Abstract
Advances in our understanding of molecular mechanisms of tumorigenesis have translated into knowledge-based therapies directed against specific oncogenic signaling targets. These therapies often induce dramatic responses in susceptible tumors. Unfortunately, most advanced cancers, including those with robust initial responses, eventually acquire resistance to targeted therapies and relapse. Even though immune-based therapies are more likely to achieve complete cures, acquired resistance remains an obstacle to their success as well. Acquired resistance is the direct consequence of pre-existing intratumor heterogeneity and ongoing diversification during therapy, which enables some tumor cells to survive treatment and facilitates the development of new therapy resistant phenotypes. In this review, we discuss the sources of intratumor heterogeneity and approaches to capture and account for it during clinical decision making. Finally, we outline potential strategies to improve therapeutic outcomes by directly targeting intratumor heterogeneity.
Tumors are composed of millions of cancer cells embedded in a microenvironment distorted by neoplastic changes. The startling heterogeneity of both cancer cells and cells composing the tumor microenvironment fuels disease progression and treatment resistance. Thus, better understanding of cellular, molecular, and spatial heterogeneity within tumors and the application of this knowledge for treatment design are essential for improving clinical outcomes. In this review we summarize sources of intratumor heterogeneity (ITH), methods to quantitatively assess ITH, and its clinical relevance. While most of the points discussed are applicable to all types of cancers, some, such as spatial and microenvironmental heterogeneity, are more relevant to solid tumors, which represent our main focus.
Sources of intratumor heterogeneity
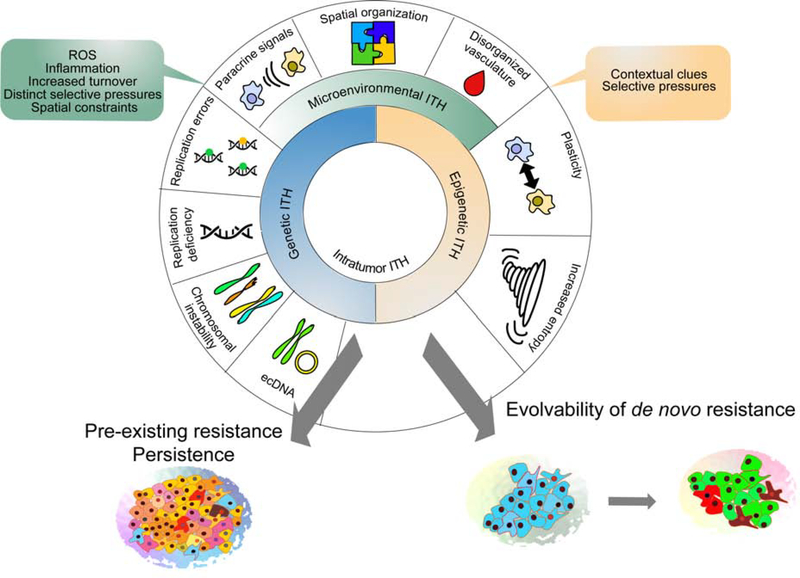
Cellular phenotypic heterogeneity within tumors is a complex, multifactorial phenomenon, which integrates genetic, epigenetic, and environmental inputs (Figure 1).
Figure 1. Sources of Intratumor heterogeneity.
Intratumor heterogeneity represents integration of inputs from genetic, phenotypic, and microenvironmental heterogeneity, in turn increasing the odds of both pre-existence of tolerant and resistant subpopulations, and the ability to evolve new adaptations.
Genetic heterogeneity is the most studied and best understood aspect of intratumor heterogeneity (ITH), although our understanding is still far from being sufficiently complete (McGranahan and Swanton, 2017). Despite mounting challenges, gene and mutation-centric focus remains at the core of molecular oncology, and the majority of cancer researchers would probably still agree with the famous statement by Bert Vogelstein: “The revolution in cancer research can be summed up in a single sentence: cancer is, in essence, a genetic disease” (Vogelstein and Kinzler, 2004). Discovering tumor-associated genetic mutations and interrogating their functional and clinical impact has been the major focus of cancer research over the last few decades, and the recent revolution in DNA sequencing technologies enabled an outpour of studies documenting startling intratumor genetic heterogeneity.
Even though eukaryotic cells replicate their DNA with astounding fidelity, the mechanism is not entirely error-free. Every time a cell divides, a few mutational errors in the form of nucleotide substitutions and small deletions are introduced even in the absence of internal and external mutagens (Zhang and Vijg, 2018). Owing to the constant turnover of tumor cells and the large size of tumor cell populations, some of these stochastic mutational hits inevitably affect genes with known cancer relevance, leading to the activation of oncogenes and inactivation of tumor suppressors. Whereas the sufficiency of these baseline replication error rates to account for carcinogenesis remains a subject of debate (Beckman and Loeb, 2006; Tomasetti and Vogelstein, 2015; Tomlinson et al., 1996), many tumors display increased mutation rates, due to either increased external or internal mutagen exposure. These include UV-related mutagenesis in skin cancers; tobacco-related mutagenesis in oral, lung, and bladder cancers; reactive oxygen species (ROS) induced mutagenesis in a wide range of malignancies, and defects in DNA repair mechanisms, which have been detected in certain tumors (e.g., deficiencies in DNA mismatch repair and APOBEC pathways) (Alexandrov et al., 2013; Fox et al., 2013; Kandoth et al., 2013).
Small-scale genetic mutations remain best understood and easiest to study, but they represent only a tip of the iceberg of genetic heterogeneity. The vast majority of spontaneous human cancers display aneuploidy, which is linked with chromosomal instability (CIN) – an increased rate of genomic mutational errors involving loss, gains, and translocations of large fragments of genomic DNA (Bakhoum and Cantley, 2018). Aneuploid cells commonly emerge following whole genome doubling (WGD) due to mitotic failure leading to tetraploidization (Dewhurst et al., 2014). WGD is an early event commonly observed across most human cancer types and it is associated with poor prognosis (Bielski et al., 2018). Compared to point mutations, these large-scale genomic events are much more likely to impact cellular phenotypes. While reduced cell fitness is the most common outcome of large-scale genomic changes, CIN enables much faster rates of genomic diversification, thus speeding up evolution during both tumor development and cancer therapies (Tang and Amon, 2013). At the same time aneuploid cells also influence their microenvironment and may trigger a specific anti-tumor immune response, which in turn may facilitate immune evasion through upregulation of immune suppressive mechanisms (Senovilla et al., 2012).
The functional relevance of genomic heterogeneity in tumor progression and therapy resistance remains poorly understood. The prevalent approach is to reduce the impact of large-scale gains to changes in copy numbers (and related expression changes) of individual “driver” genes, with known cellular functions. While in cases of complete loss of functional alleles or focal massive amplification this approach is justified, in many cases the impact of copy number alteration on the expression of an individual gene is relatively subtle. Most of these large genomic changes involve many genes, leading to significant impacts on gene regulatory networks which cannot be reduced to individual “drivers” (Tang and Amon, 2013). Furthermore, in contrast to point mutations, which are almost always irreversible, large scale DNA copy number and structural variations (with the exception of homozygous deletions) are typically much less stable, due to much higher rates of genomic mutations in aneuploid cancer cells (10−2 per chromosome per generation versus 10−7 per gene per cell division) (Lengauer et al., 1997), which might significantly complicate sub-clonal reconstruction analyses. Notably, karyotypic analyses revealed significant cell-to-cell variability of chromosomal numbers in cancer cell lines that maintained ostensibly stable average karyotypes (Li et al., 2009). The instability of copy number variations is most pertinent for extrachromosomal DNA (ecDNA), which has been shown to mediate inheritance of many “driver” genes across multiple cancers (deCarvalho et al., 2018; Turner et al., 2017; Verhaak et al., 2019). Paralleling plasmids in bacteria, this extrachromosomal inheritance enables massive focal amplification of an oncogene, while avoiding fitness penalties, associated with genomic changes in larger pieces of DNA. Whereas the exact mechanisms and inheritance patterns remain to be elucidated, even entirely stochastic distribution of ecDNA can dramatically accelerate tumor evolution and intratumor heterogeneity (Verhaak et al., 2019).
