Abstract
Liver regeneration is a fundamental biological process required for sustaining body homeostasis and restoring liver function after injury. Emerging evidence demonstrates that cytokines, growth factors, and multiple signaling pathways contribute to liver regeneration. Mammalian target of rapamycin complex 2 (mTORC2) regulates cell metabolism, proliferation and survival. The major substrates for mTORC2 are the AGC family members of kinases, including AKT, SGK, and PKC-α. We investigated the functional roles of mTORC2 during liver regeneration. Partial hepatectomy (PHx) was performed in liver-specific Rictor (the pivotal unit of mTORC2 complex) knockout (RictorLKO) and wild-type (Rictorfl/fl) mice. Rictor-deficient mice were found to be more intolerant to PHx and displayed higher mortality after PHx. Mechanistically, loss of Rictor resulted in decreased Akt phosphorylation, leading to a delay in hepatocyte proliferation and lipid droplets formation along liver regeneration. Overall, these results indicate an essential role of the mTORC2 signaling pathway during liver regeneration.
The liver is a vital organ and possesses multiple essential functions for regulating metabolism of other tissues in the body, including detoxification, decomposition, synthetization, digestion, and storage.1,2 Regeneration after injury or surgical resection is a unique capacity of the liver.3,4 It has been reported that healthy liver can regenerate back to its full size from as little as 25% of the original liver mass.5 Consequently, partial hepatectomy (PHx) and liver transplantation are widely performed among patients with various liver diseases.6 However, clinical scenarios of patients with liver dysfunction are more complicated, and regeneration processes in unhealthy livers are simultaneously affected by other complications. Indications for these procedures tightly depend on liver remnant regeneration potential. Furthermore, no therapeutic strategies to accelerate liver regeneration exist.7 Thus, a better understanding of the mechanisms whereby liver regeneration occurs would be highly beneficial for tackling these issues.
Described >80 years ago, the two-thirds PHx remains the state-of-the-art method for the in-depth analysis of signaling pathways involved in liver regeneration.8,9 After PHx, the remaining liver lobes are able to regenerate and completely replace the missing parenchyma, with consequent morphologic recovery and functional restoration.10 In this process, hepatocytes replicate first, followed by proliferation of biliary epithelia cells and sinusoidal endothelial cells. Meanwhile, newly divided cells undergo restructuring, angiogenesis, and reformation of extracellular matrix to structurally recover from PHx.11 Simultaneously, with a series of intrinsic regulations, liver function recovers to maintain the body homeostasis. In mice, it takes approximately 1 week for the liver to fully regenerate after PHx.
Several signaling pathways are involved in liver regeneration, including growth factors, cytokines, hormones, and many transcription factors. Among them, mammalian target of rapamycin (mTOR) signaling is one of the well-known pathways to be involved in liver regeneration.12 mTOR kinases include two distinct protein complexes with specific binding partners, consisting of regulatory-associated protein of mTOR (Raptor) in mTOR complex 1 (mTORC1) and rapamycin-insensitive companion of mTOR (Rictor) in mTOR complex 2 (mTORC2). mTORC2 regulates Akt, Sgk, and Pkc (AGC) kinases, which are essential for organismal development and viability in response to nutrient availability.13 Akt kinases are considered to be the major substrates for mTORC2; once activated, mTORC2 phosphorylates Akt kinases at the S473 (for Akt1) or S474 (for Akt2) position, which is required for their activation. In mammals, there are three Akt isoforms, namely, Akt1, Akt2, and Akt3. Although Akt1 and Akt2 are expressed in the liver, Akt3 is predominantly expressed in the brain. Akt2 comprises approximately 85% of total Akt in the liver and is the major Akt isoform downstream of insulin receptors in regulating glucose and fatty acid catabolism.14
Mounting evidence indicates the key roles of mTOR signaling during liver regeneration.12,15 Studies have found that the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathway is activated after PHx.16, 17, 18 mTOR phosphorylates S6K1 and is required for the expression of cyclin D1 during liver regeneration.19 Treatment with rapamycin, an mTORC1 inhibitor, after PHx leads to decreased hepatocyte proliferation and increased apoptosis.20,21 In addition, recent studies have found that ablation of Akt1 or Akt2 alone does not influence liver regeneration after PHx in mice, whereas combined loss of Akt1 and Akt2 isoforms leads to impaired regeneration and increased mortality. Intriguingly, simultaneous deletion of FoxO1 in the liver rescued the survival defects of Akt1 and Akt2 double knockout (KO) mice.22 Altogether, this body of information implies pivotal function of Akt in liver development and regeneration. However, because Akt regulates several proteins and signaling cascades, the specific requirement of mTORC2 in this process and whether Akt activation is required during liver regeneration remain poorly defined.
To better delineate the role of mTORC2 along liver regeneration, two-thirds PHx was performed in liver-specific Rictor KO (RictorLKO) mice and wild-type (WT) mice. RictorLKO mice were found to be more intolerant to PHx, having a lower survival rate than WT mice. Moreover, delayed cell proliferation and lipid droplets formation as well as cell cycle arrest were observed in RictorLKO mice. Therefore, our findings highlight the role of mTORC2 signaling in lipid formation and cell proliferation during liver regeneration.
Materials and Methods
Animals
Albumin-Cre23 and Rictorflox/flox mice24 (all in the C57BL/6 background) were purchased from Jackson Laboratory (Bar Harbor, ME) and then crossed to generate Albumin-Cre+ Rictorflox/flox (RictorLKO) and Albumin-Cre- Rictorflox/flox mice (Rictorfl/fl). Mice were sacrificed at 3, 5, and 8 weeks of age for the liver development study. For the liver regeneration study, two-thirds PHx was performed in 8-week–old male mice. Genotyping was conducted using a Mouse Direct PCR Kit (catalog number B40015; Bimake, Houston, TX) according to the standard protocol. The primers used were as follows: common reverse, 5′-CCTGCCAGCCATGGATATAA-3′; mutant forward, 5′-GAAGCAGAAGCTTAGGAAGATGG-3′; wild-type forward, 5′-GTTGTCCTTCTTTGTGCTGA-3′. Animal experiments were performed according to protocols approved by the Committee for Animal Research at the University of California, San Francisco (San Francisco, CA).
PHx Model
PHx was performed according to the technique described by Mitchell and Willenbring.25 In brief, two-thirds of the liver (consisting of median and left lobes) was removed. The two-thirds PHx and sham surgery operation were performed under isoflurane (Sigma-Aldrich, St. Louis, MO) anesthesia. After PHx, mice were sacrificed at different time points: 1 to 7 days. The sham group was used as internal control. The regenerated liver was weighed, and the liver/body weight ratio was calculated. Liver tissues and blood samples were collected and analyzed as previously described.26
Histologic and Immunohistochemical Analysis
Tissue samples were fixed with 10% neutral buffered formalin (catalog number 5701; Thermo Fisher Scientific, Waltham, MA) at 4°C overnight, followed by paraffin embedding. Tissue blocks were trimmed and cut at a thickness of 5 μm. Antigen retrieval was conducted by boiling slides for 10 minutes with sodium citrate buffer (pH 6.0). Then the slides were blocked with the goat serum/ phosphate-buffered saline (PBS) solution (1:20). Endogenous biotin, biotin receptors, and avidin-binding sites present in tissues were mocked with the Avidin/Biotin blocking kit (catalog number SP2001; Vector Laboratories, Burlingame, CA). After three washing steps in PBS, the tissue sections were then incubated with designated primary antibodies at 4°C overnight. The next day, slides were washed in PBS, and then endogenous peroxidase was blocked by 3% hydrogen peroxide at room temperature for 10 minutes. Subsequently, slides were incubated with the biotin conjugated secondary antibody (1:500; catalog number B2770; Thermo Fisher Scientific, Waltham, MA) for 30 minutes. The antibody was used according to the manufacturers and dilution for immunohistochemical analysis. After washing with PBS, the immunostaining signals were visualized using the Vectastain Elite ABC Kit (catalog number PK6100; Vector Laboratories, Burlingame, CA) and ImmPACT DAB EqV Peroxidase Substrate (catalog number SK4103; Vector Laboratories). Ki-67 and phosphorylated histone 3 (PH3) (Table 1) staining were quantified by using ImageJ software version 1.8.0 (NIH, Bethesda, MD; http://imagej.nih.gov/ij) and Image Pro Plus 7 software (Media Cybernetics, Rockville, MD).
Table 1.
Antibody Information for Immunohistochemistry
| Antibody | Concentration | Manufacturer | Catalog number |
|---|---|---|---|
| Ki-67 | 1:150 | Thermo Fisher Scientific | RM-9106-S1 |
| Phosphorylated -histone H3 (Ser10) | 1:100 | Cell Signaling Technology | 9701 |
Oil Red O and Periodic Acid-Schiff Staining
Liver tissues were embedded in optimum cutting temperature compound (Tissue-Tek, Sakura Finetek, Torrance, CA) and frozen in cold 2-methylbutane at −80°C. Frozen sections were cut at 5 μm in thickness. Oil Red O (catalog number KTORO; American MasterTech Scientific Laboratory Supplies, Lodi, CA) and periodic acid-Schiff (catalog number 22110645; Thermo Fisher Scientific) staining was performed on frozen sections according to the standard instructions.
Protein Extraction and Western Blot Analysis
Frozen mouse liver tissues were homogenized and lysed by applying M-PER Mammalian Protein Extraction Reagent (catalog number 78501; Thermo Fisher Scientific). A Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA) was used to determine protein concentration according to the protocol of the manufacturer. Protein lysates (25 μg) were denatured and separated by SDS-PAGE. Nonspecific proteins on transferred membranes were blocked with a nonfat dry milk and tris-buffered saline with Tween 20 solution (1:20) for 1 hour and incubated with specific primary antibodies (Table 2) at 4°C overnight. After washing in tris-buffered saline with Tween 20 for 10 minutes three times, anti-mouse or anti-rabbit horseradish peroxidase secondary antibody (1:5000) was applied. Protein expression was detected with Super Signal West Pico Chemiluminescent Substrate (Pierce Chemical Co., New York, NY).
Table 2.
Antibody Information Western Blotting
| Antibody | Concentration | Manufacturer | Catalog number |
|---|---|---|---|
| Rictor | 1:500 | Cell Signaling Technology | 9476 |
| p-AKT (Ser473) | 1:1000 | Cell Signaling Technology | 3787 |
| Total AKT | 1:1000 | Cell Signaling Technology | 9272 |
| p-FoxO1 (Ser256) | 1:1000 | Cell Signaling Technology | 84192 |
| p-mTOR (Ser2448) | 1:1000 | Cell Signaling Technology | 2971 |
| p-PRAS40 (Thr246) | 1:1000 | Cell Signaling Technology | 2997 |
| Acetyl-CoA carboxylase | 1:1000 | Cell Signaling Technology | 3676 |
| FASN | 1:1000 | Cell Signaling Technology | 3180 |
| p-4E-BP1 (Ser65) | 1:1000 | Cell Signaling Technology | 9451 |
| p-4E-BP1 (Thr37/46) | 1:2000 | Cell Signaling Technology | 2855 |
| 4E-BP1 | 1:2000 | Cell Signaling Technology | 9644 |
| p-S6 ribosomal protein (Ser235/236) | 1:2000 | Cell Signaling Technology | 4858 |
| p-AMPKα1 (Ser485) | 1:1000 | Cell Signaling Technology | 2537 |
| p-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | 1:1000 | Cell Signaling Technology | 4370 |
| p44/42 MAPK (Erk1/2) | 1:1000 | Cell Signaling Technology | 9102 |
| PCNA | 1:1000 | Cell Signaling Technology | 2586 |
| Cyclin D antibody | 1:1000 | Millipore | 06-137 |
| GAPDH | 1:5000 | Millipore | MAB374 |
4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; AKT, serine/threonine kinase; AMPK, 5'adenosine monophosphate–activated protein kinase; FASN, fatty acid synthase; FoxO1, forkhead box protein O1; GAPHD, glyceraldehyde-3-phosphate dehydrogenase; MAPK, mitogen-activated protein kinase; mTOR, the mammalian target of rapamycin; p-, phosphorylated; PCNA, proliferating cell nuclear antigen; RAS40, the proline-rich Akt substrate of 40 kDa.
