Abstract
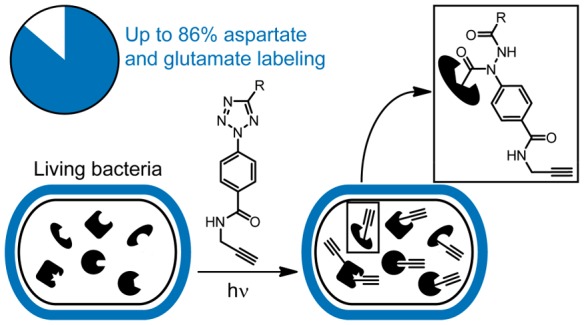
Covalent inhibitors have recently seen a resurgence of interest in drug development. Nevertheless, compounds, which do not rely on an enzymatic activity, have almost exclusively been developed to target cysteines. Expanding the scope to other amino acids would be largely facilitated by the ability to globally monitor their engagement by covalent inhibitors. Here, we present the use of light-activatable 2,5-disubstituted tetrazoles that allow quantifying 8971 aspartates and glutamates in the bacterial proteome with excellent selectivity. Using these probes, we competitively map the binding sites of two isoxazolium salts and introduce hydrazonyl chlorides as a new class of carboxylic-acid-directed covalent protein ligands. As the probes are unreactive prior to activation, they allow global profiling even in living Gram-positive and Gram-negative bacteria. Taken together, this method to monitor aspartates and glutamates proteome-wide will lay the foundation to efficiently develop covalent inhibitors targeting these amino acids.
Short abstract
2,5-Disubstituted tetrazoles were developed into light-activatable broadly reactive alkyne probes for residue-specific monitoring of aspartates and glutamates in living bacteria with high specificity.
Introduction
Covalent inhibitors have recently re-emerged as important entities in drug development.1 This is best exemplified by the approval of several kinase inhibitors for clinical use in cancer.2 Moreover, covalent inhibitors are prevalent among antibiotics. Key examples are the large class of β-lactams1 but also other antibiotics like fosfomycin,3 showdomycin,4 and optimized arylomycins.5 Nevertheless, covalent inhibitors, which do not rely on an enzymatic activity, still almost exclusively bind to cysteine residues. Targeting additional amino acids could largely help to address protein pockets that do not contain a suitable cysteine and, in this way, enlarge the scope of proteins accessible for covalent inhibitor development.
In the antibiotics field, identifying new binding sites for covalent inhibitors is urgently needed in order to efficiently treat multiresistant bacterial infections.6 Covalent inhibitors are uniquely suited to identify new targets that can be addressed with small molecules, as they allow efficient mapping of many potential binding sites in parallel using chemoproteomics.7,8 In bacteria, the almost exclusive focus on cysteine-directed covalent inhibitors raises a severe issue as cysteine is even less frequent in many bacteria (e.g., 0.6% of all amino acid residues in Staphylococcus aureus are cysteine) than in human cells (2.3%).9 Therefore, many important binding pockets in bacterial proteins lack a suitable cysteine residue. Covalent inhibitors that target other amino acid residues would thus be important for antibiotic development, and methods to broadly profile their target engagement with chemoproteomics are highly desirable.
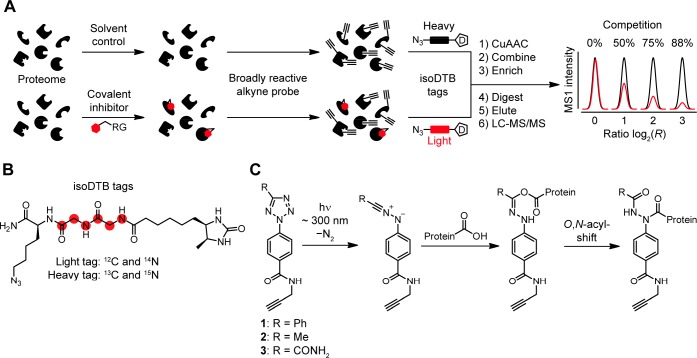
One technology that was key to facilitating the development of covalent inhibitors at cysteines is residue-specific profiling that is usually based on the isoTOP-ABPP (isotopic tandem orthogonal proteolysis activity-based protein profiling) platform (Figure 1A).7 In this technology, a proteome is split into two samples. One is treated with a covalent inhibitor and the other one with only the solvent as a control. In this way, the inhibitor will covalently bind to its target residues and block their intrinsic reactivity. In the second step, a broadly reactive alkyne probe is used to label many amino acid residues with alkynes. Thereby, binding of the covalent inhibitor is translated into a lack of alkynylation by the probe at the specific interaction sites of the covalent inhibitor. The relative degree of alkynylation in the compound- and solvent-treated samples is quantified by modification with isotopically differentiated affinity tags using copper-catalyzed azide–alkyne cycloaddition (CuAAC). Biotin tags that have an isotopically labeled linker that is cleaved by the tobacco-etch (TEV) protease are most commonly used.10 Recently, isotopically labeled desthiobiotin azide (isoDTB) tags (Figure 1B) have been introduced that obliviate the need to use a cleavable linker (isoDTB-ABPP).11 After the combination of the samples, enrichment, and proteolytic digestion, the probe-modified peptides are identified and quantified by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Because the peptides modified with the probe are directly detected, not only the target protein of the covalent inhibitor but also its exact interaction site are identified. Here, residues that are not bound by the inhibitor will show ratios of around one (R ≈ 1), whereas residues that are strongly engaged by the covalent inhibitor will show high ratios (R ≫ 1). In this way, quantitative information on the sites that are modified by the covalent inhibitor is obtained.
Figure 1.
Concept of this study. (A) Workflow of competitive residue-specific proteomics using the isotopically labeled desthiobiotin azide (isoDTB) tags.11 RG, reactive group; D, desthiobiotin. (B) Structure of the isoDTB tags.11 (C) Light-induced reactivity of 2,5-disubstituted tetrazoles 1–3 with carboxylic acids in proteins. While other nucleophiles might attack the nitrilimine, only for carboxylic acids a stable product can be formed via an O,N-acyl-shift.
