the Protein Preparation Wizard module of Maestro [88], but taking into account the HDAC reactivity in the presence of hydroxamic acids, both HDAC1 and HDAC1 were prepared with protonated histidines in positions 140 and 610, respectively.
The Glide Grid (Glide, Schrödinger, LLC, New York, NY, 2018) module was used to calculate the grid-box with a 20 Å length, using as box center the ligands JRJ and TSN for CK2 and HDAC6, respectively; and, the catalytic Zn for HDAC1, applying the versatile metal binding restriction to the zinc ion in both HDACs. Note must be taken that when using the ligand as box center, the program automatically deletes the ligand for the grid generation. The set of described ligands were prepared with the LigPrep module of Maestro (S. LigPrep, Schrödinger Release 2018-2, LLC, New York) considering a pH of 7 ± 4, and to obtain all possible hydroxamic protonation states and the metal binding site identification was also used. Ligands with a protonated hydroxamic acid were used for docking on CK2 whereas ligands with the deprotonated hydroxamate were docked in HDAC1 and HDAC6. The docking calculations were performed with Glide using the XP algorithm [89], applying van der Waals radii scaling factor of 1.0/0.8 and establishing halogen atoms as acceptors. Finally, visual inspection was applied to select the most plausible ligand orientation within the enzyme, which was further used for MD simulations.
For the MD simulations of the CK2-ligand complexes, all ligands were prepared as follows: the geometry optimization and charge distributions were calculated quantum mechanically (RHF/6-31+G**) with Gaussian 09 Revision A.1 (Gaussian, Inc., Wallingford, CT).The general AMBER force field 2 (GAFF2) was used to assign bonded and nonbonded parameters [90].
For HDAC1 and HDAC6-ligand complexes, binding metal sites and ligands were parametrized with the MCPB module embedded into the MTK++ software package in AMBER16 [91], as this methodology has been widely used to facilitate the modeling of metal effects on metalloproteins [92]. The optimized zinc and potassium metal coordination spheres were obtained by geometry optimization in the gas phase using Gaussian09 at B3LYP/6-31G* level, getting the equilibrium values of bond lengths, angles and force constants of the atoms bound to the metal.
Classical molecular dynamics (MD) simulations were performed on all complexes, with the AMBER16 program (http://ambermd.org/). The classic ff14SB AMBER force field [93] for the protein parametrization was applied together with previously assigned active site and ligand parameters. After that, systems were minimized at vacuum, to release possible undesired interactions. Complexes were then embedded in a TIP3P water octahedron of approximately 10 000 to 13 000 water molecules. System neutrality was achieved in all systems by adding sodium or chlorine ions when necessary. Water molecules and counter ions were minimized and then the embedded system was heated to 300 K for 25 ps at constant volume, keeping the protein restrained to initial positions using quadratic harmonic restraints with a constant force of 50 kcal mol−1Å−2. In all systems, the hydrogen bond lengths were kept at their equilibrium distance by means of the SHAKE algorithm [94]. Atom pair distance cutoffs were applied at 10.0 Å to compute the van der Waals interactions, while long-range electrostatics were computed by means of particle-mesh Ewald (PME) method [95]. Finally, MD simulation production was performed up to 20 ns using the thermostat NPT ensemble at 300 K, generating snapshots each 20 ps for further analysis. The trajectories of all complexes were collected and analyzed by the cpptraj module of AMBER16 [96] in order to obtain the root mean square deviation (RMSD) value of the atomic positions of the ligands.
3.3. Biological Assays
In Vitro HDAC1 and HDAC6 inhibitions were performed using Fluorometric Drug Discovery Assay Kits from Enzo Life Sciences, Inc. The reactions were prepared in HDAC assay buffer (50 mM Tris-HCl, pH = 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mg/mL BSA). 15 µL of HDAC1 (0.4 µg/well) or HDAC6 (0.5 µg/well) were incubated at 37 °C for 30 minutes with 10 µL of inhibitor diluted in 5% DMSO and 25 µL of fluorogenic substrate (0.05 µL/well of 5 mM solution). Reactions were stopped with the developer and trichostatin A, diluted in buffer without BSA, and the plate was incubated at 30 °C for 45 min, followed by measuring the fluorescence (Ex. 360 nm, Em. 450 nm, Fluoroskan, Thermo Scientific).
Recombinant human protein kinase GST-CK2 was produced in bacteria and purified as described elsewhere. The phosphorylation reactions were conducted at 37 °C for 5 min in 50 uL samples each containing 1 pmol of human recombinant CK2α, 40 mM of peptide substrate (RRRADDSDDDDD), and an appropriate concentration of the tested compounds (0.1–200 µM). The reaction buffer contained 20 µM [γ-32P] ATP (specific radioactivity 300–1000 cpm/pmol), 15 mM Mg2+, 20 mM Tris–HCl pH 7.5, and 6 mM 2-mercaptoethanol. 10 µL of the assay mixture was spotted onto a square (1 cm/1 cm) of Whatman P81 paper and allowed to dry. Each square was immersed in cold 0.5% phosphoric acid, and washed three times during 10 min, allowed to dry and transferred into a vessel containing Opti-Phase liquid (scintillation solution) and radioactivity was quantified using a scintillation counter (MicroBeta, Perkin Elmer) by Cerenkov counting. All experiments were performed in triplicate. IC50 values were calculated using GraphPad Prism (version 4.0) software.