Biochemical Assays
For glucose measurement, blood was collected in test strips and directly measured in a glucometer (Accu-Chek Active, Roche Diagnostics, Mannheim, Germany). Other blood biochemical indicators were sent and analyzed by the Zuckerberg San Francisco General Hospital Clinical Lab.
Hepatocyte Ploidy Analysis
Primary mouse hepatocytes were isolated using a two-step collagenase perfusion as previously described.27 Before perfusion, a single lobe was ligated and collected for histologic analysis. Hepatocytes were separated from nonparenchymal cells using slow spin centrifugation and passed through a 70-μm cell strainer before purification through 50% equilibrated Percoll (GE Healthcare Biosciences, Marlborough, MA). Isolated hepatocytes were resuspended in Williams E Medium (Gibco, Waltham, MA) supplemented with GlutaMAX (Gibco), nonessential amino acids (Gibco), penicillin-streptomycin (Corning Inc., Corning, NY), and fetal bovine serum. Hepatocyte ploidy staining was performed as previously described.28 Briefly, cells in suspension were stained with 15 μg/mL of Hoechst (catalog number 33342; Invitrogen, Carlsbad, CA), and 5 μmol/L of reserpine (Sigma-Aldrich) at 37°C for 30 minutes. After initial incubation, 5 μg/mL of propidium iodide was added to the cells and kept in ice. DNA content of labeled live cells was measured by flow cytometer (LSR II; BD, Franklin Lakes, NJ). Acquired data were analyzed by FACSDiva software version 8.0 (BD) and FlowJo software version 10 (BD).
RNA Sequencing and Data Analysis
Experimental design consisted of six groups: WT0D, WT2D, WT4D, KO0D, KO2D, and KO4D. WT represents WT Rictorfl/fl liver samples, whereas KO represents liver samples from RictorLKO mice. Each group had two biological replicates. RNA was isolated from liver tissues using Zymo quick-RNA miniprep kit (catalog number D7001; Zymo Research, Irvine, CA). RNA was quantified by nanodrop. RNA quality control was conducted with Bioanalyzer (Aligent, Santa Clara, CA). Library preparation and sequencing were performed by Novogene (Sacramento, CA). Gene read counts were in Ensembl ID and converted to Entrez ID using the Bioconductor Package Maintainer org.Mm.eg.db version 3.8.2 (Bioconductor, Buffalo, NY; https://www.bioconductor.org). Duplicated and unannotated genes were removed. Data were filtered with counts per million >0.1 in at least two libraries. Normalized data were obtained by subtracting WT or KO data of 2D and 4D with 0D data relatively. The Edge R package version 3.28.1 (Bioconductor) was used to normalize data, calculate fragments per kilogram of exon model per million reads mapped, draw multidimensional scaling plot, and conduct KEGG pathway analysis. Heatmap was generated by Complexheatmap. Other plots was drawn with ggplot2 (R Foundation for Statistical Computing, Vienna, Austria).29
Statistical Analysis
Prism software version 6.0 (GraphPad, San Diego, CA) was applied to analyze the data, which are presented as means ± SD. Comparisons between two groups were performed with the two-tailed unpaired t-test. Survival curves were compared by the log-rank t-test. P < 0.05 was considered statistically significant.
Results
Activation of mTORC2 Signaling after PHx in Mice
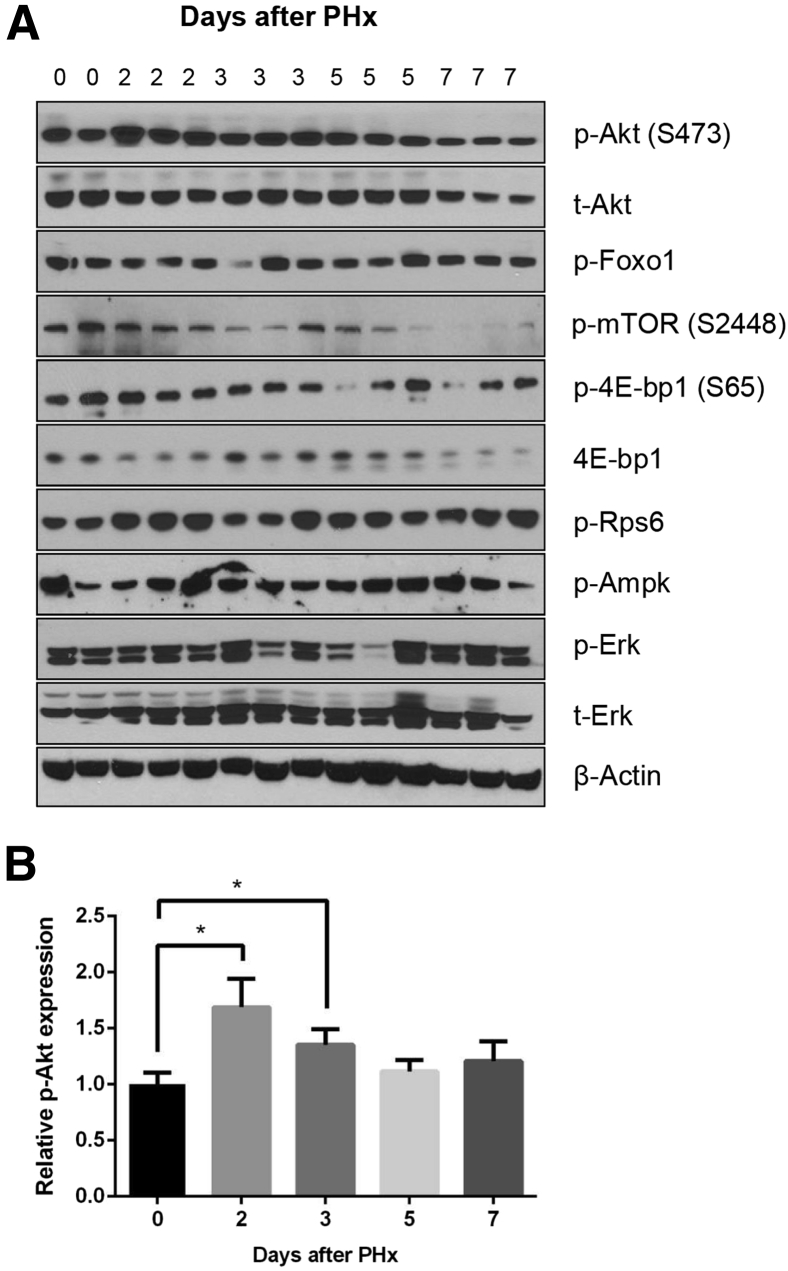
To investigate the functional contribution of mTORC2 signaling on liver regeneration, two-thirds PHx was performed in 8-week–old male WT mice (in the C57BL/6 genetic background). Because mTORC2 is responsible for phosphorylation of Akt at the Ser473 site [p-Akt (S473)], expression levels of mTORC2 signaling were determined by Western blot analysis for the expression of p-Akt (S473) during PHx (Figure 1A). The upregulation of p-Akt (S473) was observed from day 2 after PHx. The relative p-Akt expression was quantified by calculating the p-Akt (S473) to total Akt ratio. p-Akt (S473) levels peaked at 2 to 3 days and returned to the baseline levels at 5 days after PHx (Figure 1B). An increased expression of phosphorylated Rps6 (p-Rps6) expression was also observed (Figure 1A). The expression of phosphorylated Foxo1 (p-Foxo1), phosphorylated 4E-bp1 (p-4E-bp1), phosphorylated Ampk (p-Ampk), and phosphorylated Erk (p-Erk) did not significantly change during PHx (Figure 1A). Altogether, these results indicate the activation of mTORC2 signaling along liver regeneration after PHx.
Figure 1.
Liver mammalian target of rapamycin (mTOR) complex 2 pathway is activated after partial hepatectomy. A: Western blot analysis of indicated proteins in livers lysates from wild-type mice before (0 days) and after partial hepatectomy (PHx) (2 to 7 days). B: The densitometry analysis of the ratio of p-Akt (S473) to t-Akt is shown in wild-type mice before and after PHx. ∗P < 0.05. p-, phosphorylated; t-, total.
mTORC2 Deficiency in the Liver Does Not Affect Liver Development or Hepatocyte Polyploidy
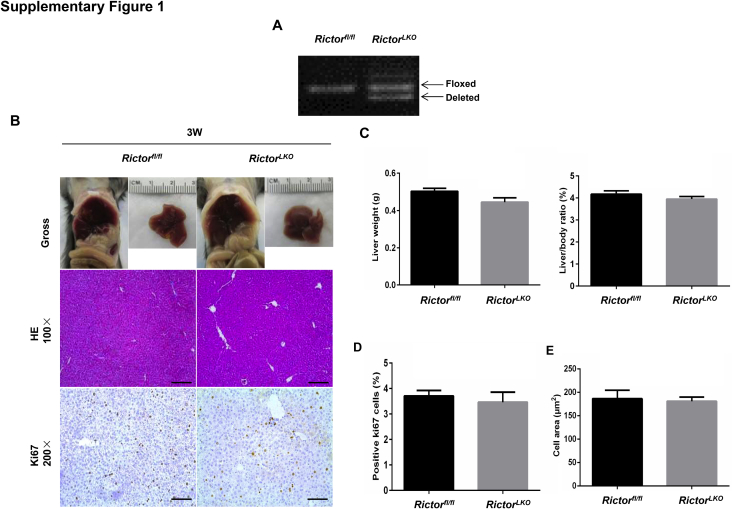
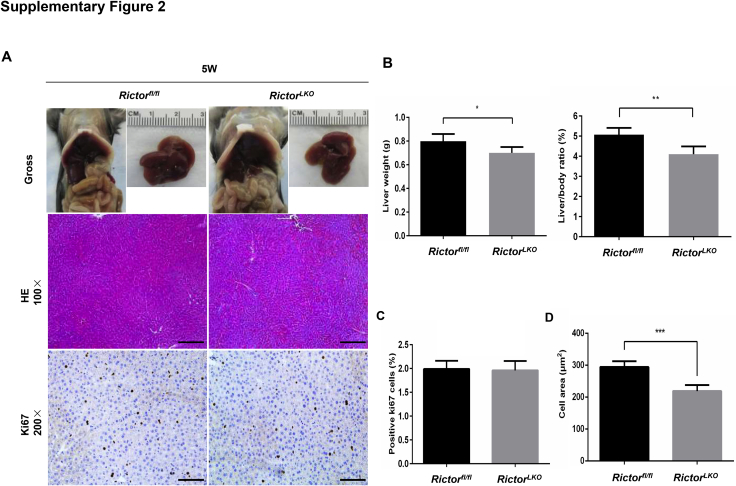
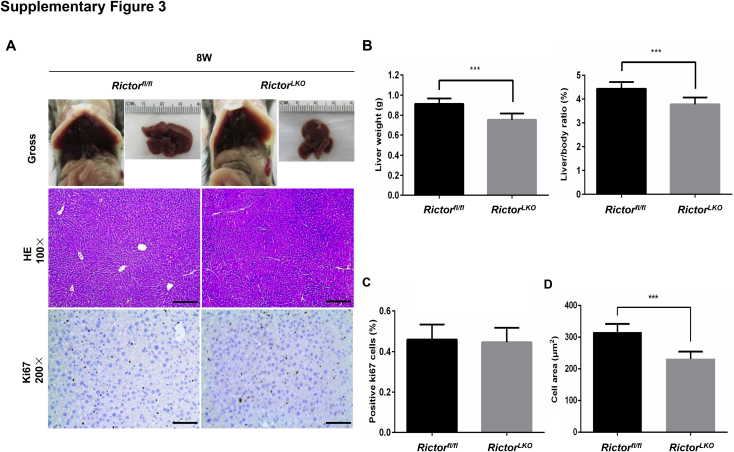
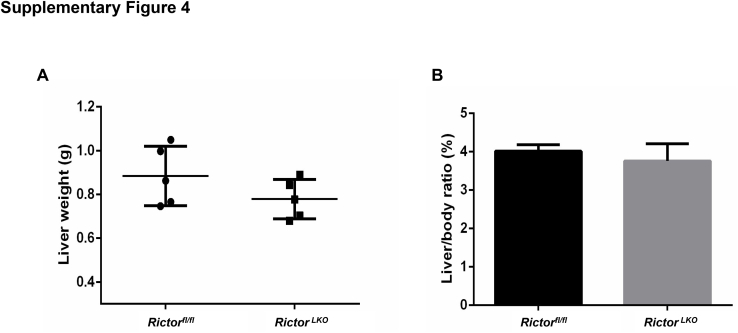
Because Rictor is the unique component of mTORC2 complex,13 the function of mTORC2 can be abolished by the deletion of Rictor. By crossing Albumin-Cre mice with Rictorfl/fl mice (all in the C57BL/6 background), RictorLKO mice were generated. Before subsequent experiments, genotyping was performed to validate depletion of Rictor in these mice (Supplemental Figure S1A). The detailed metabolic phenotype of RictorLKO mice has been previously described; specifically, it has been reported that loss of mTORC2 in the liver results in impaired glycolysis and lipogenesis on insulin stimulation or feeding, whereas otherwise these mice are rather healthy.30, 31, 32 Similar to previous reports,30, 31 RictorLKO mice were born at Mendelian ratio and developed normally. At 3 weeks of age, the liver was similar in all aspects in Rictorfl/fl mice and RictorLKO mice (Supplemental Figure S1, B–E). However, ablation of Rictor in the mouse liver resulted in a slight decrease in liver weight as well as liver weight to body weight ratio in 5-week–old mice (Supplemental Figure S2). Histologically, a significant reduction of hepatocyte area was observed in RictorLKO mice (Supplemental Figure S2). However, there was no significant difference in Ki-67–positive cells in liver tissues from WT Rictorfl/fl mice and RictorLKO littermates (Supplemental Figures S2). These phenotypes were also preserved in 8-week–old adult mice (Supplemental Figure S3). Meanwhile, no spontaneous hepatic apoptosis, necroptosis, or necrosis was observed in the liver tissues from Rictorfl/fl mice and RictorLKO mice (Supplemental Figures S3). The phenotypes observed here were similar to those that have been reported previously.30 The decreased hepatocyte area was due to the decreased glycogen and triglyceride storage in the RictorLKO liver. Indeed, in fasting state, there was no difference in liver weight and liver/body ratio between Rictorfl/fl and RictorLKO mice (Supplemental Figures S4).
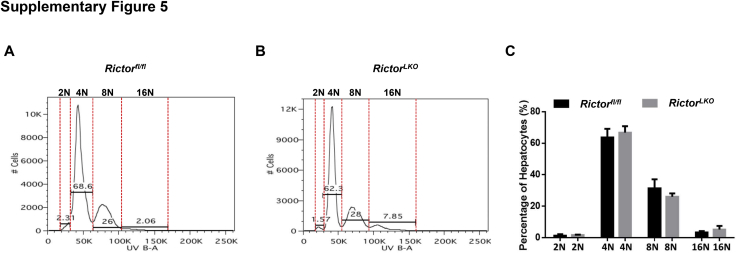
Hepatocytes are reported to be highly polyploid, which might influence how they function. Hepatocyte ploidies were analyzed using primary hepatocytes isolated from Rictorfl/fl mice and RictorLKO mice. The hepatocytes from the two cohorts of mice demonstrated similar ploidies (Supplemental Figure S5). In summary, these results are consistent with previous reports that mTORC2 is dispensable for normal liver development32 and loss of mTORC2 does not affect liver homeostasis or hepatocyte ploidies.
mTORC2 Is Required for Liver Regeneration
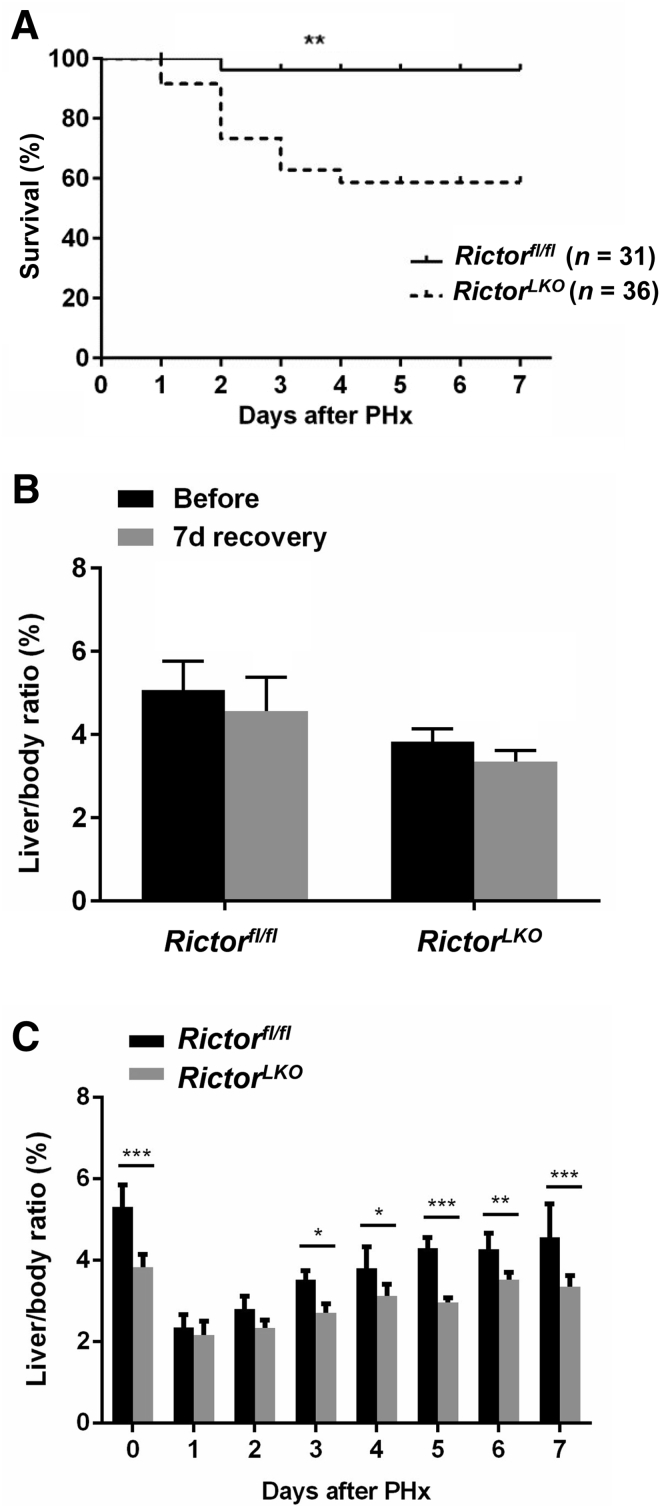
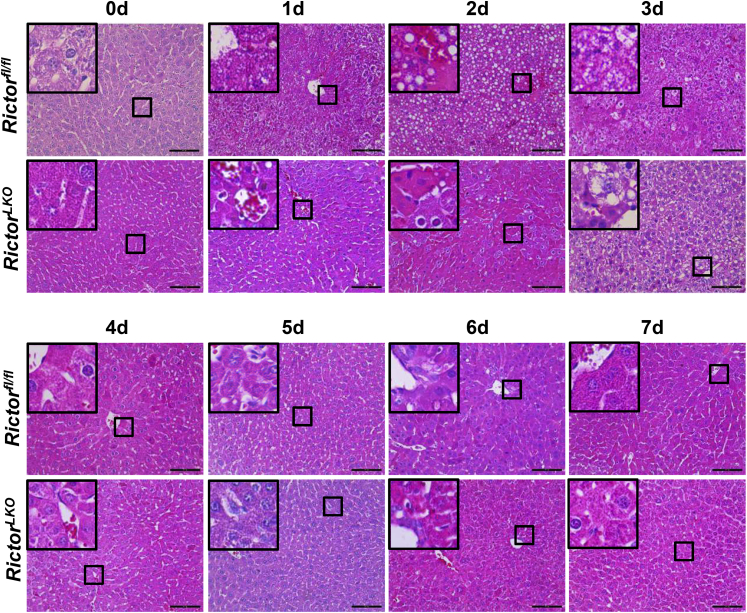
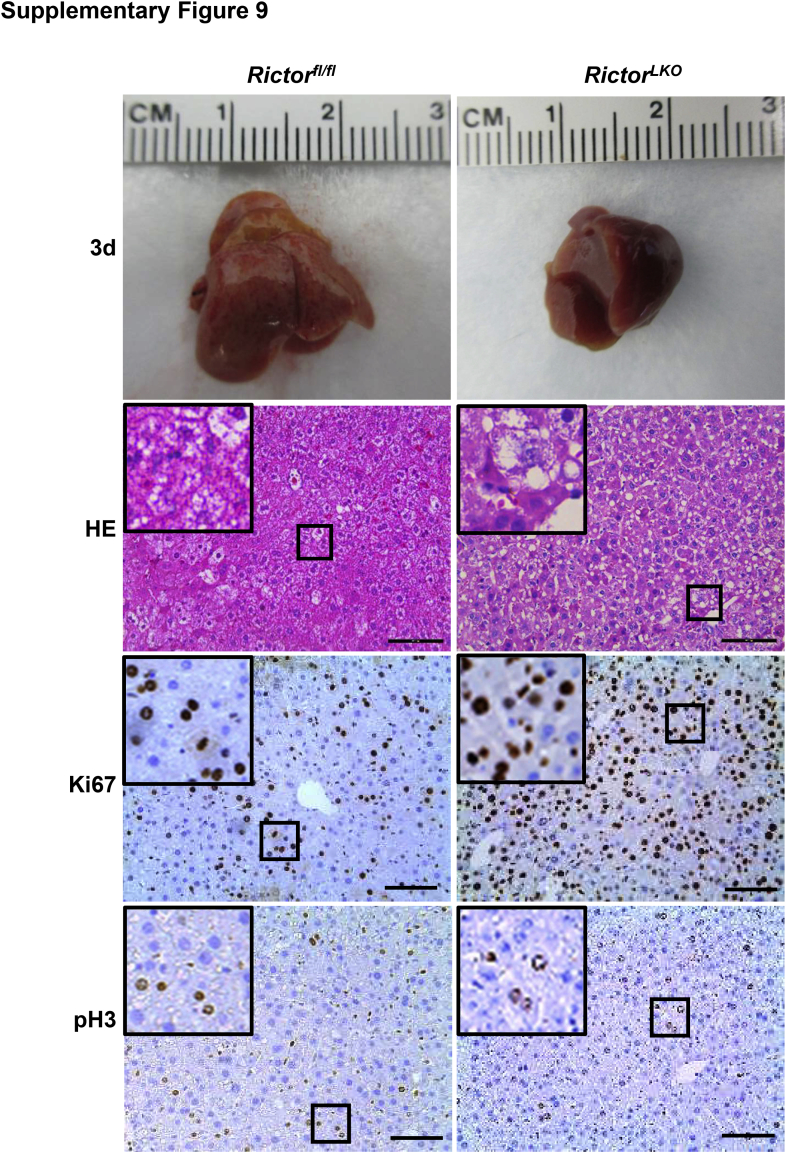
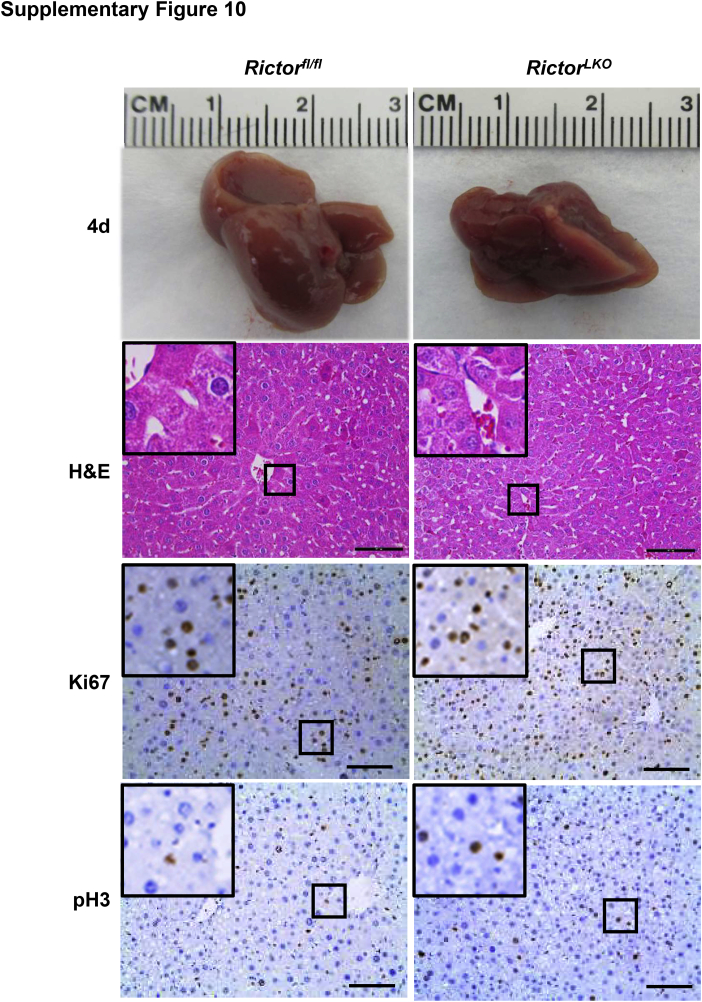
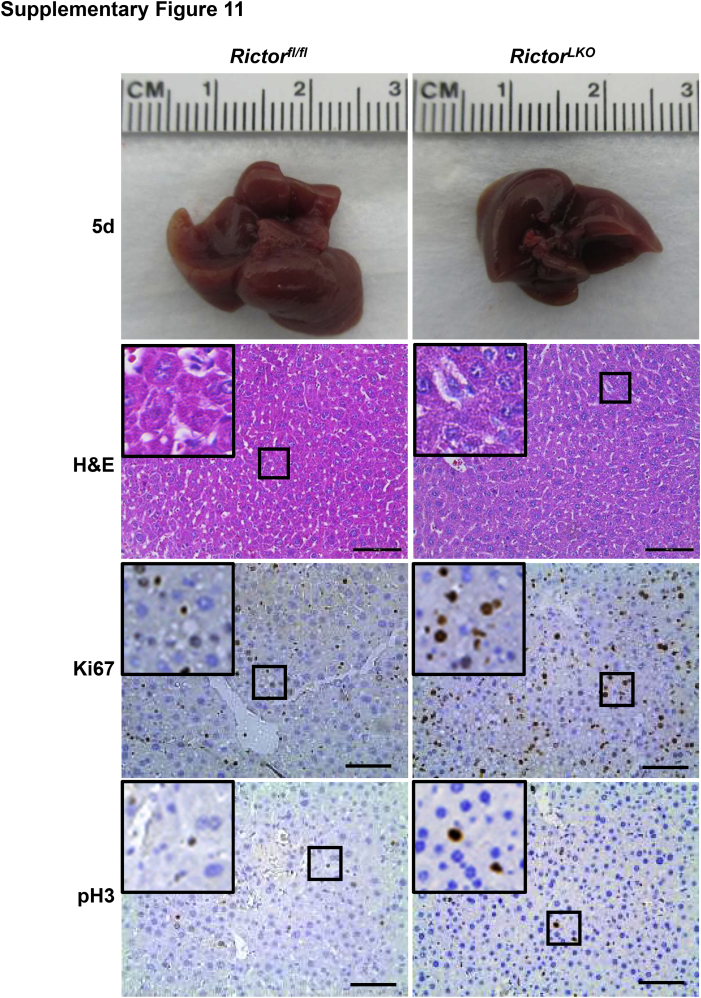
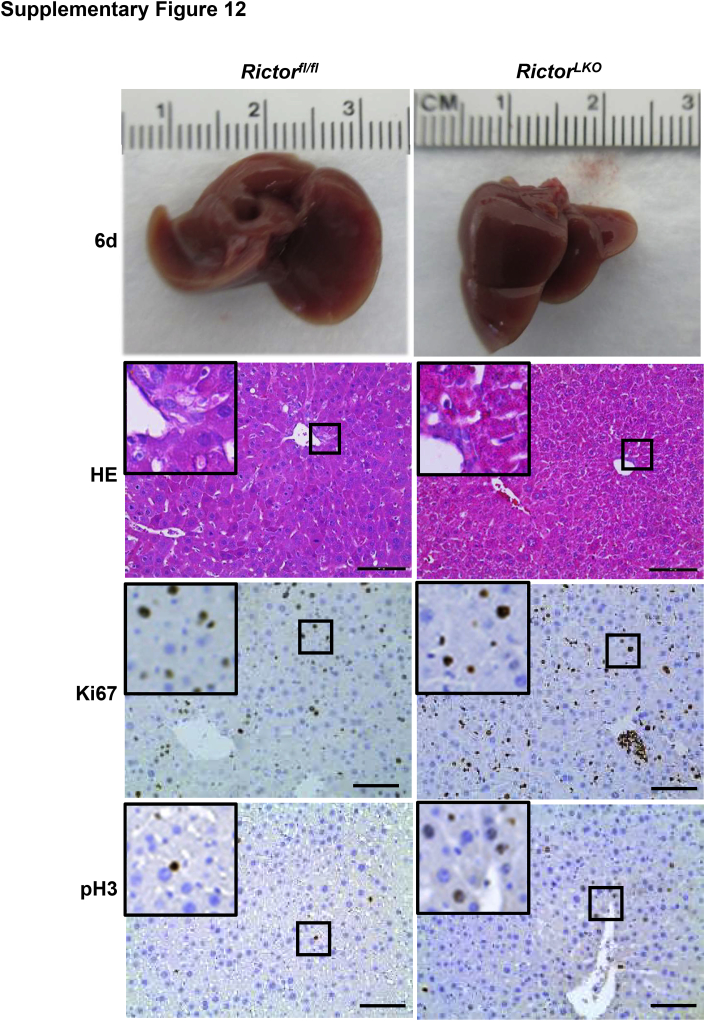
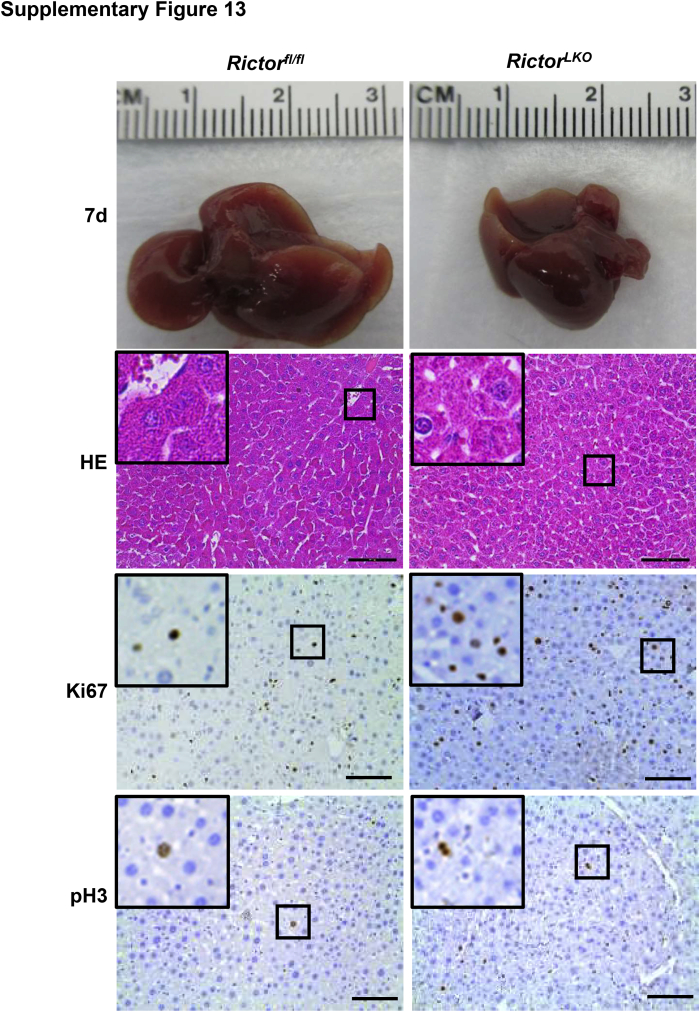
Liver-specific Akt1/2 KO in mice has been reported to impair liver regeneration and increase mortality after PHx.22 We hypothesized that loss of mTORC2, which resulted in the impairment of Akt activities, would recapitulate these phenotypes after PHx. To test this hypothesis, PHx was performed in Rictorfl/fl and RictorLKO mice. Rictor-deficient mice were found to be more intolerant to PHx and exhibited decreased survival probability compared with their WT littermates (Figure 2A). Several RictorLKO mice died from day 1 to day 4 after PHx, and the remaining mice survived (Figure 2A). After 7 days of recovery, the liver weight/body weight ratio showed no difference in the two mouse cohorts before and after PHx, suggesting that all surviving mice were able to fully recover in terms of liver regeneration (Figure 2B). Although the liver/body weight ratio was similar in Rictorfl/fl and RictorLKO mice at 1 day after PHx, liver/body weight ratio recovered at a slower pace in RictorLKO mice when compared with their WT littermates (Figure 2C). Grossly, the enlargement in liver mass was evident during liver regeneration from day 0 to day 7 after PHx (Supplemental Figures S6–S13). Histologically, comparisons between two groups were conducted by hematoxylin and eosin staining (Figure 3 and Supplemental Figures S6–S13). Inflammatory responses could be observed in both groups at day 1 to day 3 after PHx. Increased vacuolization areas accumulated at day 2 and day 3 in Rictorfl/fl mice compared with RictorLKO mice (Figure 3 and Supplemental Figures S8 and S9).
Figure 2.
Liver-specific deletion of Rictor increases mouse mortality after partial hepatectomy (PHx). A: Survival curves from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice after PHx. B: Liver weight and liver/body ratio of Rictorfl/fl and RictorLKO mice before and 7 days after PHx. C: Liver weight and liver/body ratio of Rictorfl/fl and RictorLKO mice at 0 to 7 days after PHx. n = 31 Rictorfl/fl mice (A); n = 36 RictorLKO mice (A); n = 6 Rictorfl/fl mice (B); n = 5 RictorLKO (B); n = 3 Rictorfl/fl mice (C); n = 4 RictorLKO mice (C). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus RictorLKO(Kaplan-Meier log rank test).
Figure 3.
Histologic analysis of mouse livers in response to partial hepatectomy. Representative hematoxylin and eosin–stained sections of livers isolated from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice before (0 days) and after partial hepatectomy (1 to 7 days). Boxed areas are shown at higher magnification in the insets. Original magnification: × 100 (main images); ×370 (insets). Scale bars = 200 μm.
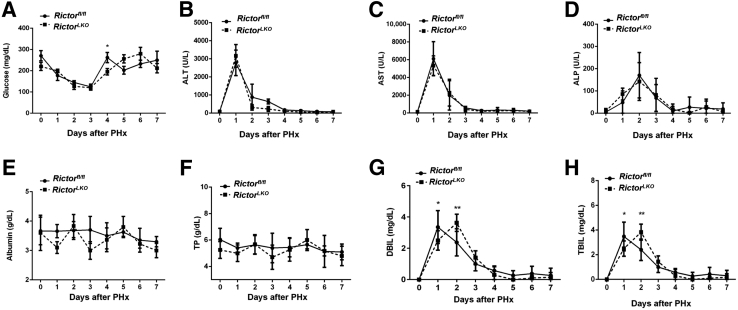
Subsequently, a series of serum biochemical markers were analyzed to evaluate liver functions among Rictorfl/fl and RictorLKO mice after PHx. No remarkable change of blood glucose levels during liver regeneration from day 0 to day 7 after PHx was detected (Figure 4A). Liver injury–related serum markers, such as alanine transaminase, aspartate transaminase, and alkaline phosphatase, markedly increased at day 1 after PHx and subsequently returned to normal, with no significant difference between the two groups of mice at each time point (Figure 4, B–D). Furthermore, no remarkable change of albumin and total protein levels was detected during liver regeneration (Figure 4, E and F). Interestingly, serum levels of direct bilirubin and total bilirubin peaked at day 1 after PHx in Rictorfl/fl mice, whereas serum levels of direct bilirubin and total bilirubin peaked at day 2 after PHx in RictorLKO mice (Figure 4, G and H), consistent with a delayed regeneration in RictorLKO mice. In summary, the present results indicated that loss of mTORC2 delays liver regeneration after PHx. The overall phenotype is similar to that observed in liver-specific Akt1/2 KO mice.
Figure 4.
Rictor deficiency in mouse liver does not significantly affect serum biochemical markers during liver regeneration. Blood glucose (A), serum alanine transaminase (ALT) (B), aspartate transaminase (AST) (C), alkaline phosphatase (ALP) (D), albumin (E), total protein (TP) (F), direct bilirubin (DBil) (G), and total bilirubin (TBil) (H) levels are shown. ∗P < 0.05, ∗∗P < 0.01 Rictorfl/fl versus RictorLKO at the same time point.
mTORC2 Deficiency Delays Hepatocyte Proliferation and Lipid Biosynthesis
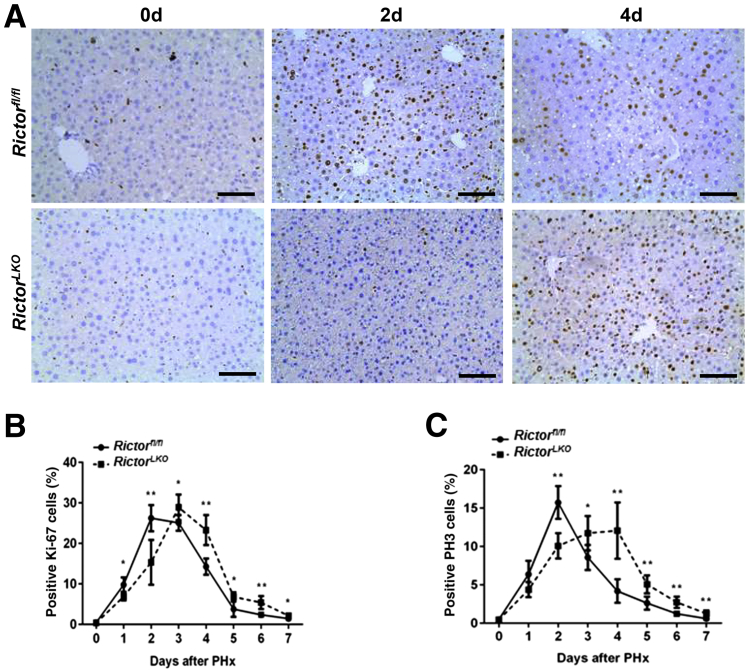
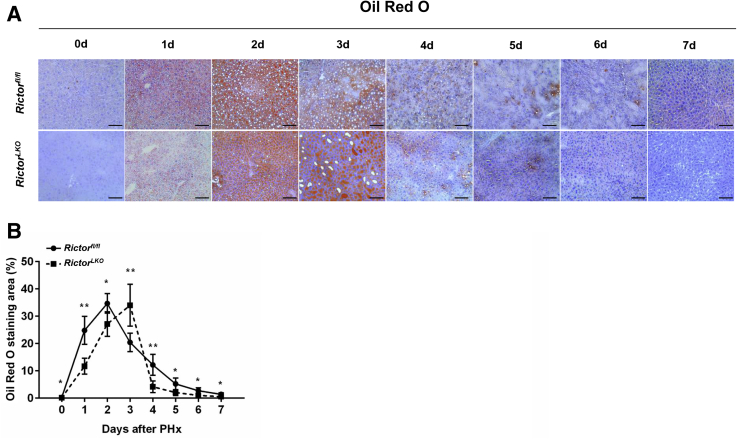
To elucidate the potential mechanisms responsible for the observed events, the hepatocyte proliferation rate was studied after PHx between Rictorfl/fl and RictorLKO mice by immunohistochemical staining of Ki-67 and PH3, two markers of proliferation. The percentage of Ki-67–positive cells markedly increased at day 1 and day 2 after PHx in Rictorfl/fl mice and decreased from day 3 after PHx. In contrast, the percentage of Ki-67–positive cells slightly increased to reach a peak at day 3 and then slowly decreased until day 7 after PHx in RictorLKO mice (Figure 5, A and B, and Supplemental Figures S6–S13). A similar trend was also observed in terms of the percentage of PH3-positive cells, showing that proliferative cells burst out during the first 2 days after PHx and markedly decreased in Rictorfl/fl mice, while the cells maintained a slow but relatively moderate level in RictorLKO mice (Figure 5C and Supplemental Figures S6–S13). Consistent with the delayed peaks of proliferation markers, delay in the lipid accumulation was also found in RictorLKO mice after PHx (Figure 6). Taken together, these results indicate that mTORC2 affects cell proliferation and lipid formation during liver regeneration.
Figure 5.
Delayed cell proliferation characterizes Rictor-deficient regenerative livers. A: Representative Ki-67 staining images in Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice livers 0 day (0d), 2 days (2d), and 4 days (4d) after partial hepatectomy (PHx). B: Quantification of positive Ki-67 nuclei/total nuclei at different time points in Rictorfl/fl and RictorLKO mice livers before and after PHx. C: Quantification of positive phosphorylated histone 3 (PH3) nuclei/total nuclei at different time points in Rictorfl/fl and RictorLKO mice before and after PHx. ∗P < 0.05, ∗∗P < 0.01 Rictorfl/fl versus RictorLKO at the same time point. Original magnification, ×200. Scale bars = 100 μm.
Figure 6.
Liver-specific Rictor deletion delays lipid droplet formation in mice after partial hepatectomy (PHx). A: Representative Oil Red O staining images in liver sections from mice before [0 days (0d)] and after PHx [1 (1d) to 7 days (7d)]. B: Quantification of Oil Red O staining area at different time points after PHx in Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice liver sections. ∗P < 0.05, ∗∗P < 0.01 Rictorfl/fl versus RictorLKO at the same time point. Original magnification, ×200. Scale bars = 100 μm.
mTORC2 Modulates Liver Regeneration via Regulation of Cell Cycle and Lipid Accumulation
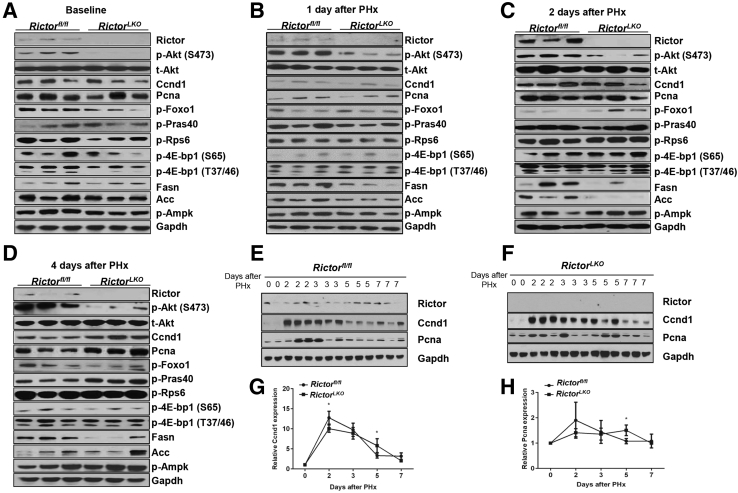
Next, the molecular differences between Rictorfl/fl and RictorLKO mice were studied after PHx. The major signaling cascades involved in liver regeneration were investigated at the baseline and 1 day, 2 days, and 4 days after PHx using Western blotting analysis (Figure 7).
Figure 7.
Delayed expression of Ccnd1 and Pcna in Rictor-deficient regenerative livers. A–D: Western blot analysis of indicated proteins in livers lysates from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 0 day (A), 1 day (B), 2 days (C), and 4 days (D) after partial hepatectomy. E and F: Western blot analysis of indicated proteins at different time points in Rictorfl/fl and RictorLKO mouse livers before and after PHx. G and H: Densitometry analysis of hepatic Ccnd1 (G) and Pcna (H) expression at different time points in Rictorfl/fl and RictorLKO mice livers before and after PHx. ∗P < 0.05 Rictorfl/fl versus RictorLKO at the same time point.
Because Akt is a major substrate of mTORC2, deletion of Rictor was accompanied by down-regulation of p-Akt (S473). Moreover, levels of p-Foxo1, p-Rps6, p-4E-bp1(S65), and p-4E-bp1 (T37/46) were also lower in RictorLKO mice at the baseline level (Figure 7A). Genes involved in hepatocyte proliferation (Ccnd1 and Pcna) and lipogenesis (Fasn, Acc and p-Ampk) were expressed at similar levels in Rictorfl/fl and RictorLKO mice (Figure 7A).
At day 1 after PHx (Figure 7B), expression of p-Akt (S473) markedly increased in Rictorfl/fl mice, whereas it remained at low levels in RictorLKO mice. However, expression of p-Rps6, p-4E-bp1(S65), and p-4E-bp1 (T37/46) was similar in the two cohorts of mice. Unexpectedly, p-FoxO1 was upregulated in RictorLKO mice when compared with Rictorfl/fl mice. Although the expression of hepatocyte proliferation genes, Ccnd1 and Pcna, was similar in these mice, the expression of lipogenic pathway genes, including Fasn and Acc, was slightly lower in RictorLKO mice at this time point. At day 2 after PHx (Figure 7C), expression of p-Akt (S473) remained at low levels in RictorLKO mice. Expression of p-Rps6, p-4E-bp1(S65), p-4E-bp1 (T37/46), and p-Foxo1, as well as Ccnd1 and Pcna, was similar to what have been observed in day 1 among Rictorfl/fl mice and RictorLKO mice. This finding coincided with the delay in lipid droplet accumulation during PHx in RictorLKO mice at this time point. At 4 days after PHx (Figure 7D), a recovery of Fasn and Acc1 levels was seen in RictorLKO mice. Intriguingly, the expression of Ccnd1 and Pcna was higher in RictorLKO mice than that detected in Rictorfl/fl mice.
Because delayed yet sustained hepatocyte proliferation was a major cellular phenotype in RictorLKO mice after PHx (Figure 5), the detailed time course analysis of Ccnd1 and Pcna expression was performed in Rictorfl/fl and RictorLKO mice after PHx (Figure 7, E and F). Ccnd1 was expressed at higher levels in Rictorfl/fl mice than in RictorLKO mice at 2 days after PHx. In addition, although Ccnd1 and Pcna levels returned to the baseline levels at 5 days PHx in Rictorfl/fl mice, they remained higher in RictorLKO mice (Figure 7, G and H). Thus, the present findings imply a critical regulatory function of mTORC2 on hepatocyte proliferation during liver regeneration.
Global RNA Sequencing Confirms the Role of mTORC2 Signaling in Regulating Cell Proliferation and Metabolism during Liver Regeneration
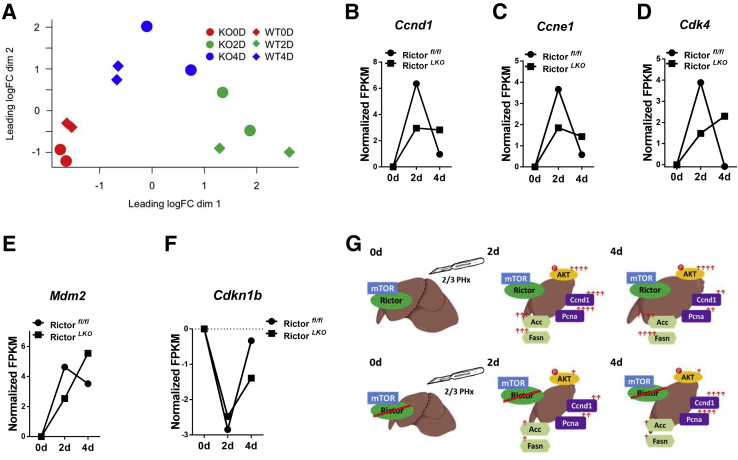
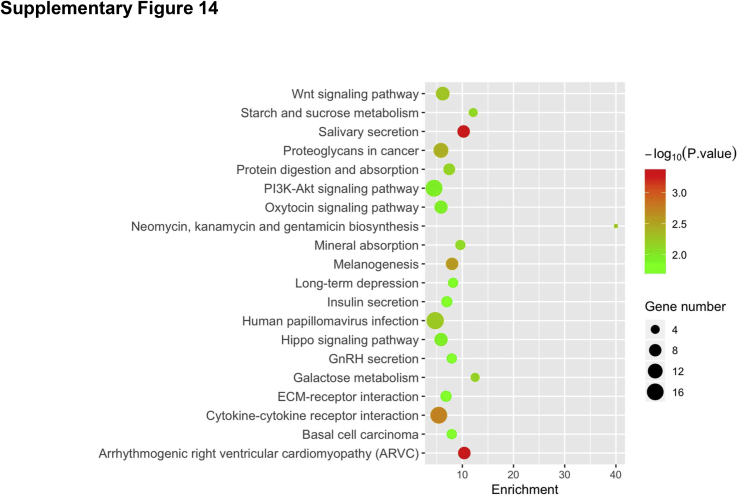
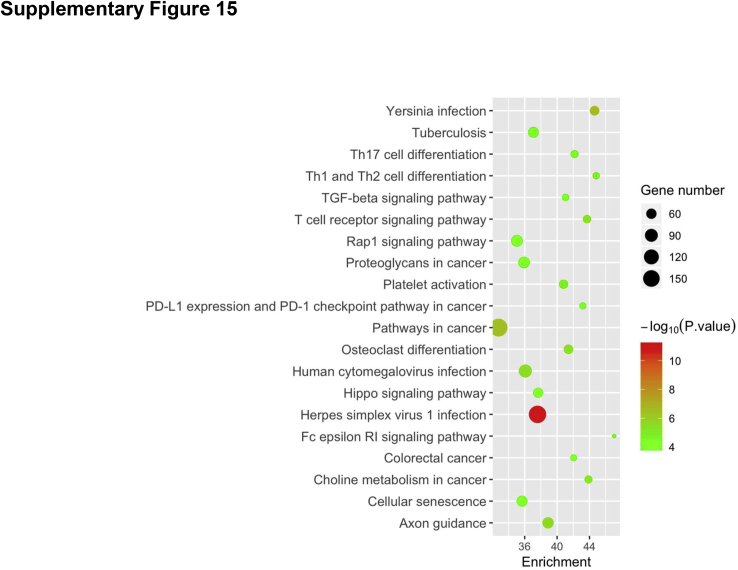
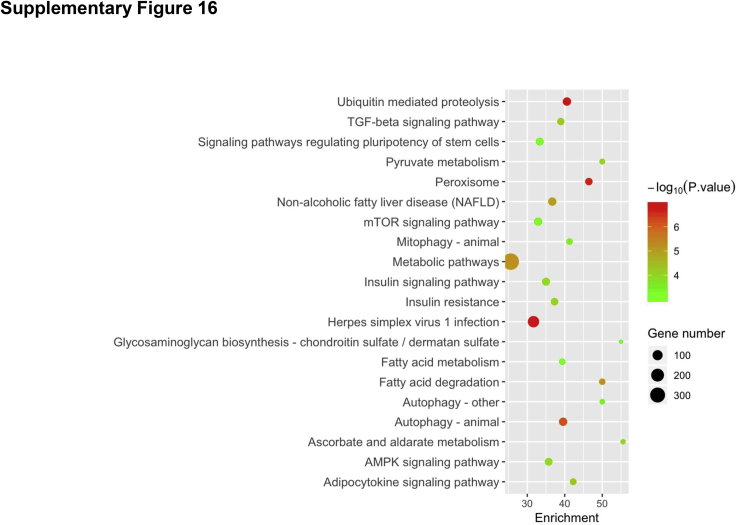
To further investigate the pathways regulated by mTORC2 during liver regeneration after PHx, RNA sequencing analysis of liver tissues was performed from Rictorfllfl and RictorLKO mice at 0 day (baseline), 2 days, and 4 days after PHx. Multidimensional scaling plot revealed relatively closer gene expression patterns of Rictorfllfl and RictorLKO mouse liver at day 0. PHx promoted significant gene expression changes in the liver. It is clear, however, that the global gene expression patterns at day 4 are closer to day 0 in Rictorfllfl mice but not in RictorLKO mice (Figure 8A). These results provide unbiased evidence that liver tissues are returning to their baseline state at day 4 in Rictorfllfl mice but not in RictorLKO mice. Because the delayed cell cycle progression is a major phenotype observed in RictorLKO mice, the major cell cycle regulators, including Ccnd1, Ccne1, Cdk4, Mdm2, and p27, were analyzed based on the RNA sequencing data. Ccnd1, Ccne1, Cdk4, and Mdm2 were expressed at high levels at day 2 in Rictorfl/fl mice, and their expression levels were lower at day 4 after PHx than those seen in day 2 after PHx. In contrast, in RictorLKO mice, these genes remained at higher levels at day 4 after PHx (Figure 8, B–E). The expression of Cdkn1d or p27, a cell cycle inhibitor, was significantly downregulated at day 2 after PHx, and it returned to the level close to the baseline at day 4 after PHx in Rictorfl/fl mice. In RictorLKO mice, p27 expression remained at a low level at day 4 after PHx (Figure 8F). In addition, KEGG pathway analysis was performed for genes differently expressed on day 0, day 2, or day 4 in Rictorfllfl and RictorLKO mouse liver tissues. Multiple pathways were found to be deregulated when mTORC2 was deleted at baseline levels as well as during liver regeneration after PHx (Supplemental Figures S14–S16). Overall, the RNA sequencing studies provide unbiased evidence and confirm that the loss of mTORC2 leads to a delayed liver regeneration after PHx.
Figure 8.
RNA sequencing results show mammalian target of rapamycin (mTOR) complex 2 signaling regulating cell proliferation and metabolism. A: Multidimensional scaling plot of the global gene expression patterns of 0 day (0D), 2 days (2D), and 4 days (4D) after partial hepatectomy (PHx) in Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice. B–F: Normalized fragments per kilogram of exon model per million reads mapped (FPKM) of indicated gene (Ccnd1, Ccne1, Cdk4, Mdm2, and Ckdn1b) expression at 0d, 2d, and 4d after PHx in Rictorfl/fl and RictorLKO mice. Dotted line indicates normalized FPKM value equals to 0. G: Graphical abstract for the study.
Discussion
With increased understanding of the biological mechanisms underlying liver regeneration, the clinical practice for curing liver diseases has improved enormously during the past decades. However, there are still challenges for more aggressive surgical resections in the setting of malignancy due to predamaged liver functions and improper regenerative processes. Thus, liver transplantation is the last choice for surgical treatment in addition to palliative treatments.33 A successful liver transplantation calls for providing an adequately sized, functioning graft for the recipient as well as maintaining a high safety profile for the donor. Complications, such as small transplantation and recipients' liver failure, still happen frequently.34 The trade-off between removal and retention remains the major challenge for surgeons. Thus, a better understanding of liver regeneration may shed light on clinical treatment strategies and availability of high-quality transplantable organs.
Numerous complex and interconnected signaling pathways have been uncovered to be involved in liver regeneration.35 mTOR signaling has been validated to be essential for regulating organismal growth, development, proliferation, and viability in response to nutrient availability.36 The mTOR kinase is considered to be a master regulator of growth that functions in two distinct complexes: mTORC1 (defined by the Raptor subunit) and mTORC2 (defined by the Rictor subunit).36 Although much is known about the inputs, outputs, and regulatory features of mTORC1, mTORC2 regulation and function remain poorly understood. In the present study, we investigated the functional role of mTORC2 during liver regeneration after PHx using RictorLKO mice. The results show that mTORC2 signaling is activated after PHx (Figure 1) and liver-specific deletion of Rictor causes significant early-stage mortality after PHx (Figure 2A). The data obtained in this study are similar to those observed in liver-specific Akt1/Akt2 double KO mice. Indeed, approximately 40% of Akt1/Akt2 KO mice did not survive after the PHx.22 These findings are consistent with the fact that Akt kinases are considered to be the major substrates of mTORC2. Previous studies have found that Pi3k and Pdk1 pathways are required for liver regeneration.17,37 Our data therefore add additional evidence to support the major role of the PI3k/Akt signaling cascade during this important physiologic process.
Mechanistically, we discovered that mTORC2 regulates hepatocyte proliferation and lipid accumulation after PHx, which is reflected by the delayed peak of Ki-67 and PH3 immunostaining as well as Oil Red O staining in RictorLKO mice. Consistently, at the biochemical levels, we found that the lipogenic pathway genes Fasn and Acc are expressed at lower levels at 2 days after PHx and higher expression of the cell proliferation genes Ccnd1 and Pcna in RictorLKO mice is seen at 4 days after PHx (Figure 8G). We compared our data with that reported in Akt1/2 double KO mice by Pauta et al,22 who found that WT mice exhibit significant lipid droplet accumulation at 2 to 4 days after PHx. The observation is consistent with our results. In contrast, Akt1/2 double KO mice failed to accumulate lipid droplets after PHx. This phenotype is different from that of RictorLKO mice. In RictorLKO mice, lipid droplet accumulation was delayed but not abolished. Of note, either deletion of Akt1/2 or ablation of mTORC2 was found to cause early-stage mortality after PHx. Our PHx surgery was performed by one expert researcher (H.W.), thus ruling out the possibility that the early mortality observed in RictorLKO mice was due to a surgical technique issue. The precise mechanisms leading to the higher mortality in RictorLKO mice or liver-specific Akt1/2 double KO mice remain to be determined. It was previously reported that there was severe hypoglycemia 12 hours after PHx in Akt1/2 KO mice, suggesting that this metabolic defect might be the major cause of the early death during PHx in these mice.22 The blood glucose levels in control and RictorLKO mice was analyzed at 24 hours after PHx, and the two cohorts of mice demonstrated the similar blood glucose levels (Figure 4A). Notably, the blood was collected from the surviving mice, and the possibility that those dead mice might have hypoglycemia cannot be ruled out. Clearly, additional studies are required to further investigate this issue. Regarding the hepatocyte proliferation, Pauta et al22 found that in WT mice, hepatocyte proliferation peaked at 2 days after PHx, decreased at 4 days, and returned to the baseline levels at 6 days. In Akt1/2 double KO mice, hepatocyte proliferation was lower throughout the time after PHx. Intriguingly, RictorLKO mice had delayed but more sustained hepatocyte proliferation after PHx. Overall, the molecular phenotypes are milder in RictorLKO mice than those seen in Akt1/2 double KO mice. The data are consistent with the finding that Akt1/2 kinase activities are regulated by multiple factors, and mTORC2 is one of the major complexes in this regulatory network.38 Loss of mTORC2 significantly decreased Akt1/2 activity, presumably without abolishing it. In addition, male mice were used in these studies for a better head to head comparison with studies by Pauta et al22 between roles of Akt1/2 and mTORC2 during liver regeneration. Although we do not expect sex difference in terms of mTORC2 regulation of liver regeneration, expanded studies with both male and female mice will be of great help to further substantiate our findings.
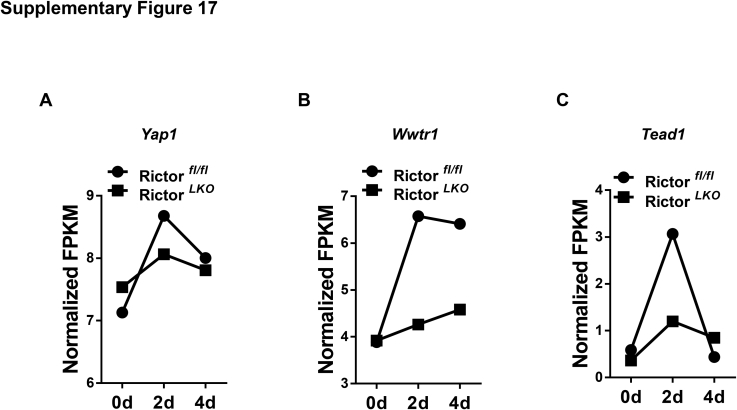
Using RNA sequencing analysis, we identified pathways that are differentially expressed in Rictorfl/fl and RictorLKO mouse liver tissues at baseline levels (Supplemental Figure S14) as well as during regeneration (Supplemental Figures S15 and S16). As expected, metabolic regulating pathways, such as PI3K-Akt signaling pathways and galactose metabolism pathways, were downregulated in RictorLKO mice at the baseline levels (Supplemental Figure S14). At day 2 after PHx, immunoregulatory signaling cascades, including Th17, Th1, and Th2 cell differentiation signaling and T-cell receptor signaling, were also significantly impaired in RictorLKO mice, indicating immunosuppression during liver regeneration in the condition of mTORC2 deficiency (Supplemental Figure S15). At day 4 after PHx, transforming growth factor-β signaling pathway as well as signals in regulating mitophagy and autophagy were identified to be down-regulated in RictorLKO mice, suggesting a potential cell proliferation brake occurring in WT mice for preventing liver overgrowth (Supplemental Figure S16). Furthermore, multiple metabolic cascades, especially fatty acid metabolic pathways, were deregulated in RictorLKO mice at this time point. The data are consistent with the key roles of these metabolic events during liver regeneration. Hippo cascade is a major signal that regulates organ size and hepatocyte proliferation.39 Yap1, Wwtr1, and Tead1, all key transcriptional factors downstream of Hippo kinases, were induced at higher levels in WT mice than those in RictorLKO mice, especially at day 2 after PHx (Supplemental Figure S17). The result suggests that mTORC2 may function to regulate liver regeneration via Hippo cascade. Clearly, additional functional studies are required to further analyze the contribution of these pathways during liver regeneration.
Our investigation uncovers the role of mTORC2 in regulating hepatocyte proliferation and lipid metabolism. mTORC2/Akt cascade regulates multiple downstream signaling pathways, including Tsc/mTORC1 and FoxOs.38,40 Tsc/mTORC1 in turns regulates p70S6k/Rps6 and 4E-bp1/Eif4e signaling pathways, which regulate lipogenesis and protein translation, respectively.41 FoxO family members are transcriptional factors, which regulate cell proliferation, metabolism, and apoptosis. FoxO1 is considered to be the major FoxO isoform in the liver.42 In Akt1//2 double KO mice, it was found that concomitant ablation of FoxO1 rescued the liver regeneration deficiency. The results suggest that FoxO1 may be the major molecule downstream of Akt during liver regeneration. However, the functional contribution of Tsc/mTORC1 in liver regeneration downstream of Akt has not been determined. It would be of high importance to further investigate these two pathways in liver regeneration, hepatocyte proliferation, and lipogenesis downstream of mTORC2. The hypothesis should be tested by generating liver-specific Tsc1/Rictor KO mice (to allow the activation of mTORC1 in the absence of mTORC2) or liver specific Foxo1/Rictor double KO mice (to simultaneously delete Foxo1 and mTORC2). The mice could be subjected to PHx, and the hepatocyte proliferation and lipogenesis after PHx could be evaluated. These studies would provide important information about the biochemical crosstalk among these pathways in PHx.
At the translational level, together with the studies by Pauta et al,22 we demonstrate that mTORC2/AKT cascade is a major signaling pathway in modulating liver regeneration after PHx. This study suggests that strategies that maintain or increase the mTORC2/AKT pathway may be useful to accelerate liver regeneration. On the other hand, mTORC2 inhibitors or AKT inhibitors may lead to delayed liver regeneration in humans. Therefore, a critical consideration and well evaluation of liver function shall be applied before hepatectomy and mTORC2 inhibitor administration.
Acknowledgments
We thank the University of California, San Francisco Liver Center Cell Biology core for the assistance with primary mouse hepatocytes isolation; Parnassus Flow Cytometry Core (PFCC) for assistance in generating the flow cytometry data; and Congyu Xu for her professional help with R language.
Footnotes
Supported by NIH grants R01CA136606 (X.C.), R01CA204586 (X.C.), P30DK026743 (University of California, San Francisco Liver Center), and K08DK101603 (B.W.); the Burroughs Wellcome Fund Career Award for Medical Scientists (B.W.); China Scholarship Council contracts 201706240075 (P.W.) and 201703170154 (R.S.); National Natural Science Foundation of China grant 81902449 (M.X.); and West China Hospital, Sichuan University, Post-Doctoral Research Project funds (H.W.); and in part by the NIH Diabetes Research Center grant P30 DK063720.
M.X. and H.W. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2019.12.010.
Contributor Information
Yong Zeng, Email: zengyong@medmail.com.cn.
Xin Chen, Email: xin.chen@ucsf.edu.
Supplemental Data
Supplemental Figure S1.
A: Representative genotyping results of Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice. B: Representative gross, hematoxylin and eosin (H&E)–, and Ki-67–stained sections of livers from 3-week–old Rictorfl/fl and RictorLKO mice. C: Liver weight and liver/body ratio of 3-week–old Rictorfl/fl and RictorLKO mice. D: Quantification of positive Ki-67 nuclei/total nuclei in 3-week–old Rictorfl/fl and RictorLKO mouse livers. E: Morphometric analysis of cell area was analyzed in 3-week–old Rictorfl/fl and RictorLKO mouse livers. Original magnification: ×100 (B, H&E); ×200 (B, Ki-67). Scale bars: 200 μm (B, H&E); 100 μm (B, Ki-67).
Supplemental Figure S2.
A: Representative gross, hematoxylin and eosin (H&E)–, and Ki-67–stained sections of livers from 5-week–old Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice. B: Liver weight and liver/body ratio of 5-week–old Rictorfl/fl and RictorLKO mice. C: Quantification of positive Ki-67 nuclei/total nuclei in 5-week–old Rictorfl/fl and RictorLKO mouse livers. D: Morphometric analysis of cell area was analyzed in 5-week–old Rictorfl/fl and RictorLKO mouse livers. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Original magnification: ×100 (A, H&E); ×200 (A, Ki-67). Scale bars: 200 μm (A, H&E); 100 μm (A, Ki-67).
Supplemental Figure S3.
A: Representative gross, hematoxylin and eosin (H&E)–, and Ki-67–stained sections of livers isolated from 8-week–old Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice. B: Liver weight and liver/body ratio of 8-week–old Rictorfl/fl and RictorLKO mice. C: Quantification of positive Ki-67 nuclei/total nuclei in 8-week–old Rictorfl/fl and RictorLKO mouse livers. D: Morphometric analysis of cell area was analyzed in 8-week–old Rictorfl/fl and RictorLKO mouse livers. ∗∗∗P < 0.001. Original magnification: ×100 (A, H&E); ×200 (A, Ki-67). Scale bars: 200 μm (A, H&E); 100 μm (A, Ki-67).
Supplemental Figure S4.
Eight-Week–old Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) male mice were fasted for 14 hours and then sacrificed. Liver weight and body weight were measured. No significant difference of liver weight (A) and liver/body ratio (B) was observed. n = 5 Rictorfl/fl mice; n = 5 RictorLKO mice.
Supplemental Figure S5.
Primary hepatocytes were isolated from 8-week–old Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) male mice. FACS analysis was conducted for evaluating the percentage of ploidy in primary hepatocyte. A: Representative percentage of 2N, 4N, 8N, and 16N in Rictorflox/flox mouse. B: Representative percentage of 2N, 4N, 8N, and 16N in RictorLKO mouse. C: Comparison of 2N, 4N, 8N, and 16N percentage among Rictorflox/flox and RictorLKO mice primary hepatocytes. No significant difference was observed. Red dotted lines indicate the peak of each polidy.
Supplemental Figure S6.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice before (0 day) partial hepatectomy. Small black boxes inside figures indicate areas of interest, and large black boxes on left upper corner denote enlarged views for better visualizing. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 μm (HE); 100 μm (Ki-67 and pH3).
Supplemental Figure S7.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 1 day after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S8.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 2 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S9.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 3 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S10.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3) –stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 4 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S11.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3) –stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 5 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S12.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 6 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S13.
Representative gross, hematoxylin and eosin (H&E)–, Ki-67–, and phosphorylated histone 3 (pH3)–stained sections of liver tissues from Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice 7 days after partial hepatectomy. Boxed areas are shown at higher magnification in the insets. Original magnification: ×100 (H&E, main image); ×200 (Ki-67 and pH3, main images); ×370 (H&E, inset); ×740 (Ki-67 and pH3, insets). Scale bars: 200 mm (HE); 100 mm (Ki-67 and pH3).
Supplemental Figure S14.
Top 20 KEGG pathways enriched in down-regulated genes among Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) livers before partial hepatectomy (baseline). The number of differentially expressed genes in each pathway is represented by the area of dot. Percentage of genes differently expressed in the pathway, or enrichment, is demonstrated with horizontal axis. The P value indicates the significance of enrichment in the listed ontological terms.
Supplemental Figure S15.
Top 20 KEGG pathways enriched in of down-regulated genes among Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice livers 2 days after partial hepatectomy. Gene expression was normalized to baseline expressions.
Supplemental Figure S16.
Top 20 KEGG pathways enriched in of down-regulated genes among Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice livers 4 days after partial hepatectomy. Gene expression was normalized to baseline expressions.
Supplemental Figure S17.
Normalized fragments per kilogram of exon model per million reads mapped (FPKM) of Yap1 (A), Wwtr1 (B), and Tead1 (C) expression at 0 day (0d), 2 days (2d), and 4 days (4d) after partial hepatectomy in Rictor wild-type (Rictorfl/fl) and Rictor knockout (RictorLKO) mice.
References
- 1.Wolf J.H., Holmes M.V., Fouraschen S., Keating B.J., Baker T., Emond J., Rader D.J., Shaked A., Olthoff K.M. Serum lipid expression correlates with function and regeneration following living donor liver transplantation. Liver Transpl. 2016;22:103–110. doi: 10.1002/lt.24220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corless J.K., Middleton H.M., 3rd Normal liver function: a basis for understanding hepatic disease. Arch Intern Med. 1983;143:2291–2294. doi: 10.1001/archinte.143.12.2291. [DOI] [PubMed] [Google Scholar]
- 3.Riehle K.J., Dan Y.Y., Campbell J.S., Fausto N. New concepts in liver regeneration. J Gastroenterol Hepatol. 2011;26 Suppl 1:203–212. doi: 10.1111/j.1440-1746.2010.06539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalopoulos G.K. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 6.Clavien P.A., Petrowsky H., DeOliveira M.L., Graf R. Strategies for safer liver surgery and partial liver transplantation. N Engl J Med. 2007;356:1545–1559. doi: 10.1056/NEJMra065156. [DOI] [PubMed] [Google Scholar]
- 7.Forbes S.J., Newsome P.N. Liver regeneration: mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol. 2016;13:473–485. doi: 10.1038/nrgastro.2016.97. [DOI] [PubMed] [Google Scholar]
- 8.Oh S.H., Swiderska-Syn M., Jewell M.L., Premont R.T., Diehl A.M. Liver regeneration requires Yap1-TGFbeta-dependent epithelial-mesenchymal transition in hepatocytes. J Hepatol. 2018;69:359–367. doi: 10.1016/j.jhep.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong B., Sun R., Huang M., Chow M.D., Zhong X.B., Xie W., Lee Y.H., Guo G.L. Fibroblast growth factor 15-dependent and bile acid-independent promotion of liver regeneration in mice. Hepatology. 2018;68:1961–1976. doi: 10.1002/hep.30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalopoulos G.K. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preziosi M.E., Monga S.P. Update on the mechanisms of liver regeneration. Semin Liver Dis. 2017;37:141–151. doi: 10.1055/s-0037-1601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panasyuk G., Patitucci C., Espeillac C., Pende M. The role of the mTOR pathway during liver regeneration and tumorigenesis. Ann Endocrinol (Paris) 2013;74:121–122. doi: 10.1016/j.ando.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Huang K., Fingar D.C. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Yu W.N., Chen X., Peng X.D., Jeon S.M., Birnbaum M.J., Guzman G., Hay N. Spontaneous hepatocellular carcinoma after the combined deletion of akt isoforms. Cancer Cell. 2016;29:523–535. doi: 10.1016/j.ccell.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X., Luo L., Chen J. Roles of mTOR signaling in tissue regeneration. Cells. 2019;8:1075. doi: 10.3390/cells8091075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haga S., Ogawa W., Inoue H., Terui K., Ogino T., Igarashi R., Takeda K., Akira S., Enosawa S., Furukawa H., Todo S., Ozaki M. Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J Hepatol. 2005;43:799–807. doi: 10.1016/j.jhep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Haga S., Ozaki M., Inoue H., Okamoto Y., Ogawa W., Takeda K., Akira S., Todo S. The survival pathways phosphatidylinositol-3 kinase (PI3-K)/phosphoinositide-dependent protein kinase 1 (PDK1)/Akt modulate liver regeneration through hepatocyte size rather than proliferation. Hepatology. 2009;49:204–214. doi: 10.1002/hep.22583. [DOI] [PubMed] [Google Scholar]
- 18.Chen P., Yan H., Chen Y., He Z. The variation of AkT/TSC1-TSC1/mTOR signal pathway in hepatocytes after partial hepatectomy in rats. Exp Mol Pathol. 2009;86:101–107. doi: 10.1016/j.yexmp.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Espeillac C., Mitchell C., Celton-Morizur S., Chauvin C., Koka V., Gillet C., Albrecht J.H., Desdouets C., Pende M. S6 kinase 1 is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. J Clin Invest. 2011;121:2821–2832. doi: 10.1172/JCI44203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y.P., Ballou L.M., Lin R.Z. Rapamycin-insensitive regulation of 4e-BP1 in regenerating rat liver. J Biol Chem. 2001;276:10943–10951. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 21.Palmes D., Zibert A., Budny T., Bahde R., Minin E., Kebschull L., Holzen J., Schmidt H., Spiegel H.U. Impact of rapamycin on liver regeneration. Virchows Arch. 2008;452:545–557. doi: 10.1007/s00428-008-0604-y. [DOI] [PubMed] [Google Scholar]
- 22.Pauta M., Rotllan N., Fernandez-Hernando A., Langhi C., Ribera J., Lu M., Boix L., Bruix J., Jimenez W., Suarez Y., Ford D.A., Baldan A., Birnbaum M.J., Morales-Ruiz M., Fernandez-Hernando C. Akt-mediated foxo1 inhibition is required for liver regeneration. Hepatology. 2016;63:1660–1674. doi: 10.1002/hep.28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., Magnuson M.A. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 24.Magee J.A., Ikenoue T., Nakada D., Lee J.Y., Guan K.L., Morrison S.J. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell C., Willenbring H. Addendum: a reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2014;9:1532. doi: 10.1038/nprot.2014.122. [DOI] [PubMed] [Google Scholar]
- 26.Song X., Liu X., Wang H., Wang J., Qiao Y., Cigliano A., Utpatel K., Ribback S., Pilo M.G., Serra M., Gordan J.D. Combined CDK4/6 and pan-mTOR inhibition is synergistic against intrahepatic cholangiocarcinoma. Clin Cancer Res. 2019;25:403–413. doi: 10.1158/1078-0432.CCR-18-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B., Zhao L., Fish M., Logan C.Y., Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan A.W., Taylor M.H., Hickey R.D., Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy M., Sorokina O., Skene N., Simonnet C., Mazzo F., Zwart R., Sher E., Smith C., Armstrong J.D., Grant S.G. Proteomic analysis of postsynaptic proteins in regions of the human neocortex. Nat Neurosci. 2018;21:130. doi: 10.1038/s41593-017-0025-9. [DOI] [PubMed] [Google Scholar]
- 30.Hagiwara A., Cornu M., Cybulski N., Polak P., Betz C., Trapani F., Terracciano L., Heim M.H., Ruegg M.A., Hall M.N. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Yuan M., Pino E., Wu L., Kacergis M., Alexander A., Soukas Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287:29579–29588. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamming D.W., Demirkan G., Boylan J.M., Mihaylova M.M., Peng T., Ferreira J., Neretti N., Salomon A., Sabatini D.M., Gruppuso P.A. Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2) FASEB J. 2014;28:300–315. doi: 10.1096/fj.13-237743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meirelles Junior R.F., Salvalaggio P., Rezende M.B., Evangelista A.S., Guardia B.D., Matielo C.E., Neves D.B., Pandullo F.L., Felga G.E., Alves J.A., Curvelo L.A., Diaz L.G., Rusi M.B., Viveiros Mde M., Almeida M.D., Pedroso P.T., Rocco R.A., Meira Filho S.P. Liver transplantation: history, outcomes and perspectives. Einstein (Sao Paulo) 2015;13:149–152. doi: 10.1590/S1679-45082015RW3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.G. A complete treatment of adult living donor liver transplantation: a review of surgical technique and current challenges to expand indication of patients. Am J Transplant. 2015;15:17–38. doi: 10.1111/ajt.12907. [DOI] [PubMed] [Google Scholar]
- 35.Valizadeh A., Majidinia M., Samadi-Kafil H., Yousefi M., Yousefi B. The roles of signaling pathways in liver repair and regeneration. J Cell Physiol. 2019;234:14966–14974. doi: 10.1002/jcp.28336. [DOI] [PubMed] [Google Scholar]
- 36.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson L.N., Larson S.D., Silva S.R., Rychahou P.G., Chen L.A., Qiu S., Rajaraman S., Evers B.M. PI3K/Akt activation is critical for early hepatic regeneration after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1401–G1410. doi: 10.1152/ajpgi.00062.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalle Pezze P., Sonntag A.G., Thien A., Prentzell M.T., Godel M., Fischer S., Neumann-Haefelin E., Huber T.B., Baumeister R., Shanley D.P., Thedieck K. A dynamic network model of mTOR signaling reveals TSC-independent mTORC2 regulation. Sci Signal. 2012;5:ra25. doi: 10.1126/scisignal.2002469. [DOI] [PubMed] [Google Scholar]
- 39.Patel S.H., Camargo F.D., Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masui K., Tanaka K., Akhavan D., Babic I., Gini B., Matsutani T., Iwanami A., Liu F., Villa G.R., Gu Y., Campos C., Zhu S., Yang H., Yong W.H., Cloughesy T.F., Mellinghoff I.K., Cavenee W.K., Shaw R.J., Mischel P.S. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C., Cigliano A., Jiang L., Li X., Fan B., Pilo M.G., Liu Y., Gui B., Sini M., Smith J.W., Dombrowski F., Calvisi D.F., Evert M., Chen X. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology. 2015;61:200–213. doi: 10.1002/hep.27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tikhanovich I., Cox J., Weinman S.A. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol. 2013;28 Suppl 1:125–131. doi: 10.1111/jgh.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]