Abstract
Anammox (anaerobic ammonium oxidation) bacteria are important for the nitrogen cycle in both natural environments and wastewater treatment plants. These bacteria have a strong tendency to grow in aggregates like biofilms and granular sludge. To understand the formation of anammox aggregates, it is required to unravel the composition of the extracellular polymeric substances (EPS), which are produced by the bacteria to develop into aggregates and granules. Here, we investigated anionic polymers in anammox granular sludge, focussing on sialic acids and sulfated glycosaminoglycans. Quantification assays and fluorescent stains indicated that sialic acids and sulfated glycosaminoglycans were present in the anammox EPS (1.6% equivalents of sialic acids and 2.4% equivalents of sulfated glycosaminoglycans). Additionally, the potential genes for the biosynthesis of sialic acids and sulfated glycosaminoglycans were analyzed in the anammox draft genomes. The finding of these components in anammox granular sludge and previously in other nonpathogenic bacteria pointed out that sialic acids and sulfated glycosaminoglycans are worth investigating in the context of a broader function in microbial communities and biofilm systems in general.
1. Introduction
In the early 1990s, the anaerobic ammonium oxidation (anammox) process in which ammonium and nitrite are converted into nitrogen gas was discovered.1 The bacteria that were found to perform this process are referred to as anammox bacteria, and they belong to a separate order named “Candidatus Brocadiales”, within the phylum Planctomycetes. They can perform the anammox process without the need of an organic carbon source or oxygen. In natural environments, anammox bacteria contribute significantly to the global nitrogen cycle.2 Moreover, the anammox process is applied in wastewater treatment to remove nitrogen from wastewater. Anammox bacteria have a strong tendency to grow in an aggregated form.3 This trait is exploited in anammox wastewater treatment plants where the bacteria are grown in the granular sludge form. Granular sludge systems provide high sludge settling velocities, ensuring a high biomass retention and utilization of relatively small reactors, costs, and footprint.
Similar to other kinds of biofilms, in granular sludge, bacteria are immobilized in a self-produced matrix. This matrix consists of a complex mixture of components, which is referred to as extracellular polymeric substances (EPS). In biofilm research, unravelling the EPS composition and function is important to move toward a comprehensive understanding and better control of biofilm formation. Due to limitations in methodology for extraction and characterization, the EPS composition is still far from fully characterized.4 Great efforts have been taken along this research line in recent years, and progress has been made with respect to solubilization of the biofilm and the discovery of new components in EPS.5−8
In our previous study, the presence of strongly anionic groups in EPS of anammox granules was indicated by means of staining with the cationic dye Alcian Blue.6 Alcian Blue stained two targets after gel electrophoresis of alkaline-extracted EPS, which could be distinguished by the molecular weight and strength of the negative charge: the high molecular-weight component (> 235 kDa) stained at pH 2.5, indicating acidic groups (i.e., carboxyl group and/or sulfate group) and a component with an apparent molecular weight of around 12 kDa stained at pH 1.0, indicating an even stronger negative charge. Because of the strong acidic character, combined with the high S-content (1.4%) measured in the extracted EPS, this strongly negatively charged component was assumed to be a polymer with sulfate groups. However, the nature and functions of these acidic components remain unknown.
Negatively charged groups are reported to play an important role in the adhesion capacity of EPS9 and protection of bacteria against environmental stresses.10 Moreover, negatively charged polysaccharides have been a target of interest in a few biofilm studies, e.g., alginates and uronic acids.11 However, the existence of polysaccharides with other negatively charged groups is also possible.
Sialic acids are a group of negatively charged nine-carbon monosaccharides, which are mostly found as the terminal residue of glycoconjugates in eukaryotes or pathogenic bacteria where they play important roles in mediating cellular recognition, adhesion processes, and protecting the underlying tissue.12,13 There are different types of sialic acids. The most known is N-acetyl neuraminic acid among many other (more than 50) derivatives. Sialic acid types were found in prokaryotes also. These are present in different isomeric forms, named after pseudaminic acid and legionaminic acid. These sialic acids are synthesized by bacteria via (partly) different pathways in comparison to their eukaryotic counterpart.14 They were also referred to as bacterial sialic acids in literature.
Sulfated polysaccharides are mostly known from the extracellular matrix of animal as sulfated glycosaminoglycans. Sulfated glycosaminoglycans are complex linear polysaccharides that can be classified into three major groups: (1) chondroitin sulfate and dermatan sulfate, (2) heparin and heparan sulfate, and (3) keratan sulfate. These molecules are associated with functions in mediating adhesion and cell signalling in mostly eukaryotes and pathogenic bacteria.15 The presence of the strongly negatively charged components in combination with the high sulfur content in our previous study, raised the question: is the strong anionic component detected in the EPS of anammox granular sludge due to the presence of components like sulfated glycosaminoglycans?
Here, we present a study in which the previously revealed anionic components in anammox EPS are further analyzed, focussing on identification, quantification, and localization of sialic acids and sulfated glycosaminoglycans. To this end, alkaline-extracted EPS was subjected to quantification assays and to mass spectrometry. In addition, specific fluorescent stains were applied to the intact anammox granules. Finally, genome database searches were performed to find potential pathways that could be involved in the formation of these anionic components by anammox bacteria.
2. Materials and Methods
2.1. Anammox Granular Sludge and EPS Extraction
Anammox granular sludge was collected from the full-scale anammox reactor in Sluisjesdijk, Rotterdam.16 The VSS (volatile suspended solids) content of the granules was 0.71 g/g granules (determined in accordance with APHA, 2005).17 The dominant anammox species in the granules was “Ca. Brocadia sapporoensis” (according to FISH, clone library analysis, and protein analysis in Boleij et al.).7 EPS extraction was performed as described in Boleij et al.6 The granules were solubilized in 0.1 M NaOH for 5 h while being stirred with a magnetic stirrer (IKA, C-MAG HS7), using a magnet with a diameter that covers the surface of the bottle, at 400 rpm. After centrifugation at 3100 g for 20 min at 4 °C, the pellet was discarded. Polymers in the supernatant were precipitated out by decreasing the pH to 5 using 1 M HCl. The precipitated polymers were collected by centrifugation at 3100 g for 20 min at 4 °C and directly lyophilized.
2.2. Sialic Acid Quantification Assay
To measure sialic acids, the sialic acid quantitation kit (Sigma-Aldrich) was used according to manufacturer instruction. The protocol was performed as described by de Graaff et al.18 The assay was applied on crushed lyophilized granules, and N-acetylneuraminic acid was used as a standard. This assay measures N-acetylneuraminic acid (NeuAc) after its release by an enzymatic cleavage using the neuraminidase. Therefore, it is suitable to measure the amount of NeuAc in either free form or in glycoproteins, cell surface glycoproteins, polysialic acids, and capsular polysaccharides.