While genetic heterogeneity within populations of tumor cells is shaped by the several mechanisms of genetic diversification described above, they are generally thought to be lacking a powerful diversification mechanism observed in natural populations throughout all taxa of life – (para)sexual recombination. However, the assumption of strict asexuality of cancers might be incorrect. Several publications, as well as our unpublished work have documented evidence of spontaneous cell fusions between genetically distinct cancer cells, or between cancer and non-cancer cells within the tumor microenvironment (Gast et al., 2018; Jacobsen et al., 2006; Lu and Kang, 2009). Genomes of hybrid cells are often unstable (Duelli et al., 2007; Storchova and Pellman, 2004), which could lead to additional diversification associated with ploidy reduction, similar to a fusion-mediated ploidy conveyor in mature hepatocytes (Duncan et al., 2010) or asexual recombination observed in C. Albicans (Zhang et al., 2015). As the mechanistic underpinning of spontaneous cell fusions remains poorly explored, it is unclear whether it is related to the cell-in-cell phenomenon, cell cannibalism and entosis that have been observed in numerous cancer types upon various stress conditions (Fais and Overholtzer, 2018). Whereas the relevance of this experimentally observed phenomenon is difficult to access in primary tumors (apart from cases of tumors in recipients of tissue transplants, such as described in case reports of a bone marrow transplant recipients) (LaBerge et al., 2017; Lazova et al., 2013), mathematical modeling suggests that fusion-mediated recombination can further enhance diversity in populations of tumor cells (ref to be provided after BioRxiv deposition).
One reason why progress has been rather slow with understanding the links between genetic heterogeneity and tumor evolution is the paucity of suitable experimental models that reproduce the ITH of human tumors. Conventional genetically engineered mouse models (GEMM) are dominated by those driven by powerful combination mutations within a single cell, providing convenient and reproducible experimental systems to study molecular mechanisms. However, these models rarely display the degree of subclonal and cellular genetic heterogeneity seen in spontaneous cancers. However, next-generation GEMM are designed to more accurately mimic human cancers, including ITH (Kersten et al., 2017). Among others, the KPC mouse model of pancreatic adenocarcinoma (PAD) driven by mutant Kras and Trp53 transgenes displays extensive subclonal heterogeneity largely driven by large-scale copy number alterations targeting genes with known functional relevance in human PAD (Niknafs et al., 2019). Another promising GEMM based approach to understand the impact of ITH is to combine low-penetrance oncogenic drivers with a source of genetic diversification, such as Sleeping Beauty transposons (Mann et al., 2016). The use of such models will enable more detailed studies assessing the functional relevance of subclonal interactions in tumor evolution, which then enable the design of improved therapeutic strategies for heterogeneous tumors.
Epigenetic heterogeneity
Studies on tumor heterogeneity have primarily focused on cancer cells genomes even though selection forces act on phenotypes rather than genotypes. Despite the critical importance of cancer-associated mutations on clinically relevant phenotypic features, such as responses to growth signaling, proliferation, and death, epigenetic (we use the term in a broader sense, covering all non-genetic phenotypic determinants) mechanisms have a greater impact on tumor cell phenotypes (Flavahan et al., 2017). For example, the gene expression and epigenetic profiles of genetically abnormal CD44+CD24- progenitor-like cancer cells isolated from primary breast tumors shows closer resemblance to the profiles of normal CD44+CD24- mammary epithelial cells than to that of more differentiated CD44-CD24+ cancer cells from the same tumor; the same is true for CD44-CD24+ cells (Shipitsin et al., 2007). These data suggest that differentiation state-related epigenetic programs have a dominant impact in shaping phenotypes compared to cancer-related genetic aberrations. Similar observations have been made in brain tumors, which represent one of the best examples of cancer stem cell-driven malignancies (Liau et al., 2017; Neftel et al., 2019; Patel et al., 2014). However, more differentiated cancer cells can give rise to stem cell-like cancer cells through epithelial-tomesenchymal transition (EMT) triggered by certain microenvironmental signals such as hypoxia (Mohlin et al., 2017) or interaction with stromal cells (Karnoub et al., 2007). Thus, tumor cells display high epigenetic and phenotypic plasticity. This phenotypic plasticity is likely to be key for the phenomena of drug-tolerant persistence, as well as the ability of cells to acquire resistance through stable non-genetic changes in gene expression, as we will elaborate below.
While epigenetic heterogeneity could be key in acquisition of traits of profound clinical importance, such as therapeutic resistance (Shaffer et al., 2017) or metastatic dissemination (Roe et al., 2017), accounting for non-genetic heterogeneity is far more challenging since, in contrast to genetic heterogeneity, the phenotypes of tumor cells can be highly plastic. Epigenetically defined phenotypic traits range from essentially hard-wired ones, which can be conceptualized as epimutations, such as silencing of key tumor suppressor genes, mediated by DNA hypermethylation on one end of the spectrum, to noise-driven cell-to-cell differences that dissipate within a few cell divisions on the other end of the spectrum (Marusyk et al., 2012). Whereas these two extremes are conceptually the easiest to deal with, the majority of non-genetic phenotypic heterogeneity, reflecting integration of microenvironmental inputs and stochastic events and differentiation status, falls in the gray zone in terms of heritability.
Studies assessing epigenetic heterogeneity within tumors has been largely focusing on DNA methylation, since this is technically less challenging to measure even at single cell level than chromatin modification (Mazor et al., 2016). Due to the reversible nature of epigenetic modifications, it was not clear if they can be used to define subclones, track tumor evolution, and assess intratumor subclonal and cellular heterogeneity. However, studies in prostate cancer (Brocks et al., 2014), glioma (Mazor et al., 2015), and chronic lymphocytic leukemia (CLL) (Gaiti et al., 2019) have demonstrated that inferring tumor evolution based on genetic and DNA methylation patterns largely overlaps. Furthermore, quantitative measures of ITH in Barrett’s esophagus based on DNA methylation and genetic mutations were both predictive of the progression to esophageal carcinoma implying that diversity is an inherent tumor characteristic and heritable epigenetic changes have similar impact on tumor evolution as genetic ones (Merlo et al., 2010).
ITH for DNA methylation has consistently been observed in regulatory regions that affect the transcription of genes relevant to the disease process. For example, in prostate cancer high ITH is observed for enhancers bound by the androgen receptor (AR) (Brocks et al., 2014), a ligand-dependent transcription factor driving the expression of many genes essential for the survival and proliferation of prostate tumor cells. Similarly, in CLL binding sites for transcription factors with known functional relevance such as NFKB1 and MYBL1 displayed lower epimutation potentially due to the protection of these sites from epimutations by transcription factor binding (Gaiti et al., 2019). Advances in technologies have enabled the single-cell multiomic profiling of tumors that allows the direct analyses between genetic and epigenetic alterations and gene expression profiles. A recent study performing single cell RNA-seq (scRNA-seq) and single cell ATAC-seq (scATAC-seq) profiling of mixed-phenotype acute leukemias (MPALs) have demonstrated common malignant signatures despite extensive heterogeneity across patients and within individual cases, and identified the RUNX1 transcription factor as a key regulator of these programs (Granja et al., 2019).
Given that tumor cells display clear parallels with differentiation hierarchy related phenotypic heterogeneity in normal tissues, the cancer stem cell framework has been, and still remains, a popular paradigm to conceptualize the ITH. However, phenotypic ITH cannot be reduced to differentiation hierarchies, as tumor cells display far greater cell-to-cell variability compared to their differentiation state normal counterparts (Jenkinson et al., 2017; Landau et al., 2014). This increased phenotypic variability likely reflects the impact of intratumor genetic heterogeneity, as well as greater variability in contextual signals provided by aberrant and less structured (compared to normal tissues) tumor microenvironments. In addition to increased plasticity and the diversifying inputs of genetic and microenvironmental heterogeneity, this increased cell-to-cell variability might also be a consequence of global epigenetic changes, generally observed in cancer cell epigenomes (such as global hypomethylation of CpG islands) (Feinberg, 2007), and can be summarized as higher entropy of the cancer epigenome (Jenkinson et al., 2017).
Microenvironmental heterogeneity
Though heterogeneity of genotypes within tumors translates into diversity of phenotypes, phenotypes are not simple functions of genotypes. Indeed, all normal cells share identical wild type genomes, which encodes highly diverse phenotypic manifestations. Phenotypic diversity in normal cells reflects developmental processes triggered by responses to microenvironmental cues. Whereas tumor cells often display aberrant responses to microenvironmental signals (e.g., suppression of death from anoikis), they still retain a large repertoire of normal responses. An extreme example is the ability of some tumor cells to become a functional part of normal tissues, when embedded in normal microenvironments. For example, human breast cancer cell lines co-injected with normal mammary epithelial cells into the mammary fat pads will become part of a functional mammary epithelium (Bussard and Smith, 2012).