In order to expand the scope of the described residue-specific proteomics workflow to amino acid residues other than cysteine, broadly reactive alkyne probes are needed that specifically address a subset of amino acids of interest. For cysteines, iodoacetamide-alkyne (IA-alkyne) is most widely used.12 For lysines, acylation reagents have been shown to efficiently label a large number of residues in the proteome.8,13 Recently, sulfonyl triazoles have successfully been applied to study tyrosines.14 Many other chemistries have been developed to target, e.g., lysines,8,13,15−17 tyrosines,18−20 methionines,21,22 histidines,23 tryptophanes,24 as well as aspartates and glutamates25−27 in proteins, but their exploration as broadly reactive alkyne probes for residue-specific proteomics is still lacking.
We were especially enticed by targeting aspartates and glutamates in the proteome as these amino acids frequently occur in the bacterial proteome (∼12% of all residues),9 tend to be in pockets or on the proteome surface due to their polarity, and could show unique reactivity over all other nucleophilic amino acids as their initially nucleophilic character can be turned into an electrophilic reactivity by suitable activators. Nevertheless, only a few chemotypes including sulfonate esters,28 diazonium salts,27 and oxazolium salts25,26 have been studied for the development of carboxylic-acid-directed covalent inhibitors. Therefore, we reasoned that the development of additional selective chemotypes could be fostered by the availability of an isoDTB-ABPP-based platform that allows investigating aspartates and glutamates proteome-wide.
Previously, isoxazolium salts based on Woodward’s reagent K have been investigated as probes for a chemoproteomic study of carboxylic acids.26 These studies led to the identification of a few target proteins, but residue-specific information could not be obtained on a global level.
Another very interesting chemotype for targeting carboxylic acids is light-activatable 2,5-disubstituted tetrazoles (Figure 1C).29 These compounds are inherently unreactive but, upon light activation, produce nitrilimines through liberation of nitrogen.29 These nitrilimines are highly reactive intermediates that were originally described as bio-orthogonal moieties to address alkenes in proteins.29 Lately, it has been shown that they can also react with a number of proteinogenic nucleophiles including carboxylic acids.30−33 This prompted us to explore their usage as broadly reactive alkyne probes. These probes would have several advantages over other strategies targeting carboxylic acids. First, by changing the substituent at the 5-position, their electronic properties can be fine-tuned, which should allow adjusting their reactivity, selectivity, and target profile.31 Second, they would form very stable 1,2-diacyl-1-arylhydrazines after reaction with carboxylic acids, which should facilitate the chemoproteomics analysis.33 Third, they are unreactive and therefore stable and potentially nontoxic before activation, which could enable their usage in living cells as has been previously shown for light-activatable cysteine-directed electrophiles.34 2,5-Disubstituted tetrazoles have been used in a proteomic context before in order to broadly identify their target proteins, but these studies have not investigated the global specificity of the probes toward certain amino acids or looked at the interactions in a residue-specific manner.33
Therefore, we set out to study the reactivity and selectivity of 2,5-disubstituted tetrazoles in a proteome-wide context. We demonstrate their ability to act as broadly reactive alkyne probes to study aspartates and glutamates in vitro and in situ with high specificity even in challenging Gram-negative bacteria. Furthermore, we study the binding of covalent ligands and introduce a new class of carboxylic-acid-directed protein ligands, namely, hydrazonyl chlorides.
Results
Synthesis of 2,5-Disubstituted Tetrazoles
In order to investigate the proteome-wide reactivity of 2,5-disubstituted tetrazoles, we set out to synthesize three different probes (1–3, Figure 1C). Due to the different effects of the substituents at the 5-position (aromatic phenyl group for 1, aliphatic methyl group for 2, and electron-withdrawing carboxamide group for 3), we reasoned that these probes should allow us to tailor their reactivity and selectivity.31 All three probes were synthesized according or similar to literature-known procedures (Scheme S1).33 For probes 1 and 3 we synthesized the diazonium salt starting from para-aminobenzoic acid and reacted it with benzaldehyde phenylsulfonylhydrazone or ethyl glyoxylate para-tosylhydrazone to give the respective tetrazoles.33 These were coupled to propargyl amine using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl) to give probe 1 and the ethyl ester precursor of probe 3. This precursor was reacted with ammonia to give the final carboxamide 3. For the synthesis of probe 2, we synthesized the diazonium salt starting from methyl para-amino-benzoate and reacted this with acetamidine hydrochloride using subsequent oxidation with iodine and potassium iodide.37 Saponification of the ester with sodium hydroxide and coupling to propargyl amine using EDC HCl yielded probe 2.33
We first studied the activation of the probes to the nitrilimine in PBS solution by UV light using LC-MS detection. For all three probes, we detected quantitative light activation followed by hydrolysis (probes 1–3) or reaction with chloride (probe 3) within 10 min of UV irradiation at 280–315 nm (Figure S1) detection indicating efficient activation.31
Aspartate and Glutamate-Specific Proteome-wide Labeling with 2,5-Disubstituted Tetrazoles
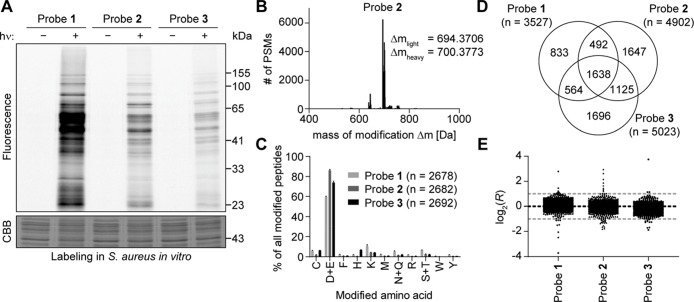
With these probes in hand, we next evaluated their proteome-wide reactivity in the Staphylococcus aureus (S. aureus) strain SH1000. After optimization of the irradiation time (Figure S2) and probe concentration (Figure S3) using a gel-based readout, we directly compared the labeling with the three probes at 100 μM with irradiation for 10 min at 280–315 nm (Figure 2A). For all three probes, we could detect strong labeling of many different proteins in the gel. All probes are therefore in principle suitable to investigate many binding sites in proteins. We observed no striking differences in the overall labeling pattern but stronger labeling with probe 1 in comparison to probe 2 and 3, which is most likely caused by its increased lipophilicity.
Figure 2.