2.3. Sialic Acid Measurement with Mass Spectrometry
The sialic acid analysis was performed following a recently established approach by Kleikamp et al.19 Mass spectrometric sialic acid analysis was performed following physical disruption and homogenization of a 2.5 mg freeze-dried granule material using a lab mortar and pestle and hydrolysis using diluted (2 M) acetic acid solution. Released sialic acids were further labeled directly from the speed vac dried lysate using DMB (1,2-diamino-4,5-methylenedioxybenzene dihydrochloride). Labeled sialic acid derivatives were further analyzed by reverse phase chromatography Orbitrap mass spectrometry (QE plus quadrupole Orbitrap, Thermo, Germany). Sialic acids were identified by comparison to commercial standards and mass.
2.4. EPS Extraction for Sulfated Glycosaminoglycans Measurement
In the previous study where anionic polymers were discovered,6 an alkaline extraction using precipitation at pH 5 was applied. For the measurement of sulfated glycosaminoglycans in this study, the same extraction method (as described in section 2.1) was used as a reference. In addition, the alkaline extraction was applied with precipitation at pH 2.5 to investigate if there are more sulfated glycosaminoglycans extracted when the precipitation was performed at a lower pH. As a control, the alkaline protocol was applied, using dialysis to remove NaOH, instead of precipitation. In theory, this would include all polymers that are solubilized, with a molecular cut-off of 3 kDa. Samples were lyophilized directly after extraction until further analysis. Hence, three extracted samples extract 1 (pH 5), extract 2 (pH 2,5), and extract 3 (dialyzed) were subjected to the sulfated glycosaminoglycan assay and the preceding treatment.
2.5. Pretreatment for Sulfated Glycosaminoglycan Assay
As a pretreatment for the glycosaminoglycan assay, samples were denatured and treated with proteinase K (Sigma Aldrich, 30 units/mg protein). For denaturation, a 4 mg/mL lyophilized sample was solubilized in a solution of 6 M urea, 25 mM NH4HCO3, and 10 mM DTT and incubated at 65 °C for 30 min. After cooling to room temperature, iodoacetamide was added to a final concentration of 40 mM, and samples were incubated for 30 min at room temperature. For the enzyme treatment, the samples were diluted four times in TRIS buffer (50 mM TRIS, 5 mM CaCl2, and 10 mM EDTA) to decrease the urea concentration, avoiding inhibition of the enzyme treatment. Proteinase K was added to a final concentration of 125 μg/mL, and the samples were incubated overnight at 37 °C.
2.6. Sulfated Glycosaminoglycan Assay
The Blyscan sulfated glycosaminoglycan assay was used to quantify sulfated glycosaminoglycans in the three extracts (described in section 2.4), as well as in the whole granules and the pellet remaining after alkaline solubilization. The assay is based on binding of 1,9-dimethyl-methylene blue (DMMB) to sulfated glycosaminoglycans at a low pH (measured pH in the DMMB solution was 1.7). The DMMB–sulfated glycosaminoglycan complex will precipitate, and subsequently, the nonbound soluble dye can be removed. Then, the bound dye is released using a dissociation reagent, and the absorbance is measured to indicate the amount of dye that formed a complex with the sulfated glycosaminoglycans. The standard that is included in the kit is bovine tracheal chondroitin 4-sulfate. The protocol was performed according to manufacturer instructions (Blyscan, Biocolor (UK)), after the pretreatment was applied as described in the previous section.
2.7. SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) and Staining with Coomassie Blue and Alcian Blue
The extracted EPS was analyzed by SDS-PAGE, as described in Boleij et al.6 SDS-PAGE was performed using NuPage Novex 4–12% bis–tris gels (Invitrogen). EPS samples were prepared in NuPAGE LDS-buffer, and DTT (dithiothreitol) was added to a final concentration of 10 mM. The proteins were denatured by incubation at 70 °C for 10 min. Subsequently, 10 μL of sample was loaded per well. The Thermo Scientific spectra multicolor broad range protein ladder was used as a molecular-weight marker. The gel electrophoresis was performed at 200 V for 35 min. The gels were stained by three different staining procedures afterward.
For visualization of proteins, the Colloidal Blue staining kit (Invitrogen) was used according to manufacturer instructions. For staining of acidic glycoconjugates, the cationic dye Alcian Blue 8GX (Fluka, Sigma Aldrich) was used. To differentiate between the relatively weaker acidic groups like carboxylate (R-COO–) and the stronger acidic group like sulfate (R-OSO3–), staining with Alcian Blue was performed at different pH values, namely, pHs 2.5 and 1.0. An adapted protocol of Møller and Poulsen20 was used. After electrophoresis, the gels were extensively washed in solution I (25% (v/v) ethanol and 10% (v/v) acetic acid, pH 2.5) for 2.5 h while refreshing the solution four times. Subsequently, the gel was stained in 0.125% (w/v) Alcian Blue in solution I (the solution was stirred overnight to dissolve the Alcian Blue and centrifuged before use) for 30 min and washed in solution I overnight. For staining of sulfated groups at pH 1.0, the same protocol was performed except that solution I was replaced by solution II (0.1 M HCl and 25% (v/v) ethanol, pH 1.0) according to Tobisawa et al.21
2.8. Lectin Staining
The glycoconjugates of the granules were studied after fluorescence lectin bar-coding and subsequent fluorescence lectin-binding analysis.22 Consequently all commercially available lectins (FITC, Fluorescein, Alexa488) were screened for binding to granule structures. Lectin staining was already described in several publications.23,24 In short, the fully hydrated, intact granules were incubated with a few droplets of lectins (0.1 mg/mL) for 20 min at room temperature in the dark and washed three times to remove the unbound lectins. The lectin-stained, hydrated, intact granules were mounted in coverwell chambers (Thermofisher) with 1 mm spacers to avoid compression of the granule. Samples were examined using a Leica SP5X instrument (Leica, Germany) equipped with an upright microscope and a super continuum light source (white laser). The setup was controlled by LAS AF software version 2.4.1. Confocal images were recorded as serial scan and a step size of 1 μm using a 25× NA 0.95 water immersion lens. Laser excitation was at 490 nm, emission was from 485 to 495 nm (reflection) and 505 to 600 nm (lectins). For improvement of resolution and contrast, some image datasets were subjected to blind deconvolution with Huygens version 18.10.0 (SVI, The Netherlands). Data were projected using Imaris version 9.2.1 (Bitplane). All image data sets were finalized in Photoshop (Adobe).