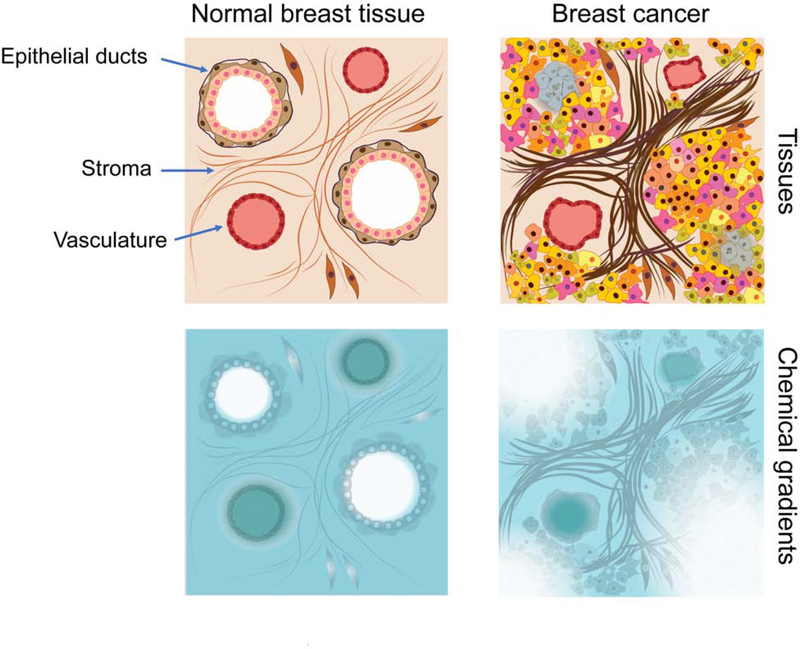
On the other hand, tumor formation entails not only genetic and epigenetic transformation of normal cells, but also highly aberrant microenvironments. Normal tissues are organized in functional and structural units with near-equal access to vasculature, which provides oxygen and nutrients while removing waste products. For example, breast epithelium is organized as two layers – basal and luminal layer, with cells facing either basal membrane, or lumen (Figure 2). As blood and lymphatic vasculature resides in sparsely populated connective tissue, both cell layers obtain nearly-equal access to fully diffusible factors (such as oxygen), while cellular access to contact signaling from the extracellular matrix (ECM) components of basal membrane, as well as to stroma-produced cytokine, is primarily confined to the basal layer. Perturbations in normal tissue architecture due to aging and chronic inflammation contribute to increased cancer risk and tumor progression (Fane and Weeraratna, 2019), and tissue organization is completely lost in invasive and metastatic tumors. These changes result in cellular and paracrine interactions not observed in healthy tissues, which contribute to selection and further diversify cancer cell phenotypes. While some tumor cells are still in contact with stroma-derived extracellular matrix, its composition is altered compared to normal tissues (Naba et al., 2012), and many tumor cells are separated from the stroma by more than one cell layer. Moreover, blood and lymphatic vasculature in tumors are disorganized with significant functional, spatial, and temporal heterogeneity (Carmeliet and Jain, 2000; Stacker et al., 2014). These perturbations in both stromal and epithelial organization leads to significant spatial and temporal variability in nutrients, oxygenation, growth factors, and pH (Korenchan and Flavell, 2019; Yuan, 2016), in turn providing diverse and abnormal contextual signals, which are absent in healthy normal tissues. As sensing environmental cues integrates a multitude of inputs, existence of different spatial and temporal gradients of microenvironmental components might lead to dramatic, combinatorial diversification of contextual signals, thus inducing phenotypic ITH, reflective of cellular responses to these contextual signals, rather than specific well-defined phenotypes (Figure 2).
Figure 2. Microenvironmental heterogeneity.
In normal tissues (such as normal breast) most epithelial cells experience similar concentrations of nutrients, oxygen, and growth factors. However, structural disorganization of epithelia, stroma, and vasculature leads to significant inequality in concentrations of these factors (one factor is illustrated). Further, differences in diffusion and consumption rates of different factors combinatorialy increase variability in microenvironmental cues directly influencing phenotypic heterogeneity and creating distinct selection forces.
In addition to diversification of contextual cues, structural and microenvironmental disorganization in carcinogenesis also creates relatively consistent new microenvironments, leading to predictable association of tumor cell phenotypes with these microenvironmental features. For example, local acidification at the tumor edge drives mesenchymal transition, mediating tissue invasion (Estrella et al., 2013). There is a striking similarity between the arrangement of cancer cells around blood vessels and vegetation around waterways in desert regions (Alfarouk et al., 2013). In addition to directly influencing tumor cell phenotypes, predictably distinct microenvironmental habitats could provide distinct selective pressures (Gatenby and Gillies, 2008). Several studies have linked genetic heterogeneity with distinct habitat locations (Gillies et al., 2018; Hoefflin et al., 2016; Lloyd et al., 2016) and have shown that spatial distribution of genetically distinct tumor cell populations correlates with poor clinical outcome (Janiszewska et al., 2015).
Microenvironmental heterogeneity also involves heterogeneity in immune cell infiltration, which is of obvious importance for immunotherapies. Leukocytes are frequently one of the most abundant cell types within tumors and their highly mobile nature can lead to rapidly changing spatial heterogeneity that can create immunologically active or silent niches. Because T cells can directly eliminate certain cancer cells or even populations of cancer cells with specific markers, it is not surprising that the frequency and location of T cells have directly been linked to subclonal heterogeneity in cancer. Furthermore, since T cells are activated by specific tumor neoantigens, many of which are generated by tumor-specific mutations, the location of T cells with specific TCRs also varies within tumors and correlates with the number and types of mutations. For example, in non-small lung cancer (NSLC) assessing mutations and TCR diversity in different regions of the same tumor demonstrated that the abundance of expanded intratumor ubiquitous TCRs is associated with the number of nonsynonymous mutations (Joshi et al., 2019). At the same time the number of expanded regional TCRs correlated with the number of regional nonsynonymous mutations.
In serous ovarian cancer metastases, higher epithelial CD8+ T cell infiltration was associated with lower tumor ITH presumably due to immune-mediated elimination of certain subclones present as minor subpopulations (Zhang et al., 2018). Subclonal neoepitopes have been shown to be more immunogenic than clonal ones (Jimenez-Sanchez et al., 2017) potentially explaining these results. In some cases, immune evasion was due to HLA LOH in the tumor cells coupled with upregulation of immune checkpoint inhibitors. Interestingly, while tumor regions with high CD8+ and CD4+ T cell densities had high TCR diversity, this was not associated with tumor cell ITH implying that T cell and tumor cell cellular heterogeneity are related but distinct features.
Thus, phenotypes of tumor cells are shaped by an integration genetic, epigenetic and microenvironmental inputs (Figure 1). Whereas we have introduced these inputs separately, they are highly interrelated. For example, microenvironmental inflammation has been linked with increased chromosomal instability and increased epigenetic plasticity (Colotta et al., 2009; Grivennikov et al., 2010; Korkaya et al., 2011). On the other hand, chromosomal instability has been linked to the induction of inflammatory signaling through cGAS-STING anti-viral pathway (Bakhoum and Cantley, 2018), thus modulating microenvironments, including immune responses. The resulting phenotypic ITH translates to heterogeneity in the sensitivity of tumor cells to anti-cancer drugs and cytotoxic immunity. Additionally, some of the heterogeneous features of the tumor microenvironment can directly modulate therapeutic responses as described below.
Quantitative assessment of intratumor heterogeneity
ITH in tissue samples
Even though all types of ITH have been linked to poor patient outcomes and therapeutic resistance in many different tumor types, assessing heterogeneity in human tissue samples is still a major challenge, which partially explains why ITH is still not commonly used to guide clinical decisions. One of the best examples for this is ITH for ERBB2 (encoding HER2) amplification in HER2+ breast cancer driven by oncogenic HER2 signaling. HER2-targeted therapies have been very successful for HER2+ tumors, but they are only effective when the tumor cells uniformly express and are dependent on HER2. However, in many cases HER2 expression and ERBB2 amplification is not homogeneous within tumors and this has been associated with shorter disease-free survival (Rye et al., 2018). Assessing HER2 status by immunohistochemistry and FISH has long been used in the clinic for the identification of patients who will most likely benefit from HER2-targeted therapies. However, due to the increasing recognition of ITH, HER2 heterogeneity, defined as HER2 positivity by FISH in > 5% and < 50% of tumor cells, has also been incorporated into the report. Considering that HER2 status is determined by automated counting of FISH signal in 50 single cells, this assay could relatively easily be expanded into a more quantitative assessment of cellular heterogeneity for HER2, and several clinical trials are ongoing (NCT02326974) to determine if this could potentially improve the clinical management of patients. Preliminary analysis of the data indicate that patients with higher pretreatment cellular heterogeneity for ERBB2 amplification are less likely to have a complete pathologic response to neoadjuvant pertuzumab and T-DM1 treatment (Filho et al., 2019).