Light-activatable 2,5-disubstituted tetrazoles allow global monitoring of aspartates and glutamates in the S. aureus proteome in vitro with high specificity. (A) Gel-based analysis of labeling with probes 1–3. S. aureus lysate was treated with 100 μM of the indicated probe, incubated for 30 min, irradiated with light (λ = 280–315 nm) for 10 min, and labeled with TAMRA-azide using CuAAC. Controls were performed without irradiation. Gel-based analysis was performed with in-gel fluorescence scanning and staining using Coomassie Brilliant Blue (CBB). (B) Analysis of the mass of modification on tryptic peptides after labeling of S. aureus lysate with 100 μM probe 2. MSFragger software35 was used to determine, which masses of modification occur in the proteomic samples labeled with probe 2 after light activation and CuAAC to the light and heavy isoDTB tags. Expected masses of modification for tryptic peptides labeled with 2 according to the reactivity shown in Figure 1C and additionally modified with light or heavy isoDTB tag, respectively, are 694.3663 and 700.3738 Da. PSM: peptide-spectrum match. (C) Analysis of the amino acid specificity of the probes. Proteomes labeled with the indicated probe after light activation and modified by CuAAC with the light and heavy isoDTB tags were analyzed with MaxQuant software36 allowing the modification on any potentially nucleophilic amino acid. Peptides were included in the analysis if the localization probability for a single residue was more than 75%. Data shows the mean ± the standard deviation. The total number of identified PSMs is given in parentheses. (D) Venn diagram of the number of quantified aspartates and glutamates with the three different probes. (E) Plot of the ratios log2(R) for aspartates and glutamates in proteomic samples, in which the heavy- and light-labeled sample were both modified with 100 μM of the indicated probe without pretreatment with an inhibitor. The expected value of log2(R) = 0 is indicated by the black line; the preferred quantification window (−1 < log2(R) < 1) is indicated by the two gray lines. Each dot represents one quantified aspartate or glutamate. All data for panels B–E originates from biologically independent duplicates of technical duplicates.
As all probes showed promising labeling using gel-based detection, we set out to investigate their reactivity and selectivity in a mass-spectrometry-based setup using the isoDTB-ABPP workflow (Figure 1A). Here, we treated both samples with the same concentration of the probe and did not pretreat with an inhibitor. As the two samples are in this case identical, the isotopic ratios R for all residues are expected to be around one, which gives an additional quality control and can be used to investigate if the method gives quantitative results.
First, we wanted to establish in an unbiased way if we detect peptides with the expected modification in the samples. For this purpose, we utilized the MSFragger software that allows an Open Search, in which peptides are identified and assigned their modification state without the need to predefine a modification (Figure 2B, and Figure S4 and Table S1).35 Indeed, in all cases we were able to mainly detect the modification with the expected mass for the 1,2-diacyl-1-aryl-hydrazine moiety of the respective probe modified with the light or heavy isoDTB tag, respectively. This indicates that the expected reactivity via the nitrilimine intermediate is directly observed using the chemoproteomic setup.
Next, we set out to investigate the amino acid specificity of the probes. For this purpose, we used MaxQuant software36 and allowed the modification with the respective probe to be on any potentially nucleophilic amino acid (C, D, E, F, H, K, M, N, Q, R, S, T, W, Y). We further analyzed all peptides with a high localization probability for a single residue (>75%, analogous to class I phosphosites) as determined by the Andromeda algorithm implemented in MaxQuant that uses a probabilistic scoring model for the localization.38 The median localization probability for all analyzed peptides across all probes was >95%. We manually verified a selection of annotated modification sites using the respective MS2 spectra (Figure S5). For all three probes (Figure 2C, Table S1), we detected that most peptides are modified on aspartates and glutamates. Interestingly, probe 2 with a methyl group at the 5-position showed an increased selectivity of more than 85% of all peptides modified at aspartates and glutamates. In order to verify this selectivity, we utilized the MSFragger software35 that uses a fragment-ion indexing algorithm for the localization of the modifications. Here, we analyzed all spectra, for which the modification was uniquely assigned to a single residue, and detected almost exactly the same amino acid selectivity compared to MaxQuant analysis (Figure S6). Probe 2 is therefore a prime candidate as a selective tool for aspartates and glutamate labeling in vitro.
In order to better understand the selectivity of probe 2, we performed experiments with amino acids protected at the Nα-position and at the α-carboxylic acid in solution. For this purpose, we treated 100 μM of probe 2 with 5 mM of the respective amino acid and investigated the reactivity after 10 min of UV irradiation using LC-MS. In PBS, we detected only very limited adduct formation and mainly hydrolysis in the presence of glutamate (Figure S7). We reasoned that the lipophilic nature of protein binding pockets increases the reactivity of the nitrilimine. We, therefore, studied the reactivity with increasing amounts of acetonitrile (Figure S7).30,32 At 75% acetonitrile, adduct formation with glutamate was detected as the main peak. Under these conditions also aspartate mainly reacted to give the expected product (Figure S8). While serine and tyrosine exclusively showed hydrolysis of the nitrilimine, some adduct was formed for lysine and histidine. For cysteine mainly products other than hydrolysis were detected. Here, besides the adduct peak, we detected a peak that had the mass of the thiohydrazide formed from probe 2. This corresponds to a formal thiolysis of the nitrilimine. We reasoned that, in analogy to the light-induced reactivity of 2-thiazolines,39 this indicates an initial adduct formation with cysteine followed by homolytic bond cleavage of the adduct. When we performed the reaction with different irradiation times (Figure S9), the activation of probe 2 was quantitative after 7 min. Next to the hydrolysis product, this mainly resulted in the formation of the cysteine adduct and the thiohydrazide. The cysteine adduct was converted to the thiohydrazide upon further irradiation. This process was almost quantitative after 20 min of irradiation. This shows that the instability of the cysteine adduct toward UV irradiation might contribute to the selectivity of probe 2 for aspartate and glutamate labeling. As the final reaction product with cysteine does not lead to a modification with an alkyne, this reactivity does not compromise the use of 2,5-disustituted tetrazoles as probes for proteome-wide monitoring of carboxylic acids. Nevertheless, for the potential development of covalent inhibitors, this reactivity will have to be carefully considered.