3.4. Cytotoxicity Studies
Human HCT-116 colon carcinoma cells, breast adenocarcinoma cells of MCF-7 line, Jurkat T-leukemia cells, leukemia cells of HL-60 cell line and its drug-resistant sublines HL-60/adr (overexpression of MRP-1) and HL-60/vinc (overexpression of P-glycoprotein) were kindly provided by Prof. Walter Berger, Institute of Cancer Research, Vienna Medical University. Human embryonal kidney cells of HEK293 line were obtained from cell culture collection of R.E. Kavetsky Institute of Experimental Pathology, Oncology and Radiobiology, National Academy of Sciences of Ukraine (Kyiv, Ukraine).
Cells were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, USA) supplemented with 10% fetal calf serum (Sigma-Aldrich, St. Louis, USA), 50 µg/mL streptomycin (Sigma-Aldrich, St. Louis, USA), and 50 units/mL penicillin (Sigma-Aldrich, St. Louis, USA) in 5% CO2 humidified atmosphere at 37 °C. Cells were seeded (5 × 105/mL for leukemia cells and 1 × 105/mL for carcinoma cells) into 24-well tissue culture plates (Greiner Bio-one, Frickenhausen, Germany) and were permitted to adhere and grow for 24 h before the experiment. Short-term (24 h) cytotoxic effect of antitumor drugs was studied under the Evolution 300 Trino microscope (Delta Optical, Nowe Osiny, Poland) after cell staining with trypan blue dye (0.1%).
Stock solutions (2 mM) of each CK2-HDAC inhibitor in DMSO (99.5% pure, Sigma, USA) were prepared, and additionally dissolved in serum-free culture medium (RPMI for leukemia cells and DMEM for carcinoma cells) prior to addition to cell culture. Final concentration of DMSO in cell culture was 0.5% or less. Cytotoxicity studies revealed no statistically significant toxicity of 0.5% DMSO solution on all cell lines, used in these studies.
3.5. Apoptosis Analysis
For cell death analyses, cells were stained with FITC-conjugated annexin V and propidium iodide (PI) using an apoptosis detection kit (BD Biosciences, San Jose, CA), according to the manufacturer’s instructions. In particular, 24 h after the addition of various concentration of CK2-HDAC inhibitors, Jurkat T-leukemia cells were centrifuged at 2,000 rpm, washed twice with 1x PBS, and incubated for 15 minutes in Annexin V binding buffer (BD Pharmingen, USA) containing 1/50 volume of FITC-conjugated Annexin V solution and PI (50 µg/mL). Then samples were diluted two times by appropriate volume of Annexin V binding buffer (BD Pharmingen, USA) and immediately measured on FL1/FL2 (FITC-PI) channels of FACScan flow cytometer (Becton Dickinson, USA). Analysis of the obtained results was carried out using the Cytomation Summit Software v3.1 (Cytomation Inc., USA).
3.6. Cell Cycle Analysis
Cell cycle of the analyzed cells was assessed according to protocol described by Walker et al. [97]. After drug treatment, 2 × 106 cells were collected, pelleted by spinning at 1,000 rpm, 4 °C for five minutes, resuspended in 1 mL of cold PBS and fixed by adding drop by drop 4 mL of −20 °C absolute ethanol. On the next day, fixed cells were centrifuged again and cell pellets were resuspended in 1 mL of PBS. Then 100 µL of 200 µg/mL DNase-free, RNase A (Invitrogen, USA) was added to cell suspension and incubated at 37 °C for 30 minutes. After this 100 µL of 1 mg/mL propidium iodide was added to samples, which were incubated at room temperature for 5–10 min and then analyzed on FACScan flow cytometer (Becton Dickinson, USA). Cell cycle analysis was carried out using the Cytomation Summit Software v3.1 (Cytomation Inc., USA).
3.7. Studies of the Functional Status of Mitochondria
Breakdown of ΔΨm mitochondrial membrane potential was determined by FACS analysis using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1). For this purpose, the Mitochondrial Membrane Potential Detection Kit (Stratagene, La Jolla, CA, USA) was used as described in the manufacturer’s instruction. Briefly, 106 Jurkat cells were treated for 1, 3, 6, 12 and 24 h with the tested drugs. After PBS washing, cells were incubated for 10 minutes in freshly prepared JC-1 solution (10 mg/mL in culture medium) at 37 °C. Spare dye was removed by PBS washing and cell-associated fluorescence was measured on FL1/FL2 (FITC-PI) channels of FACScan flow cytometer (Becton Dickinson, USA). Analysis of the obtained results was carried out using the Cytomation Summit Software v3.1 (Cytomation Inc., USA).
