SUMMARY
Telomere-led rapid chromosome movements or rapid prophase movements (rapid prophase movements) direct fundamental meiotic processes required for successful haploidization of the genome. Critical components of the machinery that generates rapid prophase movements are unknown, and the mechanism underlying rapid prophase movements remains poorly understood. We identified S. cerevisiae Mps2 as the outer nuclear membrane protein that connects the LINC complex with the cytoskeleton. We also demonstrate that the motor Myo2 works together with Mps2 to couple the telomeres to the actin cytoskeleton. Further, we show that Csm4 interacts with Mps2 and is required for perinuclear localization of Myo2, implicating Csm4 as a regulator of the Mps2-Myo2 interaction. We propose a model in which the newly identified functions of Mps2 and Myo2 cooperate with Csm4 to drive chromosome movements in meiotic prophase by coupling telomeres to the actin cytoskeleton.
Graphical Abstract
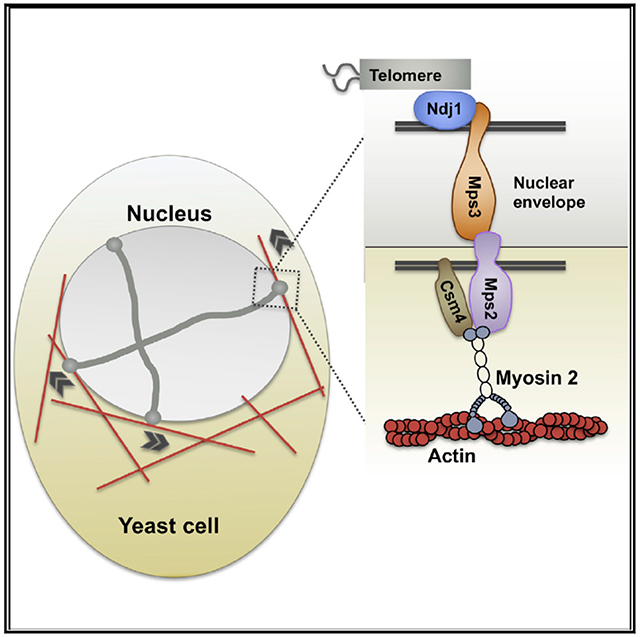
In Brief
Lee and Bisig et al. show that the extranuclear Myo2 motor protein, the nuclear-membrane-anchored LINC-like Mps2 protein, and Csm4 regulatory protein connect telomeres to the cytoskeleton and promote rapid-prophase movement in yeast.
INTRODUCTION
Meiosis involves highly regulated and coordinated chromosome movements. Homologous chromosomes must pair and recombine in the first meiotic prophase to ensure proper segregation at anaphase. Defective segregation leads to phenotypes ranging from sterility to aneuploid offspring with developmental abnormalities [1–5].
Chromosome interactions and meiotic recombination in prophase are accompanied by prominent telomere-led chromosome movements or rapid-prophase movements (rapid prophase movements) that are widely conserved in organisms from yeast to mammals [6–22]. Rapid-prophase movements have been proposed to assist homologous chromosome interactions (i.e., homologous chromosomes pairing and synapsis) [20, 23–30], reduce and/or resolve heterologous interactions [9, 16, 24, 25, 31, 32], and help resolve chromosome interlocks [33, 34]. This indicates the participation of rapid-prophase movements in critical meiotic events required for normal progression of gametogenesis.
In all organisms studied so far, meiotic prophase chromosome movements are driven by the connection of telomeres to the cytoskeleton via the LINC complex: a protein complex that bridges the intact nuclear envelope [35]. The LINC consists of inner nuclear envelope SUN (Sad1-UNC84) domain proteins and outer nuclear membrane KASH (Klarsicht, Anc-1, and Syne homology) domain proteins. Telomeres associate with the intranuclear domain of the SUN proteins, and the extranuclear portion of the KASH protein connects to the cytoskeleton (reviewed in [36–38]).
In S. cerevisiae, it is indicated that rapid-prophase movements are dependent on the actin cytoskeleton [17, 39] and require the proteins Ndj1, Mps3, and Csm4 at the nuclear membrane. Although we know Ndj1 is a meiosis-specific telomere protein that binds the N terminus (nucleoplasmic domain) of the inner nuclear envelope SUN domain protein Mps3 [16, 40–42], a KASH5-like protein and motor proteins connecting Ndj1-Mps3 with cytoskeleton components have not been identified [43, 44]. In this work, we show evidence that the Mps2 protein acts as a meiotic KASH-like protein. Mps2 interacts with Mps3 [44] and localizes at the end of chromosomes (this study) in cytological preparation of yeast chromosome spread in a manner that supports its role as a KASH protein in meiosis. Deletion of the KASH-equivalent domain of Mps2 weakens its interaction with Mps3 and interferes with rapid-prophase movements. We have also identified the type V myosin Myo2 as the cytoplasmic motor that interacts with Mps2 and contributes to rapid-prophase movements.
In S. cerevisiae, Csm4 is tail anchored to the nuclear envelope and its deletion impairs rapid-prophase movements [16, 45–48]. However, the mechanism of Csm4 function within the LINC complex is not understood. We found that Csm4 physically interacts with Mps2, is dependent on Mps2 for association with end of chromosomes via nuclear envelope, and is required for the perinuclear localization of Myo2. In sum, we propose a model in which Mps2, Csm4, and Myo2 cooperate to provide the primary engine for chromosome movements in meiotic prophase in budding yeast by coupling telomeres to the actin cytoskeleton via a LINC complex.
RESULTS
Mps2 Is Membrane Associated and Its Localization to the End of the Chromosomes Requires an Intact LINC
To uncover the LINC complex protein that connects to the cytoskeleton, we examined proteins known to bind Mps3. The SUN domain of Mps3 interacts with the C terminus of Mps2 in mitotic cells, where this Mps3-Mps2 interaction is required for spindle pole body duplication and viability [44]. Mps2 is a single pass integral membrane protein of the nuclear envelope, oriented with residues 1–311 in the cytoplasm and residues 327–387 in the lumen of the nuclear envelope [44, 49, 50] (Figure S1). Mps2, like Mps3, is highly concentrated at the spindle pole body and, at lower levels, in the nuclear membrane [43, 44]. KASH proteins within the LINC complex are typically tail-anchored membrane proteins, containing a transmembrane domain near the C terminus generally followed by 30–40 amino acids [28]. Mps2 does not contain a typical KASH domain based on its amino acid sequence, but it has a 60-amino-acid domain distal to the transmembrane domain. This is sufficiently long to allow Mps2 to be inserted co-translationally and exceeds that expected for a tail-anchored protein [45, 51, 52] (Figure S1 and schematic, Figure 1A). Together, these data suggest that Mps2 could function as a transmembrane protein as part of the LINC complex, connecting Mps3 with the cytoskeleton to generate rapid-prophase movements.
Figure 1. Mps2, a KASH-like Protein, Localizes at Chromosome Ends.
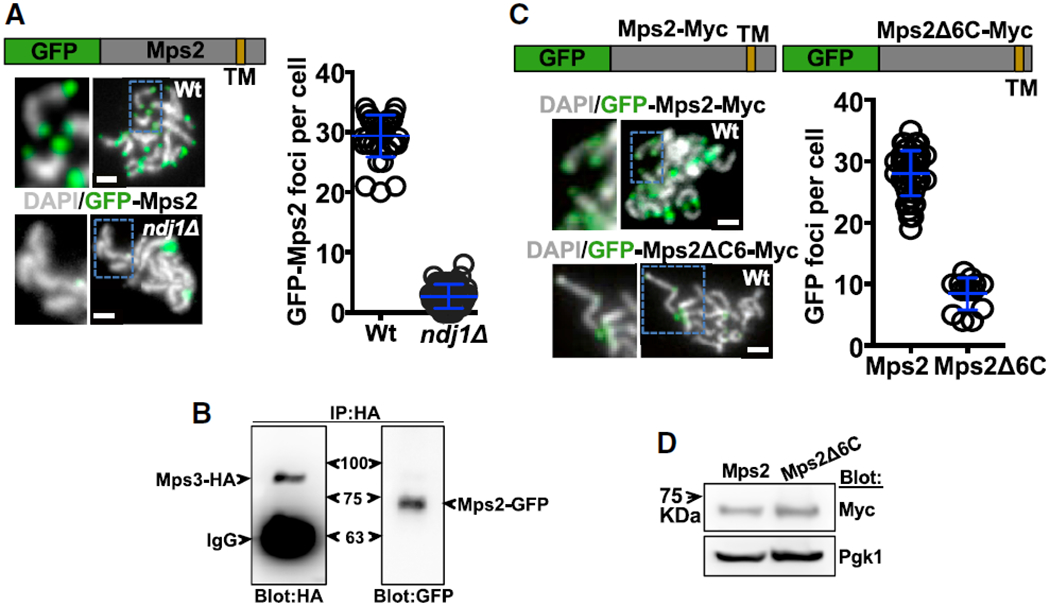
(A) Schematic: GFP-Mps2 fusion protein; TM represents the transmembrane domain (see Figure S1). Examples of nuclei at pachytene as determined by the level of chromosome condensation are shown (white: DNA labeled with DAPI; green: GFP-Mps2 fusion protein; Wt: wild-type strain; ndj1Δ: ndj1 mutant strain). Quantitation of GFP-Mps2 signal at the end of chromosomes per cell is also shown. GFP-Mps2 foci in wild-type (mean ± standard deviation; 29.4 ± 3.5; n = 36) versus ndj1Δ is shown (2.7 ± 1.9; n = 51; t test p < 0.0001).