Quantitative assessment of cell-to-cell variation in the expression of a therapeutic target at the protein level is much harder to evaluate, as it is usually measured using immunostaining and manually scored by pathologists. Scoring discordance between individuals and variation between different batches of staining contribute to limited reporting of the observed heterogeneity. However, technological advances in multiplex immunostaining techniques, digital pathology, and machine learning are paving the way to more accurate reporting of ITH.
Decreased expression of the target is only one of the many mechanisms by which tumors evade therapy. Many known resistance mechanisms are related to genetic alterations that are identified based on unbiased genome-wide analyses. Several computational approaches have been developed to use next-generation sequencing read frequency to assess ITH and infer clonal evolution of a tumor. Multiregional sequencing of renal carcinoma, glioblastoma and lung cancer have all showed significant divergence between distinct areas of the same tumor (Gerlinger et al., 2012; Thrane et al., 2018; Yates et al., 2015; Zhang et al., 2014). While evolutionary trajectories can be drawn between distinct clones at different sites, clinical benefit of these analyses remains to be determined. The relatively high cost of exome-sequencing makes it challenging to perform these analyses for all patients. A large clinical trial (NCT01888601) was initiated in 2014 for non-small cell lung cancer (NSCLC) patients, aiming at multiregional and longitudinal sampling of primary and recurrent tumors to determine the impact of ITH on clinical outcomes. Initial results from this study show that ITH for copy number alterations and genomic instability are associated with increased risk of recurrence or death (Jamal-Hanjani et al., 2017). Moreover, immune cell heterogeneity within tumors point to a correlation between T cell clonality and tumor antigen diversity in different regions of the same tumor (Reuben et al., 2017). Advances leading to decrease of sequencing costs and streamlined bioinformatic analysis will be required for the more wide-spread use of genome-wide approaches for routine ITH assessment for patient prognostication.
Regional sampling of heterogeneous tumors remains one of the largest challenges in clinical diagnostics. In certain tumor types, such as lung or breast cancer, biopsies from several distinct regions may be taken. However, in glioblastoma and pancreatic cancer, additional sample collection is associated with higher risk for the patient. Multiregional sequencing has invariably showed regional heterogeneity in different tumor types, raising many questions. How representative is a single biopsy? How many biopsies would we need to collect to cover the extent of heterogeneity of a large highly aggressive tumor? An important tool in addressing these seemingly impossible issues is “liquid biopsy”: sampling blood and other body fluids.
ITH in liquid biopsies
Blood drawn from cancer patients contains circulating tumor cells (CTC) and circulating tumor DNA (ctDNA) (Rossi and Ignatiadis, 2019). Liquid biopsies can easily be repeated weekly or bi-weekly, allowing for unprecedented longitudinal monitoring of cancer. Analysis of genomic alterations present in CTCs or ctDNA has been successfully used to follow therapy response over time and identify resistance in breast (Aceto et al., 2014; Dawson et al., 2013), colon (Misale et al., 2012; Siravegna et al., 2015), and lung (Anagnostou et al., 2019) cancer. Moreover, blood samples can also be used to assess the heterogeneity of the T cell antigen receptor repertoire, which could help the design of more effective adoptive T cell immunotherapies and monitoring their efficacy. The liquid biopsy paradigm was also applied to cerebrospinal fluid (CSF) of brain tumor patients. While not as easy as blood collection, ctDNA in CSF is associated with glioma progression and shorter survival. A recent study has shown that CSF ctDNA recapitulated the genotype of tumor biopsy and could thus be used for genotype-directed therapy monitoring (Miller et al., 2019). Yet, shedding of CTCs and ctDNA into the bloodstream is neither fully understood, nor uniform in all patients. Additional studies are required to dissect the biology of CTC and tumor DNA shedding and to determine how well ctDNA represent tumor profiles in order to validate liquid biopsy as a safe and robust way to monitor changes in ITH during treatment.
Single cell profiling
In recent years the development of single-cell sequencing technologies has enabled studies of ITH at increased resolution. Thousands of cells can now be barcoded and profiled, revealing phenotypically or genetically distinct subpopulations present in a tumor. Phenotypic heterogeneity can also be assessed by high multiplexing capacity of Cytometry-Time-of-Flight (CyTOF) (Spitzer and Nolan, 2016), multiplexed ion beam imaging (MIBI) (Angelo et al., 2014), and cyclic immunofluorescence (cycIF) (Lin et al., 2015) techniques that allow for the quantification of multiple protein expression in individual cells. The advantage of MIBI and cycIF is that they are performed in intact tissue samples maintaining tumor topology and cellular context. While all these techniques are very useful in identifying distinct subpopulations of cells, especially in the tumor microenvironment, it is much harder to clearly deconvolute complex phenotypes of tumors. In the case of brain tumors, single-cell profiling revealed four major states, associated with distinct developmental pathways hijacked by cancer cells (Neftel et al., 2019; Patel et al., 2014). Yet, in many other tumor types these distinctions are not as clear. However, phenotypic diversity may provide more insight into the genetic and epigenetic changes associated with treatment, as therapy has a more immediate effect on cellular phenotypes.
Since drug resistance can occur via different mechanisms due to genetic, epigenetic, and phenotypic heterogeneity (Figure 3), single-cell multi-omics and parallel deconvolution of all the traits of individual cancer cells would be needed to integrate all the variables. First approaches of integration of genetic and epigenetic measures at single-cell resolution show that transcriptional and epigenetic plasticity of certain subpopulations may be associated with their lineage history and underlying driver mutation (Gaiti et al., 2019). Multi-omics approaches will certainly lead to better understanding of the complex biology of the tumor ecosystem. However, more targeted approaches to identify clinically relevant variables will likely be needed as well.
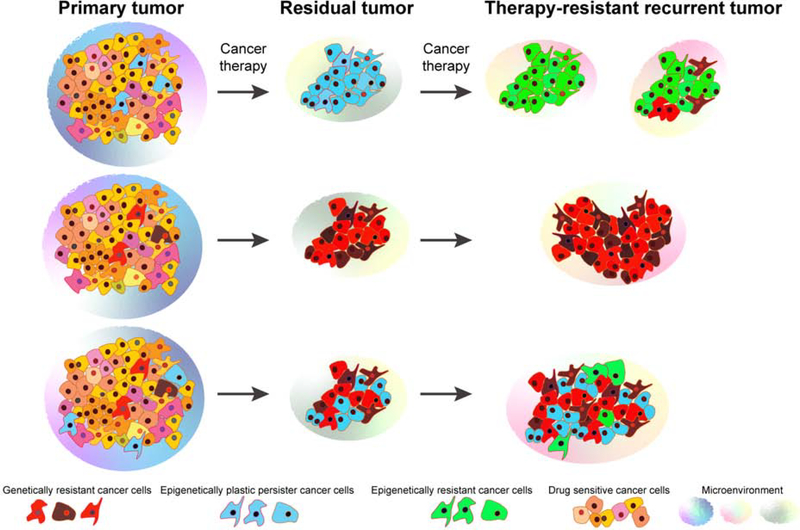
Figure 3. Evolution of therapeutic resistance.
Heterogeneous primary tumors likely contain subpopulations of cancer cells with pre-existing resistance to therapy due to genetic or epigenetic mechanisms. Genetically resistant cancer cells can also outgrow from epigenetically plastic persisters in part due to therapy-induced alterations. Interaction with the microenvironment also contributes to phenotypic heterogeneity and associated therapeutic resistance within tumors. In most advanced-stage tumors both genetic and epigenetic resistance likely to be present leading to multiple different resistance mechanisms.
Heterogeneity in therapy resistance
The majority of advanced, metastatic cancers remain incurable, even in those cases when available therapies are capable of eliminating the vast majority of tumor cells. Pre-existing intratumor heterogeneity increases the odds of at least some tumor cells to survive therapy-induced elimination, while ongoing diversification of tumor cell phenotypes during treatment enable tumor cells to adapt to therapy-imposed selective pressures, leading to the de novo resistance and eventual relapse. Here, we discuss the impact of intratumor heterogeneity on resistance to the two most promising and effective (when properly matched) types of therapies: targeted therapies and immunotherapies, both of which are commonly combined with chemotherapy. ITH has been shown to be associated with poor outcome and decreased response to cancer treatment by multiple groups in multiple human cancer types implying a universal role in therapeutic resistance (Almendro et al., 2014; Morris et al., 2016).