3.8. DCFDA and DHE Assays
Cellular ROS contents were measured by incubating control or drug-treated cells with fluorescent dyes dihydrodichlorofluorescein diacetate (DCFDA, detecting mainly H2O2) and dihydroethidium (DHE, O2●−-specific) in concentrations of 10 μM at 37 °C for 30 minutes. After incubation with the dyes, cells were washed with PBS and immediately analyzed at FL1 (DCFDA) or FL2 channel (DHE) of FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
3.9. Statistical Analyses
All experiments were performed in triplicate and repeated three times. Statistical analysis of data was conducted in GraphPad Prism 7.0 Software (GraphPad Software, Inc.) using Student’s t test. Statistical significance was set at P ≤ 0.05. Evaluation of LC50 values of each compound was done by built-in functions of GraphPad Prism (non-linear regression, curve fit, [Inhibitor] vs. normalized response - Variable slope).
4. Conclusions
In this study we demonstrate that a multitarget approach can provide interesting anticancer lead compounds. In silico studies showed plausible predicted binding modes for all the designed compounds in spite of their different dynamic behaviors. The best result for a dual CK2/HDAC1 inhibitor in the enzymatic assays was found for 11b (CK2 IC50 = 1.46 µM; HDAC1 IC50 = 3.67 µM), a DMAT derivative with a five-carbon atom chain connecting the hydroxamate moiety. Furthermore, this compound was found to be even more active against HDAC6 (IC50 = 0.86 µM).
This compound showed a promising in vitro antiproliferative activity, with LC50 in the order of 4–23 µM in several tumor cell lines. Studies on the mechanism of cell death induction by 11b showed apoptotic activity and G1/S growth arrest in Jurkat T-leukemia cells. This compound was also able to circumvent multidrug resistance of human leukemia cells, caused by overexpression of MRP-1 and P-glycoprotein, and this phenomenon was accompanied by an increase in superoxide production both in sensitive and MDR cell lines.
Compound 11d, showing lower enzymatic activity against the three protein targets (CK2 IC50 = 5.89 µM; HDAC1 IC50 = 13.7 µM; HDAC6 IC50 = 8.98 µM), unexpectedly provided the best cellular activity with LC50 in 3–8 µM range. This difference in cellular behavior may be attributed to the increase in lipophilicity brought about by the longer seven-membered chain of 11d compared to the five carbon linker of 11b. Moreover, this inhibitor presented the highest cytotoxic activity, proapoptotic capability, and the best mitochondria-targeting and MDR circumventing properties, thus being the most promising drug candidate for further in vivo studies.
Altogether, this work further demonstrates that our multitarget HDAC/CK2 approach is a valid strategy for drug discovery in the field of anticancer drug development.
Acknowledgments
We thank Tamara Ruiz-Zorrilla for her contribution at the initial steps of this work.
Abbreviations
| ATP | adenosine triphosphate |
| CK2 | Protein kinase 2 |
| BSA | Bovine Serum Albumin |
| CuAAC | Cu (I)-catalyzed alkyne azide cycloaddition |
| DCFDA | 2′,7′ –dichlorofluorescin diacetate |
| DCM | dichloromethane |
| DMAT | 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole |
| DMF | dimethylformamide |
| DMSO | dimethyl sulfoxide |
| EDCI | 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| EGFR | epidermal growth factor receptor |
| FACS | Fluorescence-activated cell sorting |
| FITC | Fluorescein isothiocyanate |
| HAT | histone acetyl transferase |
| HDAC | histone deacetylase |
| HER2 | human epidermal growth factor receptor 2 |
| HOBt | 1-hydroxybenzotriazole |
| JAK2 | Janus kinase 2 |
| LC50 | lethal concentration 50 |
| LMD | molecular dynamics |
| MDR | multi-drug resistance |
| MTD | multitarget drugs |
| MW | microwave |
| NMM | N-Methylmorpholine |
| OTHP | 2-tetrahydropyranyl |
| PI3K | osfoinositol 3-quinasa |
| RMSD | root-mean-square deviation |
| ROS | reactive oxygen species |
| RT | room temperature |
| SAR | structure activity relationship |
| SRM | surface recognition moiety |
| TBB | tetrabromobenzotriazole |
| TBI | tetrabromobenzimidazole |
| TBTA | Tris(benzyltriazolyl)methylamine |
| THF | Tetrahydrofuran |
| TsOH | p-Toluenesulfonic acid |
| ZBG | Zinc binding group |
Supplementary Materials
The following are available online, HPLC data; 13C NMR and 1H NMR of spectra of compounds 11a–d, 7a–d, 15a and 19. Figures S1–S7, Tables S1–S3 and Figure S8.