2.9. Heparin Red Staining
The fluorescent probe Heparin Red (RedProbes, Münster, Germany) was employed for staining negatively charged macromolecules (such as Heparin) in the granular matrix. For staining, the protocol of the supplier data sheet was followed. In short, 8.8 μL of Heparin Red and 1 mL of enhancer solution were mixed and added to the fully hydrated, intact granules for 1 h. The Heparin Red-stained, fully hydrated, intact granules were immediately examined by CLSM using the instrument described above. Image data sets were recorded as single or serial scan (step size 1, 0.5, or 0.17 μm) using a 63× NA 1.2 water or a 100× NA 1.4 oil immersion lens at various zoom settings. Laser excitation was at 480 and 567 nm, emission was from 470 to 490 nm (reflection) and 590 to 650 nm (Heparin Red). Image data handling was already explained above. In addition, Heparin Red data sets were loaded in Fiji (https://fiji.sc/) and color coded with the lookup table called “rainbow RGB”. For the better color separation of pixel intensities, the contrast was set to auto. By this treatment, three pixel intensities are color coded as red = strong signal, green = intermediate signal, and blue = weak signal.
2.10. BLAST (Basic Local Alignment Search Tool) Analysis of Pathways for Syntheses of Sialic Acid Types and Sulfated Glycosaminoglycans
BLAST analysis was applied to find homologous biosynthesis pathways of sialic acids and sulfated glycosaminoglycans. The BLAST tool at the NCBI website was used to BLAST reference proteins against the draft genomes included in the database of “Candidatus Brocadiales” (taxid:1127829). Matches were viewed as significantly similar when E-values were below 1E-20.
3. Results
3.1. Sialic Acids in Anammox Granular Sludge
Previously, a high amount of proteins (∼60%) was measured in the extracted EPS of anammox granules, and various glycoconjugates were found.6 Glycoconjugate modifications with acidic groups such as sulfate (sulfation) and/or sialic acid (sialylation) on the polysaccharide part are common phenomena in the extracellular matrix of eukaryotes.25 In this previous study, the presence of polyanionic groups in glycoconjugates was indicated by staining with a cationic dye (Alcian Blue). Here, analysis is performed on strongly negatively charged polyanionic components such as sulfates and sialic acids.
Sialic acids were measured in the anammox granules, using a sialic acid quantitation kit, which is based on the release of N-acetylneuramic acid (NeuAc) by an enzymatic treatment. According to this assay, the anammox granules contain 1.6% sialic acids (weight % of gram of N-acetylneuraminic acid equivalents per gram VSS of granular sludge). Hence, the quantification assay data indicates the presence of sialic acids. To identify which kinds of sialic acids are present in anammox granules, mass spectrometry (MS) was applied. With MS, sialic acids were detected in the form of N-acetyl neuraminic acid (NeuAc), deaminated neuraminic acid (KDN), and pseudaminic acid/legionaminic acid (Pse/Leg, which have the same molecular weights) (see Table S1). Hence, there are three different kinds of sialic acids widely distributed in anammox granules.
The draft genome of “Ca. Brocadiales” was analyzed for genes that could code for the pathways for the biosynthesis of sialic acids, reference pathways from known sialic acid producers were taken (see Figure S1). The genes of the required enzymes were aligned with the draft genomes of species in the order “Ca. Brocadiales” that are available in the database of NCBI. Because Campylobacter jejuni has characterized pathways for biosyntheses of NeuAc, Leg, and Pse, these pathways were used as reference genes.26 Those pathways were partly matching with proteins in the “Ca. Brocadiales” database (Tables S2 and S3). Many of the genes of Brocadiales that matched the legionaminic acid synthase were in common with the genes that matched with pseudamiminc acid synthase and the N-acetylneuraminic acid synthase. Even though these pathways are distinct, the enzymes are homologous.
In the halophile archaea Halorubrum sp. PV6, a legionaminic acid was reported to be attached to its surface layer (S-layer) glycoprotein.27 Therefore, the reported genes of Halorubrum sp. PV6 were also used as a reference to BLAST the draft genome of “Ca. Brocadiales”. All enzymes for biosynthesis of legionaminic acid were found to have a significant match with a database protein in the order of “Ca. Brocadiales” (Table 1 and Table S4). Hence, the biosynthesis pathway of Halorubrum matched better with anammox than the synthesis pathways in Campylobacter jejuni.
Table 1. Blast Search for the Pathway for Synthesis of Legionaminic Acida.
| reference gene/protein | accession number best match | name protein and species | identity (%) | E-value |
|---|---|---|---|---|
| HrrPV6_1047 AYD49518.1 (LegB) | WP_070067840.1 | NAD-dependent epimerase/dehydratase family protein [Candidatus Brocadia sapporoensis] | 38.0 | 1.8E-55 |
| HrrPV6_1046 AYD49517.1 (LegC) | WP_070067943.1 | DegT/DnrJ/EryC1/StrS family aminotransferase [Candidatus Brocadia sapporoensis] | 35.4 | 1.2E-66 |
| HrrPV6_1014 AYD49510.1 (LegH) | WP_070066607.1 | hypothetical protein [Candidatus Brocadia sapporoensis] | 32.7 | 3.1E-51 |
| HrrPV6_1049 AYD49520.1 (LegG) | TFG47122.1 | UDP-N-acetylglucosamine 2-epimerase (hydrolyzing) [Candidatus Brocadiae bacterium]* | 37.5 | 2.3E-72 |
| HrrPV6_1048 AYD49519.1 (LegF) | TLD41229.1 | N-acetylneuraminate cytidylyltransferase [Candidatus Jettenia ecosi]* | 34.6 | 8.0E-42 |
| HrrPV6_1053 AYD49523.1 (LegI) | TFG47123.1 | N-acetylneuraminate synthase [Candidatus Brocadiae bacterium]* | 50.1 | 1.5E-109 |
The reference pathway is from halophile Halorubrum sp. PV6.27 The best match shown is the best match with “Ca. Brocadia sapporoensis”. *When there was no match with “Ca. Brocadia sapporoensis”, the best match of any “Ca. Brocadiales” is shown.
3.2. Quantification of Sulfated Glycosaminoglycans
To investigate the strongly anionic polymers, the presence of sulfated glycosaminoglycans was investigated by using the Blyscan sulfated glycosaminoglycan assay. This assay is based on a dye-binding method in which 1,9-dimethylmetylene blue (DMMB) is used to estimate the amount of sulfated glycosaminoglycans. DMMB can bind free sulfated glycosaminoglycans or sulfated polysaccharides that are bound to a protein backbone (proteoglycans).
In the EPS extraction process, the anammox granules were first solubilized in NaOH. Afterward, the EPS was recovered from this alkaline solution by three methods in parallel: precipitation with acid at pH 5 (extract 1), precipitation with acid at pH 2.5 (extract 2), and with dialysis (extract 3). The different recovery methods yielded 24, 22, and 31% of the initial VSS amount, respectively (Table 2). The recovered materials were subjected to the sulfated glycosaminoglycan assay. In addition, the whole granules and the pellet that remained undissolved after alkaline treatment were analyzed.
