We have realized that in our original work, the upstream and downstream coordinates of the nucleotides in the nucleotide plots were switched in the computational program, and thus the nucleotide plots should be mirror images of those presented in the version of this article initially published. In Fig. 1 in this Addendum, the nucleotide plots in Fig. 3a–d, f–k of the original article are modified to show the corrected upstream and downstream coordinates.
Fig. 1 |. Identity and sequence contexts of rNMP incorporation in S. cerevisiae genome.
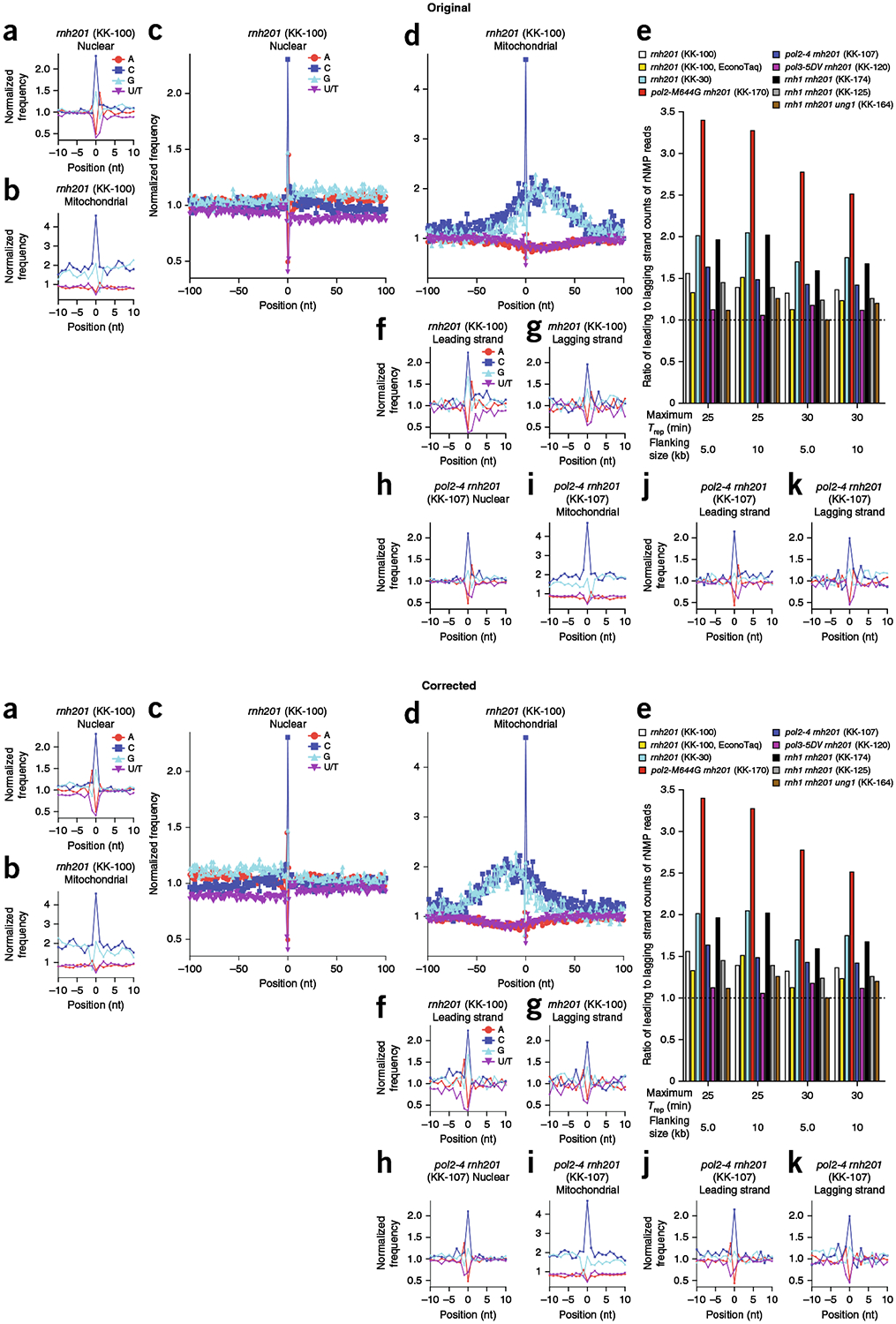
a,b, Normalized nucleotide frequencies relative to mapped positions of sequences from Ribose-seq library of rnh201Δ (KK-100) cells. Position 0 is the rNMP. c,d, Zoom-out of frequencies. e, Ratios of rNMPs on newly synthesized leading to lagging strand for all ribose-seq libraries. Early-firing ARSs selected by their replication timing (Trep) were investigated for two different flanking sizes. f,g, Normalized nucleotide frequencies relative to mapped sequence positions in leading (f) and lagging (g) strands from the rnh201Δ (KK-100) library. ARSs with a Trep of no longer than 25 min were selected with flanking size of 10 kb. h–k, Normalized nucleotide frequencies relative to mapped sequence positions from a pol2-4 rnh201Δ (KK-107) library. Reads were mapped to (a,c,h) nuclear genome, (b,d,i) mitochondrial genome, (f,j) leading strand or (g,k) lagging strand.
Some corrections to the Supplementary Information were also necessary. A revised Supplementary Information PDF including final, correct versions of all Supplementary Figures and Tables has been published with this Addendum. Nucleotide plots in Supplementary Figs. 5, 6 and 8 have been modified to show corrected upstream and downstream coordinates. In Supplementary Table 4, the position 3′ was changed to 5′ in the title of the table, coordinate +1 position was changed to −1 in the right column title, and the word “downstream” was changed to “upstream” in the legend.
Finally, some corrections are required in the text. First, in the sentence in the abstract reading “Ribonucleotides were primarily incorporated on the newly synthesized leading strand of nuclear DNA and were present upstream of (G+C)-rich tracts in the mitochondrial genome,” the word “upstream” should be “downstream”: “Ribonucleotides were primarily incorporated on the newly synthesized leading strand of nuclear DNA and were present downstream of (G+C)-rich tracts in the mitochondrial genome.”
Throughout the first paragraph of the Results subsection “Pattern of sequences flanking rNMPs in S. cerevisiae DNA,” the word “downstream” must be changed to “upstream,” all the “+1” positions should be “–1”, and three of the four 3′ positions should be 5′. In addition, the last sentence of the paragraph needs to be reworded. The complete correct paragraph is as follows:
“Upstream of incorporated rNMPs, we found that the −1 position was most frequently dA and least frequently dG both in the nuclear and in the mitochondrial genomes, with 42–52% dA and 6–16% dG among all four deoxyribonucleotides (dA, dC, dG and dT) (Supplementary Table 4). At the −1 position, dT was also frequent (31–40%) in the mitochondrial genome. In mitochondrial DNA, the high level of dA or dT at the −1 position 5′ from the rNMP could reflect the high A+T content in the mitochondrial genome. It is also possible that the dA in the −1 position influences rNMP incorporation by DNA polymerases. Alternatively, we speculate that dA in the −1 position might stabilize incorporated rNMPs, possibly by affecting base stacking and preventing its repair by mechanisms other than ribonucleotide excision repair. It will be useful to determine the nearest-neighbor thermodynamic parameters for single rNMPs in DNA duplex and, in particular, the stability trend for the base pair 5′ of the rNMP sites. We recently showed that single rGMPs embedded in a short DNA duplex have a marked effect on the elastic properties of DNA by altering the DNA structure at the site encompassing the rNMP and the nucleotide 3′ to it18. Thus, it is reasonable to think that the −1 position 5′ from the rNMP, rather than being a site for signaling for an altered structure, is the most critical site for stabilizing the incorporation of an rNMP in DNA.”
In the second paragraph of the same Results subsection, we amended the text “rNMP sites in mitochondrial DNA were primarily upstream of (G+C)-rich regions” by changing the word “upstream” to “downstream”: “rNMP sites in mitochondrial DNA were primarily downstream of (G+C)-rich regions.”
In the Discussion, the sentence “rCMP and rGMP were more abundant than rAMP and rUMP, and there was frequently a dA downstream of the rNMPs” should be changed to “rCMP and rGMP were more abundant than rAMP and rUMP, and there was frequently a dA upstream of the rNMPs.”
Finally, in the Supplementary Software mentioned in the “Code availability” section, we made changes in the script. Line 125 of the script nuc_frequencies.py (https://github.com/hesselberthlab/modmap/blob/snake/modmap/nuc_frequencies.py) was changed from
“
nuc_counts[offset][nucs] += row.count
” to
if (revcomp_strand)
:
nuc_counts[-offset][nucs] += row.count
else
:
nuc_counts[offset][nucs] += row.count
Supplementary Material
Footnotes
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-019-0505-9.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