Author Contributions
Chemical synthesis, purification and characterization of the compounds: R.M., J.M.Z. and M.P., supervized by A.R.; Molecular Modelling: B.D.G. and C.C. supervized by B.d.P.-T.; Enzymatic inhibitory evaluation: J.M.Z. (HDAC) and M.M. (CK2); In Vitro evaluation: R.P. and N.S.; Manuscript conceptualization, writing and editing: R.M. supervized jointly by A.R. and B.d.P.-T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Spanish MICIU/FEDER, UE grant number RTI2018-093539-B-I00. B. di G, thanks Fundación Universitaria San Pablo CEU for FPI fellowship and the Spanish MINECO for FPU fellowship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sawyers C.L. Mixing cocktails. Nature. 2007;449:993–996. doi: 10.1038/449993a. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann G.R., Lehár J., Keith C.T. Multi-target therapeutics: When the whole is greater than the sum of the parts. Drug Discov. Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Kwak E.L., Clark J.W., Chabner B. Targeted Agents: The Rules of Combination. Clin. Cancer Res. 2007;13:5232–5237. doi: 10.1158/1078-0432.CCR-07-1385. [DOI] [PubMed] [Google Scholar]
- 4.Simon F. The trouble with making combination drugs. Nat. Rev. Drug Discov. 2006;5:881–882. doi: 10.1038/nrd2188. [DOI] [PubMed] [Google Scholar]
- 5.Morphy R., Rankovic Z. Designed Multiple Ligands. An Emerging Drug Discovery Paradigm. J. Med. Chem. 2005;48:6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 6.Boyle N.M.O., Meegan M.J. Designed Multiple Ligands for Cancer Therapy. Curr. Med. Chem. 2011;18:4722–4737. doi: 10.2174/092986711797535344. [DOI] [PubMed] [Google Scholar]
- 7.Proschak E., Stark H., Merk D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J. Med. Chem. 2019;62:420–444. doi: 10.1021/acs.jmedchem.8b00760. [DOI] [PubMed] [Google Scholar]
- 8.Jonathan Fray M., Bish G., Brown A.D., Fish P.V., Stobie A., Wakenhut F., Whitlock G.A. N-(1,2-Diphenylethyl)piperazines: A new class of dual serotonin/noradrenaline reuptake inhibitor. Bioorg. Med. Chem. Lett. 2006;16:4345–4348. doi: 10.1016/j.bmcl.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 9.Neumeyer J.L., Peng X., Knapp B.I., Bidlack J.M., Lazarus L.H., Salvadori S., Trapella C., Balboni G. New Opioid Designed Multiple Ligand from Dmt-Tic and Morphinan Pharmacophores. J. Med. Chem. 2006;49:5640–5643. doi: 10.1021/jm0605785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bornot A., Bauer U., Brown A., Firth M., Hellawell C., Engkvist O. Systematic Exploration of Dual-Acting Modulators from a Combined Medicinal Chemistry and Biology Perspective. J. Med. Chem. 2013;56:1197–1210. doi: 10.1021/jm301653f. [DOI] [PubMed] [Google Scholar]
- 11.Costantino L., Barlocco D. Designed Multiple Ligands: Basic Research vs. Clinical Outcomes. Curr. Med. Chem. 2012;19:3353–3387. doi: 10.2174/092986712801215883. [DOI] [PubMed] [Google Scholar]
- 12.Cavalli A., Bolognesi M.L., Minarini A., Rosini M., Tumiatti V., Recanatini M., Melchiorre C. Multi-target-Directed Ligands To Combat Neurodegenerative Diseases. J. Med. Chem. 2008;51:347–372. doi: 10.1021/jm7009364. [DOI] [PubMed] [Google Scholar]
- 13.Bolognesi M.L. Harnessing Polypharmacology with Medicinal Chemistry. ACS Med. Chem. Lett. 2019;10:273–275. doi: 10.1021/acsmedchemlett.9b00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lolli G., Pinna L.A., Battistutta R. Structural Determinants of Protein Kinase CK2 Regulation by Autoinhibitory Polymerization. ACS Chem. Biol. 2012;7:1158–1163. doi: 10.1021/cb300054n. [DOI] [PubMed] [Google Scholar]
- 15.Litchfield D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/bj20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cozza G., Meggio F., Moro S. The Dark Side of Protein Kinase CK2 Inhibition. Curr. Med. Chem. 2011;18:2867–2884. doi: 10.2174/092986711796150423. [DOI] [PubMed] [Google Scholar]
- 17.St-Denis N.A., Litchfield D.W. Protein Kinase CK2 in Health and Disease. Cell. Mol. Life Sci. 2009;66:1817–1829. doi: 10.1007/s00018-009-9150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad K.A., Wang G., Unger G., Slaton J., Ahmed K. Protein kinase CK2—A key suppressor of apoptosis. Adv. Enzym. Regul. 2008;48:179–187. doi: 10.1016/j.advenzreg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra B., Issinger O.G. Protein Kinase CK2 in Human Diseases. Curr. Med. Chem. 2008;15:1870–1886. doi: 10.2174/092986708785132933. [DOI] [PubMed] [Google Scholar]