Table 2. Measured Amount of Sulfated Glycosaminoglycans (GAG) (Chondroitin Sulfate Equivalents), Compared to the Amount that Was Recovered from the Granular Sludgea.
| sample | extraction yield (VSS EPS/granules) | sulfated GAG equivalents measured (%) | sulfated GAG in granules (calculated) |
|---|---|---|---|
| extract 1 (pH 5) | 24% | 5.4% | 1.3% |
| extract 2 (pH 2.5) | 22% | 6.9% | 1.5% |
| extract 3 (dialyzed) | 31% | 5.1% | 1.6% |
| granules | 2.4% | 2.4% | |
| pellet | 0.9% |
The percentages of sulfated GAG equivalents are based on the measured amount of VSS in samples before pretreatment.
For application of the glycosaminoglycan assay, the recovered EPS was solubilized in TRIS buffer. When the solution of the extracted EPS was directly applied to the assay, 1.2% of the VSS fraction was measured as sulfated glycosaminoglycans. It was noticed that a huge amount of precipitate formed once the EPS solution was mixed with the dye solution presumably due to the high protein content in combination with the low pH of the dye solution. Formation of protein precipitation was suspected to hinder the binding of DMMB to the sulfated glycosaminoglycans. To avoid the interference of proteins, a pretreatment was applied in which the EPS was denatured and treated with proteinase K. After pretreatment, most of the proteins were successfully removed (no more binding of Coomassie blue in SDS-PAGE, see Figure S2). The 12 kDa band in the SDS-PAGE gel was still stained by Alcian Blue after pretreatment.
The pretreatment significantly increased the outcome of the quantification assay. The sulfated glycosaminoglycans measured in extracts 1, 2, and 3 were 5.4, 6.9, and 5.1% of the VSS fraction of the extracted EPS, respectively (see Table 2). When whole anammox granules were subjected to the pretreatment and quantification assay, instead of the extracted EPS, 2.4% of the VSS fraction was measured as sulfated glycosaminoglycans.
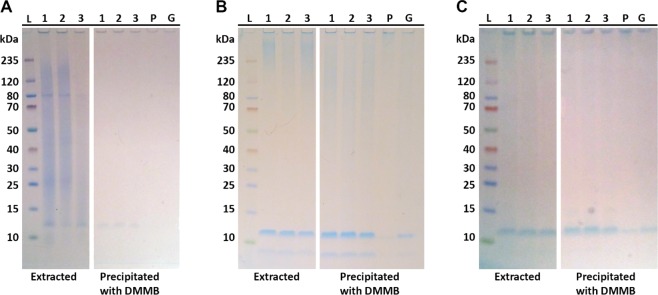
Since the quantification assay is based on precipitation of the DMMB–sulfated glycosaminoglycan complex, it also served as a method for sulfated glycosaminoglycans isolation. The precipitated DMMB–sulfated glycosaminoglycan complex was collected, resolubilized in the NuPAGE LDS-buffer and DTT (dithiothreitol), and applied on SDS-PAGE to check what components were precipitated in the DMMB complex. The profiles are shown in Figure 1. The target component was the band at 12 kDa. In Figure 1, it can be seen that the 12 kDa band is indeed present in the precipitated DMMB complex. Hence, the DMMB assay targets the 12 kDa band.
Figure 1.
Images of SDS-PAGE gels with extracts 1, 2, and 3; the pellet (P); and the whole granules (G). Lane L contains the molecular-weight ladder. Gels were stained with (A) Coomassie Blue, (B) Alcian Blue at pH 2.5, and (C) Alcian Blue at pH 1.0. In all panels, the right part contains the profile of the material after EPS extraction, and the left part is the fraction that formed a complex with the DMMB dye in the glycosaminoglycan assay. (Note that before pretreatment, the pellet and the whole granules cannot be applied on a gel since they are not soluble.).
3.3. Analysis of Anammox Genome for Synthesis of Sulfated Glycosaminoglycans
Sulfotransferases are the enzymes that transfer sulfo groups to polysaccharides.28 For sulfated glycosaminoglycan sulfotransferases, there is little literature available about the encoding genes in bacteria. Searches in the NCBI database to chondroitin, heparan, and keratan sulfotransferase resulted in bacterial genes only for the first two types. Examples for known genes are the chondroitin 4-O-sulfotransferase gene in Pseudomonas fluorescensF113 and heparan sulfate glucosamine 3-O-sulfotransferase in bacteria Sinorhizobium frediiNRG234.(29) The corresponding protein sequences were blasted against the “Ca. Brocadiales” database. For chondroitin 4-O-sulfotransferase (accession number WP_014336261.1) no significant similarity was found (i.e., E-values were higher than 2E-05). Interestingly, blasting against the heparan sulfate glucosamine 3-O-sulfotransferase (accession number YP_002823420.1) resulted in significant similarity with several proteins, shown in Table 3, which means there is a higher chance for anammox bacteria to produce heparan sulfate.
Table 3. List with Significant Matches (E-value < 1E-20) of Blast Search against Heparan Sulfate Glucosamine 3-O-Sulfotransferase (Accession Number YP_002823420.1).
| accession number | name protein and species | identity (%) | E-value |
|---|---|---|---|
| RZV93996.1 | hypothetical protein EX341_03835 [Candidatus Scalindua sp. SCAELEC01] | 31.8 | 6.04E-33 |
| RZV93978.1 | hypothetical protein EX341_03745 [Candidatus Scalindua sp. SCAELEC01] | 29.81 | 4.76E-31 |
| GAN32649.1 | hypothetical protein BROSI_A1164 [Candidatus Brocadia sinica JPN1] | 30.22 | 9.45E-31 |
| WP_082059063.1 | hypothetical protein [Candidatus Brocadia sinica] | 30.2 | 2.72E-30 |
| KXK32771.1 | sulfotransferase [Candidatus Brocadia sinica] | 28.9 | 7.13E-29 |
| KXK32773.1 | sulfotransferase [Candidatus Brocadia sinica] | 28.8 | 3.62E-28 |
| WP_052562786.1 | hypothetical protein [Candidatus Brocadia sinica] | 29.6 | 1.95E-27 |
| TLD42416.1 | sulfotransferase [Candidatus Jettenia ecosi] | 27.3 | 3.66E-24 |
| WP_099326166.1 | sulfotransferase [Candidatus Kuenenia stuttgartiensis] | 29.0 | 6.33E-24 |
| WP_007223162.1 | sulfotransferase [Candidatus Jettenia caeni] | 26.4 | 2.60E-23 |
| KHE93325.1 | sulfotransferase [Candidatus Scalindua brodae] | 27.1 | 1.80E-22 |
| TLD42415.1 | sulfotransferase [Candidatus Jettenia ecosi] | 28.4 | 1.26E-21 |
| KXK28276.1 | sulfotransferase [Candidatus Brocadia sinica] | 25.8 | 1.30E-21 |
| ODS30602.1 | sulfotransferase [Candidatus Scalindua rubra] | 27.3 | 4.78E-21 |
| OQZ03151.1 | hypothetical protein B6D35_00200 [Candidatus Brocadia sp. UTAMX2] | 24.8 | 6.16E-20 |
| KXK28277.1 | sulfotransferase [Candidatus Brocadia sinica] | 29.1 | 7.05E-20 |
3.4. Lectin Staining and Heparin Red Staining
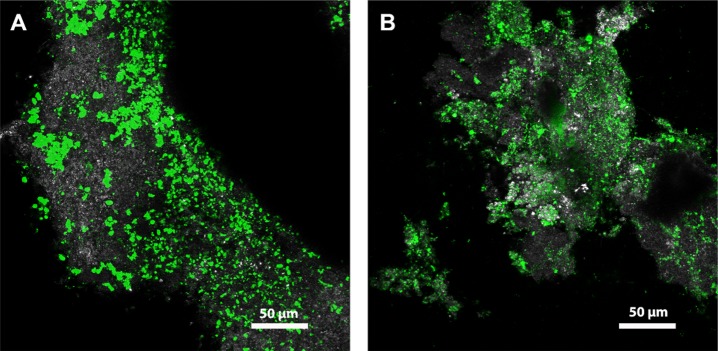
To localize the distribution of sialic acids in granules, lectin staining was applied. A screening with 70 lectins was performed, and several that bind to sialic acid were found to bind to the matrix of the anammox granules. In Figure 2, images with anammox granules stained by fluorescent lectins Wheat germ agglutinin (WGA) and Helix aspersa agglutinin (HAA), respectively, are shown. WGA binds sialic acids (N-acetyl neuraminic acid) and N-acetyl glucosamine (GlcNAc).30 HAA binds to O-linked glycans composed of GalNAc.31 The visualization of the fluorescent signal of these lectins provides the possible distribution of sialic acids in the granules.
Figure 2.
Confocal laser scanning microscopy of fully hydrated, intact anammox granules stained with fluorescently labeled lectins shown as maximum intensity projections. (A) WGA (Wheat germ agglutinin, 40 optical sections) and (B) HAA (Helix aspersa agglutinin, 66 optical sections). Color allocation: grey: reflection signal, green: lectin staining.
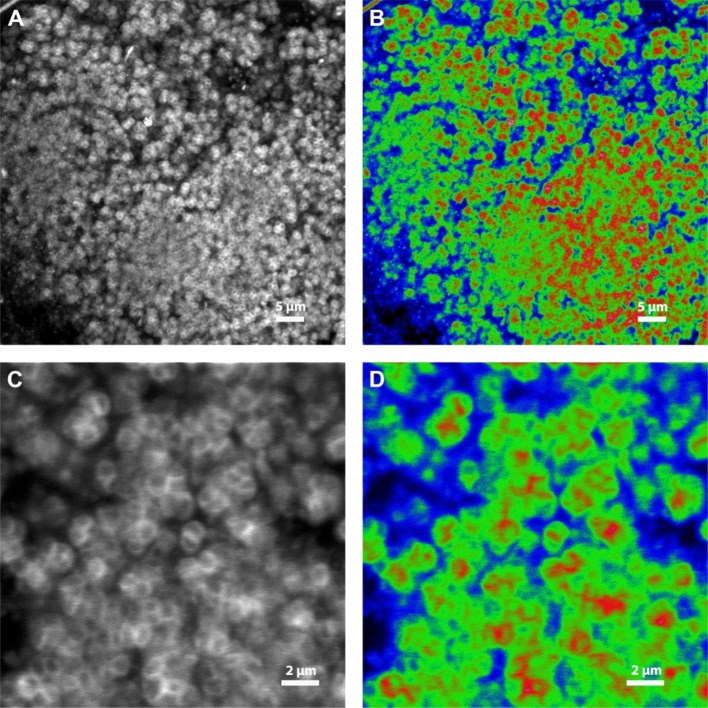
Figure 3 shows anammox granules that were stained with Heparin Red, which is a fluorescent molecular probe. It is used for direct detection of Heparins in blood plasma. Heparin Red is polycationic. It forms complex aggregations with polyanionic components and was used to localize polyanionic components. When Heparin Red is bound to polyanionic components, which have a charge density more negative than 0.81 per monosaccharide, it emits a fluorescent signal. On top of that, the fluorescence intensity of Heparin Red is quenched when it forms stable aggregates on the polyanionic chains. The higher the charge density, the more quenched the fluorescent signal is.32 Since both heparin (1.8–2.4 sulfate per disaccharide) and heparan sulfate (0.8–1.8 sulfate per disaccharide) are highly negatively charged macromolecules,33,34 their binding to Heparin Red results to a quenched signal. Figure 3 shows the cells in the granules, after Heparin Red staining (Figure 3C,D shows higher magnifications). Figure 3A,C is in white and black. In comparison, Figure 3B,D is the same image data set as Figure 3A,C, respectively, but using a look up table in which the pixel intensities are color coded as: very low pixel intensities in blue: no binding of Heparin Red, high pixel intensities in red: binding of Heparin Red, and intermediate pixel intensities in green: strong binding of Heparin Red due to quenching. In Figure S3, the same images, subjected to deconvolution, are shown. The black and white images show that after staining the measured signal looks like the shape of the cells. This suggests that the polyanionic components are located around the cell wall or in the capsule of the bacteria.
Figure 3.
Confocal laser scanning microscopy of Heparin Red, bound to components in the fully hydrated, intact anammox granule. (A) and (C) are greyscale images of the fluorescent signal. (A) and (B) represent an image series of 45 optical sections at 0.17 μm step size shown as maximum intensity projection, (C) and (D) represent a single image. (B) and (D) are the images of (A) and (C), respectively, converted to color images with three pixel intensities: red = binding of Heparin Red, green = quenched signal, implying high negative charge density, and blue = no binding of Heparin Red (see detailed explanation in the Results section).
4. Discussion
In EPS of anammox and other biofilm forming bacteria, the presence of proteins/glycoproteins and polyanionic groups are reported.18,35 As both sulfate groups and sialic acids are anionic and both sulfation and sialylation are common modifications of (glyco)proteins and polysaccharides in the extracellular matrix of eukaryotes, the polyanionic components of anammox EPS were studied, with a focus on these two acidic modifications. Looking at the function of similar structures in known systems can direct us in finding potential functionalities of sulfation and sialylation in biofilm systems.
With the colorimetric quantification assays, around 1.6% of the VSS was measured as sialic acids (NeuAc equivalents) and 2.4% of the VSS was measured as sulfated glycosaminoglycans (chondroitin equivalents). Since the samples that we analyzed are not well-defined mixtures containing possibly interfering components, and we do not have the information to know the exact appropriate standard, these assays are not valid for an absolute quantification in EPS of anammox granules. Possible biases in these assays were not evaluated. In addition, the applied treatments might not release all target components. The lectin and heparin staining intensity appeared very intense in comparison with the amount that was measured with the assays. However, the assays did indicate the presence of sialic acids and sulfated glycosaminoglycans, respectively, and therefore, they are worth to further look into.
In anammox granules, the presence of sialic acids in the form of NeuAc, KDN, and Pse/Leg was confirmed with MS measurements. The gene-encoding enzymes for the synthesis of sialic acids are partly present in “Ca. Brocadia sapporoensis” genomes. The enzymes that are necessary for sialic acids synthesis but could not be found particularly in the “Ca. Brocadia sapporoensis” genome were found in other “Ca. Brocadiales” species. There are more than 50 different variants of sialic acids. In animals, sialic acids are often found as terminal residue of (mucin-)glycoprotein, which have a function in protection against proteases, cell signalling/recognition, and adhesion processes.36 In bacteria, various nine-carbon sugars like NeuAc are known, e.g., Pse and Leg. These are mostly reported in pathogenic bacteria and are thought to mimic the host with the purpose of invading the host.37 However, this image may be biased due to the increased focus of studies on those species that are involved in host–microbe interactions. Recent examples of nonpathogenic organisms carrying sialic acid derivatives are, e.g., seawater-adapted aerobic granular sludge, dominated by “Candidatus Accumulibacter”18 or the surface layer (S-layer) glycoprotein of haloarchaea Halorubrum sp. PV6, which has a sialic acid as a terminal residue.27
Anammox bacteria are known to produce S-layer glycoproteins also.38 Specifically, the anammox granular sludge used in this study contains a putative S-layer glycoprotein, visualized with SDS-PAGE at 80 kDa.6 The glycan structure of this glycoprotein was determined to be composed of a methylated N-acetyl hexosamine (HexNAc) backbone, substituted with a pentose, and a dideoxyhexose residue, and carrying an unknown terminal residue of 350 Da. Hence, the S-layer glycoprotein is a possible source of the measured sialic acids in anammox EPS. However, the band at 80 kDa was not stained by Alcian Blue. If the terminal residue would be a derivative of sialic acid, the overall negative charge of the molecule is not strong enough for it to be stained by Alcian Blue.
The smear at >235 kDa was only stained by Alcian Blue at pH 2.5, indicating carboxylic groups. The profile resembles high molecular-weight polysaccharides or glycoproteins. It is also possible that both the S-layer proteins and other high molecular-weight polysaccharides or glycoproteins are differently sialylated. For example in Campylobacter jejuni, different NeuB genes were found to be involved in biosynthesis of lipo-oligosaccharides and flagella.39 Based on the results, sialic acids are confirmed to be present, but the exact molecular location remains to be determined. Determining the saccharide sequence that the sialic acids are attached to would reveal more about the nature of these components. In known eukaryotic systems, there are many biological roles for sialic acids of which cellular recognition is a very important one.12,40 In granular sludge and biofilms, sialic acids might also play a role in masking bacterial cells to be protected against invaders.
The other targeted components in this study were sulfated glycosaminoglycans. To determine the exact type of sulfated polysaccharide (or glycoprotein), the component should be isolated and the molecular structure should be determined. From the analyses presented here, the staining with Alcian Blue at pH 1.0, the complexing with DMMB, and the staining with Heparin Red, the component resembles sulfated glycosaminoglycans. Regarding the homology with the sulfotransferases in anammox genomes, it fits best with heparan sulfate glucosamine 3-O-sulfotransferase. Heparin Red staining indicated that the strongly polyanionic components, suggested to be sulfated glycosaminoglycans, are located at the outside of the cell walls. In comparison, the sulfated glycosaminoglycan content in aerobic granular sludge is around 3.1% of the VSS. However, the negatively charged macromolecules, which were stained by Heparin Red, seem to be differently distributed. While in the aerobic granular sludge, the signal was observed in the space in between capsules within the microcolony and also in the extracellular matrix in between the colonies;41 in the anammox granules, it appears as the shape of the cells, indicating the strongly polyanionic components are located around the cell walls or in the capsules of the anammox cells.
In the extracellular matrix of eukaryotes, heparan sulfates are located at the outside of cell surfaces, with the function of cellular recognition and adhesion to extracellular matrix components.42 Interestingly, heparan sulfate is the most ancient of all known glycosaminoglycans.43 With this in mind, it may be suggested that heparan sulfate might have a similar function in anammox granules also. To confirm what kind of sulfated polysaccharides are present, the monosaccharide sequence should be determined. Knowing the exact structure would aid in predicting the function of these molecules by comparing with known systems. In addition, since bacteria do not have an endoplasmatic reticulum and Golgi system as used to synthesize sulfated glycosaminoglycans in eukaryotes, the machinery that is needed for the synthesis remains to be determined.
Currently, both sialic acids and sulfated glycosaminoglycans that are found in prokaryotes are mostly related to the microbe–host interaction in the context of mimicking host extracellular matrix components to bypass its immune system.15,44 However, recently, they are also found in prokaryotic, nonpathogenic organisms,19,35,45,46 as well as in this study. This indicates they could be more broadly present, with a differential functionality in microbial communities and biofilms in general. Proposed functions of these negatively charged components include protecting the cells, cell–cell or cell–matrix adhesion, scavenging of other components, and involvement in biomineralization. Looking into the question if these polymers could also have similar roles in the biofilm extracellular matrix as in the multicellular eukaryotic cells will improve the understanding of the composition and functioning of the biofilm matrix. In addition, analyzing the more complex components of the biofilm matrix paves the way to the production of biopolymers that currently need to be extracted from higher organisms. (e.g., heparin, which is used as an anticoagulant, is extracted from porcine mucosa.)
Acknowledgments
This research was funded by the SIAM Gravitation Grant 024.002.002, the Netherlands Organization for Scientific Research. The authors acknowledge Stef Broenink (TU Delft) for his help with the development of the pretreatment method and sulfated glycosaminoglycan assay measurements. The help of Ute Kuhlicke (Helmholtz Centre for Environmental Research, Magdeburg) in laser microscopy and image analysis is highly appreciated.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.9b07207.
The authors declare no competing financial interest.
Supplementary Material
References
- Mulder A.; van de Graaf A. A.; Robertson L. A.; Kuenen J. G. Anaerobic Ammonium Oxidation Discovered in a Denitrifying Fluidized Bed Reactor. FEMS Microbiol. Ecol. 1995, 16, 177–184. 10.1111/j.1574-6941.1995.tb00281.x. [DOI] [Google Scholar]
- Kuenen J. G. Anammox Bacteria: From Discovery to Application. Nat. Rev. Microbiol. 2008, 6, 320–326. 10.1038/nrmicro1857. [DOI] [PubMed] [Google Scholar]
- Peeters S. H.; van Niftrik L. Trending Topics and Open Questions in Anaerobic Ammonium Oxidation. Curr. Opin. Chem. Biol. 2019, 49, 45–52. 10.1016/j.cbpa.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Seviour T.; Derlon N.; Dueholm M. S.; Flemming H.-C.; Girbal-Neuhauser E.; Horn H.; Kjelleberg S.; van Loosdrecht M. C. M.; Lotti T.; Malpei M. F.; et al. Extracellular Polymeric Substances of Biofilms: Suffering from an Identity Crisis. Water Res. 2019, 151, 1–7. 10.1016/j.watres.2018.11.020. [DOI] [PubMed] [Google Scholar]
- Pronk M.; Neu T. R.; van Loosdrecht M. C. M.; Lin Y. M. The Acid Soluble Extracellular Polymeric Substance of Aerobic Granular Sludge Dominated by Defluviicoccus Sp. Water Res. 2017, 122, 148–158. 10.1016/j.watres.2017.05.068. [DOI] [PubMed] [Google Scholar]
- Boleij M.; Pabst M.; Neu T. R.; Van Loosdrecht M. C. M.; Lin Y. Identification of Glycoproteins Isolated from Extracellular Polymeric Substances of Full-Scale Anammox Granular Sludge. Environ. Sci. Technol. 2018, 52, 13127–13135. 10.1021/acs.est.8b03180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij M.; Seviour T.; Wong L. L.; van Loosdrecht M. C. M.; Lin Y. Solubilization and Characterization of Extracellular Proteins from Anammox Granular Sludge. Water Res. 2019, 164, 114952. 10.1016/j.watres.2019.114952. [DOI] [PubMed] [Google Scholar]
- Lin Y.; Reino C.; Carrera J.; Pérez J.; van Loosdrecht M. C. M. Glycosylated Amyloid-like Proteins in the Structural Extracellular Polymers of Aerobic Granular Sludge Enriched with Ammonium-Oxidizing Bacteria. Microbiology 2018, e00616. 10.1002/mbo3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats N.; De Winder B.; Stal L. J.; Mur L. R. Isolation and Characterization of Extracellular Polysaccharides from the Epipelic Diatoms Cylindrotheca Closterium and Navicula Salinarum. Eur. J. Phycol. 1999, 34, 161–169. 10.1080/09670269910001736212. [DOI] [Google Scholar]
- Roberts I. S. The Biochemistry and Genetics of Capsular Polysaccharide Production in Bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C.; Wingender J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Tanner M. E. The Enzymes of Sialic Acid Biosynthesis. Bioorg. Chem. 2005, 33, 216–228. 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Varki A. Biological Roles of Glycans. Glycobiology 2016, 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J. J.; Marzaioli A. M.; De Castro C.; Hall R. M. 5,7-Di-N-Acetyl-Acinetaminic Acid: A Novel Non-2-Ulosonic Acid Found in the Capsule of an Acinetobacter Baumannii Isolate. Glycobiology 2015, 25, 644–654. 10.1093/glycob/cwv007. [DOI] [PubMed] [Google Scholar]
- DeAngelis P. L. Evolution of Glycosaminoglycans and Their Glycosyltransferases: Implications for the Extracellular Matrices of Animals and the Capsules of Pathogenic Bacteria. Anat. Rec. 2002, 268, 317–326. 10.1002/ar.10163. [DOI] [PubMed] [Google Scholar]
- van der Star W. R. L.; Abma W. R.; Blommers D.; Mulder J.-W.; Tokutomi T.; Strous M.; Picioreanu C.; van Loosdrecht M. C. M. Startup of Reactors for Anoxic Ammonium Oxidation: Experiences from the First Full-Scale Anammox Reactor in Rotterdam. Water Res. 2007, 41, 4149–4163. 10.1016/j.watres.2007.03.044. [DOI] [PubMed] [Google Scholar]
- American Public Health Association (APHA) , Standard Methods for the Examination of Water and Wastewater; American Public Health Association (APHA): 2005, 21st edition,Washington, DC. [Google Scholar]
- de Graaff D. R.; Felz S.; Neu T. R.; Pronk M.; van Loosdrecht M. C. M.; Lin Y. Sialic Acids in the Extracellular Polymeric Substances of Seawater-Adapted Aerobic Granular Sludge. Water Res. 2019, 155, 343–351. 10.1016/j.watres.2019.02.040. [DOI] [PubMed] [Google Scholar]
- Kleikamp H. B. C.; Lin Y. M.; McMillan D. G. G.; Geelhoed J. S.; Naus-Wiezer S. N. H.; van Baarlen P.; Saha C.; Louwen R.; Sorokin D. Y.; van Loosdrecht M. C. M.; et al. Tackling the Chemical Diversity of Microbial Nonulosonic Acids – a Universal Large-Scale Survey Approach. Chem. Sci. 2020, 11, 3074–3080. 10.1039/C9SC06406K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller H. J.; Poulsen J. H.. Staining of Glycoproteins/proteoglycans on SDS-Gels. In The Protein Protocols Handbook; Springer Science & Business Media, 2009; pp 773–777. [Google Scholar]
- Tobisawa Y.; Imai Y.; Fukuda M.; Kawashima H. Sulfation of Colonic Mucins by N-Acetylglucosamine 6-O-Sulfotransferase-2 and Its Protective Function in Experimental Colitis in Mice. J. Biol. Chem. 2010, 285, 6750–6760. 10.1074/jbc.M109.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu T.; Kuhlicke U. Fluorescence Lectin Bar-Coding of Glycoconjugates in the Extracellular Matrix of Biofilm and Bioaggregate Forming Microorganisms. Microorganisms 2017, 5, 5. 10.3390/microorganisms5010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu T. R.; Lawrence J. R. [10] Lectin-Binding Analysis in Biofilm Systems. Methods Enzymol. 1999, 310, 145–152. 10.1016/S0076-6879(99)10012-0. [DOI] [PubMed] [Google Scholar]
- Neu T. R.; Lawrence J. R.. In Situ Characterization of Extracellular Polymeric Substances (EPS) in Biofilm Systems. In Microbial Extracellular Polymeric Substances: Characterization, Structure and Function; Wingender J., Neu T. R., Flemming H.-C., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 1999; pp 21–47. [Google Scholar]