Finally, we used MaxQuant software to also quantify the modified aspartates and glutamates.36 For this purpose, we allowed modification only at these residues. We were able to quantify more than 3500 aspartates and glutamates with each of the three probes (Figure 2D, Table S1). Probes 2 and 3 even quantified around 5000 residues, each. As the heavy and light samples were mixed at a ratio of 1:1, all residues are expected to be quantified with an R value close to one (log2(R) ≈ 0). In all cases, the number of peptides that were outside of the preferred window of −1 < log2(R) < 1 was ≤1% indicating that the probes allow reliable residue-specific quantification (Figure 2E, Table S1). There is a significant overlap between the residues detected with each probe (Figure 2D), but there are also many residues that are exclusively detected with one of the probes. Furthermore, digestion with chymotrypsin instead of trypsin in an additional experiment led to an additional increase in the total number of quantified residues for probe 2 to 6136 (Figure S10 and Table S1). By combining the data from the three probes and the additional experiments with chymotrypsin digestion for probe 2, we were able to quantify a total of 8971 aspartates and glutamates in the proteome of S. aureus. It is striking that probe 1 allowed quantification of fewer aspartates and glutamates, while showing the most intense labeling by gel-based experiments (Figure 2A). Due to the lower specificity of the probe, it can be speculated that this number is lowered by the higher number of peptides modified at other sites as the total amount of detected modified peptides is similar between all probes (Figure 2C). In order to obtain an estimate for the sensitivity of the method, we added bovine serum albumin (BSA) at different concentrations to the S. aureus lysate and performed isoDTB-ABPP with probe 2. We were able to detect BSA down to a concentration of 10 nM (Table S1). In summary, probe 2 is an ideal tool for the global investigation of carboxylic acids in the bacterial proteome in vitro.
Residue-Specific Profiling of Covalent Protein Ligands Targeting Aspartates and Glutamates
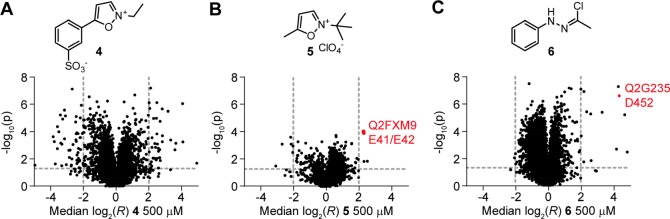
With probe 2 as an optimized probe in hand, we next wanted to take first steps toward globally investigating carboxylic-acid-directed protein ligands in the proteome of S. aureus. For this purpose, we initially investigated established carboxylic-acid-directed chemistry. Here, isoxazolium salts have been used as protein ligands in various instances.25,26 We decided to investigate Woodward’s reagent K (4, Figure 3A) and isoxazolium salt 5 (Figure 3B). By performing the chemoproteomic workflow with these ligands at 500 μM (and additionally at 200 μM for 4, Figure S11 and Table S2) and probe 2 as an optimized broadly reactive alkyne probe, we were able to identify 44 aspartates and glutamates that are able to interact with these compounds in the S. aureus proteome. These hits, e.g., include the interaction of 5 with the residues E41/E42 in the ATP-binding site of the essential protein pyruvate kinase (UniProt code, Q2FXM9; Figure 3B, Table S2). For compound 4, we also detected 50 residues that increase in labeling upon compound treatment. For eight of these residues, we detected another aspartate or glutamate in the same protein that is engaged by at least 50% indicating that this engagement might lead to a structural change causing an increased labeling. Put together, these competitive studies show that our method allows target engagement studies at aspartates and glutamates in the whole proteome and could verify that isoxazolium salts engage these residues also in bacterial proteins.
Figure 3.
Probe 2 reveals the targeted residues of carboxylic-acid-directed covalent protein ligands proteome-wide using isoDTB-ABPP in vitro. (A–C) Volcano plots of isoDTB-ABPP experiments comparing samples pretreated with 500 μM of the indicated covalent ligand to a solvent control. Plotted are the ratio (log2(R)) between the heavy (solvent-treated, 1% HCl for 4, DMSO for 5, DMF for 6) and light (compound-treated) labeled samples and the probability in a one-sample t test that R is equal to one (−log10(p)). The targeted E41/E42 of pyruvate kinase (UniProt code: Q2FXM9) and D452 of nicotinate phosphoribosyltransferase (UniProt code: Q2G235) are highlighted in red. All data originates from two (B and C) or three (A) biologically independent experiments; performed in technical duplicates (A and B) or triplicates (C).
Enticed by the possibility to address aspartates and glutamates in the proteome with nitrilimines, we next explored if the reactive nitrilimine could also be generated without the use of light and therefore used as a covalent protein ligand. Previous studies that used nitrilimines for conjugation to alkenes have utilized hydrazonyl chlorides for this purpose, but to the best of our knowledge these chemotypes have not been used as carboxylic-acid-directed protein ligands.40 We therefore synthesized compound 6 (Scheme S2), which can liberate a nitrilimine through elimination of hydrochloric acid. Using 6 at 500 μM in competitive experiments with probe 2, we identified 13 residues that interact with 6 in the S. aureus proteome (Figure 3C, Table S2). These peptides include the residue D452 of the essential protein nicotinate phosphoribosyl-transferase (Npt) (UniProt code: Q2G235). In order to prove this interaction, we recombinantly expressed and purified this protein. Upon treatment with 6in vitro, we were able to detect quantitative modification of Npt and a shift in the mass of the protein that corresponds to the modification with one molecule of the nitrilimine formed from 6 (Figure S12). To a lower degree, we were also able to detect double and triple modification. As the quite reactive hydrazonyl chloride was used at a high concentration on an isolated protein, this points to the fact that multiple carboxylic acid residues can be modified by 6 in this setup.
In order to verify the binding sites on Npt and quantify their engagement, we used a recently established protocol based on reductive dimethyl labeling for quantification.8 After treatment of recombinant Npt with 6, we were able to detect the 6-modified peptide with modification at the expected residue D452 and quantified the target engagement to approximately 50% (Figure S13 and Table S2), which is the highest engagement among all quantified aspartates and glutamates in Npt. Nevertheless, we were also able to detect modified peptides at other sites, which points to the fact that other residues are also reactive when compound 6 is used on an isolated protein at this high concentration.
Despite the considerable target engagement at D452, we could detect no effect on enzyme activity (Figure S14), pointing toward the fact that Npt is not inhibited by covalent modification of this residue by 6. Nevertheless, this experiment shows that hydrazonyl chlorides are interesting warheads for targeting carboxylic acids in proteomes and that the product of the reaction with carboxylic acids in proteins is a 1,2-diacyl-1-arylhydrazine, which is in agreement with the formation of an intermediate nitrilimine.
Through compiling of the proteome-wide isoDTB-ABPP data for all three competitors (4–6), we detected a total of 56 aspartates and glutamates in 48 different proteins that are targeted by at least one of these compounds in the proteome of S. aureus. These residues show a slightly increased probability to be at functional sites (5.4%) compared to other quantified residues (4.0%) or the genomic background (2.7%, Figure S15). This suggests that in functional sites higher residue reactivity or additional noncovalent interactions might facilitate high occupancy binding of the ligands. Due to the limited number of identified liganded residues with the used three competitors, this observation will need to be verified once a larger data set of ligandable residues is available. Interestingly, residues in essential proteins are strongly enriched among the quantified residues compared to the genomic background (Figure S16). Concerning the functional classes of proteins, the liganded residues and other quantified residues show a very similar distribution to each other, in which enzymes and proteins involved in gene expression are enriched, and receptors, transporters, and channels are depleted in comparison to the whole genome (Figure S17). These facts point to the chance to functionally target diverse important proteins with these compounds. Comparing the specific interactions of the three compounds, one can see that most residues are exclusively targeted by one of the compounds indicating some specificity even with these very small compounds at high concentrations (Figure S18). Chemoproteomic profiling of proteomes with probe 2 is therefore a very promising strategy for the optimization of carboxylic-acid-directed protein ligands.
Global Investigation of Aspartates and Glutamates in Living Bacteria
Having a working platform in hand that allows quantifying many aspartates and glutamates in vitro, we next wanted to investigate if this technology can also be transferred to monitor these amino acids in living bacteria. As the probes are nonreactive before light activation, we reasoned that they could be nontoxic in bacteria. We therefore measured the minimum inhibitory concentration (MIC) for all three probes for the growth of S. aureus (Figure S19). All three compounds had no MIC or effect on the optical density of the culture up to a concentration of 200 μM. The probes can therefore be used in living bacteria without affecting their viability.
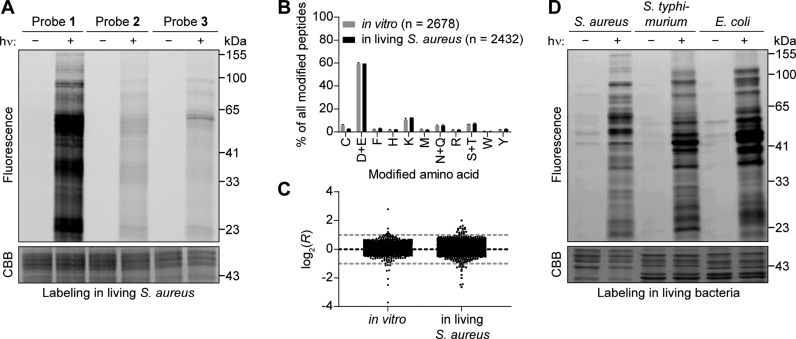
We next checked for labeling in situ using a gel-based readout. Here, we pretreated living S. aureus cells with the probes and then labeled the aspartates and glutamates using UV irradiation. After optimization of the pretreatment (Figure S20) and irradiation time (Figure S21) using probe 1, we directly compared all three probes (Figure 4A). We could detect that there is only very weak labeling with probes 2 and 3 in comparison to probe 1. The difference is much larger than in vitro (Figure 2A). Probe 1 therefore seems to be taken up into cells much more efficiently than the other probes. We, therefore, decided to utilize probe 1 for in situ experiments, although the selectivity for aspartates and glutamates is not quite as high, because of the observed striking difference in labeling intensity.
Figure 4.
Specific labeling of aspartates and glutamates with 2,5-disubstituted tetrazoles in living bacteria. (A) Probe 1 efficiently labels proteins in living S. aureus. Living bacteria were treated with the indicated probe for 1 h and labeled by irradiation with light (λ = 280–315 nm) for 10 min. After lysis, TAMRA azide was attached using CuAAC, and labeling was analyzed using gel electrophoresis with in-gel fluorescence scanning and Coomassie Brilliant Blue (CBB) staining. Controls were performed without irradiation. (B) Analysis of the amino acid specificity of probe 1in vitro and in living S. aureus. Samples were analyzed with MaxQuant software36 allowing the modification with probe 1 and the heavy or light isoDTB tag to be on any potentially nucleophilic amino acid. Peptides were further analyzed if the localization probability for a single residue was more than 75%. The total number of identified modification sites is given in parentheses. Data shows the mean ± the standard deviation. (C) Plot of the ratios log2(R) of samples, in which the heavy- and light-labeled sample were both modified with the same concentration of the indicated probe in living S. aureus without pretreatment with an inhibitor. The expected value of log2(R) = 0 is indicated by the black line; the preferred quantification window (−1 < log2(R) < 1) is indicated by the two gray lines. One data point in living bacteria at log2(R) = −8.9 is not shown for clarity. All data for panels B and C originates from biologically independent duplicates of technical triplicates for data in living bacteria and from biologically independent duplicates of technical duplicates for data in vitro. (D) Probe 1 efficiently labels proteins in the living Gram-negative bacteria S. typhimurium and E. coli. The experiment was performed in the indicated bacteria as described for S. aureus in part A.
Next, we performed a mass-spectrometry-based experiment as described above, in which two samples of living bacteria were individually treated with probe 1, irradiated, lysed, clicked to the light and heavy isoDTB tags, respectively, and mixed at a ratio of 1:1 before analysis. We could verify the same selectivity for aspartates and glutamates as seen in vitro for probe 1 (Figure 4B, Table S1). We were able to quantify 3928 residues with the number of incorrectly quantified peptides again being ≤1% (Figure 4C, Table S1). The method is therefore applicable to also quantitatively monitor aspartates and glutamates in living S. aureus.
Encouraged by this result, we also investigated the Gram-negative bacteria Escherichia coli (E. coli) and Salmonella typhimurium (S. typhimurium). Gram-negative bacteria possess a very strong outer barrier consisting of the cell wall and two cell membranes as well as efficient efflux mechanisms. This renders the development of inhibitors and probes that efficiently engage proteins in these cells very challenging.6 After testing the three probes by gel-based experiments and demonstrating that probe 1 again gives the strongest labeling (Figures S22 and S23), we directly compared the labeling in living S. aureus, E. coli, and S. typhimurium using probe 1 (Figure 4D). Probe 1 labeled the proteomes of the Gram-negative bacteria to a similar degree as in S. aureus, which indicates that this probe is efficiently taken up also into these most challenging cells. Performing a mass-spectrometry-based experiment in E. coli, we were able to quantify 1905 residues with good selectivity (Figure S24 and Table S1). Our platform therefore allows global profiling of aspartates and glutamates also in challenging Gram-negative bacteria, which will have important implications for drug discovery of covalent inhibitors through target engagement studies.
Discussion
We here describe the first method to globally map aspartates and glutamates in a residue-specific fashion. We synthesized three light-activatable 2,5-disubstituted tetrazole probes and investigated them as broadly reactive alkyne probes to study the proteome-wide interactions of covalent, carboxylic-acid-directed protein ligands using isoDTB-ABPP.
Initially, we optimized the labeling conditions in vitro in the proteome of S. aureus and could show that probe 2 exhibits the highest selectivity with >85% of all peptides being labeled at aspartates and glutamates. In this way, we were able to quantify more than 6000 residues using this probe and, in aggregate, more than 8500 residues across all probes. This corresponds to 8.9% of all aspartates and glutamates encoded in the S. aureus genome. Furthermore, we quantify 398 residues at functional sites including 174 residues at functional sites of essential proteins. Residues at functional sites of essential proteins (Figure S5) include, e.g., E293 at the ATP binding site of phosphoglycerate kinase (UniProt code: Q2G031), E179 at the active site of the GMP synthase GuaA (UniProt code: Q2G0Y6), E197 at a magnesium binding site of succinate-CoA ligase (UniProt code: Q2FZ37), D233 at the active site of FabH (UniProt code: Q2FZS0), E330 at the ATP binding site of the glycine-tRNA ligase GlyQS (UniProt code: Q2FY08), and E453 at the substrate binding site of 6-phosphogluconate dehydrogenase (UniProt code: Q2FY60). In this way, the method bears great potential to identify functionally important effects on carboxylic acids in the bacterial proteome. Future studies should be able to further increase this coverage of carboxylic acids and especially functional sites. For this purpose, the proteome can be prefractionated using, e.g., strong cation exchange or high pH fractionation in order to decrease the complexity of the samples in individual MS experiments. Furthermore, it is tempting to speculate that more chemically diverse 2,5-disubstituted tetrazoles can be synthesized to cover more functional sites by specific interactions. The 2,5-disubstituted tetrazole probe 2 offers great specificity and high coverage for aspartate and glutamate residues in lysates. Thus, it allows for the first time a broad study of effects on aspartates and glutamates in the proteome.
Next, we used this technology to map the interactions of carboxylic-acid-directed protein ligands. Here, we first profiled two isoxazolium salts (4 and 5) and identified several residues that are modified by these compounds. These include many residues at functional sites and in essential proteins. Furthermore, we proposed, synthesized, and evaluated hydrazonyl chlorides as a new class of carboxylic-acid-directed protein ligands. This design was based on the fact that they can liberate nitrilimines upon elimination of hydrochloric acid that are similar to those resulting from our light-activatable probes. We synthesized one member of this compound class (6) and found 13 targeted binding sites in the proteome of S. aureus. We verified the interaction of 6 with the recombinant, essential protein nicotinate phosphoribosyltransferase in vitro and verified the expected binding site and mass of the modification on the protein. Nevertheless, due to the high reactivity and concentration of 6, some additional labeling of other sites was also observed indicating that further optimization of this chemotype will be necessary. Taken together, this shows that our method gives residue-specific target engagement information for carboxylic-acid-directed protein ligands in the bacterial proteome.
So far, all three tested competitors did not show antibacterial activity up to 500 μM (Figure S25). Compound 6 is nevertheless a most promising candidate for further optimization into a specific carboxylic-acid-directed covalent inhibitor with biological activity as it shows specific engagement of several residues. Besides varying the substituents of the molecule in order to introduce binding to specific proteins and to tailor its reactivity, it would be especially interesting to investigate various leaving groups instead of chloride in order to further tune the stability, reactivity, and selectivity of these compounds as has recently been shown for targeting tyrosines.14 Furthermore, future development will have to carefully investigate the possibility to avoid reactivity with cysteine residues in order to develop inhibitors that can be used in biologically relevant settings. The presented method will be instrumental in evaluating these compounds in a proteome-wide sense and using them to generate a more complete map of aspartates and glutamates that can be addressed with covalent ligands.
Finally, we could show that probe 1 can be used to monitor close to 4000 aspartates and glutamates with good selectivity when it is used in living S. aureus cells. The probe even allows obtaining a broad map of these residues in living Gram-negative bacteria, which are notoriously hard to penetrate with chemical probes. In this way, the methodology will allow obtaining important insights into the behavior of aspartates and glutamates in this biologically relevant setting.
Taken all of this together, we present a method that for the first time allows to globally and residue-specifically monitor aspartates and glutamates in the bacterial proteome in vitro and in living bacteria. The method furthermore allows the identification of sites that can be addressed with covalent ligands in the proteome. In this way, we are convinced that this methodology will have important implications for the design of new chemotypes for covalent inhibitors that target carboxylic acids as well as to broadly understand the target engagement at these amino acid residues in order to more efficiently develop new antibiotics with a covalent mechanism-of-action.
While this manuscript was under consideration, 2H-azirines were reported as additional probes to residue-specifically monitor aspartates and glutamates in the proteome.41 We are convinced that with these two complementary probe technologies using constitutive and light-activatable electrophiles, respectively, important progress will be made in the design of specific carboxylic-acid-directed covalent inhibitors.
Acknowledgments
We gratefully acknowledge Prof. Dr. Stephan A. Sieber and his group for their generous support.
Glossary
Abbreviations
- ABPP
activity-based protein profiling
- CBB
Coomassie Brilliant Blue
- CuAAC
copper-catalyzed azide–alkyne cycloaddition
- DMF
dimethylformamide
- DMSO
dimethyl sulfoxide
- E. coli
Escherichia coli
- EDC·HCl
N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride
- HCl
hydrochloric acid
- IA-alkyne
iodoacetamide alkyne
- isoDTB tags
isotopically labeled desthiobiotin azide tags
- isoDTB-ABPP
isotopically labeled desthiobiotin azide tag-based activity-based protein profiling
- isoTOP-ABPP
isotopic tandem orthogonal proteolysis activity-based protein profiling
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- MIC
minimum inhibitory concentration
- MS
mass spectrometry
- PSM
peptide-spectrum match
- R
ratio between the heavy and the light channel in isoDTB-ABPP experiments
- S. aureus
Staphylococcus aureus
- S. typhimurium
Salmonella typhimurium, TEV, tobacco-etch virus
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01268.
Additional text, figures, schemes, and tables presenting chemoproteomic data, biochemical results, synthetic procedures, experimental conditions, and 1H and 13C NMR spectra (PDF)
Table S1: isoDTB-ABPP data for the evaluation of the mass of modification, selectivity, quantification, and sensitivity for probes 1–3in vitro in S. aureus, in living S. aureus, and in living E. coli. (XLSX)
Table S2: Competitive isoDTB-ABPP data for the competitors 4–6in vitro in S. aureus as well as of reductive dimethyl labeling experiments to establish and quantify the site of labeling on recombinant nicotinate phosphoribosyltransferase (XLSX)
Table S8: Annotation database for all aspartates and glutamates in the genome of S. aureus SH1000 (XLSX)
Author Present Address
† K.B.: Institute of Organic Chemistry and Biochemistry of the Czech Academy of Science, Flemingovo n. 2, 16610 Prague, Czech Republic
Author Present Address
‡ B.L.H.B.: Division of Drug Discovery and Safety, Leiden Academic Centre for Drug Research, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands
Author Contributions
§ K.B., B.L.H.B., and P.R.A.Z. contributed equally to this work. K.B. and B.L.H.B. performed MS- and gel-based experiments. K.B. cloned and expressed nicotinate phosphoribosyltransferase and performed biochemical experiments. B.L.H.B. and P.R.A.Z. synthesized compounds. P.R.A.Z. performed reactivity assays with amino acids using LC-MS. K.B., B.L.H.B., and S.M.H. acquired and analyzed MS data. S.M.H. conceived and supervised the project. S.M.H. wrote the manuscript with contribution from all authors.
S.M.H. and P.R.A.Z. acknowledge funding by the Fonds der Chemischen Industrie through a Liebig Fellowship and a Ph.D. fellowship. We acknowledge financial support by the TUM Junior Fellow Fund.
The authors declare no competing financial interest.
Supplementary Material
References
- Singh J.; Petter R. C.; Baillie T. A.; Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discovery 2011, 10, 307–317. 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Chaikuad A.; Koch P.; Laufer S. A.; Knapp S. The Cysteinome of Protein Kinases as a Target in Drug Development. Angew. Chem., Int. Ed. 2018, 57, 4372–4385. 10.1002/anie.201707875. [DOI] [PubMed] [Google Scholar]
- Hendlin D.; Stapley E. O.; Jackson M.; Wallick H.; Miller A. K.; Wolf F. J.; Miller T. W.; Chaiet L.; Kahan F. M.; Foltz E. L.; Woodruff H. B.; Mata J. M.; Hernandez S.; Mochales S. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 1969, 166, 122–123. 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- Nishimura H.; Mayama M.; Komatsu Y.; Kato H.; Shimaoka N.; Tanaka Y. Showdomycin, a New Antibiotic from a Streptomyces Sp. J. Antibiot. 1964, 17, 148–155. [PubMed] [Google Scholar]
- Smith P. A.; Koehler M. F. T.; Girgis H. S.; Yan D.; Chen Y.; Chen Y.; Crawford J. J.; Durk M. R.; Higuchi R. I.; Kang J.; Murray J.; Paraselli P.; Park S.; Phung W.; Quinn J. G.; Roberts T. C.; Rouge L.; Schwarz J. B.; Skippington E.; Wai J.; Xu M.; Yu Z.; Zhang H.; Tan M. W.; Heise C. E. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 2018, 561, 189–194. 10.1038/s41586-018-0483-6. [DOI] [PubMed] [Google Scholar]
- Lakemeyer M.; Zhao W.; Mandl F. A.; Hammann P.; Sieber S. A. Thinking Outside the Box-Novel Antibacterials To Tackle the Resistance Crisis. Angew. Chem., Int. Ed. 2018, 57, 14440–14475. 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
- Backus K. M.; Correia B. E.; Lum K. M.; Forli S.; Horning B. D.; Gonzalez-Paez G. E.; Chatterjee S.; Lanning B. R.; Teijaro J. R.; Olson A. J.; Wolan D. W.; Cravatt B. F. Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534, 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker S. M.; Backus K. M.; Lazear M. R.; Forli S.; Correia B. E.; Cravatt B. F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017, 9, 1181–1190. 10.1038/nchem.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium . UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E.; Wang C.; Simon G. M.; Richter F.; Khare S.; Dillon M. B.; Bachovchin D. A.; Mowen K.; Baker D.; Cravatt B. F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanon P. R. A.; Lewald L.; Hacker S. M. Isotopically Labeled Desthiobiotin Azide (isoDTB) Tags Enable Global Profiling of the Bacterial Cysteinome. Angew. Chem., Int. Ed. 2020, 59, 2829–2836. 10.1002/anie.201912075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers A. E.; Cravatt B. F. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 2005, 127, 10018–10019. 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C. C.; Kleinman J. I.; Nomura D. K. NHS-Esters As Versatile Reactivity-Based Probes for Mapping Proteome-Wide Ligandable Hotspots. ACS Chem. Biol. 2017, 12, 1478–1483. 10.1021/acschembio.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm H. S.; Toroitich E. K.; Borne A. L.; Brulet J. W.; Libby A. H.; Yuan K.; Ware T. B.; McCloud R. L.; Ciancone A. M.; Hsu K.-L. Global targeting of functional tyrosines using sulfur-triazole exchange chemistry. Nat. Chem. Biol. 2020, 16, 150–159. 10.1038/s41589-019-0404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B. K.; Loveridge C. J.; Thyssen S.; Wormer G. J.; Nielsen A. D.; Palmfeldt J.; Johannsen M.; Poulsen T. B. STEFs: Activated Vinylogous Protein-Reactive Electrophiles. Angew. Chem., Int. Ed. 2019, 58, 3533–3537. 10.1002/anie.201814073. [DOI] [PubMed] [Google Scholar]
- Tamura T.; Ueda T.; Goto T.; Tsukidate T.; Shapira Y.; Nishikawa Y.; Fujisawa A.; Hamachi I. Rapid labelling and covalent inhibition of intracellular native proteins using ligand-directed N-acyl-N-alkyl sulfonamide. Nat. Commun. 2018, 9, 1870. 10.1038/s41467-018-04343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akcay G.; Belmonte M. A.; Aquila B.; Chuaqui C.; Hird A. W.; Lamb M. L.; Rawlins P. B.; Su N.; Tentarelli S.; Grimster N. P.; Su Q. Inhibition of Mcl-1 through covalent modification of a noncatalytic lysine side chain. Nat. Chem. Biol. 2016, 12, 931–936. 10.1038/nchembio.2174. [DOI] [PubMed] [Google Scholar]
- Ban H.; Gavrilyuk J.; Barbas C. F. Tyrosine bioconjugation through aqueous ene-type reactions: a click-like reaction for tyrosine. J. Am. Chem. Soc. 2010, 132, 1523–1525. 10.1021/ja909062q. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Ma D.; Du D.; Xi Z.; Yi L. An efficient reagent for covalent introduction of alkynes into proteins. Org. Biomol. Chem. 2014, 12, 9528–9531. 10.1039/C4OB01873G. [DOI] [PubMed] [Google Scholar]
- Narayanan A.; Jones L. H. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 2015, 6, 2650–2659. 10.1039/C5SC00408J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Yang X.; Jia S.; Weeks A. M.; Hornsby M.; Lee P. S.; Nichiporuk R. V.; Iavarone A. T.; Wells J. A.; Toste F. D.; Chang C. J. Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355, 597–602. 10.1126/science.aal3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. T.; Nelson J. E.; Suero M. G.; Gaunt M. J. A protein functionalization platform based on selective reactions at methionine residues. Nature 2018, 562, 563–568. 10.1038/s41586-018-0608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S.; He D.; Chang C. J. Bioinspired Thiophosphorodichloridate Reagents for Chemoselective Histidine Bioconjugation. J. Am. Chem. Soc. 2019, 141, 7294–7301. 10.1021/jacs.8b11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y.; Ishiyama T.; Sasaki D.; Abe J.; Sohma Y.; Oisaki K.; Kanai M. Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins. J. Am. Chem. Soc. 2016, 138, 10798–10801. 10.1021/jacs.6b06692. [DOI] [PubMed] [Google Scholar]
- Martin-Gago P.; Fansa E. K.; Winzker M.; Murarka S.; Janning P.; Schultz-Fademrecht C.; Baumann M.; Wittinghofer A.; Waldmann H. Covalent Protein Labeling at Glutamic Acids. Cell Chem. Biol. 2017, 24, 589–597. 10.1016/j.chembiol.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Qian Y.; Schurmann M.; Janning P.; Hedberg C.; Waldmann H. Activity-Based Proteome Profiling Probes Based on Woodward’s Reagent K with Distinct Target Selectivity. Angew. Chem., Int. Ed. 2016, 55, 7766–7771. 10.1002/anie.201602666. [DOI] [PubMed] [Google Scholar]
- Mix K. A.; Raines R. T. Optimized diazo scaffold for protein esterification. Org. Lett. 2015, 17, 2358–2361. 10.1021/acs.orglett.5b00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E.; Simon G. M.; Cravatt B. F. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008, 4, 405–407. 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herner A.; Lin Q. Photo-Triggered Click Chemistry for Biological Applications. Top. Curr. Chem. 2016, 374, 1. 10.1007/s41061-015-0002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Dai J.; Hu M.; Liu C.; Meng R.; Liu X.; Wang C.; Luo T. Photo-induced coupling reactions of tetrazoles with carboxylic acids in aqueous solution: application in protein labelling. Chem. Commun. 2016, 52, 4702–4705. 10.1039/C5CC10445A. [DOI] [PubMed] [Google Scholar]
- Herner A.; Marjanovic J.; Lewandowski T. M.; Marin V.; Patterson M.; Miesbauer L.; Ready D.; Williams J.; Vasudevan A.; Lin Q. 2-Aryl-5-carboxytetrazole as a New Photoaffinity Label for Drug Target Identification. J. Am. Chem. Soc. 2016, 138, 14609–14615. 10.1021/jacs.6b06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Qian L.; Li L.; Bernhammer J. C.; Huynh H. V.; Lee J. S.; Yao S. Q. Tetrazole Photoclick Chemistry: Reinvestigating Its Suitability as a Bioorthogonal Reaction and Potential Applications. Angew. Chem., Int. Ed. 2016, 55, 2002–2006. 10.1002/anie.201508104. [DOI] [PubMed] [Google Scholar]
- Cheng K.; Lee J. S.; Hao P.; Yao S. Q.; Ding K.; Li Z. Tetrazole-Based Probes for Integrated Phenotypic Screening, Affinity-Based Proteome Profiling, and Sensitive Detection of a Cancer Biomarker. Angew. Chem., Int. Ed. 2017, 56, 15044–15048. 10.1002/anie.201709584. [DOI] [PubMed] [Google Scholar]
- Abo M.; Weerapana E. A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J. Am. Chem. Soc. 2015, 137, 7087–7090. 10.1021/jacs.5b04350. [DOI] [PubMed] [Google Scholar]
- Kong A. T.; Leprevost F. V.; Avtonomov D. M.; Mellacheruvu D.; Nesvizhskii A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Ramanathan M.; Wang Y. H.; Liu S. T. One-Pot Reactions for Synthesis of 2,5-Substituted Tetrazoles from Aryldiazonium Salts and Amidines. Org. Lett. 2015, 17, 5886–5889. 10.1021/acs.orglett.5b03068. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; Blagoev B.; Gnad F.; Macek B.; Kumar C.; Mortensen P.; Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Matsuura T.; Ito Y. Photoinduced reactions—LXXIX: Photochemistry of 2-thiazolines. Tetrahedron 1975, 31, 1245–1250. 10.1016/0040-4020(75)80164-5. [DOI] [Google Scholar]
- Zengeya T. T.; Garlick J. M.; Kulkarni R. A.; Miley M.; Roberts A. M.; Yang Y.; Crooks D. R.; Sourbier C.; Linehan W. M.; Meier J. L. Co-opting a Bioorthogonal Reaction for Oncometabolite Detection. J. Am. Chem. Soc. 2016, 138, 15813–15816. 10.1021/jacs.6b09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N.; Hu J.; Zhang Z.-M.; Liu W.; Huang M.; Fan Y.; Yin X.; Wang J.; Ding K.; Ye W.; Li Z. 2H-Azirine-Based Reagents for Chemoselective Bioconjugation at Carboxyl Residues Inside Live Cells. J. Am. Chem. Soc. 2020, 142, 6051–6059. 10.1021/jacs.9b12116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.