Heterogeneity in therapy resistance targeted therapy
Small molecule inhibitors, directed against abnormal signaling of mutated kinases, are commonly used as frontline therapies for cancers with druggable driver lesions, such as EGFR and ALK in EGFR and ALK mutant lung cancers, or BRAF in melanoma (Zhang et al., 2009). Despite the often-spectacular initial tumor responses, and continuous development of more efficient inhibitors, resistance emerges with near inevitability in advanced, metastatic cancers. In many cases of clinical relapse, resistance is associated with specific genetic mutations, such as genomic amplification of the therapy target, point mutations that allosterically reduce the ability of drug to block enzymatic activity, or amplification/mutation of other genes, enabling tumor cells to sustain oncogenic signaling (Lovly and Shaw, 2014). Most of these mutational changes are considered to be sufficient to confer full resistance, underlying clinical relapse. In this case, probability of pre-existing or de novo resistance could be inferred using mathematical models, involving target population size (numbers of tumor cells capable of self-renewal), mutational probability and proliferation/death rates (Bozic et al., 2013; Foo and Michor, 2014; Wodarz and Komarova, 2005). Assessing tumors at different stages of progression and building mathematical models based on these data could also be used to predict probable tumor evolutionary paths towards resistance (Angelova et al., 2018; Khan et al., 2018). The reality, however, might be more complicated, as development of resistance might be multifactorial (Hong et al., 2019; Shaffer et al., 2017; Vander Velde et al., 2019), in which case preexisting diversity and ongoing diversification, which fuel natural selection, rather than presence of specific mutations, might be essential for the evolution of resistance (Merlo et al., 2006). The relationship between genetic diversity and evolution is more complicated for copy number variations due to CIN: while providing a powerful source of diversification, the majority of large-scale chromosomal changes are disadvantageous (Tang and Amon, 2013). Both experimental studies and analysis of clinical data support the existence of a “sweet spot” of CIN-related diversity, where intermediate levels, which, balance evolvability with ability to maintain fitness of tumor cell populations, are most dangerous (Andor et al., 2016; Godek et al., 2016). This constraint most likely does not apply to copy number changes mediated by extrachromosomal DNA, which avoids fitness cost of large-scale chromosomal changes, while providing a highly dynamic focused diversification mechanism, enabling populations of tumor cells to quickly find optimum under therapy-imposed selection pressures (Nathanson et al., 2014).
Despite the clear evidence of genetic mechanisms of resistance, in many cases no known genetic “drivers” could be identified. Whereas it is formally possible that resistance in these cases could be attributed to uncharacterized mutations, commonality of long remission times and a growing body of experimental evidence suggests that resistance can arise through epigenetic mechanisms, involving semi-stable changes in gene expression mediated by chromatin remodeling (Hinohara et al., 2018; Knoechel et al., 2014; Liau et al., 2017; Risom et al., 2018; Sharma et al., 2010; Shu et al., 2016). A seminal paper by Sharma and colleagues (Sharma et al., 2010) implied stochastic, reversible epigenetic transition into a distinct drug-tolerant phenotypic state, enabling tumor cells to survive therapy, but incapable of supporting robust net positive growth rates to drive tumor relapse. A number of subsequent studies demonstrated the relevance of this phenomenon toward multiple therapeutic modalities across many cancer types including breast and brain tumors (Hinohara et al., 2018; Hong et al., 2019; Knoechel et al., 2014; Liau et al., 2017; Risom et al., 2018; Sharma et al., 2010; Shu et al., 2016). At least in some cases, phenotypic transition towards drug tolerance might be induced by the treatment (Goldman et al., 2015; Pisco and Huang, 2015), although the ability to undergo phenotypic transition is likely to be limited to only certain sub-populations of tumor cells (Hong et al., 2019; Shaffer et al., 2017).
Drug tolerance is often thought as a distinct, well-defined phenotypic state that might be defined by similar molecular underpinnings across multiple cancer types such as dependency on certain metabolic pathways (Hangauer et al., 2017). However, our lineage tracing-based studies revealed that tolerance towards ALK inhibitors develops from heterogeneous pre-existent subpopulations which differ in their ability to survive or grow under different inhibitors (Vander Velde et al., 2019). Likewise, functional phenotypic heterogeneity was recently implied in chemo-resistance in pancreatic cancers (Seth et al., 2019), suggesting that tolerance might reflect ITH in general, rather than a well-defined phenotypic state. At the same time, certain epigenetic states may confer “universal” drug resistance. For example, high expression and activity of the KDM5 family of histone demethylases has been linked to multiple different types of therapeutic resistance in multiple different cancer. In lung cancer high KDM5A expression is associated with EGFR inhibitor resistance (Sharma et al., 2010), in melanoma quiescent cells required for tumor propagation that also play a role in resistance to chemotherapeutic agents and BRAF inhibitors have high KDM5B levels (Roesch et al., 2013). A recent study in breast cancer investigating mechanisms by which high KDM5B activity leads to endocrine resistance determined that higher KDM5B levels are associated with higher cell-to-cell transcriptomic heterogeneity (Hinohara et al., 2018). Thus, mutant and abnormally expressed epigenetic regulators may influence tumor progression and therapeutic responses via their effect on cellular epigenetic and phenotypic heterogeneity, and therefore, may represent good combination agents even if their single agent efficacy is modest.
Whereas drug tolerance is likely responsible for the ability of tumor cells to survive therapy, leading to minimal residual disease, clinical relapse requires acquisition of stronger resistance phenotypes, capable of maintaining net positive tumor growth in the presence of treatment. Bona fide resistance can develop from drug-tolerant cells through acquisition of new genetic mutations, thus requiring new genetic diversification (Figure 3). Notably, acquisition of EGFR T790M gatekeeper mutations by tolerant cells leads to more robust tumor cell phenotypes. These cells are less sensitive to T790M targeting third generation EGFR inhibitors compared to T790M mutations on the background of therapy naïve cells (Hata et al., 2016). By maintaining a reservoir of viable tumor cells, tolerance enables acquisition of multiple distinct resistance mechanisms, thus supporting more heterogeneous relapse, which reduces chances of second line therapy success. Similarly, in colorectal carcinomas treatment with EGFR and BRAF inhibitors downregulates DNA repair pathways and upregulates error-prone polymerases in drug-tolerant persister cells, therefore increasing mutation rates and the likelihood of resistance (Russo et al., 2019).
On the other hand, full resistance can also develop from weakly resistant sub-populations through non-genetic mechanisms. A case in point is a recent study on acquisition of resistance to BRAF inhibitors, where transient expression heterogeneity in resistance associated genes, such as AXL and EGFR, enabled survival of subpopulations of tumor cells, but development of fully resistant phenotypes involved acquisition of multiple, partially coordinated gene expression changes (Shaffer et al., 2017). A similar phenomenon has been described in breast and lung cancers (Hong et al., 2019; Vander Velde et al., 2019). These studies shed a new light on the association between “stemness” and resistance. While EMT and stemness are often considered to be proximal causes of therapy resistance (Holohan et al., 2013; Singh and Settleman, 2010), phenotypic plasticity per se might be the true culprit, by enabling cancer cells to access resistance phenotypes by rewiring of gene regulatory networks (Pisco and Huang, 2015).
Whereas the majority of studies focus on cell-autonomous resistance mechanisms, a growing body of evidence suggests that cell-to-cell and microenvironmental interactions could dramatically impact drug sensitivity. For example, growth factors that can be produced by both tumor cells and normal cells within tumor microenvironment, have been shown to partially or completely de-sensitize tumor cells to many types of Tyrosine Kinase Inhibitors (TKIs), across numerous cancer types (Wilson et al., 2012). Similarly, drug sensitivity could be dramatically reduced by signaling, mediated by interaction of tumor cells with the extracellular matrix (ECM) (Hirata et al., 2015). Whereas in typical in vitro studies, every tumor cell can be equally impacted, microenvironmental heterogeneity in vivo likely leads to heterogeneity in environmentally mediated therapy resistance, where only some of the tumor cells can evade the impact of therapy (Marusyk et al., 2016). While the impact of microenvironmentally mediated resistance on relapse remains unclear (as tumor cells remain sensitive outside of the borders of protective niches), the phenomenon likely contributes to residual disease, providing reservoir of tumor cells capable of developing cell-autonomous resistance mechanisms (Hirata and Sahai, 2017; Meads et al., 2009). The link between microenvironmental heterogeneity is not limited to paracrine signals, cell-to-cell, and cell-ECM contact. Given the well documented impact of hypoxia, acidity, and nutrient status on therapy sensitivity (Gillies et al., 2012), metabolic microenvironmental heterogeneity likely provides an important modulator of tumor responses to therapies, a notion supported by in silico modeling (Robertson-Tessi et al., 2015).