(B) Co-immunoprecipitation of Mps3-HA and Mps2-GFP from total meiotic yeast extract using anti-HA antibodies for pull-down. This yeast strain co-expressed Mps3-HA and Mps2-GFP, and blots were probed with anti-HA (left panel) and anti-GFP (right panel) antibodies.
(C) Schematics: GFP-Myc-Mps2 fusion protein and GFP-Myc-Mps2Δ6C, which lacks 6 amino acids from the Mps2 C terminus. Examples of nuclei at pachytene stage show GFP-Myc-Mps2 and GFP-Myc-Mps2Δ6C localization at the end of chromosomes in the wild-type strain. Antibodies against GFP were used to reveal the protein fusions in chromosome spreads. Quantitation of GFP fluorescent signal at the end of the chromosomes per cell is also shown. GFP-Myc-Mps2 versus GFP-Myc-Mps2Δ6C foci 28.1 ± 3.7, n = 56 versus 8.4 ± 2.6, n = 14; unpaired t test; p < 0.0001. (D) The western blot shows similar expression of strains expressing GFP-Myc-Mps2 and GFP-Myc-Mps2Δ6C. Myc antibodies were used to visualize both Mps2 and Mps2Δ6C, which contain Myc tags. Pgk1 was used as loading control.
In meiotic cells, Mps3 localizes to telomeres via interactions with Ndj1, the meiosis-specifc telomere protein [40]. We predicted that, if Mps2 interacts with Mps3 within the LINC complex, then Mps2 should also be localized to chromosome ends via its interaction with Mps3. To investigate this, we performed chromosome spread in which condensed meiotic chromosomes are spread with chemical/mechanical forces and probed with antibodies to study the proteins of interest. As yeast conducts meiosis with an intact nuclear envelope, ends of chromosomes in chromosome spreads contain part of the nuclear envelope. Proteins associated with nuclear envelope, especially those capable of accumulating at high concentration, can easily been identified using this method and appear as localized at the end of the chromosomes. Indeed, in strains containing Mps2-GFP, we observed an average of 28–31 GFP foci localized at the end of pachytene chromosomes (Figure 1A). This is near to the number expected for the total number of bivalent chromosomes (32) in wild-type and similarly to that observed for Ndj1, Mps3, and Csm4 [16, 40, 41] (Figure 1A). The localization of Mps2 at the end of chromosomes required Ndj1 (Figure 1A), presumably through Mps3 at the inner nuclear envelope. We confirmed that Mps3 and Mps2 form part of the same complex in meiotic cells by immunoprecipitation. We used a yeast strain expressing both Mps2-GFP and Mps3-hemagglutinin (HA) after 4 h in sporulation media. Using HA antibodies, we observed that Mps3-HA was able to pull down itself and Mps2-GFP (Figure 1B).
The C Terminus of Mps2 Is Required for Its Connection to End of Chromosomes via LINC Complex (Mps3) and for Normal Rapid-Prophase Movements and Meiosis
If Mps2 is part of the machinery generating rapid-prophase movements, disturbing Mps2 localization and/or interaction with the LINC complex is expected to result in deficient rapid-prophase movements. To test this hypothesis, we deleted six amino acids from the C terminus of Mps2, which are required for Mps2 to interact with the SUN domain of Mps3 in vitro [44]. Indeed, a wild-type strain expressing GFP-Mps2ΔC6 (Mps2 amino acids 1–381) displayed only weak GFP signals at a few chromosome ends in meiotic chromosome spreads compared to control expressing GFP-Mps2 (Figure 1C), suggesting that disruption of the Mps2-Mps3 interaction precludes the association of Mps2 with the end of the chromosomes. GFP-Mps2 and GFP-Mps2ΔC6 show similar expression level monitored by western blots in total cell extract after 4 h in sporulation media (Figure 1D). Our data suggest that Mps2 is a LINC complex component that localizes to the end of the chromosomes via Mps3.
To investigate the physiological function of Mps2 as a LINC complex component, we measured rapid-prophase movements in an mps2ΔC6 strain. Specifically, we analyzed time-lapse images of GFP-lacI bound to an array of lacO sequences integrated near the right telomere of chromosome IV [16] and measured the maximum speed, average speed, and bias (this is ~0 for random movement, <0 for tendency to remain in place, and >0 for tendency to move away from the starting position). The bias term is adapted from measures of bacterial motility of both unpaired and paired telomeres. Detailed methods regarding through-focus microscopy and semi-automatic quantification and analyses were published in [16]. csm4 deletion mutant served as a control for near-absent telomere movement. We found that telomeres moved significantly slower in wild-type compared to the mps2ΔC6 strain in maximum and average speed analyses at 4 h (Video S1) and 5 h (Figures 2A–2C and S2) after cells were transferred to sporulation media. Similarly, measurement of bias (Figure S2), which describes the ability for a spot to move away from its starting point, is reduced respect to wild-type. Meiotic progression was impaired in mps2ΔC6 strain. Sporulation and spore viability were also reduced in the mps2ΔC6 strain, and progression through meiotic prophase is affected (Figure S3). These findings are consistent with a deficiency in rapid-prophase movements, though they could also reflect defective spindle body duplication, as observed in mps2ΔC6 mitotic cells [44].
Figure 2. Mps2 Promotes Rapid-Prophase Movements in Budding Yeast.
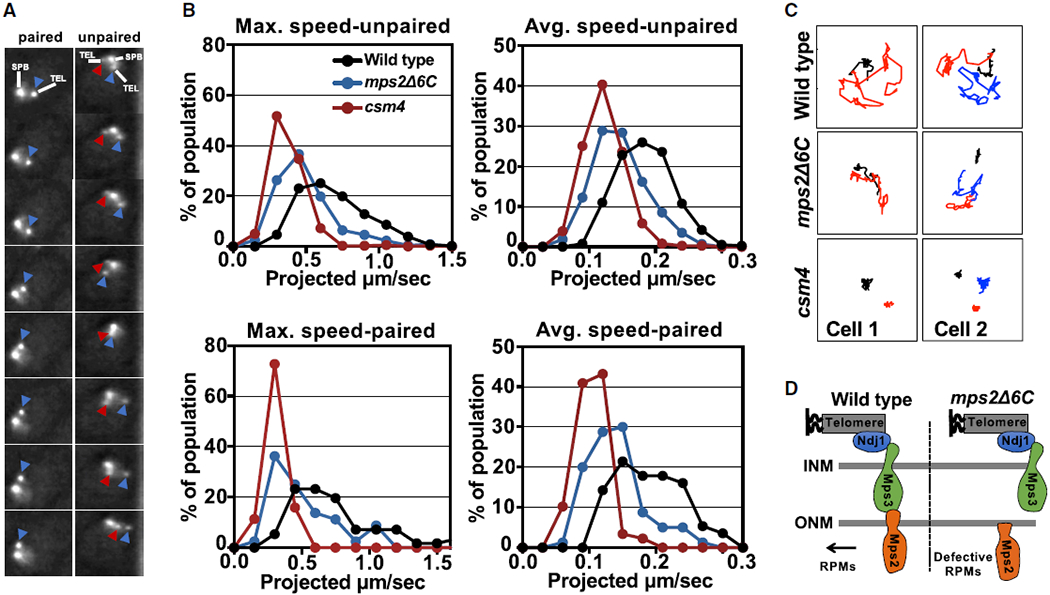
(A) The image frames in the pictures provide an illustration of how speed was measured in cells with paired (left) and unpaired (right) chromosomes 4 right telomeres. These images were acquired with through-focus microscopy, every 1 s, for a total of 61 frames (detailed description for through-focus microscopy and its quantification and semi-auto analyses were published in [16]; see Video S1). In different frames, the same color arrows mark the same lacO256/lacI-GFP spots.
(B) Histograms display meiotic rapid-prophase movements activity in zygotene by measures of maximum and average speed for unpaired and paired chromosomes (see Figure S2) in mps2Δ6C mutant, wild-type, and csm4Δ strains. In our strain background, for strains that are not delayed in meiotic progression, 4 h post-shift into sporulation medium roughly correlates with zygotene and 5 h post-shift correlates with pachytene (see Figure S3). For csm4Δ, a mutant known for prolonged delay, 4.5 h post-shift roughly correlates with zygotene [16], is used as a control for near absence of rapid-prophase movements. All measurements are for lacO256/lacI-GFP spots adjacent to the 4R telomere, and each profile is the result of a total of three independent experiments (250–300 cells scored per strain), where there are 2 spots (unpaired, top) or 1 spot (paired, lower panel).
(C) Example of traces (spots in 61-frame time-lapse series) of paired and unpaired telomere movement in wild-type, mps2Δ6C, and csm4Δ in zygotene cells. (Black line marks the spindle pole body movement. Figures with red line only mark paired telomere and with both red and blue lines mark unpaired telomeres.) Represented are projection images of telomere movement in 61 s. Cell 1 and cell 2 are examples of cells with paired and unpaired telomeres, respectively.
(D) Schematic of wild-type Mps2 and Mps2Δ6C association to other components of the LINC complex.
Overall, our data suggest that the impaired interaction of an Mps2 carboxyl terminal mutant and Mps3 [44] affects the integrity of the LINC complex, leading to defective rapid-prophase movements (Figure 2D). Thus, Mps2 fulfills structural and functional predictions of a KASH-like protein within the LINC complex, bridging the Mps3/Ndj1/telomere complex to cytoplasm and cytoskeleton.
Mps2 Interacts with Myo2 Cytoplasmic Molecular Motor
According to our model, the Mps2 C terminus contacts Mps3 within the lumen of the nuclear envelope, whereas the N terminus is located in the cytoplasm (Figure 2D). To identify proteins that interact with the cytoplasmic domain of Mps2, we performed a yeast two-hybrid screen, anticipating that we would find interactions with cytoskeletal elements. Specifically, we used amino acids 1–310 of Mps2 as bait to screen a prey library of yeast genomic DNA fragments [53]. We isolated clones expressing previously reported Mps2 interactors, including Yaf9, Spc24, and Bbp1 [49, 54], validating the screen (Table S1).
Intriguingly, we identified Mps2-interacting clones that encode amino acids 943–1,574 of myosin 2 (Myo2), an essential type V myosin that was not previously linked with Mps2/LINC. This region of Myo2 contains the cargo binding region, which in mitotic cells interacts with adaptor proteins on the vacuole, secretory vesicles, peroxisomes, mitochondria, and astral microtubules to promote their transport along actin cables [55]. Additional yeast two-hybrid analyses revealed that the interaction with Myo2 required amino acids 125–310 of Mps2, a region that contains two predicted coiled-coil domains (Figure 3A). The cargo-binding domain of Myo2 (1,087–1,574 amino acids) comprises a complex fold of 15 antiparallel helices [55, 56] and contains a number of coiled-coil domains that could interact with similar domains on Mps2. To confirm the interaction, we expressed Myo2-HA and Mps2-Myc fusion proteins in yeast and performed co-immunoprecipitation. Antibodies specific for HA immunoprecipitated Myo2-HA and Mps2-Myc from wild-type cells undergoing meiosis (Figures 3B and S4). However, Myc antibodies did not co-immunoprecipitate Myo2-HA with Mps2-Myc, possibly due to competition from other Myo2 interacting partners. Overall, our data suggest that Mps2 interacts with the cytoplasmic molecular motor Myo2.
Figure 3. Myo2 Interacts with Mps2 and Is Required for Myo2 Localization at the Nuclear Envelope.
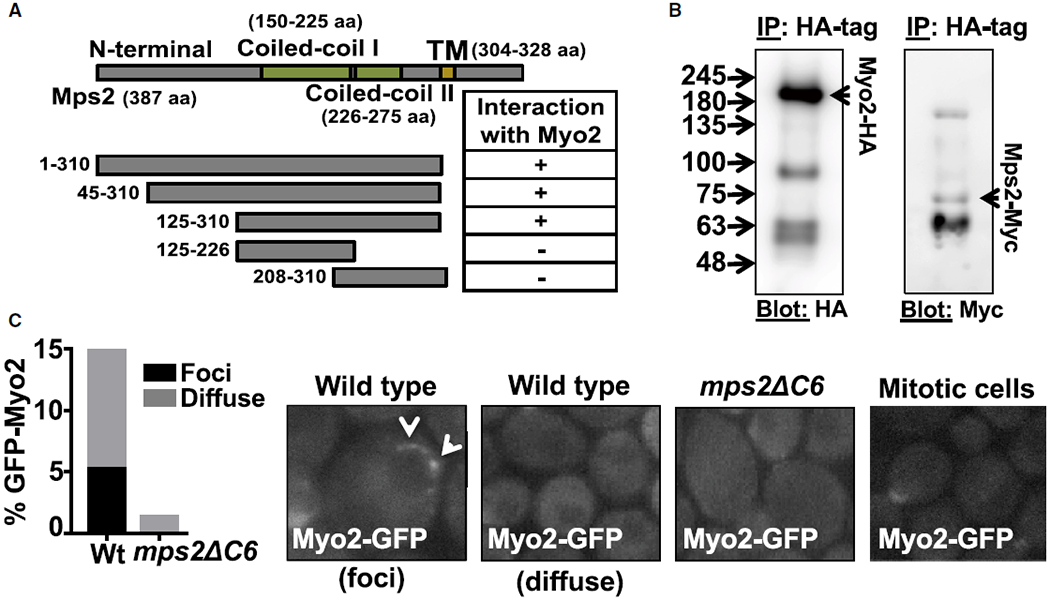
(A) Schematic of predicted functional domains of Mps2 and Mps2 truncation mutants. Absence (−) or presence (+) of a two-hybrid interaction with Myo2 amino acids 943–1,574 is indicated (see Table S1 for proteins that interact with the cytoplasmic domain of Mps2).
(B) Co-immunoprecipitation of Myo2-HA and Mps2-Myc from total yeast extract using anti-HA. This yeast strain co-expressed Myo2-HA and Mps2-Myc from endogenous promoters, and blots were probed with anti-HA (left panel) and anti-Myc (right panel) antibodies (see Figure S4).
(C) Examples of GFP-Myo2 localization in vivo wild-type, mps2Δ6C nuclei, and mitotic dividing cells. Two patterns of Myo2-GFP signal identified as “foci” or “diffuse” (see Video S2). Percentage of nuclei with the indicated perinuclear GFP-Myo2 patterns is also shown (n = 100 nuclei scored in each mutant). The describe pattern of Myo2-GFP in wild-type and absence of perinuclear signal in mps2ΔC6 is highly reproducible.
Myo2 Interacts with the LINC Complex and Requires Mps2 for Perinuclear Localization
We hypothesized that the myosin motor protein acts at telomeres to promote rapid-prophase movements via Mps2. We analyzed the cellular localization of various myosin family members by GFP fusion to Myo1, Myo2, Myo4, and Myo5 in yeast meiotic chromosome spreads. We detected significant accumulation of Myo1-GFP, Myo2-GFP, and Myo4-GFP, but not Myo5-GFP, at the end of chromosomes (Figure S5A). Not every telomere in a given spread was labeled, suggesting that the interactions may be transient.
In mitotic cells, Myo2 is concentrated at the tip of the emerging bud and at the bud neck of large buds as well as at the shmoo tip of mating cells [57, 58]. In contrast, live imaging of meiotic whole cells revealed that Myo2-GFP can concentrate as foci on the nuclear envelope, which has not been reported for mitotic cells (Figure 3C). We also observed a diffuse distribution of Myo2 around meiotic nuclei (Figure 3C, “diffuse” versus “foci”). The perinuclear Myo2-GFP foci moved rapidly around the nucleus (Video S2), in a manner comparable to the behavior of Mps3-GFP [16]. Notably, the “diffused” and “foci” perinuclear pattern of Myo2-GFP were absent in mps2∆C6 (Figure 3C; Video S2). These data suggest that Mps2 is required to recruit Myo2 to the nuclear envelope during meiosis.
Expression of a Dominant-Negative Allele of Myo2 (Myo2ΔN) Delocalizes Myo2 from Nuclear Envelope, Perturbs Rapid-Prophase Movements, and Delays Meiotic Progression
We investigated the potential roles of myosin family proteins in rapid prophase movements and meiotic progression. Myo2 is an essential gene, so we constructed an inducible dominant-negative allele, MYO2ΔN. The Myo2 cargo-binding domain (amino acids 1,087–1,574, without motor domain) was placed under the control of the GAL1 promoter [58], in a strain expressing a fusion of the human estrogen receptor to the yeast Gal4 transcription factor [59]. Expression of Myo2 cargo-binding domain protein serves as dominant negative and competes with endogenous Myo2 for Mps2 binding. We induced Myo2ΔN in meiotic cells by adding β-estradiol 2 h after transferring to sporulation medium. Expression of Myo2ΔN disrupted the perinuclear localization of Myo2-GFP (Figure 4A) and led to a significant reduction in rapid-prophase movements, to levels slightly better than those observed in csm4 deletion (Figures 4B and 4C; Video S3). Anaphase I was delayed and spore viability reduced in Myo2ΔN with or without estradiol, and sporulation levels were affected in Myo2ΔN upon induction (Figure S6).
Figure 4. Myo2 Interacts with Mps2 and Is Required for Rapid-Prophase Movements in Budding Yeast.
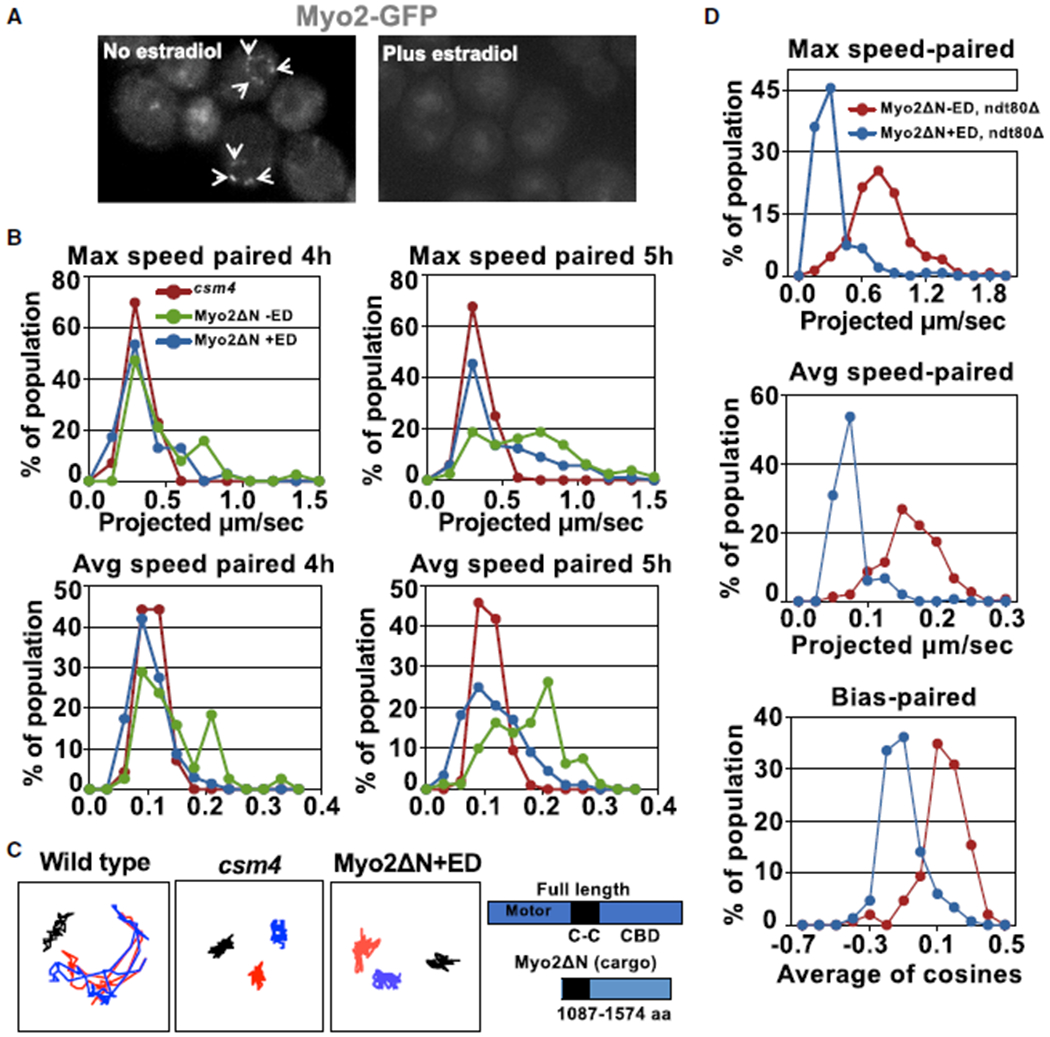
(A) Live-image examples of nuclei showing distribution of Myo2-GFP in strain (MYO2-GFP, Pgal-Myo2ΔN) carrying inducible Myo2ΔN treated with and without estradiol. Expression of Myo2ΔN by adding estradiol competes away Myo2-GFP at the nuclear envelope.
(B) Histograms display rapid-prophase movements at early (4 h post-shift/zygotene in Myo2ΔN with and without estradiol and 4.5 h post-shift/zygotene in csm4Δ) and late (5 h post-shift/zygotene in Myo2ΔN with and without estradiol and 8.5 h post-shift/zygotene in csm4Δ) prophase in maximum and average speed analyses for paired telomeres in different strains, as indicated (see Figures S5 and S6). All measurements are for lacO256/lacI-GFP spots adjacent to the 4R telomere, and each profile is the result of three independent experiments (a total of 250–300 cells scored per strain; see Videos S3 and S4).
(C) Traces represent projection images of independent telomere movements in 61-frame time-lapse series (unpaired telomeres are represented in red and blue; black marks the spindle pole body movement) in zygotene from WT, csm4Δ, and Myo2ΔN with estradiol. Schematics indicate the functional domains within full-length Myo2 and Myo2ΔN, lacking the motor domain at the N terminus. +ED, estradiol; CBD, cargo-binding domain.
(D) Histograms display rapid-prophase movements characteristics maximum speed, average speed, and bias for paired telomeres in the indicated yeast strain at 6 h after cells were transferred to sporulation media. ED, estradiol.
To confirm the role of Myo2 in rapid-prophase movements, we created a strain carrying Myo2ΔN in a ndt80Δ background. The ndt80Δ allele block cells in pachytene with complete synaptonemal complexes [60]. We showed in a previous study [16] that rapid-prophase movements sustain a 0.9 μm/s maximum speed in ndt80Δ mutants. This system allows rapid-prophase movements measurements after longer periods of Myo2ΔN expression and competition to wild-type Mps2. Again, in these conditions, Myo2ΔN expression in early meiotic prophase severely impaired rapid prophase movements (Figure 4D). We speculate that, although rapid prophase movements are strongly inhibited, the residual rapid-prophase movements, perhaps supported by additional Myo proteins, are sufficient to achieve a level of pairing and recombination. Myo1 and Myo4 associate to a portion of chromosome ends (Figure S5A), and the rapid-prophase movement histograms for maximum and average speed for unpaired and paired chromosomes in wild-type versus myo1, myo4, and myo 3/5 mutants are similar (Figure S5B; Video S4).
Nevertheless, the dominant-negative Myo2ΔN delocalizes Myo2 and impairs rapid-prophase movements, showing that Myo2 plays a critical function in supporting rapid-prophase movements.
Csm4 Interacts with Mps2, Is Required for Myo2 Nuclear Localization, and Regulates Mps2-Myo2 Rapid-Prophase Movements Functions
Given our new perspective on the roles of Mps2 and Myo2, we next investigated the role of Csm4 in rapid-prophase movements. Csm4 is a meiosis-specific, tail-anchored membrane protein [45] that localizes to the end of the chromosomes in chromosome spreads, and it is essential for rapid-prophase movements and normal meiotic progression [16, 45–48]. Csm4 has been proposed to be a meiotic KASH protein that partners with Mps3 within the LINC complex. Consistent with this hypothesis, deletion of the Csm4 transmembrane domain creates a null allele [47] and significantly impairs rapid prophase movements and homologous chromosome pairing [48]. However, only a minimal region of Csm4 is predicted to project into the lumen of the nuclear envelope. In contrast, our data suggest that Mps2 interacts with Mps3 within the lumen of the nuclear envelope, as a functional and structural LINC complex component.
We considered that Csm4 is not a structural LINC component but instead functions primarily as an accessory protein to Mps2 to regulate LINC complex interactions with cytoskeleton components. We found that Csm4, unlike Ndj1, was not required for the localization of Mps2 to the end of the chromosomes in chromosome spread analyses, suggesting that Csm4 does not influence the Mps2-Mps3 interaction (Figure 5A). In addition, expression of Mps2ΔC6 abrogated the localization of Csm4 to the end of the chromosomes, but not of Ndj1 or Mps3 (Figure 5B), suggesting that Csm4 requires Mps2 for association to the end of the chromosomes. To determine whether Mps2 and Csm4 interact, we co-expressed Mps2-GFP-Myc and Csm4-HA-tagged proteins in yeast. Csm4-HA and Mps2-GFP-Myc co-immunoprecipitated from total meiotic yeast extract (Figures 5C, S4B, and S4C), suggesting that Mps2 and Csm4 form part of the same complex during meiosis. We previously demonstrated that Ndj1 and Mps3 are required for Csm4 to localize to the end of the chromosomes in chromosome spread analyses [16], presumably reflecting localization of Csm4 to the nuclear envelope. Our current results suggest that Csm4 is recruited to the end of the chromosomes via Mps2, with both proteins positioned in the outer nuclear membrane.
Figure 5. Csm4 Forms a Complex with Mps2, and Csm4 Is Required for the Nuclear Localization of Myo2.
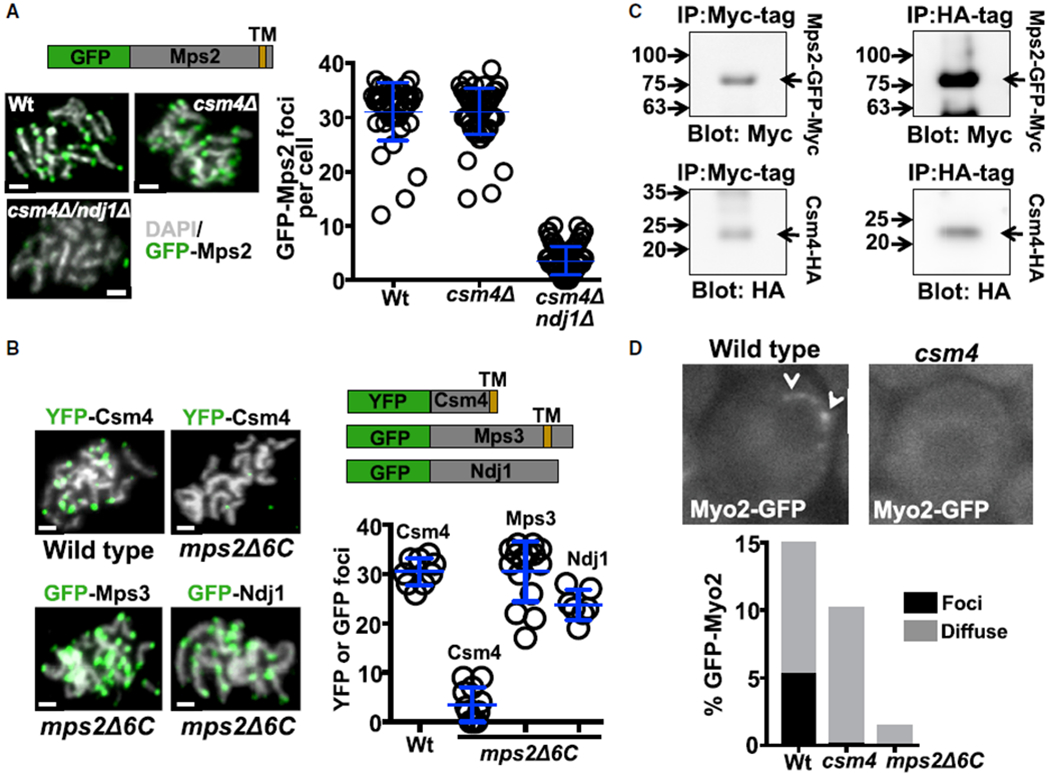
(A) Examples of nuclei at pachytene stage. White: DNA labeled with DAPI; green: GFP-Mps2 fusion protein. csm4Δ, Csm4 mutant strain; csm4Δ/ndj1Δ, double mutant strain for csm4 and ndj1; Wt, wild-type strain. Schematic depicts the domains of the Mps2-GFP-fusion protein. Quantitation of GFP-Mps2 signal at the end of the chromosomes per cell is also shown. GFP-Mps2 foci in wild-type obtained from three independent experiments (mean ± standard deviation; 31.14 ± 5.4; n = 42) compared to csm4Δ (31.15 ± 4.3; n = 14; unpaired t test; p = 0.99) and csm4Δ/Ndj1Δ (3.5 ± 2.6; n = 108; unpaired t test; p < 0.0001).
(B) Examples of nuclei at pachytene stage (white: DNA labeled with DAPI; green: YFP and GFP fusion proteins, as indicated). mps2Δ6C, mutant strain for mps2Δ6C, which lacks 6 amino acids from the Mps2 C terminus is shown. Schematic depicts the domains of the Csm4-YFP fusion, Mps3-GFP, and Ndj1-GFP proteins. TM represents the predicted transmembrane domain of Csm4 and Mps3. Quantitation of YFP-Csm4 (in wild-type, 30.4 ± 2.7, n = 9 and mps2Δ6 3.5 ± 3.4, n = 12, unpaired t test, p < 0.0001), GFP-Mps3 (30.5 ± 6.1; n = 15), and GFP-Ndj1 (23.7 ± 3.1; n = 7) signal at the end of the chromosomes per cell is also shown.
(C) Co-immunoprecipitation of Mps2-GFP-Myc and Csm4-HA from total yeast extract, using anti-Myc or anti-HA, as indicated. Yeast co-expressed Mps2-GFP-Myc and Csm4-HA, and blots were probed with anti-HA (lower panels) and anti-Myc antibodies (upper panels) as indicated (see Figure S4).
(D) Examples of Myo2-GFP localization in vivo for wild-type and csm4Δ cells. Quantitation of Myo2 associated with nuclei is also shown.
Notably, csm4Δ cells lacked perinuclear Myo2-GFP foci (Figure 5D; Video S2). The formation of Myo2 foci at the nuclear envelope depends on both Mps2 and Csm4. We propose that Csm4 promotes rapid-prophase movements by recruiting Myo2 and/or by stabilizing the Mps2-Myo2 interaction.
DISCUSSION
Mps2 May Connect the LINC Complex to Myo2 on Actin and Cooperate with Csm4 to Mediate Dynamic Chromosome Movements in Yeast Meiosis
Understanding the role and regulation of rapid-prophase movements has been limited by a lack of insight into the machinery that generates rapid-prophase movements. Using complementary genetic, biochemical, and functional approaches, we obtained evidence that Mps2 may function as a LINC component, interacting with the cytoplasmic type V myosin motor (Myo2). Mps2 and Myo2 are required to generate rapid-prophase movements, suggesting that Myo2 is a primary engine of rapid prophase movements in budding yeast (Figure 6).
Figure 6.
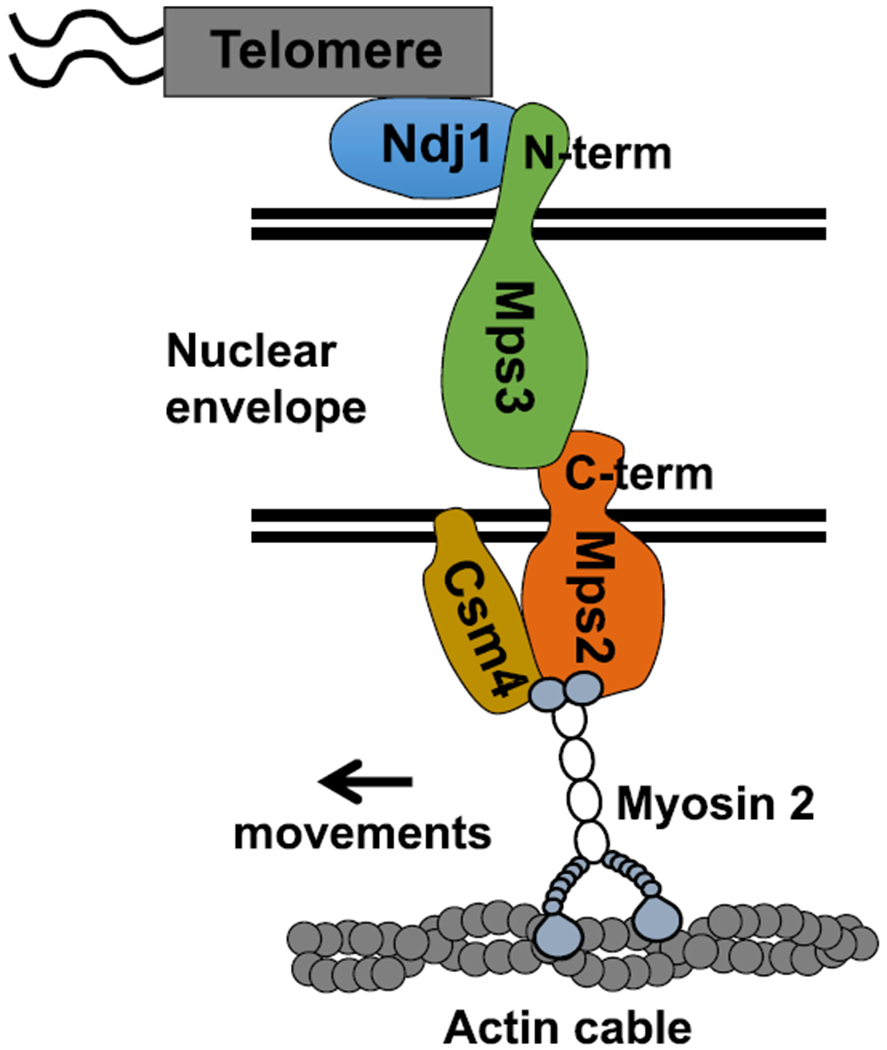
Tentative Model for Meiotic Chromosome Telomere-Nuclear Envelope Attachment and Connection to Cytoskeleton Components in Yeast
Our model proposes that Mps3 and Mps2 are core components of the LINC complex connecting telomeres to the cytoskeleton in meiotic budding yeast (Figure 6). Although Mps2 lacks obvious amino acid sequence homology to KASH proteins, it appears to function as a KASH protein partner to the SUN protein Mps3 within the budding yeast LINC complex.
On the other hand, although Csm4 is an essential regulator of rapid-prophase movements, Csm4 appears not to be a structural component of the LINC complex. Importantly, Csm4 and Mps2 interact and both are required to recruit Myo2 to the nuclear envelope. Our model considers that Mps2 recruits Myo2 via direct interaction and that Csm4 could act to stabilize the interaction between Mps2 and Myo2. Alternatively, Csm4 could recruit actin cables or promote actin polymerization at the nuclear envelope, which would indirectly recruit Myo2 and other components of the actin cytoskeleton. The evidence we present regarding local function of Myo2 at telomeres does not rule out the possibility that Myo2 dominant-negative overexpression or deletion of other Myosin family members may affect other cellular functions relevant to rapid-prophase movements, for example, on the role that different myosins play in stabilizing the cytoskeletal network.
Based on observations that individual telomeres move in association with actin cables, it was suggested that telomeres move via actin treadmilling [17, 47]. We show that Myo2 might generate the force driving rapid-prophase movements, perhaps in addition to actin treadmilling. Combining both activities could modulate chromosome movements and, for example, generate back and forth motions that might help resolve chromosome interlocks [17, 47].
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Roberto J. Pezza (Roberto-Pezza@omrf.org). For yeast strains generated in this study and further information about the reagents please contact Roberto J. Pezza (Roberto-Pezza@omrf.org). All yeast strains are available for sharing.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast culture
Diploids were grown 24 h in 5 mL YPDA at 30°C. For synchronous meiosis cells were grown to 5 × 107 cells/ml in YPAcetate for 16 h, then shifted to 1% potassium acetate at 108 cells/ml for 5 h to reach zygotene stage.
METHOD DETAILS
Yeast strains
Strain list and corresponding background can be found in STAR Methods. CSM4 alleles: csm4::LEU2 was constructed by replacing the PstI-PmeI fragment from the CSM4 coding sequence with a PstI-SmaI fragment containing TRP1 [61]. CSM4 was epitope-tagged at the N terminus by inserting EcoRV and EcoRI site immediately after the start codon in CSM4. A Smal-EcoRI 3x HA fragment was PCR amplified from pFA-3x HA-TRP1 [62] and ligated into the EcoRV-EcoRI sites. The 3xHA-CSM4 constructs was cloned onto MCB917, a derivative of pRS304 [63] that carries a fragment of chromosome 16 in the NotI-SacI sites. The 3xHA-CSM4 was integrated into csm4::LEU2 strains by digestion with BglII. Mps2ΔC: The deletion of six codons from the C terminus of Mps2 was accomplished by transformation with a PCR product [62]. GFP-MPS2 was introduced by transformation with plasmid p30 [50]. mps3Δ240-430: partial deletion alleles of MPS3 deleted residues 240–430, was constructed by PCR and sequenced before use. The mutant alleles were cloned into the URA3 vector YIplac211 and integrated at the genomic site of MPS3. Successful replacements of MPS3 with the deletion alleles were identified by PCR screening of 5-FOAR colonies [33].
Yeast two-hybrid analysis
Bait plasmids were constructed in pBGKT7 (Clontech) and transformed into yeast strain AH109. Bait transformants were transformed with a library of random sheared yeast genomic DNA in pACTII and interaction was selected on minus adenine, histidine, leucine, and tryptophan medium. Approximately 1x106 transformants were screened. Library plasmids were recovered and rescreened.
Measuring rapid prophase movements in budding yeast
For time-lapse acquisitions, cells from sporulating cultures were concentrated, spread across polyethyleneimine-treated coverslips, then covered with a thin 1%agarose pad to anchor the cells to the coverslip [64]. The coverslip was then inverted over a silicone rubber gasket attached to a glass slide, to provide an air space and to prevent drying while imaging. In some experiments, following image acquisition the cell preparations were placed in a humid chamber at 30°C overnight to determine the effects of image acquisition on final levels of sporulation. All time-lapse experiments employed an extended depth-of-focus method to acquire fluorescence images [65]. [16] Images were made at 27°C using an upright Axioplan 2ie microscope fitted with a 100 ×, NA1.4 plan-Apo objective (Carl Zeiss MicroImaging), a high-speed switching DG-5 xenon illuminator (Sutter), a CoolSNAP HQ digital camera (Photometrics), and a BNC555 pulse generator (Berkeley Nucleonics) to synchronize camera exposure with focusing movements and illumination. Each image in a time-lapse series employed the full camera frame (1392 × 1040 6.45 μm square pixels) to acquire 12-bit images in 250 ms exposures while focus traveled through 10 μm (5 μm on either side of the mid-focal plane). Longer travel and exposure times provide better resolved final images, but these conditions clearly revealed the spots of interest and minimized fading of the fluorescent signals. Fine sampling in the plane of focus (0.0645 μm/pixel) was found to provide better spot discrimination than lower levels of resolution. Following image deconvolution, cells were examined individually to determine the positions of the spots in each time-lapse series. The position of each spot was defined as the pixel coordinate nearest the centroid of the brightest local pixels. Positions were assigned automatically by software then edited manually to remove spurious assignments and to correct for overlapping spots; assignment is aided by the summation of intensity that occurs when spots coincide (the images essentially are summed intensity projections, rather than the more familiar maximum intensity projections). Image acquisition, deconvolution, viewing, and quantification were all carried out using custom-written software [16].
Co-immunoprecipitation
To prepare cell lysate, cells were washed with ddH2O supplemented with 0.2 mM PMSF and resuspended in 1 mL/g sample of buffer H (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.1% IGPAL, 150 mM KCl) with protease inhibitors cocktail (Price) plus 3.6 g/g sample of Zirconium Silicate beads (Next Advance). Cells were homogenized in the Blue Bullet Blender at speed 10 for 15 min in a cold room, and the lysate was cleared by centrifugation at 70000 g for 30 min at 4°C. For immunoprecipitation, 15 μg/g sample of anti-HA or 12 μg/g sample of anti-Myc antibodies were added to the lysates, incubated overnight at 4°C in rotation followed by incubation with 30 μL/g sample of Dynabeads M-280 Sheep Anti-Rabbit or Anti-Mouse IgG for 1 h at 4°C with rotation. Beads were washed four times with ice-cold IP buffer, and bound proteins were eluted by boiling for 5 min with SDS-PAGE sample buffer. Proteins were separated by 4%–15% gradient SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. The blots were probed with individual primary antibodies as indicated, and then incubated with HRP-conjugated donkey anti-rabbit or anti-mouse antibodies. In all blots, proteins were visualized by enhanced chemiluminescence.
Spreads/localizations in budding yeast
Spread meiotic nuclei were prepared from sporulating cells by spheroplasting, fixing, and drying the resulting preparations on slides. Spheroplasting was carried out by removing 5-10 mL samples, washing the cells in 2% wt/vol potassium acetate/0.8M sorbitol/ pH7.0, adding dithiothreitol to a final concentration of 10mM for 10 min at 30°C and then adding Zymolyase 100T (ICN Biomedicals, Irvine, CA) to 25 μg/ml final concentration. The percent of cells spheroplasted at a given time was determined by counting samples diluted into potassium acetate-sorbitol with versus without 2% wt/vol Sarkosyl; only the spheroplasts lyse in 2% Sarkosyl. At 70%–90% spheroplasting, the cells were washed once with ice-cold 0.1 M 2-(N-morpholino) ethane sulfonic acid/1 M sorbitol/1 mM EDTA/0.5 mM MgCI2/pH 6.4 and kept on ice as a pellet with the supernatant removed. Spheroplasts in the pellet were mixed with 0.1 M 2- (N-morpholino)ethanesulfonic acid/l mM EDTA/0.5 mM MgCI2/pH 6.4 at a volume ratio of 1 part spheroplasts:25 parts buffer. Paraformaldehyde (4% wt/vol)/pH 7.0 (unbuffered) was added at a volume ratio of 1 part suspensions parts paraformaldehyde, and 0.4ml of the mixture was placed on a glass microscope slide precoated with poly-L-lysine. After 10 min at room temperature, the slides were gently drained and 0.35 mL more of 4% wt/vol paraformaldehyde/pH 7.0 was added for another 5 min. The final fixative was gently drained off, the surface of the slide rinsed with 5 mL of 0.4% vol/vol PhotoFlo200 (Eastman Kodak, Rochester, NY), and the preparations allowed to air dry. GFP antibodies were used for fluorescent localization of GFP tagged proteins.
QUANTIFICATION AND STATISTICAL ANALYSIS
Prism was used for statistical analysis of media, standard deviation, and t test for significance. Statistical details of experiments, including number of cells, what n represents, mean and standard deviation of the analyzed populations, and test of significance when comparing two samples, can be found in Results and in the Figure legends.
DATA AND CODE AVAILABILITY
This study did not generate/analyze data/code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Dynabeads M-280 Sheep Anti-Rabbit IgG | Invitrogen | #11203D; RRID: AB_2783009 |
| Dynabeads M-280 Sheep Anti-Mouse IgG | Invitrogen | #11202D; RRID: AB_2783640 |
| Anti-HA | Millipore | #05-904; RRID: AB_11213751 |
| Anti-cMyc | EMD Millipore | #05-724; RRID: AB_11211891 |
| Anti-GFP | Abcam | #ab290; RRID: AB_303395 |
| Experimental Models: Organisms/Strains | ||
| Library of random sheared yeast genomic DNA in pACTII | This paper | N/A |
| Y, a, mps3Δ240-430 | This paper | MCY 1456 |
| X, alpha, csm4::LEU2 | This paper | MCY 1536 |
| Y, a, csm4::LEU2 | This paper | MCY 1539 |
| Y, a, mps2ΔC6-TRP1 | This paper | MCY 1885 |
| X, alpha, mps2ΔC6-TRP1 | This paper | MCY1886 |
| X, alpha, MYO5-GFP-TRP1 | This paper | MCY 2037 |
| Y, a, MYO5-GFP-TRP1 | This paper | MCY 2038 |
| X, alpha, myo3::TRP1 myo5::TRP1 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 | This paper | MCY 2083 |
| Y, a, myo3::TRP1 myo5::TRP1 leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1- GFP/lacI at LYS2 | This paper | MCY 2092 |
| X, a, MYO1-GFP-TRP1 | This paper | MCY 2134 |
| Y, alpha, MYO1-GFP-TRP1 | This paper | MCY 2135 |
| Y, a, kanMX-CLB2-Ubi-MYO1 | This paper | MCY 2146 |
| X, alpha, kanMX-CLB2-Ubi-MYO1 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MCY 2147 |
| X, a, GFP-myc-MPS2-URA3 csm4::LEU2 | This paper | MCY 2213 |
| Y, alpha, GFP-myc-MPS2-URA3 csm4::LEU2 | This paper | MCY 2214 |
| X, a, GFP-myc-MPS2-URA3 ndj1::TRP1 | This paper | MCY 2215 |
| Y, alpha, GFP-myc-MPS2-URA3 ndj1::TRP1 | This paper | MCY 2216 |
| X, alpha, GFP-myc-MPS2-URA3 csm4::LEU2 ndj1::LEU2 | This paper | MCY 2217 |
| Y, a, GFP-myc-MPS2-URA3 csm4::LEU2 ndj1::LEU2 | This paper | MCY 2218 |
| X, alpha, GFP-myc-MPS2-URA3 | This paper | MCY 2219 |
| Y, a, GFP-myc-MPS2-URA3 | This paper | MCY 2220 |
| Y, a, myo4::TRP1 leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MCY 2241 |
| X, alpha, myo4::TRP1 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MCY 2244 |
| X, alpha, GFP-myc-mps2ΔC6-TRP1 | This paper | MCY 2249 |
| Y, a, GFP-myc-mps2ΔC6-TRP1 | This paper | MCY 2251 |
| Y a kanmx-GAL-HA-MYO3 | This paper | MCY 2387 |
| X, alpha, kanmx-GAL-HA-MYO3 | This paper | MCY 2388 |
| X, alpha, MYO4-GFP-TRP1 | This paper | MCY 2443 |
| Y, a, MYO4-GFP-TRP1 | This paper | MCY 2444 |
| Y, alpha, mps3Δ240-430 mps2ΔC6-TRP1 | This paper | MCY 2484 |
| X, a, mps3Δ240-430 mps2ΔC6-TRP1 | This paper | MCY 2486 |
| X, alpha, GAL-MYO2-DN-LEU2 GAL4-ER-URA3 | This paper | MCY 2541 |
| Y, a, GAL-MYO2-DN-LEU2 GAL4-ER-URA3 | This paper | MCY 2543 |
| Y, a, mps2ΔC6-TRP1 leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MCY 2722 |
| X, alpha, mps2ΔC6-TRP1 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MCY 2728 |
| Y, alpha, GAL4-ER-URA3 GAL-GST-MYO2-TRP1 leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MCY 3190 |
| X, a, GAL4-ER-URA3 GAL-GST-MYO2-TRP1 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MCY 3192 |
| Y, alpha, GAL4-ER-URA3 GAL-MYO2-DN-TRP1 ndt80::HPH leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MCY 3245 |
| X, a, GAL4-ER-URA3 GAL-MYO2-DN-TRP1 ndt80::HPH Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MCY 3247 |
| X, a, ade5°canR CYHs his7-2 leu2 lys2-1° met13-d trp1–63 tyr1-1(−2?) ura3-13(PER1+) GAL+ (wild-type) | This paper | MDY 1480 |
| Y, alpha, ade5°canR CYHs his7-2 leu2 lys2-1° met13-d trp1–63 tyr1-1(−2?) ura3-13 (PER1+) GAL+ (wild-type) | This paper | MDY 1508 |
| Y, alpha, leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MDY 1567 |
| X, a, Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MDY 2426 |
| X, a, csm4::LEU2 Pdmc1-I/GFP-URA3 at Ura3, Pcyc1-I/GFP-URA3 atCyc1, Laco256-LEU2 at Chr4telo | This paper | MDY 2609 |
| X, alpha, mps3Δ240-430 | This paper | MDY 2661 |
| Y, alpha, csm4::LEU2 leu1 TUB1/GFP lacO256-LEU2-IVRtelo P-DMC1-GFP/lacI at LYS2 | This paper | MDY 2778 |
| X, a, Myo2-GFP-TRP1 mps2ΔC6-TRP1 | This paper | MDY 3554 |
| Y, alpha, Myo2-GFP-TRP1 mps2ΔC6-TRP1 | This paper | MDY 3557 |
| X, alpha, GFP-myc-MPS2-URA3 Mps3-HA-TRP1 | This paper | MDY 4027 |
| Y, a, GFP-myc-MPS2-URA3 Mps3-HA-TRP1 | This paper | MDY 4030 |
| Software and Algorithms | ||
| ImageJ | [66] | https://imagej.nih.gov/ij/ |
| AxioVision | Zeiss | https://www.zeiss.com/ |
| Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| OMRFQUANT | Our laboratory | N/A |
Highlights.
Mps2 connects the LINC complex to the cytoskeleton
Myo2 interacts with Mps2 and connects the telomer to cytoskeleton actin filaments
Csm4 works together with Mps2 and Myo2 to regulate rapid-prophase movements
ACKNOWLEDGMENTS
We thank Robin Harris for assistance in the yeast two-hybrid assays. We thank Dean Dawson for advice and discussion. We thank Angela Andersen from Life Science Editors editorial services for her assistance in manuscript writing. R.J.P. received financial support from NIH/NIGMS (R01 GM125803), and C.G.B. was supported by Fulbrigth-Conicet.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.01.054.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hassold T, Hunt PA, and Sherman S (1993). Trisomy in humans: incidence, origin and etiology. Curr. Opin. Genet. Dev 3, 398–403. [DOI] [PubMed] [Google Scholar]
- 2.Hassold T, Sherman S, and Hunt PA (1995). The origin of trisomy in humans. Prog. Clin. Biol. Res 393, 1–12. [PubMed] [Google Scholar]
- 3.Hassold T, and Hunt P (2001). To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet 2, 280–291. [DOI] [PubMed] [Google Scholar]
- 4.Hassold T, Hall H, and Hunt P (2007). The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet 16, R203–R208. [DOI] [PubMed] [Google Scholar]
- 5.Hunt PA, and Hassold TJ (2008). Human female meiosis: what makes a good egg go bad? Trends Genet. 24, 86–93. [DOI] [PubMed] [Google Scholar]
- 6.Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang KT, et al. (2013). The LINC complex is essential for hearing. J. Clin. Invest 123, 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alleva B, and Smolikove S (2017). Moving and stopping: regulation of chromosome movement to promote meiotic chromosome pairing and synapsis. Nucleus 8, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiraoka Y, and Dernburg AF (2009). The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605. [DOI] [PubMed] [Google Scholar]
- 9.Koszul R, and Kleckner N (2009). Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 19, 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao KT, and Ellingson DJ (1969). Observations on nuclear rotation and oscillation in Chinese hamster germinal cells in vitro. Exp. Cell Res 55, 39–42. [DOI] [PubMed] [Google Scholar]
- 11.Lee CY, Horn HF, Stewart CL, Burke B, Bolcun-Filas E, Schimenti JC, Dresser ME, and Pezza RJ (2015). Mechanism and regulation of rapid telomere prophase movements in mouse meiotic chromosomes. Cell Rep. 11, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibuya H, Morimoto A, and Watanabe Y (2014). The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 10, e1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheehan MJ, and Pawlowski WP (2009). Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc. Natl. Acad. Sci. USA 106, 20989–20994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trelles-Sticken E, Dresser ME, and Scherthan H (2000). Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol 151, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, and Kaback DB (2007). Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104, 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, and Dresser ME (2008). Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133, 1175–1187. [DOI] [PubMed] [Google Scholar]
- 17.Koszul R, Kim KP, Prentiss M, Kleckner N, and Kameoka S (2008). Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133, 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christophorou N, Rubin T, Bonnet I, Piolot T, Arnaud M, and Huynh JR (2015). Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol 17, 1388–1400. [DOI] [PubMed] [Google Scholar]
- 19.Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, and Hiraoka Y (1994). Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273. [DOI] [PubMed] [Google Scholar]
- 20.Wynne DJ, Rog O, Carlton PM, and Dernburg AF (2012). Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J. Cell Biol 196, 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, and Jantsch V (2010). Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 6, e1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickards GK (1975). Prophase chromosome movements in living house cricket spermatocytes and their relationship to prometaphase, anaphase and granule movements. Chromosoma 49, 407–455. [DOI] [PubMed] [Google Scholar]
- 23.Long J, Huang C, Chen Y, Zhang Y, Shi S, Wu L, Liu Y, Liu C,Wu J, and Lei M (2017). Telomeric TERB1-TRF1 interaction is crucial for male meiosis. Nat. Struct. Mol. Biol 24, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rog O, and Dernburg AF (2015). Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 10, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, and Jantsch V (2007). The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12, 873–885. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya H, Ishiguro K, and Watanabe Y (2014). The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol 16, 145–156. [DOI] [PubMed] [Google Scholar]
- 27.Shibuya H, Hernández-Hernández A, Morimoto A, Negishi L, Höög C, and Watanabe Y (2015). MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163, 1252–1266. [DOI] [PubMed] [Google Scholar]
- 28.Horn HF, Kim DI, Wright GD, Wong ES, Stewart CL, Burke B, and Roux KJ (2013). A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol 202, 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, and Watanabe Y (2012). A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol 198, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, and Dernburg AF (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrador L, Barroso C, Lightfoot J, Müller-Reichert T, Flibotte S, Taylor J, Moerman DG, Villeneuve AM, and Martinez-Perez E. (2013). Chromosome movements promoted by the mitochondrial protein SPD-3 are required for homology search during Caenorhabditis elegans meiosis. PLoS Genet. 9, e1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labella S, Woglar A, Jantsch V, and Zetka M (2011). Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell 21, 948–958. [DOI] [PubMed] [Google Scholar]
- 33.Lee CY, Conrad MN, and Dresser ME (2012). Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 8, e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Garcia M, Schubert V, Osman K, Darbyshire A, Sanchez-Moran E, and Franklin FCH (2018). TOPII and chromosome movement help remove interlocks between entangled chromosomes during meiosis. J. Cell Biol 217, 4070–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, and Hodzic D (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fridkin A, Penkner A, Jantsch V, and Gruenbaum Y (2009). SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell. Mol. Life Sci 66, 1518–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razafsky D, and Hodzic D (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol 186, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr DA, and Fridolfsson HN (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol 26, 421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trelles-Sticken E, Adelfalk C, Loidl J, and Scherthan H (2005). Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol 170, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad MN, Lee CY, Wilkerson JL, and Dresser ME (2007). MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104, 8863–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad MN, Dominguez AM, and Dresser ME (1997). Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276, 1252–1255. [DOI] [PubMed] [Google Scholar]
- 42.Chua PR, and Roeder GS (1997). Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11, 1786–1800. [DOI] [PubMed] [Google Scholar]
- 43.Starr DA, and Fischer JA (2005). KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136–1146. [DOI] [PubMed] [Google Scholar]
- 44.Jaspersen SL, Martin AE, Glazko G, Giddings TH Jr., Morgan G, Mushegian A, and Winey M (2006). The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol 174, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beilharz T, Egan B, Silver PA, Hofmann K, and Lithgow T (2003). Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem 278, 8219–8223. [DOI] [PubMed] [Google Scholar]
- 46.Kosaka H, Shinohara M, and Shinohara A (2008). Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 4, e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, and Alani E (2008). Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4, e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonntag Brown M, Zanders S, and Alani E (2011). Sustained and rapid chromosome movements are critical for chromosome pairing and meiotic progression in budding yeast. Genetics 188, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schramm C, Elliott S, Shevchenko A, and Schiebel E (2000). The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muñoz-Centeno MC, McBratney S, Monterrosa A, Byers B, Mann C, and Winey M (1999). Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell 10, 2393–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kutay U, Hartmann E, and Rapoport TA (1993). A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72–75. [DOI] [PubMed] [Google Scholar]
- 52.Wattenberg B, and Lithgow T (2001). Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2, 66–71. [DOI] [PubMed] [Google Scholar]
- 53.Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, and Kodadek T (1997). DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Masson I, Saveanu C, Chevalier A, Namane A, Gobin R, Fromont-Racine M, Jacquier A, and Mann C (2002). Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol 43, 1431–1443. [DOI] [PubMed] [Google Scholar]
- 55.Eves PT, Jin Y, Brunner M, and Weisman LS (2012). Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J. Cell Biol 198, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pashkova N, Jin Y, Ramaswamy S, and Weisman LS (2006). Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 25, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lillie SH, and Brown SS (1994). Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol 125, 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reck-Peterson SL, Novick PJ, and Mooseker MS (1999). The tail of a yeast class V myosin, myo2p, functions as a localization domain. Mol. Biol. Cell 10, 1001–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamin KR, Zhang C, Shokat KM, and Herskowitz I (2003). Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17, 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Ajimura M, Padmore R, Klein C, and Kleckner N (1995). NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol 15, 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berben G, Dumont J, Gilliquet V, Bolle PA, and Hilger F (1991). The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7, 475–477. [DOI] [PubMed] [Google Scholar]
- 62.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 63.Sikorski RS, and Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yumura S, Mori H, and Fukui Y (1984). Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol 99, 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conchello JA, and Dresser ME (2007). Extended depth-of-focus microscopy via constrained deconvolution. J. Biomed. Opt 12, 064026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze data/code.