- 20.Chua M.M., Ortega E.C., Sheikh A., Lee M., Abdul-Rassoul H., Hartshorn L.K., Dominguez I. CK2 in Cancer: Cellular and Biochemical Mechanisms and Potential Therapeutic Target. Pharmaceuticals. 2017;10:18. doi: 10.3390/ph10010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unger G.M., Davis A.T., Ahmed J.W., Ahmed K. Protein Kinase CK2 as Regulator of Cell Survival: Implications for Cancer Therapy. Curr. Cancer Drug Targets. 2004;4:77–84. doi: 10.2174/1568009043481687. [DOI] [PubMed] [Google Scholar]
- 22.Cozza G., Moro S. Kinase CK2 Inhibition: An Update. Curr. Med. Chem. 2013;20:671–693. doi: 10.2174/092986713804999312. [DOI] [PubMed] [Google Scholar]
- 23.Cozza G., Girardi C., Ranchio A., Lolli G., Sarno S., Orzeszko A., Kazimierczuk Z., Battistutta R., Ruzzene M., Pinna L.A. Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: Structural features and pharmacological potential. Cell. Mol. Life Sci. 2014;71:3173–3185. doi: 10.1007/s00018-013-1552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cozza G. The Development of CK2 Inhibitors: From Traditional Pharmacology to in Silico Rational Drug Design. Pharmaceuticals. 2017;10:26. doi: 10.3390/ph10010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Battistutta R. Protein Kinase CK2 in Health and Disease. Cell. Mol. Life Sci. 2009;66:1868–1889. doi: 10.1007/s00018-009-9155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarno S., Moro S., Meggio F., Zagotto G., Dal Ben D., Ghisellini P., Battistutta R., Zanotti G., Pinna L.A. Toward the rational design of protein kinase casein kinase-2 inhibitors. Pharmacol. Therapeut. 2002;93:159–168. doi: 10.1016/S0163-7258(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 27.Raaf J., Guerra B., Neundorf I., Bopp B., Issinger O.-G., Jose J., Pietsch M., Niefind K. First Structure of Protein Kinase CK2 Catalytic Subunit with an Effective CK2β-Competitive Ligand. ACS Chem. Biol. 2013;8:901–907. doi: 10.1021/cb3007133. [DOI] [PubMed] [Google Scholar]
- 28.Prudent R., Cochet C. New Protein Kinase CK2 Inhibitors: Jumping out of the Catalytic Box. Chem. Biol. 2009;16:112–120. doi: 10.1016/j.chembiol.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Swider R., Masłyk M., Zapico J.M., Coderch C., Panchuk R., Skorokhyd N., Schnitzler A., Niefind K., de Pascual-Teresa B., Ramos A. Synthesis, biological activity and structural study of new benzotriazole-based protein kinase CK2 inhibitors. RSC Adv. 2015;5:72482–72494. doi: 10.1039/C5RA12114K. [DOI] [Google Scholar]
- 30.Purwin M., Hernández-Toribio J., Coderch C., Panchuk R., Skorokhyd N., Filipiak K., de Pascual-Teresa B., Ramos A. Design and synthesis of novel dual-target agents for HDAC1 and CK2 inhibition. RSC Adv. 2016;6:66595–66608. doi: 10.1039/C6RA09717K. [DOI] [Google Scholar]
- 31.Pastor M., Zapico J.M., Coderch C., Maslyk M., Panchuk R., de Pascual-Teresa B., Ramos A. From a MMP2/CK2 multitarget approach to the identification of potent and selective MMP13 inhibitors. Org. Biomol. Chem. 2019;17:916–929. doi: 10.1039/C8OB02990C. [DOI] [PubMed] [Google Scholar]
- 32.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 33.Sekiguchi Y., Nakaniwa T., Kinoshita T., Nakanishi I., Kitaura K., Hirasawa A., Tsujimoto G., Tada T. Structural insight into human CK2alpha in complex with the potent inhibitor ellagic acid. Bioorgan. Med. Chem. Lett. 2009;19:2920–2923. doi: 10.1016/j.bmcl.2009.04.076. [DOI] [PubMed] [Google Scholar]
- 34.Pierre F., Chua P.C., O’Brien S.E., Siddiqui-Jain A., Bourbon P., Haddach M., Michaux J., Nagasawa J., Schwaebe M.K., Stefan E., et al. Discovery and SAR of 5-(3-Chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic Acid (CX-4945), the First Clinical Stage Inhibitor of Protein Kinase CK2 for the Treatment of Cancer. J. Med. Chem. 2011;54:635–654. doi: 10.1021/jm101251q. [DOI] [PubMed] [Google Scholar]
- 35.Battistutta R., Cozza G., Pierre F., Papinutto E., Lolli G., Sarno S., O’Brien S.E., Siddiqui-Jain A., Haddach M., Anderes K., et al. Unprecedented Selectivity and Structural Determinants of a New Class of Protein Kinase CK2 Inhibitors in Clinical Trials for the Treatment of Cancer. Biochemistry. 2011;50:8478–8488. doi: 10.1021/bi2008382. [DOI] [PubMed] [Google Scholar]
- 36.Witt O., Deubzer H.E., Milde T., Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Marks P.A., Rifkind R.A., Richon V.M., Breslow R., Miller T., Kelly W.K. Histone deacetylases and cancer: Causes and therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 38.Gräff J., Tsai L.-H. The Potential of HDAC Inhibitors as Cognitive Enhancers. Annu. Rev. Pharmacol. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- 39.Andrews K.T., Haque A., Jones M.K. HDAC inhibitors in parasitic diseases. Immunol. Cell Biol. 2012;90:66–77. doi: 10.1038/icb.2011.97. [DOI] [PubMed] [Google Scholar]
- 40.Rotili D., Simonetti G., Savarino A., Palamara A.T., Migliaccio A.R., Mai A. Non-Cancer Uses of Histone Deacetylase Inhibitors: Effects on Infectious Diseases and β-Hemoglobinopathies+ Curr. Top. Med. Chem. 2009;9:272–291. doi: 10.2174/156802609788085296. [DOI] [PubMed] [Google Scholar]
- 41.Gluckman P.D., Hanson M.A., Buklijas T., Low F.M., Beedle A.S. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat. Rev. Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 42.Gregoretti I., Lee Y.-M., Goodson H.V. Molecular Evolution of the Histone Deacetylase Family: Functional Implications of Phylogenetic Analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehrmann H., Pritchard L.L., Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 45.Lane A.A., Chabner B.A. Histone Deacetylase Inhibitors in Cancer Therapy. J. Clin. Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 46.Minucci S., Pelicci P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 47.Pontiki E., Hadjipavlou-Litina D. Histone deacetylase inhibitors (HDACIs). Structure—Activity relationships: History and new QSAR perspectives. Med. Res. Rev. 2012;32:1–165. doi: 10.1002/med.20200. [DOI] [PubMed] [Google Scholar]
- 48.Paris M., Porcelloni M., Binaschi M., Fattori D. Histone Deacetylase Inhibitors: From Bench to Clinic. J. Med. Chem. 2008;51:1505–1529. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 49.Schobert R., Biersack B. Multimodal HDAC Inhibitors with Improved Anticancer Activity. Curr. Cancer Drug Targets. 2018;18:39–56. doi: 10.2174/1568009617666170206102613. [DOI] [PubMed] [Google Scholar]
- 50.Hesham H., Lasheen D., Abouzid K. Chimeric HDAC inhibitors: Comprehensive review on the HDAC-based strategies developed to combat cancer. Med. Res. Rev. 2018;38:2058–2109. doi: 10.1002/med.21505. [DOI] [PubMed] [Google Scholar]
- 51.Tang C., Li C., Zhang S., Hu Z., Wu J., Dong C., Huang J., Zhou H.-B. Novel Bioactive Hybrid Compound Dual Targeting Estrogen Receptor and Histone Deacetylase for the Treatment of Breast Cancer. J. Med. Chem. 2015;58:4550–4572. doi: 10.1021/acs.jmedchem.5b00099. [DOI] [PubMed] [Google Scholar]
- 52.Duan W., Li J., Inks E.S., Chou C.J., Jia Y., Chu X., Li X., Xu W., Zhang Y. Design, Synthesis, and Antitumor Evaluation of Novel Histone Deacetylase Inhibitors Equipped with a Phenylsulfonylfuroxan Module as a Nitric Oxide Donor. J. Med. Chem. 2015;58:4325–4338. doi: 10.1021/acs.jmedchem.5b00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ai T., Cui H., Chen L. Multi-Targeted Histone Deacetylase Inhibitors in Cancer Therapy. Curr. Med. Chem. 2012;19:475–487. doi: 10.2174/092986712798918842. [DOI] [PubMed] [Google Scholar]
- 54.Seo S.-Y. Multi-targeted hybrids based on HDAC inhibitors for anti-cancer drug discovery. Arch. Pharm. Res. 2012;35:197–200. doi: 10.1007/s12272-012-0221-9. [DOI] [PubMed] [Google Scholar]
- 55.Guerrant W., Patil V., Canzoneri J.C., Oyelere A.K. Dual Targeting of Histone Deacetylase and Topoisomerase II with Novel Bifunctional Inhibitors. J. Med. Chem. 2012;55:1465–1477. doi: 10.1021/jm200799p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L., Petrelli R., Gao G., Wilson D.J., McLean G.T., Jayaram H.N., Sham Y.Y., Pankiewicz K.W. Dual inhibitors of inosine monophosphate dehydrogenase and histone deacetylase based on a cinnamic hydroxamic acid core structure. Bioorgan. Med. Chem. 2010;18:5950–5964. doi: 10.1016/j.bmc.2010.06.081. [DOI] [PubMed] [Google Scholar]
- 57.Tavera-Mendoza L.E., Quach T.D., Dabbas B., Hudon J., Liao X., Palijan A., Gleason J.L., White J.H. Incorporation of histone deacetylase inhibition into the structure of a nuclear receptor agonist. Proc. Nat. Acad. Sci. USA. 2008;105:8250–8255. doi: 10.1073/pnas.0709279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishiguchi G.A., Rico A., Tanner H., Aversa R.J., Taft B.R., Subramanian S., Setti L., Burger M.T., Wan L., Tamez V., et al. Design and Discovery of N-(2-Methyl-5′-morpholino-6′-((tetrahydro-2H-pyran-4-yl)oxy)-[3′,-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers. J. Med. Chem. 2017;60:4869–4881. doi: 10.1021/acs.jmedchem.6b01862. [DOI] [PubMed] [Google Scholar]
- 59.Ling Y., Liu J., Qian J., Meng C., Guo J., Gao W., Xiong B., Ling C., Zhang Y. Recent Advances in Multi-target Drugs Targeting Protein Kinases and Histone Deacetylases in Cancer Therapy. Curr. Med. Chem. 2020;27:1–24. doi: 10.2174/0929867327666200102115720. [DOI] [PubMed] [Google Scholar]