A standard approach for a biomedical research paper entails reducing a phenomenon, such as therapy resistance, to a single mechanism. However, resistance might entail a combined impact of multiple mechanisms operating both within single cells (Hong et al., 2019; Shaffer et al., 2017; Vander Velde et al., 2019) and across different functionally distinct subpopulations (Chabon et al., 2016; Piotrowska et al., 2015). Thus, the development of clinical resistance is likely to involve not only different sources of pre-existing ITH, but also different inputs for additional diversification. For example, resistance to EGFR inhibitors in a cell line model of EGFR-mutant lung cancer can develop from both pre-existing resistance-conferring mutations, but also from drug tolerant cells, acquiring diverse resistance mechanisms (Hata et al., 2016; Ramirez et al., 2016). While the role of microenvironmental diversification remains poorly explored, its inputs likely shape both genetic and epigenetic diversification. Sufficiently complete understanding of resistance cannot be accomplished exclusively with mainstream reductionistic methodological approaches, but instead requires integrative studies and development of new tools.
Heterogeneity in therapy resistance – immunotherapy
Immune surveillance is one of the key microenvironmental constrains that all tumors must overcome in order to progress and grow. Immunoediting refers to the process by which the immune system impacts tumor evolution and it is classified into three phases: elimination, equilibrium, and escape (Dunn et al., 2004). Studies in experimental models and patients with compromised immune system have demonstrated that most neoplastic cells are eliminated by the immune system before they become clinically relevant. However, tumor cells can have variable expression of neoantigens and may display differences in signaling pathways related to immunity, thus, ITH for these traits will limit the efficacy of antitumor immune responses. As a consequence, all clinically-relevant tumors have already escaped immune surveillance by various mechanisms.
Immunotherapy is thought to be relatively unaffected by ITH, since tumor neoantigens are usually not functional cancer drivers, thus, they are not expected to be impacted by cancer therapy. However, recent data demonstrates that ITH influences the success of immunotherapy as well. Acquired resistance to immunotherapy can develop due to ITH for neoantigens, antigen presentation, and interferon signaling (Havel et al., 2019; Kalbasi and Ribas, 2019). Tumor mutational burden (TMB) defined as the number of somatic mutations within tumors, is one of the strongest predictors of response to immune checkpoint inhibitors with high TMB in general associated with better response (Ribas and Wolchok, 2018). However, higher TMB can also increase ITH and recent data in both experimental models and patients show that tumors with high subclonal ITH are less likely to respond to immune checkpoint inhibitors than more homogeneous tumors (Anagnostou et al., 2017; Gejman et al., 2018; McGranahan et al., 2016; Milo et al., 2018; Wolf et al., 2019). Resistance to immunotherapy due to ITH could be due to multiple mechanisms. Higher ITH is associated with higher risk of having a resistant clone. At the same time, subclonal mutations may not trigger such an effective immune response as clonal ones (Anagnostou et al., 2017; Gejman et al., 2018; McGranahan et al., 2016; Milo et al., 2018; Wolf et al., 2019), although there is also data suggesting that subclonal neoantigens are more immunogenic (Jimenez-Sanchez et al., 2017). Thus, ITH has prognostic and predictive value for immunotherapy as well.
Microenvironmental and spatial heterogeneity are of key importance for immune therapies as well. For example, attraction of immune cells is mediated by paracrine action of chemokines and cytokines, key suppressors of immune responses CD73 and IDO1 act in a paracrine manner, and hypoxia (Noman et al., 2015) and acidification (Pilon-Thomas et al., 2016) can potently suppress cytotoxic immunity. The location of immune cells within tumors is also highly heterogeneous, which is likely influenced by stromal cells and is associated with probability of response to immune checkpoint inhibitors (Hirata and Sahai, 2017). Based on the frequency and location of immune cells tumors can be classified as immune cold (immune desert), peritumoral, stromal-restricted, and inflamed (Gruosso et al., 2019; Keren et al., 2018; Rosenthal et al., 2019). While this classification was applied to tumors in different patients reflecting intertumor heterogeneity, even within the same tumor there is significant spatial heterogeneity for TIL frequency with some regions being fully infiltrated and others devoid of leukocytes. In metastatic urothelial cancer a fibroblast TGFβ signature correlated with immune exclusion and resistance to immune checkpoint inhibitors, both of which was improved by combined treatment with TGFBR inhibitors and anti-PD-L1 antibody (Mariathasan et al., 2018). Thus, decreasing tumor cell and microenvironmental ITH would improve the efficacy of immunotherapy as well.
Accounting for heterogeneity in therapeutic decision making
Given the profound importance of ITH shaping therapy responses, it could be beneficial to explore its utility as a predictive marker. Perhaps more importantly, considering strategies to reduce ITH might lead to improvement of long-term therapeutic responses, prolonging remission and increasing the odds of driving tumors toward extinction. Given the current focus on specific “drivers” of resistance, the main effort is directed towards the detection of resistance conferring mutations in tissue and liquid biopsies, with the idea that this data should shed light on the emergence of resistance, potentially informing therapeutic interventions to intercept the relapse. However, given the well-established link between ITH and evolvability of tumors, complementary consideration of ITH could provide an independent predictive marker, which could also be factored in when formulating the initial treatment strategy, as well as informing decisions on changing therapies during remission.
For example, in HER2+ breast cancer tumors with higher ITH for HER2 are less likely to respond to treatment (Rye et al., 2018). Therapeutic resistance is likely caused by cancer cells that lack HER2 amplification and overexpression making them resistant to HER2-targeted therapies. Thus, the combination of HER2-targeting and non-targeted agents (e.g., chemotherapy and PI3K/AKT inhibitors) is predicted to be the most beneficial in tumors with high ITH. Indeed, a recent clinical trial (NCT03248492) testing trastuzumab deruxtecan (DS-8201), an anti-HER2 antibody linked to topoisomerase inhibitor, in metastatic HER2+ breast cancer patients who failed prior treatment has given positive results (Modi et al., 2019). While DS-8201 is also a HER2-targeting antibody, its antitumor effects do not require HER2 dependency, since it simply serves as a chemotherapy delivery agent. Thus, it is also efficacious against tumor cells that lack HER2 amplicon but still have HER2 expression, which makes it more efficacious in heterogeneous HER2+ tumors. This example clearly shows benefits of understanding the cancer cell dependencies and their combinations in improving efficacy of treatments for heterogeneous tumors. Mathematical modeling can also help with predicting optimal drug combinations and treatment schedules that most likely to work in heterogeneous tumors (Chakrabarti and Michor, 2017; Gallaher et al., 2018; Leder et al., 2014) 31026176. Indeed, several clinical trials have been designed based on these models and following evolutionary principles and some have shown promising results (Gallaher et al., 2018).
In addition to guiding therapeutic decision-making, many aspects of ITH could be considered therapeutic targets (Figure 4). Whereas little can be done to prevent stochastic mutations occurring during replication, maximizing tumor debulking through surgery or irradiation should reduce ITH, stemming from all of the inputs. Moreover, at least some types of ITH could be targeted more directly. Given the links between inflammation, CIN, and phenotypic plasticity (Colotta et al., 2009; Grivennikov et al., 2010; Korkaya et al., 2011), use of anti-inflammatory agents as therapy explicitly directed against ITH might warrant exploration. Similarly, many types of epigenetic drugs, such as histone deacetylase (HDAC), histone demethylase, and bromodomain inhibitors might reduce phenotypic plasticity and ITH. Phenotypic plasticity underlying tolerance and resistance can be disrupted by epigenetic inhibitors, such as those targeting histone deacetylases (Sharma et al., 2010), BET bromodomain proteins (Knoechel et al., 2014; Risom et al., 2018) or histone demethylases (Hinohara and Polyak, 2019; Hinohara et al., 2018; Liau et al., 2017). While current strategies primarily focus on short-term cytotoxic/cytostatic effects of the drugs, consideration of longer-term effects, mediated by reduction of ITH might be warranted, even when the agents are not effective in the short term. Reducing microenvironmental heterogeneity might also be beneficial, both in the short and long term. While anti-angiogenic agents did not live up to the initial promise of eliminating tumors by blocking their access to oxygen/nutrients, the reverse idea – normalizing blood vasculature using lower doses of anti-angiogenic drugs, might still prove to be clinically useful (Jain, 2014). In addition to improving drug delivery, which is currently the main rationale behind the strategy, normalization of blood vasculature could also reduce microenvironmental heterogeneity, while also inhibiting selection for more invasive and aggressive sub-populations (Robertson-Tessi et al., 2015).
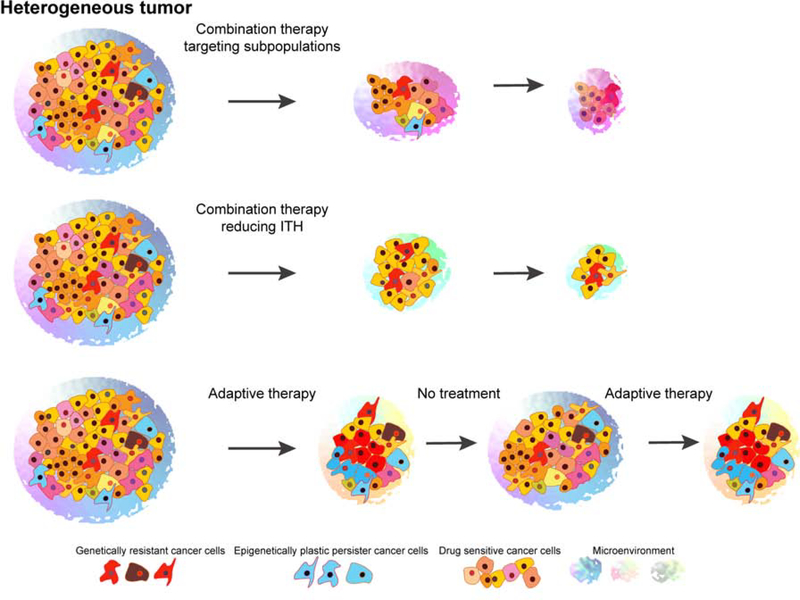
Figure 4. Optimal therapies for heterogeneous tumors.
The effective treatment of heterogeneous tumors requires optimized therapy to minimize the evolution of therapeutic resistance. There are three general approaches by which this can be achieved: (1) combination therapies targeting different tumor subpopulations, different dependencies, or both cell-autonomous and non-cell-autonomous functions; (2) therapies combining tumor-targeting agents with compounds that reduce ITH such as HDAC or BET inhibitors; and (3) adaptive therapy when ITH is maintained and the competition of drug-resistant and drug-sensitive cells is limited by alternating therapy and no treatment.
Concluding remarks
Despite the recent explosion of interest toward cancer evolution and ITH, our knowledge in this area remains rudimentary. Moving from observational studies toward deeper understanding of ITH and using this knowledge to improve clinical outcomes would require not only the development of novel methodologies to quantify different types of ITH, but also new frameworks to conceptualize and integrate this knowledge. Achieving these goals would necessitate moving from reductionistic approaches of molecular oncology toward multidisciplinary studies, involving meaningful integration of clinical observations, bioinformatical analyses, and systems biology approaches with experimental and mathematical modeling.
Acknowledgements
We thank Dr. Daria Miroshnychenko for her help with designing Figure 2. This work was supported by the National Cancer Institute R35CA197623 (K.P.), R00CA201606 (M.J.), Susan G. Komen Breast Cancer Foundation CCR17481976 (A.M.), and the Breast Cancer Research Foundation (K.P.).
Footnotes
Declaration of Interests
K.P. is a member of the scientific advisory boards of Farcast Biosciences and Acrivon Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarouk KO, Ibrahim ME, Gatenby RA, and Brown JS (2013). Riparian ecosystems in human cancers. Evol Appl 6, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, Gonzalez-Farre X, Munoz M, Russnes HG, Helland A, et al. (2014). Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 6, 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, Marrone K, Sivakumar IKA, Bruhm DC, Rosner S, et al. (2019). Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res 79, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, et al. (2017). Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 7, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, and Maley CC (2016). Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. (2014). Multiplexed ion beam imaging of human breast tumors. Nat Med 20, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova M, Mlecnik B, Vasaturo A, Bindea G, Fredriksen T, Lafontaine L, Buttard B, Morgand E, Bruni D, Jouret-Mourin A, et al. (2018). Evolution of Metastases in Space and Time under Immune Selection. Cell 175, 751–765 e716. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, and Cantley LC (2018). The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 174, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman RA, and Loeb LA (2006). Efficiency of carcinogenesis with and without a mutator mutation. Proc Natl Acad Sci U S A 103, 14140–14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, Chang MT, Schram AM, Jonsson P, Bandlamudi C, et al. (2018). Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 50, 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, Moon YS, Yaqubie A, Kelly N, Le DT, et al. (2013). Evolutionary dynamics of cancer in response to targeted combination therapy. Elife 2, e00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, Koop C, Oakes C, Zucknick M, Lipka DB, Weischenfeldt J, et al. (2014). Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep 8, 798–806. [DOI] [PubMed] [Google Scholar]
- Bussard KM, and Smith GH (2012). Human breast cancer cells are redirected to mammary epithelial cells upon interaction with the regenerating mammary gland microenvironment in-vivo. PLoS One 7, e49221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, and Jain RK (2000). Angiogenesis in cancer and other diseases. Nature 407, 249–257. [DOI] [PubMed] [Google Scholar]
- Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA, et al. (2016). Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7, 11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, and Michor F (2017). Pharmacokinetics and Drug Interactions Determine Optimum Combination Strategies in Computational Models of Cancer Evolution. Cancer Res 77, 3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F, Allavena P, Sica A, Garlanda C, and Mantovani A (2009). Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. (2013). Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209. [DOI] [PubMed] [Google Scholar]
- deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, Koeman J, Seth S, Protopopov A, Felicella M, et al. (2018). Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 50, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Grönroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. (2014). Tolerance of whole- genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discovery 4, 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, and Lazebnik Y (2007). A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol 17, 431–437. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, and Grompe M (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, and Schreiber RD (2004). The three Es of cancer immunoediting. Annu Rev Immunol 22, 329–360. [DOI] [PubMed] [Google Scholar]
- Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, et al. (2013). Acidity generated by the tumor microenvironment drives local invasion. Cancer Res 73, 1524–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S, and Overholtzer M (2018). Cell-in-cell phenomena in cancer. Nat Rev Cancer 18, 758–766. [DOI] [PubMed] [Google Scholar]
- Fane M, and Weeraratna AT (2019). How the ageing microenvironment influences tumour progression. Nat Rev Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP (2007). Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–440. [DOI] [PubMed] [Google Scholar]
- Filho OM, Viale G, Trippa L, Li T, Yardley DA, Mayer IA, Abramson VG, Arteaga CL, Spring L, Waks AG, et al. (2019). HER2 heterogeneity as a predictor of response to neoadjuvant T-DM1 plus pertuzumab: Results from a prospective clinical trial. Journal of Clinical Oncology 37, 502–502. [Google Scholar]
- Flavahan WA, Gaskell E, and Bernstein BE (2017). Epigenetic plasticity and the hallmarks of cancer. Science 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo J, and Michor F (2014). Evolution of acquired resistance to anti-cancer therapy. J Theor Biol 355, 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EJ, Prindle MJ, and Loeb LA (2013). Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev 32, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F, Chaligne R, Gu H, Brand RM, Kothen-Hill S, Schulman RC, Grigorev K, Risso D, Kim KT, Pastore A, et al. (2019). Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature 569, 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher JA, Enriquez-Navas PM, Luddy KA, Gatenby RA, and Anderson ARA (2018). Spatial Heterogeneity and Evolutionary Dynamics Modulate Time to Recurrence in Continuous and Adaptive Cancer Therapies. Cancer Res 78, 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast CE, Silk AD, Zarour L, Riegler L, Burkhart JG, Gustafson KT, Parappilly MS, Roh-Johnson M, Goodman JR, Olson B, et al. (2018). Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv 4, eaat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, and Gillies RJ (2008). A microenvironmental model of carcinogenesis. Nat Rev Cancer 8, 56–61. [DOI] [PubMed] [Google Scholar]
- Gejman RS, Chang AY, Jones HF, DiKun K, Hakimi AA, Schietinger A, and Scheinberg DA (2018). Rejection of immunogenic tumor clones is limited by clonal fraction. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies RJ, Brown JS, Anderson ARA, and Gatenby RA (2018). Eco-evolutionary causes and consequences of temporal changes in intratumoural blood flow. Nat Rev Cancer 18, 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies RJ, Verduzco D, and Gatenby RA (2012). Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer 12, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godek KM, Venere M, Wu Q, Mills KD, Hickey WF, Rich JN, and Compton DA (2016). Chromosomal Instability Affects the Tumorigenicity of Glioblastoma Tumor-Initiating Cells. Cancer Discov 6, 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A, Majumder B, Dhawan A, Ravi S, Goldman D, Kohandel M, Majumder PK, and Sengupta S (2015). Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun 6, 6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, Parks B, Gars E, Liedtke M, Zheng GXY, et al. (2019). Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol 37, 1458–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruosso T, Gigoux M, Manem VSK, Bertos N, Zuo D, Perlitch I, Saleh SMI, Zhao H, Souleimanova M, Johnson RM, et al. (2019). Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest 129, 1785–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, et al. (2017). Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, et al. (2016). Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 22, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel JJ, Chowell D, and Chan TA (2019). The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 19, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinohara K, and Polyak K (2019). Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol 29, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinohara K, Wu HJ, Vigneau S, McDonald TO, Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, Papanastasiou M, et al. (2018). KDM5 Histone Demethylase Activity Links Cellular Transcriptomic Heterogeneity to Therapeutic Resistance. Cancer Cell 34, 939–953 e939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M, Larkin J, Marais R, and Sahai E (2015). Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer Cell 27, 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata E, and Sahai E (2017). Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb Perspect Med 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefflin R, Lahrmann B, Warsow G, Hubschmann D, Spath C, Walter B, Chen X, Hofer L, Macher-Goeppinger S, Tolstov Y, et al. (2016). Spatial niche formation but not malignant progression is a driving force for intratumoural heterogeneity. Nat Commun 7, ncomms11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, and Johnston PG (2013). Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13, 714–726. [DOI] [PubMed] [Google Scholar]
- Hong SP, Chan TE, Lombardo Y, Corleone G, Rotmensz N, Bravaccini S, Rocca A, Pruneri G, McEwen KR, Coombes RC, et al. (2019). Single-cell transcriptomics reveals multistep adaptations to endocrine therapy. Nat Commun 10, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen BM, Harrell JC, Jedlicka P, Borges VF, Varella-Garcia M, and Horwitz KB (2006). Spontaneous fusion with, and transformation of mouse stroma by, malignant human breast cancer epithelium. Cancer Res 66, 8274–8279. [DOI] [PubMed] [Google Scholar]
- Jain RK (2014). Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell 26, 605–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. (2017). Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 376, 2109–2121. [DOI] [PubMed] [Google Scholar]
- Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA, et al. (2015). In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 47, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson G, Pujadas E, Goutsias J, and Feinberg AP (2017). Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat Genet 49, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez A, Memon D, Pourpe S, Veeraraghavan H, Li Y, Vargas HA, Gill MB, Park KJ, Zivanovic O, Konner J, et al. (2017). Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 170, 927–938 e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K, Robert de Massy M, Ismail M, Reading JL, Uddin I, Woolston A, Hatipoglu E, Oakes T, Rosenthal R, Peacock T, et al. (2019). Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat Med 25, 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbasi A, and Ribas A (2019). Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, and Weinberg RA (2007). Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563. [DOI] [PubMed] [Google Scholar]
- Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang SR, Kurian A, Van Valen D, West R, et al. (2018). A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten K, de Visser KE, van Miltenburg MH, and Jonkers J (2017). Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 9, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, Mateos JF, Vatsiou A, Lampis A, Damavandi MD, et al. (2018). Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer Discov 8, 1270–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. (2014). An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 46, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenchan DE, and Flavell RR (2019). Spatiotemporal pH Heterogeneity as a Promoter of Cancer Progression and Therapeutic Resistance. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Liu S, and Wicha MS (2011). Regulation of cancer stem cells by cytokine networks: attacking cancer’s inflammatory roots. Clin Cancer Res 17, 6125–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBerge GS, Duvall E, Grasmick Z, Haedicke K, and Pawelek J (2017). A Melanoma Lymph Node Metastasis with a Donor-Patient Hybrid Genome following Bone Marrow Transplantation: A Second Case of Leucocyte-Tumor Cell Hybridization in Cancer Metastasis. PLoS One 12, e0168581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. (2014). Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazova R, Laberge GS, Duvall E, Spoelstra N, Klump V, Sznol M, Cooper D, Spritz RA, Chang JT, and Pawelek JM (2013). A Melanoma Brain Metastasis with a Donor-Patient Hybrid Genome following Bone Marrow Transplantation: First Evidence for Fusion in Human Cancer. PLoS One 8, e66731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder K, Pitter K, Laplant Q, Hambardzumyan D, Ross BD, Chan TA, Holland EC, and Michor F (2014). Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell 156, 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, and Vogelstein B (1997). Genetic instability in colorectal cancers. Nature 386, 623–627. [DOI] [PubMed] [Google Scholar]
- Li L, McCormack AA, Nicholson JM, Fabarius A, Hehlmann R, Sachs RK, and Duesberg PH (2009). Cancer-causing karyotypes: chromosomal equilibria between destabilizing aneuploidy and stabilizing selection for oncogenic function. Cancer Genet Cytogenet 188, 1–25. [DOI] [PubMed] [Google Scholar]
- Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD, Rodig SJ, et al. (2017). Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell 20, 233–246 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Fallahi-Sichani M, and Sorger PK (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun 6, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, and Gatenby RA (2016). Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer Res 76, 3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, and Shaw AT (2014). Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res 20, 2249–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, and Kang Y (2009). Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc Natl Acad Sci U S A 106, 9385–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KM, Newberg JY, Black MA, Jones DJ, Amaya-Manzanares F, Guzman-Rojas L, Kodama T, Ward JM, Rust AG, van der Weyden L, et al. (2016). Analyzing tumor heterogeneity and driver genes in single myeloid leukemia cells with SBCapSeq. Nat Biotechnol 34, 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, and Polyak K (2012). Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12, 323–334. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Tabassum DP, Janiszewska M, Place AE, Trinh A, Rozhok AI, Pyne S, Guerriero JL, Shu S, Ekram M, et al. (2016). Spatial Proximity to Fibroblasts Impacts Molecular Features and Therapeutic Sensitivity of Breast Cancer Cells Influencing Clinical Outcomes. Cancer Res 76, 6495–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJA, Smirnov IV, Reis GF, Phillips JJ, Barnes MJ, et al. (2015). DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell 28, 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Song JS, and Costello JF (2016). Intratumoral Heterogeneity of the Epigenome. Cancer Cell 29, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, and Swanton C (2017). Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 168, 613–628. [DOI] [PubMed] [Google Scholar]
- Meads MB, Gatenby RA, and Dalton WS (2009). Environment-mediated drug resistance: a major contributor to minimal residual disease. Nature reviews Cancer 9, 665–674. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, and Maley CC (2006). Cancer as an evolutionary and ecological process. Nat Rev Cancer 6, 924–935. [DOI] [PubMed] [Google Scholar]
- Merlo LMF, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, and Maley CC (2010). A Comprehensive Survey of Clonal Diversity Measures in Barrett’s Esophagus as Biomarkers of Progression to Esophageal Adenocarcinoma. Cancer Prevention Research 3, 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, Zheng Y, Skakodub A, Mehta SA, Campos C, et al. (2019). Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565, 654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo I, Bedora-Faure M, Garcia Z, Thibaut R, Perie L, Shakhar G, Deriano L, and Bousso P (2018). The immune system profoundly restricts intratumor genetic heterogeneity. Sci Immunol 3. [DOI] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. (2012). Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, Andre F, Iwata H, Ito Y, Tsurutani J, et al. (2019). Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N Engl J Med. [Google Scholar]
- Mohlin S, Wigerup C, Jogi A, and Pahlman S (2017). Hypoxia, pseudohypoxia and cellular differentiation. Exp Cell Res 356, 192–196. [DOI] [PubMed] [Google Scholar]
- Morris LG, Riaz N, Desrichard A, Senbabaoglu Y, Hakimi AA, Makarov V, Reis-Filho JS, and Chan TA (2016). Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7, 10051–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, and Hynes RO (2012). The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 11, M111 014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. (2014). Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849 e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknafs N, Zhong Y, Moral JA, Zhang L, Shao MX, Lo A, Makohon-Moore A, IacobuzioDonahue CA, and Karchin R (2019). Characterization of genetic subclonal evolution in pancreatic cancer mouse models. Nat Commun 10, 5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman MZ, Hasmim M, Messai Y, Terry S, Kieda C, Janji B, and Chouaib S (2015). Hypoxia: a key player in antitumor immune response. A Review in the Theme: Cellular Responses to Hypoxia. Am J Physiol Cell Physiol 309, C569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, Damaghi M, Wojtkowiak JW, Mule JJ, Ibrahim-Hashim A, and Gillies RJ (2016). Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res 76, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska Z, Niederst MJ, Karlovich CA, Wakelee HA, Neal JW, Mino-Kenudson M, Fulton L, