Abstract
The histone acetyltransferase Elp3 (Elongator Protein 3) is the catalytic subunit of the highly conserved Elongator complex. Elp3 is essential for the complex functions of Elongator in both the nucleus and cytoplasm of neurons, including the epigenetic control of neuronal motility genes and the acetylation of α-tubulin that affects axonal branching and cortical neuron migration. Accordingly, misregulation of Elp3 has been implicated in human disorders that specifically affect neuronal function, including Familial Dysautonomia (FD), a disease characterized by degeneration of the sensory and autonomic nervous system, and the motor neuron degenerative disorder amyotrophic lateral sclerosis (ALS). These studies underscore the importance of Elp3 in neurodevelopment and disease, and the need to further characterize the multiple nuclear and cytoplasmic based roles of ELP3 required for neurogenesis in animal models, in vivo. In this report, we investigate the behavioral and morphological consequences that result from targeted reduction of Elp3 HAT levels specifically in the developing Drosophila nervous system. We demonstrate that loss of Elp3 during neurodevelopment leads to a hyperactive phenotype and sleep loss in the adult flies, a significant expansion in synaptic bouton number and axonal length and branching in the larval neuromuscular junction as well as the misregulation of certain genes known to be involved in these processes. Our results uncover a novel role for Elp3 in the regulation of synaptic bouton expansion during neurogenesis that may be linked with a requirement for sleep.
Introduction
The generation of complex synaptic regulatory networks and diverse cell types during neurogenesis is achieved, in large part, by precisely coordinated and tightly controlled gene expression profiles distinct for each neuronal cell lineage (Orphanides & Reinberg 2002). Maintenance of such differential gene control is largely dependent on the way that DNA and its associated histone proteins are packaged into a highly organized chromatin structure in the nucleus of eukaryotic cells. (Orphanides & Reinberg 2002, Reik 2007, Kiefer 2007, Elefant et al. 2000b). Activation of gene expression profiles requires that chromatin condensation be disrupted to allow for transcription factor binding and RNA polymerase assembly and passage. Control of such chromatin reorganization is achieved in large part by specific patterns of covalent modifications on the N-terminal tails of histone proteins that include acetylation, methylation, and phosphorylation (Berger 2001, Elefant et al. 2000a, Fischle et al. 2003, Jenuwein & Allis 2001, Strahl & Allis 2000). These distinct histone modification motifs serve as signals for the recruitment of chromatin organizational proteins, which influence chromatin structure and subsequent epigenetic gene control (Jenuwein & Allis 2001, Strahl & Allis 2000, Felsenfeld & Groudine 2003, Berger 2002, Fischle et al. 2003, Margueron et al. 2005).
Histone acetylation is one of the best characterized of the histone modifications and is carried out by a family of enzymes termed histone acetyltransferases (HATs). The HAT Elp3 (Elongator Protein 3) is the catalytic subunit of the highly conserved Elongator complex, which consists of six subunits, ELP1–6. Elp3 contains conserved motifs characteristic of the GNAT family of HATs and acetylates histone H3 both in vitro and in vivo. The Elongator complex was initially identified as a mutisubunit complex that copurifies with the hyperphosphorylated form of the RNA polymerase II (RNAPII) holoenzyme in yeast and human cells (Hawkes et al. 2002, Kim et al. 2002, Winkler et al. 2001). Support for a direct role for Elp3 in transcriptional regulation includes genetic studies revealing defective phenotypes for yeast elp3 nulls including slow activation of certain genes, defects in global histone H3 acetylation patterns essential for gene activation (Winkler et al. 2002, Kristjuhan et al. 2002, Kristjuhan & Svejstrup 2004), and the finding that Elp3 is essential for the association of Elongator with nascent RNA in vivo (Petrakis et al. 2004, Svejstrup 2007). The Elongator complex has also been reported to play a variety of different roles in distinct regions of the cell in addition to its nuclear role in transcriptional elongation including the formation of modified wobble uridines in tRNAs in the cell cytoplasm (Esberg et al. 2006, Huang et al. 2005), and polarized exocytosis (Rahl et al. 2005).
Misregulation of Elp3 has been implicated in a number of human disorders that specifically affect neuronal function, including Familial dysautonomia (FD), an autosomal recessive neurodevelopmental disease characterized by degeneration of the sensory and autonomic nervous system (Slaugenhaupt & Gusella 2002, Axelrod 2004, Gardiner et al. 2007, Simpson et al. 2009), and the motor neuron degenerative disorder amyotrophic lateral sclerosis (ALS) (Wallis et al. 2008). Accordingly, studies in mammalian cells reveal that Elp3 is essential for promoting histone H3 acetylation throughout the coding regions of certain neuronal cell motility genes that is linked to their transcriptional activation (Close et al. 2006), supporting a nuclear-based epigenetic role for Elp3 in neuronal gene regulation. However, the role of Elp3 in neuronal function is complex, as more recent studies support a cytoplasm-based role for Elp3 in the acetylation of α-tubulin required for the migration and differentiation of projection neurons in cultured mouse cortical neurons (Gardiner et al. 2007, Wynshaw-Boris 2009, Creppe et al. 2009). and in motor neuron axonal branching and length in zebra fish (Simpson et al. 2009). Taken together, these studies underscore the importance of Elp3 in neurogenesis and disease, and the need to further characterize the multiple nuclear and cytoplasm-based roles of ELP3 in the developing nervous system using effective in vivo multicellular model systems.
Here, we explore the behavioral and morphological consequences of targeting Elp3 HAT reduction specifically in the developing Drosophila nervous system, in vivo. We demonstrate that loss of ELP3 HAT activity during neurodevelopment leads to a hyperactive phenotype and sleep loss in adult flies, a significant expansion in synaptic bouton number and axonal length and branching in the larval neuromuscular junction as well as the misregulation of certain genes known to be involved in these processes. Our results provide insight into a novel role for Elp3 in the regulation of synaptic bouton formation during neurogenesis that may be associated with a requirement for sleep.
Results
Ubiquitous reduction of ELP3 in Drosophila results in lethality.
We previously cloned the human homologue of ELP3 in Drosophila, referred to as Dmel\ELP3 (FlyBase 2009, Zhu et al. 2007). To explore developmental ELP3 function in vivo, we generated Dmel\ELP3 knockdown flies to assess potential phenotypes resulting from ELP3 loss. GAL4 targeted RNAi knockdown technology (Brand & Perrimon 1993) was used to create transgenic flies capable of inducible reduction of endogenous Dmel\ELP3 in specific cell and tissue-types of choice. This strategy has been successfully used for functional analysis of numerous genes in Drosophila (Rushton et al. 2009, Cerrato et al. 2006, Zhu et al. 2007). The Dmel\ELP3/RNAi construct was created by selecting a 650 bp RNAi non-conserved target sequence specific for Dmel\ELP3 (Figure 1A and B) shown by BLAST searches to exhibit non-redundancy within the genome. Three independently derived transgenic fly lines with insertions for this construct were chosen for use. Importantly, the insertions were homozygous viable, and did not cause any observable aberrant phenotypes in the absence of GAL4 induction.
FIGURE 1. Expression of Dmel\ELP3 RNAi reduces endogenous Dmel\ELP3 levels.
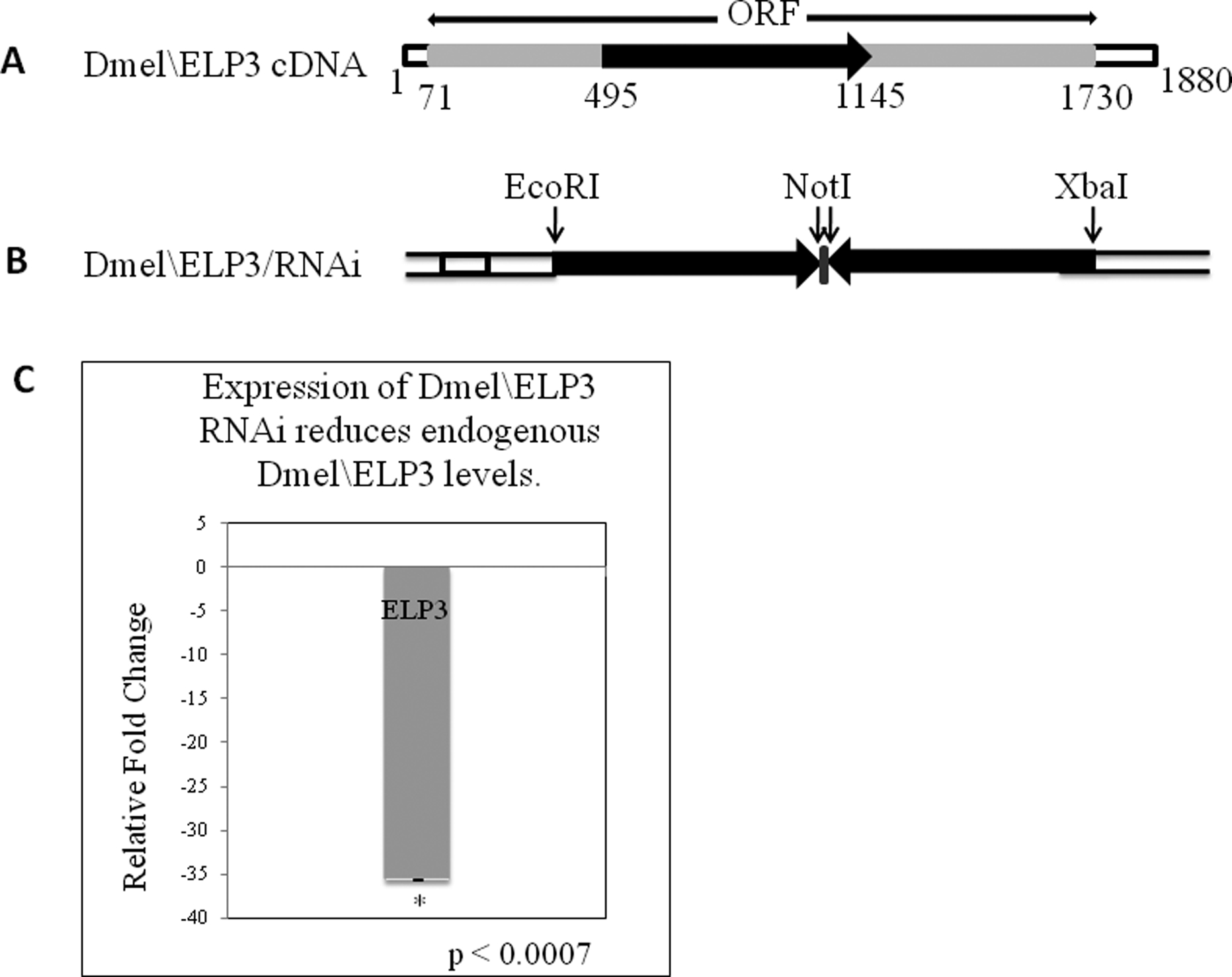
(A) Schematic of the Dmel\ELP3 ORF. Solid arrow represents the location of the 650bp RNAi non-conserved target sequence chosen for use in creating the Dmel\ELP3/RNAi construct. (B) Schematic of the Dmel\ELP3/RNAi construct. The 650bp RNAi target cDNA sequence was amplified by PCR using the cDNA Dmel\ELP3 clone as template and cloned into a sense–antisense inverted gene arrangement in the pUAST inducible expression vector, under the control of GAL4–UAS-binding sites. The inverted cDNA fragments are joined by a PCR generated short polylinker sequence and common NotI restriction sites, serving as the hinge region of the RNAi hairpin. (C) Shown is a histogram depicting qPCR analysis of Dmel\Elp3 expression between Dmel\ELP3/RNAi and control w1118 samples. Progeny resulting from a cross between homozygous Dmel\TIP60/RNAi line B and daughterless w*; P{GAL4-da.G32}UH1 were allowed to develop to the early pupal stage. The relative fold change in mRNA expression level was measured using the comparative Ct method with rp49 as the internal control gene. Error bars represent standard deviation. Asterisks (*) indicates significant fold change in relation to control where p < 0.0007. Error bar depicts standard deviation of the mean.
To test for efficient ELP3 knockdown and to determine whether ELP3 is essential for multicellular development, we induced Dmel\ELP3/RNAi expression in the fly using the robust ubiquitous GAL4 daughterless (da) driver (Bloomington stock # 5460). Higher levels of GAL4 induction can be obtained at 29°C when compared with 25°C, presumably due to higher activity of the yeast GAL4 transcription factor. Induction of RNAi at 29°C is commonly and successfully used for functional analysis of many genes in Drosophila (Duffy 2002, Fortier 2000). Induction of Dmel\ELP3/RNAi using the da-GAL4 driver for each of the three independently derived Dmel\ELP3/RNAi insertion lines at 29°C (Supplemental Table 1), but not 25°C (data not shown), revealed that fly viability was significantly reduced for each of the three independent lines tested. In each case, no defects in development were observed until the late pupal stage, which is the stage at which the majority of lethality occurred. The variation in lethality between the Dmel\ELP3/RNAi line A when compared to lines B and C is likely due to position effect variegation on expression due to random transgene insertion.
To verify that endogenous Dmel\ELP3 was down-regulated, progeny resulting from a cross between Dmel\ELP3/RNAi line B or w1118 control fly lines and the da GAL4 line were allowed to develop to the pre-pupal stage and the fold change in gene expression between the two lines was assessed using real-time PCR, before lethality in the late pupal stage occurred. Analysis of RNAi induced Elp3 knockdown using real-time PCR revealed significant reduction of ELP3 (Figure 1C) at 29°C but not at 25°C (data not shown), supporting a role for ELP3 in fly viability and making it necessary to maintain flies at 29°C for efficient ELP3 knockdown. Our results revealed that at 29°C, endogenous Dmel\ELP3 transcript levels were significantly reduced in the Dmel\ELP3/RNAi line B sample by 45 fold (Figure 1C), indicating that GAL4-induced Dmel\ELP3/RNAi expression is robustly silencing endogenous Dmel\ELP3 expression. These results were confirmed using a second ubiquitous GAL4 driver 337 (data not shown). Taken together, our results demonstrate that Dmel\ELP3 is essential for Drosophila multicellular development, and support our system as a valuable in vivo model for the functional analysis of Dmel\ELP3 during development.
Targeted reduction of Dmel\ELP3 in the nervous system causes an increase in climbing and locomotor activities and a loss of sleep in flies.
Analysis of temporal and spatial patterns of Dmel\ELP3 expression during embryogenesis utilizing in situ hybridization experiments (our unpublished data; BDGP gene expression report – accession # CG15433) reveal that high levels of Dmel\ELP3 are detected in the nervous tissues, and specifically in the central nervous system of the fly. Moreover, reduction in ELP3 production has been implicated in neuronal degeneration associated with amyotrophic lateral sclerosis (ALS) (Simpson et al. 2009) and familial dysautonomia (Slaugenhaupt & Gusella 2002, Axelrod 2004, Gardiner et al. 2007). To investigate Dmel\ELP3 in the nervous system of the fly, we targeted silencing of endogenous Dmel\ELP3 using GAL4 fly line y1w*;P{GawB}60IIA, shown by our laboratory and others (Chan & Kravitz 2007) to induce GAL4 preferentially in the brain and central nervous system (CNS). Induction of our two strongest independent Dmel\ELP3/RNAi fly lines B and C at 29°C resulted in no significant reduction of viability when compared to control progeny from a w1118 and 60IIA-GAL4 cross. However, visual assessment of the adult Dmel\ELP3/RNAi flies from independent lines B and C revealed that they were far more active than control flies in that they exhibited continuous movement, including circling the top of the vial and rapid jumping. To further explore this phenotype, we utilized the climbing assay, commonly used to validate and quantitatively assess behavioral manifestation of nervous system dysfunction. Wild-type Drosophila display a negative geotactic response such that when they are tapped to the bottom of a vial, they immediately climb to the top and remain there (Feany & Bender 2000). This natural response is compromised as a result of aging and defects in neurological and muscle processes, and thus climbing ability has been successfully used to monitor and quantitate the progression of severity in Drosophila age-related neurological disease models (Chen & Feany 2005, Chan & Bonini 2000). Dmel\ELP3/RNAi male and female flies consistently climbed more rapidly than the control w1118 flies after day 15 to end of period tested (Supplemental Figure 1 A and B). Induction of Dmel\ELP3/RNAi using the well characterized pan-neural GAL4 driver elavC155 also resulted in a similar hyperactive phenotype (data not shown). Our results indicate that loss of Dmel\ELP3 in all neurons, and specifically in the brain and CNS of the fly, results in the flies having an accelerated rate of climbing in later adulthood.
Another characteristic of flies expressing Dmel\ELP3/RNAi preferentially in the brain and central nervous system using GAL4 driver 60IIA and elavC155 was defects in their locomotor ability. Dmel\ELP3/RNAi flies that climbed to the top of the vials continuously circled the upper portion of the tube, and often executed rapid high jumps back down below the 9cm mark of the vial side shortly after climbing past it. To determine whether this observed hyperactivity of the Dmel\ELP3/RNAi flies was significant, we monitored their locomotor activity utilizing the Drosophila Activity Monitoring System (DAMS), a powerful assay used to study and quantify gross activity. The results showed that the Dmel\ELP3/RNAi flies broke the beam a significant total number of times more than the control flies, indicating that loss of Dmel\ELP3 in the brain and CNS caused a significant increase in the total activity of the flies (Figure 2). Our results demonstrate that loss of ELP3 in the nervous system, and specifically in the brain and CNS results in a significant increase in gross locomotor activity.
FIGURE 2. Expression of Dmel\ELP3/RNAi in the nervous system results in an increased locomotor activity in adult flies.

Flies homozygous for Dmel\ELP3/RNAi line B or control w1118 were crossed to flies homozygous for CNS GAL4 driver 60IIA. Staged 4 day old male or female progeny from this cross were each placed in separate DAMS glass tubes (32 males and 32 females total per genotype) and the number of times the flies broke the infrared beam was recorded in 30 minute intervals over a 24 hour period with 12 hour light and dark cycles. Shown is a histogram depicting the total activity for adult flies measured over a 24-hour period. Asterisks (*) indicates significant fold change in relation to control where p ≤ 0.0004. Error bars depict standard deviation of the mean.
The hyperactivity of the Dmel\ELP3/RNAi flies prompted us to ask whether loss of Dmel\ELP3 in the nervous system also leads specifically to loss of sleep in these flies. The DAMS assay is a powerful tool to investigate and quantify changes in gross locomotor activity for different fly genotypes, though it has certain limitations for specifically studying sleep. Such limitations include insensitivity to small fly movements which occur outside of the path of the infrared beam which affects the identification of actual quiescent sleep behavior, as non-detection of beam breaks in the DAMS assay may not be associated with sleep but rather with the fly not being in the vicinity of the infrared beam path (Zimmerman et al. 2008b, Zimmerman et al. 2008a). To overcome these issues we used digital video analysis to determine whether loss of ELP3 also induced a lack of sleep in the flies. Single, staged, 4-day-old Dmel\ELP3/RNAi and control w1118 progeny from a GAL4–60IIA cross were transferred to 6cm glass tubes and behavior recordings of fly sleep carried at 5 second intervals over a 72 hour total, 12:12 hour light/dark time course. We quantified total sleep, sleep bout number and mean sleep bout duration. Female Dmel\ELP3/RNAi flies slept significantly less that the control flies both during the day and nighttime periods (Figure 3A). Additionally, female Dmel/ELP3/RNAi flies have significantly shorter sleep bouts (Figure 3B). Male flies do not have significantly different total sleep (Figure 3A) but do show a small but significant increase in bout duration (Figure 3B). Neither male nor female Dmel\ELP3/RNAi flies had significantly different sleep bout numbers than the appropriate gender control flies (data not shown). In certain strains, male Drosophila melanogaster sleep more than female flies, having pronounced daytime sleep (Shaw et al. 2000, Huber et al. 2004, Andretic & Shaw 2005, Zimmerman et al. 2008b), and have been shown to respond to sleep deprivation much less strongly than females of the same strain (Shaw et al. 2002, Huber et al. 2004, Andretic & Shaw 2005, Zimmerman et al. 2008a). Indeed, males have been shown to have increased wakefulness after 6 hours of sleep deprivation, which is opposite the response of females (Huber et al. 2004, Hendricks et al. 2003, Shaw et al. 2000). Therefore, having a sex dependent sleep phenotype in response to the induction of the ELP3 RNAi is not without precedent. In addition, both male and female Dmel\ELP3/RNAi flies show significantly greater distance moved per movement compared to control flies by video which confirms the DAMS data and explains the greater number of beam breaks observed (data not shown). Similar sleep loss results were obtained using the elavC155 pan-neuronal GAL4 driver (data not shown). Taken together, our results demonstrate that loss of Dmel\ELP3 in the nervous system reduces the amount of sleep and increases the amount of activity in the fly, implicating Dmel\ELP3 in sleep and activity related neuronal pathways.
FIGURE 3. Expression of Dmel\ELP3/RNAi in the nervous system results in sleep loss in 5 and 6 day old adult flies.

Single staged 4 day old Dmel\ELP3/RNAi and control w1118 progeny from a GAL4–60IIA cross were transferred to 6cm glass tubes and behavior recordings of fly sleep carried at 5 second intervals over a 72 hour total, 12:12 hour light/dark time course on days 5 and 6 dark periods only (n=32). (A) shown is a histogram depicting the averaged total sleep activity for adult male and female flies measured on days 5 and 6. (B) shown is a histogram depicting the mean sleep bout duration for males and females measured on days 5 and 6. Asterisks (*) indicates significant fold change in relation to control where single asterisks indicate p ≤ 0.05 and double asterisks indicate p ≤ 0.001. Error bars depict standard deviation of the mean.
Knockdown of Dmel\ELP3 results in the misregulation of genes involved in sleep, vesicle transport and fusion, and protein chaperone activity.
ELP3 is implicated in facilitating the transcriptional activation of genes (Svejstrup et al., 2007). To investigate a potential molecular basis underlying the increase in activity and loss of sleep phenotypes in the Dmel\ELP3/RNAi flies, we asked whether loss of Dmel\ELP3 leads to misexpression of genes known to be associated with such phenotypes. For this analysis, we induced ubiquitous silencing of Dmel\ELP3 using the da-GAL4 driver as we had demonstrated that this driver induces robust levels of Dmel\ELP3 knockdown in flies (Figure 1C). mRNA was prepared from early pupae, directly before the lethal pupal stage (Suplemental Table 1), to ensure potential gene changes were not due to cell death. The mRNA levels of 6 specific genes from the progeny of a Dmel\ELP3/RNAi or control w1118 with da-GAL4 was assessed by quantitative real-time PCR and the fold change in gene expression levels between the two fly lines was determined (Figure 4). The putative target genes chosen for this analysis were: heat shock chaperone genes HSC70–3 (homolog of the mammalian endoplasmic reticulum chaperone BiP) and the cytoplasmic resident chaperone HSC70–4 (Elefant & Palter 1999), selected based on their involvement of the stress response linked with sleep loss (Naidoo et al. 2007), the synaptobrevin (SYB) gene (VAMP2 homolog), an ADHD candidate gene and a vesicle-associated membrane protein (VAMP) that is part of the SNAP-receptor (SNARE) complex and mediates exocytotic vesicle fusion by interacting with specific plasma membrane proteins that allow for either cell growth or fusion of neurotransmitter containing vesicles required for neuronal firing (Wallis et al. 2008, Davids et al. 2003, Bhattacharya et al. 2002), SLEEPLESS, chosen for its role as a sleep-promoting factor (Koh et al. 2008), gelsolin, a cytoskeleton modulator found to be downregulated in FD patient fibroblasts (Close et al. 2006), and superoxide dismutase (SOD1) in which particular mutations result in ALS cases with a family history (Simpson et al. 2009). The results of our analysis demonstrated that out of the 6 genes assessed, three of the genes (HSC3/BiP, SYB and SLEEPLESS) were significantly affected, with a marked increase in mRNA levels for BiP and SYB and a marked decrease in mRNA levels for SLEEPLESS (Figure 4). Consistent with findings that ALS is linked to Elp3 loss in a SOD1 independent manner (Simpson et al. 2009), we observed that SOD1 levels were unaffected in response to ELP3 loss and internal control RP49 levels were also unaffected. That only certain genes were affected indicate that the gene changes we observed were specific. Surprisingly, loss of Dmel\ELP3 resulted in the up-regulation of certain genes which is not consistent with the recognized role of ELP3 as a transcriptional activator, suggesting that these genes are either indirect ELP3 targets, or that in some instances, ELP3, like other HATS, is also involved in repression of certain genes. Notably, each of the affected genes was associated with the increase in activity and decrease in sleep phenotypes we observed in our Dmel\ELP3/RNAi flies. Induction of BIP is shown to be associated with sleep loss and up-regulation of the SYB gene has been shown to be associated with attention deficit hyperactivity disorder (ADHD). Finally, SLEEPLESS was specifically identified in a screen for genes associated with sleep and downregulation of SLEEPLESS (Koh et al. 2008) results in a reduction of sleep in flies. To ask whether these gene changes correlated with the behavioral changes we observed for adult ELP3/RNAi flies (Supplemental Figure 1, Figures 2 and 3), we carried out qPCR analysis using RNA isolated from the heads of 15 day old control flies or flies expressing Dmel\ELP3/RNAi specifically in the CNS using GAL4 driver 60IIA (Chan & Kravitz 2007). Day 15 was chosen as it corresponds to the beginning of the period of significantly increased climbing ability of Dmel\ELP3/RNAi flies (Supplemental Figure 1). The results revealed that although the trend in gene expression profile levels for SYB, HSC3 and SLEEPLESS in these adult heads was similar to that of the da GAL4 induced ELP3/RNAi pre-pupae staged flies, the changes were not statistically significant (Supplementary Figure 2). The differences in gene expression profiles between the two samples may reflect the more restricted and less robust expression pattern of the CNS GAL4 driver 60IIA than the da GAL4 driver, leading to dilution of significant gene expression changes in the whole adult heads assayed or that significant gene misregulation in response to ELP3 loss occurs earlier in development, leading to early physiological changes in the nervous system that are maintained into adulthood. Taken together, these results support an essential role for Dmel\ELP3 in the regulation of genes in the early pupal stage of the fly that may be involved in the behavioral phenotypes we observe in the adult flies.
FIGURE 4. Specific genes associated with Elp3 phenotypes are misregulated in response to ubiquitous Elp3 loss in early pupal stages of the fly.

Three female Dmel\ELP3/RNAi/B or w1118 flies were crossed to three da-GAL4 male flies and RNA was isolated from staged early progeny pupae. Shown is a histogram depicting qPCR analysis of the expression of the indicated genes in these staged early Elp3/RNAi or control pupal progeny samples. The relative fold change in mRNA expression levels were measured using the comparative Ct method with rp49 as the internal control gene. Error bars represent standard deviation. Asterisks (*) indicates significant fold changes between Dmel/Elp3 RNAi and control flies with values of p < 0.009. Error bars depict standard deviation of the mean.
Older Dmel\ELP3/RNAi flies respond normally to sleep deprivation.
A P-element insertion in SLEEPLESS has been shown to have no effect upon normal sleep but demonstrates a significant loss of the homeotic sleep drive, i.e. this mutant is severely impaired in the ability to recover from lost sleep (Koh et al., 2008). Therefore, we examined the sleep phenotype of 15 and 16 day old control and Dmel\ELP3/RNAi females from a CNS GAL4 60IIA cross, which correspond to the beginning of the period of significantly increased climbing ability of Dmel\ELP3/RNAi flies (Supplemental figures 1A and B). Females of this age show a significant loss of total sleep and have significantly shorter sleep bouts (Supplemental Figures 3A and B). We sleep deprived ELP3 RNAi flies and controls by gentle handling for 6 hours beginning at ZT 18. The flies were then allowed to recover for 24 hours. Unhandled controls were left undisturbed during the time of the deprivation. Sleep deprived ELP3 RNAi flies showed no significant differences in the amount of recovery sleep when compared to control flies deprived for the same amount of time in either the first 4 hours (p=0.975) or second 4 hours (p=0.907) following deprivation (Supplemental Figure 4). The ELP3 RNAi and control sleep deprived groups slept significantly more than the respective control groups left undisturbed in both the first 4 hour period (ZT0 to ZT4) (p>0.001 and p>0.001, respectively) and second 4 hour period (ZT4 to ZT8) (p>0.001 and p=0.004, respectively) following the end of deprivation. These results show that loss of Dmel\ELP3 specifically in the CNS of 15 and 16 day old adult flies does not appear to effect recovery sleep after sleep deprivation.
Loss of Dmel\ELP3 in the nervous system leads to an expansion of synaptic boutons in the larval fly neuromuscular junction.
Chemical synapses transmit information directionally from a presynaptic cell to a target postsynaptic cell via the release of neurotransmitters from the presynaptic terminal or synaptic bouton. Such firing of distinct neuronal connections either initiates the muscle contractions associated with movement and activity or directly influences learning and behavior. Recent studies have demonstrated that sleep loss is associated with synaptic bouton expansion (Gilestro et al. 2009). Thus, changes in synaptic density, or synaptic plasticity, affect activity, sleep, learning, and memory processes. These studies prompted us to ask whether there was an expansion in synaptic bouton formation resulting from Dmel\ELP3 loss, which would provide a potential mechanism underlying the behavioral changes we observed in the adult Dmel\ELP3/RNAi flies. To explore this possibility, we examined bouton morphology in the fly larval neuromuscular junction, as this system is extremely advantageous to the study of synaptic plasticity in that it is very well characterized and shows striking conservation of numerous key synaptic molecules identified in mammals (Collins & DiAntonio 2007, Koh et al. 2000, Broadie & Bate 1993). Dmel\ELP3/RNAi and control w1118 flies were crossed to the elavC155 pan-neuronal GAL4 driver, and third instar progeny larvae were collected. Of note, we demonstrated that knockdown of Dmel\ELP3 using this driver results in an increased activity and reduction of sleep of the adult flies. To examine bouton morphology, boutons at muscles 6 and 7 at abdominal segment A4 were stained with anti-HRP that labels the entire presynaptic membrane, cysteine string protein (CSP) that is a specific marker of the presynaptic vesicles within boutons, and Phalloidin, a toxin that stains muscles, to identify and measure the surface area of the appropriate muscle groups and abdominal segments. The degree of bouton expansion at the NMJ was determined by counting the number of synaptic boutons. Remarkably, there was a dramatic increase (182%) of the total number of synaptic boutons in the Dmel\ELP3/RNAi larvae when compared with the wild type control (Figures 5A, B and C). Of note, there are two types of boutons that are found within larval NMJ muscles 6 and 7. These boutons are classified as type I small (Is) and type I big (Ib) by size. Type-Is boutons have larger stimulation thresholds and excitatory junctional currents of larger amplitude while type-Ib boutons exhibit more pronounced short-term facilitation (Koh et al. 2000). Intriguingly, although both type-Is and type-Ib boutons in the Dmel\ELP3/RNAi lines were significantly increased when compared to the wild-type control, there was a substantially larger expansion of type-Is boutons when directly compared to Ib (329% increase of type Is to 129% increase of type Ib), supporting partial specificity in Dmel\ELP3 function in certain bouton types (Figures 5A, B and C). In support of this concept, “satellite” bouton budding, a process that involves the budding of bouton(s) from one central bouton on the main branch, was indistinguishable in the Dmel\ELP3/RNAi flies when compared to the wild-type control, suggesting that Dmel\ELP3 does not affect this process. Of note, we also observed an increase in axonal arbor area in terms of length and branching relative to muscle surface area in the ELP3 RNA flies when compared to controls (Figure 5D). Taken together, our results indicate that Dmel\ELP3 plays a role in controlling the degree of axonal arbor length and branching and synaptic bouton expansion, and displays at least some specificity in preferentially controlling type Is bouton formation.
FIGURE 5. Loss of Dmel\ELP3 in the nervous system leads to an expansion of synaptic boutons and axonal arbor area in the larval NMJ.

Flies homozygous for either Dmel\ELP3/RNAi line B or control w1118 were crossed to flies homozygous for the nervous system elavC155 pan-neuronal GAL4 driver, and staged third instar progeny larvae were collected. Confocal imaging analysis of boutons at muscles 6/7 at abdominal segment 4 immunohistochemical stained with anti-HRP (in green) that labels the entire presynaptic membrane, CSP (in red) that is a specific marker of the presynaptic vesicles within boutons, or merged HRP and CSP (in yellow). (A) larva expressing Dmel\RNAi/B; i) is at 40x magnification and ii) is at 60x magnification (B) control larva; i) is at 40x magnification and ii) is 60x magnification (C) Histogram depicts quantitative analysis of Type Is and Ib bouton number at muscles 6 and 7 at abdominal segment 4. (P < 0.000001, n ≤ 12) (D) Muscles 6/7 at abdominal segment 4 were stained with Phalloidin, a toxin that stains muscles, to identify and measure their surface area. Histogram represents synaptic bouton arbor area relative to the muscle area in Elp3/RNAi and control larva. In the analyses, each genotype is represented by 13 larval preparations (n=13). Asterisks (*) indicates significant fold change in relation to control where single asterisks indicate p ≤ 0.000001 and double asterisks indicate p ≤ 0.00004. All error bars depict standard deviation of the mean.
Discussion
In this report, we investigate the behavioral and morphological phenotypes that result from targeted reduction of Elp3 HAT levels both ubiquitously and specifically in the developing Drosophila nervous system, in vivo. We demonstrate that targeted reduction of Elp3 in all tissues of the fly results in a significant reduction in fly viability (Suplemental Table 1). Studies of Elp3 function in mammalian cell types indicate that loss of Elp3 indeed leads to severe defects in certain specific cellular processes. For example, loss of ELP3 in human HeLa cell lines leads to repression of certain genes that encode proteins required for cell motility and cell migration. Accordingly, Elp3 depleted neuronal cells and fibroblasts from FD patients both display a significant reduction in cell motility (Close et al. 2006). More recently, studies using mouse cortical neuronal cells directly implicate Elp3 in the acetylation of α-tubulin that controls the migration and differentiation of cortical neurons (Gardiner et al. 2007, Creppe et al. 2009, Wynshaw-Boris 2009). Moreover, while this work was in progress, Simpson et al. identified a fly strain containing a lethal transposon that molecularly mapped to the ELP3 gene, confirming our findings that knockdown of ELP3 in all tissues has lethal consequences in the fly (Simpson et al. 2009). Based on these findings, we speculate that as multicellular development in our Dmel\ELP3/RNAi flies proceeds, disruption of the cell specific processes that require non-redundant Dmel\ELP3 functions accrue over time, ultimately culminating in the phenotypes we observe.
We found that loss of ELP3 in the nervous system of Drosophila results in a significant increase in gross locomotor activity and a significant reduction in the amount of time the flies sleep. Consistent with these hyperactive behavioral phenotypes, our immunohistochemical staining of the larval neuromuscular junction (NMJ) using antibodies to synaptic markers HRP, that stains neuronal membranes and cysteine string protein, a pre-synaptic marker protein that regulates Ca2+ channels and is essential for vesicle exocytosis and neurotransmitter release (Zinsmaier et al. 1994, Dawson-Scully et al. 2000, Bronk et al. 2005), show that flies depleted for ELP3 in the nervous system also display a significant increase in synaptic bouton number in the larval NMJ. Importantly, while our studies are the first to demonstrate a role for ELP3 in synaptic bouton formation, while this work was in progress, Simpson et al demonstrated that flies containing a mutation in the GCN5-related acetyltransferase (GNAT) domain of ELP3 result in a disruption of photoreceptor projections into the fly medulla, indicative of defects in neuronal communication that may arise at least in part from altered axonal targeting and synaptic development (Simpson et al. 2009). Importantly, although the actual bouton number in our ELP3 depleted flies is significantly higher than the wild-type controls, the actual boutons themselves exhibit a typical HRP and CSP staining pattern, suggesting that these boutons are functional for neuronal firing. Accordingly, in ELP3 depleted flies we observe a significant upregulation of synaptobrevin, a vesicle-associated membrane protein (VAMP) that is part of the SNAP-receptor (SNARE) complex and mediates exocytotic vesicle fusion that allows for either cell growth or fusion of neurotransmitter containing vesicles required for neuronal firing (Wallis et al. 2008, Davids et al. 2003, Bhattacharya et al. 2002). Moreover, we observe down-regulation of sleepless (sss), a brain-enriched glycosyl-phosphatidylinositol (GPI)- anchored membrane protein proposed to enhance K+ channel activity in restoring resting neuronal membrane potential, thus reducing neuronal excitability and inducing sleep (Koh et al. 2008). Together, a reduction in sleepless expression in conjunction with an increase in synaptic bouton number and concomitant increases in both CSP and synaptobrevin levels in ELP3 depleted flies suggest that neuronal firing may be occurring more readily, providing a potential molecular mechanism underlying their hyperactive behavioral phenotype.
Several important studies have recently been published on sleep processes in the fly, supporting a link between sleep need and synaptic bouton formation and function. Gilestro et al. demonstrate that levels of several synaptic structural and secretory machinery protein markers in the fly increase during periods of wakefulness and decrease after sleep, with CSP showing a significant increase after 12 hours of continuous waking (Gilestro et al. 2009). Moreover, Donlea et al. show that the number of synaptic terminals in the brain of the fly after periods of social enrichment increases and that these numbers decrease after long bouts of sleep following social enrichment (Donlea et al. 2009). Taken together, these studies support the synaptic homeostasis hypothesis, which claims that sleep is required to downscale synapse formation in the brain (Tononi & Cirelli 2006). According to this model, potentiation of the synapses occurs while organisms are awake and increases during long durations of wakefulness. Downscaling of synapses during sleep may be necessary to lower energy consumption, free up space for synapses to grow during the next waking period, and decrease cellular stress caused by the synthesis and delivery of neurotransmitter containing synaptic vesicles. The significant increase in synaptic boutons, upregulation of BIP (Elefant & Palter 1999) that potentially counteracts the cellular stress associated with long bouts of wakefulness ((Mackiewicz et al. 2008, Naidoo et al. 2007) and the reduction of total sleep we observe for the CNS expressing Dmel\ELP3/RNAi flies support the synaptic homeostasis hypothesis, although our data may also suggest that reduction in synaptic strength is a consequence rather than a drive for sleep in the fly.
How does ELP3 play an active role in the control of bouton formation and sleep in the fly? One possible explanation may be directly due to the recently discovered cytoplasmic-based function for ELP3 in the acetylation of α-tubulin that allows for their stable polymerization into microtubules. These studies demonstrate that loss of ELP3 in cultured projection neuronal cells leads to severe defects in axonal branching (Creppe et al. 2009, Wynshaw-Boris 2009, Gardiner et al. 2007). Moreover, these researchers demonstrate that purified ELP3 promotes α-tubulin acetylation of microtubules while specific blockage of this acetylation using a dominant negative version of a-tubulin that cannot be acetylated leads to axonal branching defects similar to those resulting from ELP3 loss, suggesting that Elp3-induced acetylation of α-tubulin is required for appropriate axonal branching. Transition to new axonal growth and branch formation has been shown to be accompanied by splaying of looped microtubules and formation of short microtubules that interact with the actin cytoskeleton to invade the lamellipodium (Dent & Gertler 2003, Dent & Kalil 2001). Similarly, formation of synaptic boutons at axon terminals is achieved via looping of microtubules that invades and promotes bouton budding at the plasma membrane (Colon-Ramos 2009, Collins & DiAntonio 2007). Such budding is believed to rely on destabilization of the cytoskeleton as indicated by studies demonstrating that disruption of dynamic actin filaments leads to blockage of long-term potentiation (LTP), a cellular correlate for memory formation indicative of synaptic bouton formation and function (Halpain et al. 1998, Ramachandran & Frey 2009). Based on these observations, one hypothesis may be that loss of ELP3 promotes destabilization of microtubules via reduction of α-tubulin acetylation, leading to an increase in microtubule looping and splaying as well as destabilization of the cytoskeleton, ultimately enhancing axonal branching, length, and synaptic bouton budding. Alternatively, bouton expansion may arise from the epigenetic role of ELP3 in directly regulating sleepless gene expression. ELP3-induced disruption of the transcriptional activation of the sleepless gene in the nucleus might, according to the synaptic homeostasis hypothesis, lead to long bouts of wakefulness and enhanced neuronal firing, thus triggering expansion of synaptic bouton formation via downstream pathways (Tononi & Cirelli 2006).
We found that loss of Dmel\ELP3 in the CNS of 15 and 16 day old adult flies appeared to have no effect on recovery sleep after sleep deprivation (Supplementary Figure 4). Our finding is in contrast to the lack of sleep homeostasis seen in a hypomorphic SLEEPLESS mutant which has a similar reduced sleep phenotype (Kho et al., 2008) to our ELP3/RNAi flies (Fig. 3A,B and Supplemental Figs. 3A and B) and less of a response than seen in transgenic flies over expressing BiP under the control of a heat shock promoter (Naidoo et al., 2007). This normal homeostatic response could be attributable either to the contrasting effects of the concurrent changes of both of the SLEEPLESS and BiP genes or possibly to the more restricted expression of the CNS GAL4 60IIA promoter versus the global effect of the P element insertions in the SLEEPLESS gene and the heat shock promoter used in the BiP experiments. Additionally, as Dmel/ELP3 expression levels change during fly development (Zhu et al., 2007), it is possible that this may represent a changing role for ELP3 regulation upon these genes with increasing age of the fly, consistent with our observation that the climbing ability of Dmel/ELP3\RNAi flies is more significantly affected after day 15.
The identification of a number of neurological disorders that result from HAT misregulation underscores a crucial role for acetylation in neurogenesis. Missense mutations in the CBP and p300 genes or loss of a CBP allele cause Rubinstein-Taybi syndrome (RTS), a human disease that displays complex phenotypic abnormalities including mental retardation and neoplasia. Memory loss associated with RTS is shown to be due to lack of CBP HAT activity, and treatment of an RTS mouse model with histone deacetylase inhibitors (HDACi) rescues RTS-associated memory deficits, indicating that appropriate histone acetylation is critical for long-term potentiation, learning, and memory (Alarcon et al. 2004, Korzus et al. 2004). Recent studies support the importance of selective HDACi design, as only specific HDACs appear to affect synaptic plasticity and memory formation (Guan et al. 2009). For example, neuron-specific overexpression of HDAC2 and not HDAC1 in mice results in a decrease of synapse number and memory enhancement, and deficiency of HDAC2 in mice displays the converse of these effects. Importantly, WT-151, an HDACi that selectively inhibits HDAC6, a class II HDAC known to target K40 acetylation of α-tubulin does not increase memory formation in mice, inferring that stable ELP3-induced α-tubulin acetylation does not increase synaptic bouton formation (Guan et al. 2009). Consistent with this finding, here we show for the first time that reduction of ELP3 HAT activity in the fly nervous system results in enhanced synaptic bouton formation and a decrease in sleep activity, supporting an active role for this HAT in the control of synaptic bouton formation. Further understanding of the molecular mechanisms underlying ELP3 in this process has the potential to pave the way for the design of selective epigenetic-based therapeutics for treatment of diseases affecting synaptic plasticity and degeneration.
Materials and Methods
Behavioral Assays:
Activity Assay:
Individual progeny from crosses Dmel\ELP3/RNAi/B x y1w*; P{GawB}60IIA (RNAi) and w1118 x y1w*;P{GawB}60IIA (control) were collected upon eclosion and allowed to acclimate to a 12:12 hour light/dark cycle at 29°C for 4 days after eclosion. Locomotor activity was monitored with the Drosophila Activity Monitoring System (Trikinetics) at 29°C, as per manufacturer’s instructions. Activity counts were recorded every 30 minutes for 24 hours. The significant difference observed between ELP3/RNAi and control groups for total activity was determined using a Student’s t- tests for each time point (n=32). The experiment was carried out three independent times with consistent results.
Digital Video Monitoring:
Individual progeny from crosses Dmel\ELP3/RNAi/B x y1w*; P{GawB}60IIA (RNAi) and w1118 x y1w*;P{GawB}60IIA (control) were collected upon eclosion and allowed to acclimate to a 12:12 hour light/dark cycle at 29°C for 72 hours, beginning 4 days after eclosion. At day 3, individual flies were anesthetized and transferred to Corning Pyrex Glass tubes (65mm length, 5mm diameter) containing Drosophila media at one end. Movements were monitored at 29°C and recorded every 5 seconds by use of digital video recording as previously described (Zimmerman et al. 2008b).
Analysis of video data:
Total sleep, sleep bout number and mean sleep bout duration were calculated from video data using custom software as previously described (Zimmerman et al. 2008b). The significance of differences observed between RNAi and control groups (see above) for total sleep, sleep bout number and mean sleep bout duration for 24 hours was determined using Student’s t-tests for each sex (5 and 6 day old flies, n=32) or for each day (15 and 16 day old females, n=56).
Real-time RT-PCR:
Total RNA was isolated from early pupae of the following crosses in quadruplicate: Dmel\ELP3/RNAi/B x da-GAL4, w1118 x da-GAL4 using TRIzol (Invitrogen) and treated twice with DNA-free (Ambion) to digest DNA. Total RNA was also isolated from 25 heads of 15 day old flies from either a Dmel/ELP3/RNAi/B or w1118 x GAL4 60IIA cross. cDNA was prepared using SuperScript II reverse transcriptase (Invitrogen) according to manufacturer’s instructions with 1 μg total RNA and 0.2μg/mL random hexamer primers (Roche Applied Science). PCR reactions were performed in a 20 μL reaction volume containing 1:4 dilution of 10ng cDNA, 1x Power SYBR® Green PCR Master Mix (Applied Biosystems), and 10μM both forward and reverse primers (primer pairs available upon request). PCR was performed using ABI 7500 Real-Time PCR system (Applied Biosystems) following the manufacturer’s instructions. Fold change in mRNA expression were determined by the ΔΔCt method (Livak & Schmittgen 2001, Yuan et al. 2006). Cycling parameters: 95°C for 3 min, 45–50 cycles of: 95°C for 15 sec, 60°C for 45 sec, followed by dissociation curve step.
Larval NMJ Preparations:
3rdinstar larvae were filleted in HL-3 saline, pH7.2 and pinned out on Sylgard dishes with guts removed. Preps were then fixed in 3.5% paraformaldehyde 1° antibody csp (1.5 μg/μl) incubation overnight, 4°C. Washed 6 times in PBS-T (1x phosphate buffered saline +0.1% Triton), incubated in 2° antibody for 1 hour, washed twice in 1x PBS-T, then once in 1x PBS, then mounted onto slides in Vectashield antifade mounting media. Confocal microscopy was performed using Olympus Microscope with fluoview software. Synaptic boutons were manually counted. In the analyses, each genotype is represented by 13 larval preparations (n=13). The significant difference observed in total bouton number and muscle surface area between ELP3/RNAi and control groups was determined using Student’s t-tests.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Dr. Daniela Zarnescu and Patty Estes for larval NMJ preparation training and Drs. Joseph Bentz, Daniel Marenda, Aleister Saunders, and Jeffery Twiss for valuable discussion of our work. This work was supported by NIH grant R21NS055821 to J.Z, additional support to J.Z. from NIH grant AG17628 and NIH grant R01HD057939 to F.E.
References:
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER and Barco A (2004) Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron, 42, 947–959. [DOI] [PubMed] [Google Scholar]
- Andretic R and Shaw PJ (2005) Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol, 393, 759–772. [DOI] [PubMed] [Google Scholar]
- Axelrod FB (2004) Familial dysautonomia. Muscle Nerve, 29, 352–363. [DOI] [PubMed] [Google Scholar]
- Berger SL (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene, 20, 3007–3013. [DOI] [PubMed] [Google Scholar]
- Berger SL (2002) Histone modifications in transcriptional regulation. Curr Opin Genet Dev, 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Stewart BA, Niemeyer BA, Burgess RW, McCabe BD, Lin P, Boulianne G, O’Kane CJ and Schwarz TL (2002) Members of the synaptobrevin/vesicle-associated membrane protein (VAMP) family in Drosophila are functionally interchangeable in vivo for neurotransmitter release and cell viability. Proc Natl Acad Sci U S A, 99, 13867–13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH and Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Broadie KS and Bate M (1993) Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J Neurosci, 13, 144–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronk P, Nie Z, Klose MK, Dawson-Scully K, Zhang J, Robertson RM, Atwood HL and Zinsmaier KE (2005) The multiple functions of cysteine-string protein analyzed at Drosophila nerve terminals. J Neurosci, 25, 2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrato A, Parisi M, Santa Anna S et al. (2006) Genetic interactions between Drosophila melanogaster menin and Jun/Fos. Dev Biol, 298, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY and Bonini NM (2000) Drosophila models of human neurodegenerative disease. Cell Death Differ, 7, 1075–1080. [DOI] [PubMed] [Google Scholar]
- Chan YB and Kravitz EA (2007) Specific subgroups of FruM neurons control sexually dimorphic patterns of aggression in Drosophila melanogaster. Proc Natl Acad Sci U S A, 104, 19577–19582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L and Feany MB (2005) Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci, 8, 657–663. [DOI] [PubMed] [Google Scholar]
- Close P, Hawkes N, Cornez I et al. (2006) Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell, 22, 521–531. [DOI] [PubMed] [Google Scholar]
- Collins CA and DiAntonio A (2007) Synaptic development: insights from Drosophila. Curr Opin Neurobiol, 17, 35–42. [DOI] [PubMed] [Google Scholar]
- Colon-Ramos DA (2009) Synapse formation in developing neural circuits. Curr Top Dev Biol, 87, 53–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML et al. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell, 136, 551–564. [DOI] [PubMed] [Google Scholar]
- Davids E, Zhang K, Tarazi FI and Baldessarini RJ (2003) Animal models of attention-deficit hyperactivity disorder. Brain Res Brain Res Rev, 42, 1–21. [DOI] [PubMed] [Google Scholar]
- Dawson-Scully K, Bronk P, Atwood HL and Zinsmaier KE (2000) Cysteine-string protein increases the calcium sensitivity of neurotransmitter exocytosis in Drosophila. J Neurosci, 20, 6039–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW and Gertler FB (2003) Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron, 40, 209–227. [DOI] [PubMed] [Google Scholar]
- Dent EW and Kalil K (2001) Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci, 21, 9757–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N and Shaw PJ (2009) Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science, 324, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB (2002) GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis, 34, 1–15. [DOI] [PubMed] [Google Scholar]
- Elefant F, Cooke NE and Liebhaber SA (2000a) Targeted recruitment of histone acetyltransferase activity to a locus control region. J Biol Chem, 275, 13827–13834. [DOI] [PubMed] [Google Scholar]
- Elefant F and Palter KB (1999) Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol Biol Cell, 10, 2101–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefant F, Su Y, Liebhaber SA and Cooke NE (2000b) Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. Embo J, 19, 6814–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ and Bystrom AS (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell, 24, 139–148. [DOI] [PubMed] [Google Scholar]
- Feany MB and Bender WW (2000) A Drosophila model of Parkinson’s disease. Nature, 404, 394–398. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G and Groudine M (2003) Controlling the double helix. Nature, 421, 448–453. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y and Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol, 15, 172–183. [DOI] [PubMed] [Google Scholar]
- FlyBase (2009) Gene Dmel\ELP3.
- Fortier E, Belote JM (2000) Temperature-Dependent Gene Silencing by an Expressed Inverted Repreat in Drosophila. Genesis, 26, 240–244. [DOI] [PubMed] [Google Scholar]
- Gardiner J, Barton D, Marc J and Overall R (2007) Potential role of tubulin acetylation and microtubule-based protein trafficking in familial dysautonomia. Traffic, 8, 1145–1149. [DOI] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G and Cirelli C (2009) Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science, 324, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E et al. (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature, 459, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpain S, Hipolito A and Saffer L (1998) Regulation of F-actin stability in dendritic spines by glutamate receptors and calcineurin. J Neurosci, 18, 9835–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes NA, Otero G, Winkler GS et al. (2002) Purification and characterization of the human elongator complex. J Biol Chem, 277, 3047–3052. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Lu S, Kume K, Yin JC, Yang Z and Sehgal A (2003) Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms, 18, 12–25. [DOI] [PubMed] [Google Scholar]
- Huang B, Johansson MJ and Bystrom AS (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA, 11, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G and Cirelli C (2004) Sleep homeostasis in Drosophila melanogaster. Sleep, 27, 628–639. [DOI] [PubMed] [Google Scholar]
- Jenuwein T and Allis CD (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kiefer JC (2007) Epigenetics in development. Dev Dyn, 236, 1144–1156. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lane WS and Reinberg D (2002) Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A, 99, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ and Sehgal A (2008) Identification of SLEEPLESS, a sleep-promoting factor. Science, 321, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YH, Gramates LS and Budnik V (2000) Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity. Microsc Res Tech, 49, 14–25. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG and Mayford M (2004) CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron, 42, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A and Svejstrup JQ (2004) Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J, 23, 4243–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A, Walker J, Suka N, Grunstein M, Roberts D, Cairns BR and Svejstrup JQ (2002) Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol Cell, 10, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Naidoo N, Zimmerman JE and Pack AI (2008) Molecular mechanisms of sleep and wakefulness. Ann N Y Acad Sci, 1129, 335–349. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P and Reinberg D (2005) The key to development: interpreting the histone code? Curr Opin Genet Dev, 15, 163–176. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J and Pack AI (2007) A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep, 30, 557–565. [DOI] [PubMed] [Google Scholar]
- Orphanides G and Reinberg D (2002) A unified theory of gene expression. Cell, 108, 439–451. [DOI] [PubMed] [Google Scholar]
- Petrakis TG, Wittschieben BO and Svejstrup JQ (2004) Molecular architecture, structure-function relationship, and importance of the Elp3 subunit for the RNA binding of holo-elongator. J Biol Chem, 279, 32087–32092. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ and Collins RN (2005) Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell, 17, 841–853. [DOI] [PubMed] [Google Scholar]
- Ramachandran B and Frey JU (2009) Interfering with the actin network and its effect on long-term potentiation and synaptic tagging in hippocampal CA1 neurons in slices in vitro. J Neurosci, 29, 12167–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature, 447, 425–432. [DOI] [PubMed] [Google Scholar]
- Rushton E, Rohrbough J and Broadie K (2009) Presynaptic secretion of mind-the-gap organizes the synaptic extracellular matrix-integrin interface and postsynaptic environments. Dev Dyn, 238, 554–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ and Tononi G (2000) Correlates of sleep and waking in Drosophila melanogaster. Science, 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ and Robinson DF (2002) Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature, 417, 287–291. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Lemmens R, Miskiewicz K et al. (2009) Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet, 18, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA and Gusella JF (2002) Familial dysautonomia. Curr Opin Genet Dev, 12, 307–311. [DOI] [PubMed] [Google Scholar]
- Strahl BD and Allis CD (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ (2007) Elongator complex: how many roles does it play? Curr Opin Cell Biol, 19, 331–336. [DOI] [PubMed] [Google Scholar]
- Tononi G and Cirelli C (2006) Sleep function and synaptic homeostasis. Sleep Med Rev, 10, 49–62. [DOI] [PubMed] [Google Scholar]
- Wallis D, Russell HF and Muenke M (2008) Review: Genetics of attention deficit/hyperactivity disorder. J Pediatr Psychol, 33, 1085–1099. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P and Svejstrup JQ (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci U S A, 99, 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P and Svejstrup JQ (2001) RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem, 276, 32743–32749. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A (2009) Elongator bridges tubulin acetylation and neuronal migration. Cell, 136, 393–394. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F and Stewart CN Jr. (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics, 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Singh N, Donnelly C, Boimel P and Elefant F (2007) The cloning and characterization of the histone acetyltransferase human homolog Dmel\TIP60 in Drosophila melanogaster: Dmel\TIP60 is essential for multicellular development. Genetics, 175, 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Naidoo N, Raizen DM and Pack AI (2008a) Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci, 31, 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Raizen DM, Maycock MH, Maislin G and Pack AI (2008b) A video method to study Drosophila sleep. Sleep, 31, 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier KE, Eberle KK, Buchner E, Walter N and Benzer S (1994) Paralysis and early death in cysteine string protein mutants of Drosophila. Science, 263, 977–980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