- 60.Chu-Farseeva Y.Y., Mustafa N., Poulsen A., Tan E.C., Yen J.J.Y., Chng W.J., Dymock B.W. Design and synthesis of potent dual inhibitors of JAK2 and HDAC based on fusing the pharmacophores of XL019 and vorinostat. Eur. J. Med. Chem. 2018;158:593–619. doi: 10.1016/j.ejmech.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 61.Luan Y., Li J., Bernatchez J.A., Li R. Kinase and Histone Deacetylase Hybrid Inhibitors for Cancer Therapy. J. Med. Chem. 2018;62:3171–3183. doi: 10.1021/acs.jmedchem.8b00189. [DOI] [PubMed] [Google Scholar]
- 62.Cai X., Zhai H.-X., Wang J., Forrester J., Qu H., Yin L., Lai C.-J., Bao R., Qian C. Discovery of 7-(4-(3-Ethynylphenylamino)-7-methoxyquinazolin-6-yloxy)-N-hydroxyheptanamide (CUDC-101) as a Potent Multi-Acting HDAC, EGFR, and HER2 Inhibitor for the Treatment of Cancer. J. Med. Chem. 2010;53:2000–2009. doi: 10.1021/jm901453q. [DOI] [PubMed] [Google Scholar]
- 63.Mahboobi S., Dove S., Sellmer A., Winkler M., Eichhorn E., Pongratz H., Ciossek T., Baer T., Maier T., Beckers T. Design of Chimeric Histone Deacetylase- and Tyrosine Kinase-Inhibitors: A Series of Imatinib Hybrides as Potent Inhibitors of Wild-Type and Mutant BCR-ABL, PDGF-Rβ, and Histone Deacetylases. J. Med. Chem. 2009;52:2265–2279. doi: 10.1021/jm800988r. [DOI] [PubMed] [Google Scholar]
- 64.Khan D.H., He S., Yu J., Winter S., Cao W., Seiser C., Davie J.R. Protein kinase CK2 regulates the dimerization of histone deacetylase 1 (HDAC1) and HDAC2 during mitosis. J. Biol. Chem. 2013;288:16518–16528. doi: 10.1074/jbc.M112.440446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pluemsampant S., Safronova O.S., Nakahama K.-I., Morita I. Protein kinase CK2 is a key activator of histone deacetylase in hypoxia-associated tumors. Int. J. Cancer. 2008;122:333–341. doi: 10.1002/ijc.23094. [DOI] [PubMed] [Google Scholar]
- 66.Thirumurugan P., Matosiuk D., Jozwiak K. Click Chemistry for Drug Development and Diverse Chemical–Biology Applications. Chem. Rev. 2013;113:4905–4979. doi: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- 67.Andrzejewska M., Pagano M.A., Meggio F., Brunati A.M., Kazimierczuk Z. Polyhalogenobenzimida-zoles: Synthesis and Their inhibitory activity against casein kinases. Bioorgan. Med. Chem. 2003;11:3997–4002. doi: 10.1016/S0968-0896(03)00403-6. [DOI] [PubMed] [Google Scholar]
- 68.Świder R., Masłyk M., Martín-Santamaría S., Ramos A., de Pascual-Teresa B. Multisite-directed inhibitors of protein kinase CK2: New challenges. Mol. Cell. Biochem. 2011;356:117–119. doi: 10.1007/s11010-011-0962-7. [DOI] [PubMed] [Google Scholar]
- 69.Porter N.J., Osko J.D., Diedrich D., Kurz T., Hooker J.M., Hansen F.K., Christianson D.W. Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity. J. Med. Chem. 2018;61:8054–8060. doi: 10.1021/acs.jmedchem.8b01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan Y.-G., Peng Q.-L., Gurunathan S. Combination of palladium nanoparticles and tubastatin-A potentiates apoptosis in human breast cancer cells: A novel therapeutic approach for cancer. Int. J. Nanomed. 2017;12:6503–6520. doi: 10.2147/IJN.S136142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sellmer A., Stangl H., Beyer M., Grünstein E., Leonhardt M., Pongratz H., Eichhorn E., Elz S., Striegl B., Jenei-Lanzl Z., et al. Marbostat-100 Defines a New Class of Potent and Selective Antiinflammatory and Antirheumatic Histone Deacetylase 6 Inhibitors. J. Med. Chem. 2018;61:3454–3477. doi: 10.1021/acs.jmedchem.7b01593. [DOI] [PubMed] [Google Scholar]
- 72.Huang Y., Dong G., Li H., Liu N., Zhang W., Sheng C. Discovery of Janus Kinase 2 (JAK2) and Histone Deacetylase (HDAC) Dual Inhibitors as a Novel Strategy for the Combinational Treatment of Leukemia and Invasive Fungal Infections. J. Med. Chem. 2018;61:6056–6074. doi: 10.1021/acs.jmedchem.8b00393. [DOI] [PubMed] [Google Scholar]
- 73.Wiley R.H., Hussung K.F. Halogenated benzotriazoles. J. Am. Chem. Soc. 1957;79:4395–4400. doi: 10.1021/ja01573a042. [DOI] [Google Scholar]
- 74.Pagano M.A., Andrzejewska M., Ruzzene M., Sarno S., Cesaro L., Bain J., Elliott M., Meggio F., Kazimierczuk Z., Pinna L.A. Optimization of Protein Kinase CK2 Inhibitors Derived from 4,5,6,7-Tetrabromobenzimidazole. J. Med. Chem. 2004;47:6239–6247. doi: 10.1021/jm049854a. [DOI] [PubMed] [Google Scholar]
- 75.Battistutta R., De Moliner E., Sarno S., Zanotti G., Pinna L.A. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 2001;10:2200–2206. doi: 10.1110/ps.19601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Battistutta R., Mazzorana M., Cendron L., Bortolato A., Sarno S., Kazimierczuk Z., Zanotti G., Moro S., Pinna L.A. The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules. Chembiochem. 2007;8:1804–1809. doi: 10.1002/cbic.200700307. [DOI] [PubMed] [Google Scholar]
- 77.Battistutta R., Mazzorana M., Sarno S., Kazimierczuk Z., Zanotti G., Pinna L.A. Inspecting the structure-activity relationship of protein kinase CK2 inhibitors derived from tetrabromo-benzimidazole. Chem. Biol. 2005;12:1211–1219. doi: 10.1016/j.chembiol.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 78.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernhart E., Stuendl N., Kaltenegger H., Windpassinger C., Donohue N., Leithner A., Lohberger B. Histone deacetylase inhibitors vorinostat and panobinostat induce G1 cell cycle arrest and apoptosis in multidrug resistant sarcoma cell lines. Oncotarget. 2017;8:77254–77267. doi: 10.18632/oncotarget.20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garraway L.A., Jänne P.A. Circumventing Cancer Drug Resistance in the Era of Personalized Medicine. Cancer Discov. 2012;2:214. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 81.Borgo C., Ruzzene M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Canc. Res. 2019;38:287. doi: 10.1186/s13046-019-1292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pizzi M., Piazza F., Agostinelli C., Fuligni F., Benvenuti P., Mandato E., Casellato A., Rugge M., Semenzato G., Pileri S.A. Protein kinase CK2 is widely expressed in follicular, Burkitt and diffuse large B-cell lymphomas and propels malignant B-cell growth. Oncotarget. 2015;6:6544–6552. doi: 10.18632/oncotarget.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korynevska A., Heffeter P., Matselyukh B., Elbling L., Micksche M., Stoika R., Berger W. Mechanisms underlying the anticancer activities of the angucycline landomycin E. Biochem. Pharmacol. 2007;74:1713–1726. doi: 10.1016/j.bcp.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 84.Ruiz-Ojeda F.J., Gomez-Llorente C., Aguilera C.M., Gil A., Rupérez A.I. Impact of 3-Amino-1,2,4-Triazole (3-AT)-Derived Increase in Hydrogen Peroxide Levels on Inflammation and Metabolism in Human Differentiated Adipocytes. PLoS ONE. 2016;11:e0152550. doi: 10.1371/journal.pone.0152550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson P.J., Millard C.J., Riley A.M., Robertson N.S., Wright L.C., Godage H.Y., Cowley S.M., Jamieson A.G., Potter B.V.L., Schwabe J.W.R. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 2016;7:11262. doi: 10.1038/ncomms11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hai Y., Christianson D.W. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat. Chem. Biol. 2016;12:741. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen K., Zhang X., Wu Y.-D., Wiest O. Inhibition and Mechanism of HDAC8 Revisited. J. Am. Chem. Soc. 2014;136:11636–11643. doi: 10.1021/ja501548p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 89.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 90.Case D.A., Betz R.M., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Homeyer N., et al. AMBER 2016. University of California; San Francisco, CA, USA: 2016. [Google Scholar]
- 91.Li P., Merz K.M. MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016;56:599–604. doi: 10.1021/acs.jcim.5b00674. [DOI] [PubMed] [Google Scholar]
- 92.Torras J., Maccarrone M., Dainese E. Molecular dynamics study on the Apo- and Holo-forms of 5-lipoxygenase. Biotechnol. Appl. Bioc. 2018;65:54–61. doi: 10.1002/bab.1583. [DOI] [PubMed] [Google Scholar]
- 93.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoneya M., Berendsen H.J.C., Hirasawa K. A Non-Iterative Matrix Method for Constraint Molecular Dynamics Simulations. Mol. Simulat. 1994;13:395–405. doi: 10.1080/08927029408022001. [DOI] [Google Scholar]
- 95.Cheatham T.E., III, Miller J.L., Fox T., Darden T.A., Kollman P.A. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. J. Am. Chem. Soc. 1995;117:4193–4194. doi: 10.1021/ja00119a045. [DOI] [Google Scholar]
- 96.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 97.Walker P.R., Kwast-Welfeld J., Gourdeau H., Leblanc J., Neugebauer W., Sikorska M. Relationship between Apoptosis and the Cell Cycle in Lymphocytes: Roles of Protein Kinase C, Tyrosine Phosphorylation, and AP1. Exp. Cell Res. 1993;207:142–151. doi: 10.1006/excr.1993.1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.