- Muthana S. M.; Campbell C. T.; Gildersleeve J. C. Modifications of Glycans: Biological Significance and Therapeutic Opportunities. ACS Chem. Biol. 2012, 7, 31–43. 10.1021/cb2004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhofen I. C.; Vinogradov E.; Whitfield D. M.; Brisson J. R.; Logan S. M. The CMP-Legionaminic Acid Pathway in Campylobacter: Biosynthesis Involving Novel GDP-Linked Precursors. Glycobiology 2009, 19, 715–725. 10.1093/glycob/cwp039. [DOI] [PubMed] [Google Scholar]
- Zaretsky M.; Roine E.; Eichler J. Sialic Acid-like Sugars in Archaea: Legionaminic Acid Biosynthesis in the Halophile Halorubrum Sp. PV6. Front. Microbiol. 2018, 9, 2133. 10.3389/fmicb.2018.02133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusche-Gullberg M.; Kjellén L. Sulfotransferases in Glycosaminoglycan Biosynthesis. Curr. Opin. Struct. Biol. 2003, 13, 605–611. 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Bhotmange D. U.; Singhal R. S. Identification of Chondroitin-like Molecules from Biofilm Isolates Exiguobacterium Indicum A11 and Lysinibacillus Sp. C13. J. Appl. Microbiol. 2015, 119, 1046–1056. 10.1111/jam.12914. [DOI] [PubMed] [Google Scholar]
- Monsigny M.; Roche A.-C.; Sene C.; Maget-Dana R.; Delmotte F. Sugar-Lectin Interactions: How Does Wheat-Germ Agglutinin Bind Sialoglycoconjugates?. Eur. J. Biochem. 1980, 104, 147–153. 10.1111/j.1432-1033.1980.tb04410.x. [DOI] [PubMed] [Google Scholar]
- Moore J. S.; Kulhavy R.; Tomana M.; Moldoveanu Z.; Suzuki H.; Brown R.; Hall S.; Kilian M.; Poulsen K.; Mestecky J.; et al. Reactivities of N-Acetylgalactosamine-Specific Lectins with Human IgA1 Proteins. Mol. Immunol. 2007, 44, 2598–2604. 10.1016/j.molimm.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warttinger U.; Giese C.; Harenberg J.; Holmer E.; Krämer R. A Fluorescent Probe Assay (Heparin Red) for Direct Detection of Heparins in Human Plasma. Anal. Bioanal. Chem. 2016, 408, 8241–8251. 10.1007/s00216-016-9940-y. [DOI] [PubMed] [Google Scholar]
- Capila I.; Gunay N. S.; Shriver Z.; Venkataraman G. Methods for Structural Analysis of Heparin and Heparan Sulfate. Chem. Biol. Heparin Heparan Sulfate 2005, 55–77. 10.1016/B978-008044859-6/50004-6. [DOI] [Google Scholar]
- Stick R. V.; Williams S. J. Chapter 11- Glycoproteins and Proteoglycans. Carbohydrates Essent. Mol. Life 2009, 369–412. 10.1016/B978-0-240-52118-3.00011-9. [DOI] [Google Scholar]
- Bourven I.; Bachellerie G.; Costa G.; Guibaud G. Evidence of Glycoproteins and Sulphated Proteoglycan-like Presence in Extracellular Polymeric Substance from Anaerobic Granular Sludge. Environ. Technol. 2015, 36, 2428–2435. 10.1080/09593330.2015.1034186. [DOI] [PubMed] [Google Scholar]
- Van Klinken B. J.; Dekker J.; Büller H. A.; Einerhand A. W. Mucin Gene Structure and Expression: Protection vs Adhesion. Am. J. Physiol.-Gastrointest. Liver Physiol. 1995, 269, G613–G627. 10.1152/ajpgi.1995.269.5.G613. [DOI] [PubMed] [Google Scholar]
- Lewis A. L.; Desa N.; Hansen E. E.; Knirel Y. A.; Gordon J. I.; Gagneux P.; Nizet V.; Varki A. Innovations in Host and Microbial Sialic Acid Biosynthesis Revealed by Phylogenomic Prediction of Nonulosonic Acid Structure. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13552–13557. 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling M. C. F.; de Almeida N. M.; Klingl A.; Speth D. R.; Op den Camp H. J. M.; Rachel R.; Jetten M. S. M.; van Niftrik L. A New Addition to the Cell Plan of Anammox bacteria: “Candidatus Kuenenia Stuttgartiensis” has a Protein Surface Layer as the Outermost Layer of the Cell. J. Bacteriol. 2014, 196, 80–89. 10.1128/JB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D.; Karlyshev A. V.; Hitchen P. G.; Morris H. R.; Dell A.; Gregson N. A.; Wren B. W. Multiple N-Acetyl Neuraminic Acid Synthetase (neuB) Genes in Campylobacter Jejuni: Identification and Characterization of the Gene Involved in Sialylation of Lipo-Oligosaccharide. Mol. Microbiol. 2000, 35, 1120–1134. 10.1046/j.1365-2958.2000.01780.x. [DOI] [PubMed] [Google Scholar]
- Schauer R. Sialic Acids and Their Role as Biological Masks. Trends Biochem. Sci. 1985, 10, 357–360. 10.1016/0968-0004(85)90112-4. [DOI] [Google Scholar]
- Felz S.; Neu T. R.; van Loosdrecht M. C. M.; Lin Y. Aerobic Granular Sludge Contains Hyaluronic Acid-like and Sulfated Glycosaminoglycans-like Polymers. Water Res. 2020, 169, 115291. 10.1016/j.watres.2019.115291. [DOI] [PubMed] [Google Scholar]
- Bernfield M.; Götte M.; Park P. W.; Reizes O.; Fitzgerald M. L.; Lincecum J.; Zako M. Functions of Cell Surface Heparan Sulfate Proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Yamada S.; Sugahara K.; Özbek S. Evolution of Glycosaminoglycans: Comparative Biochemical Study. Commun. Integr. Biol. 2011, 4, 150–158. 10.4161/cib.4.2.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R.; Kalivoda K. A.; Deszo E. L.; Steenbergen S. M. Diversity of Microbial Sialic Acid Metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–153. 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T.; Varki A. Chemical Diversity in the Sialic Acids and Related α-Keto Acids: An Evolutionary Perspective. Chem. Rev. 2002, 102, 439–470. 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- de Graaff M. S.; Temmink H.; Zeeman G.; van Loosdrecht M. C. M.; Buisman C. J. N. Autotrophic Nitrogen Removal from Black Water: Calcisum Addition as a Requirement for Settleability. Water Res. 2011, 45, 63–74. 10.1016/j.watres.2010.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.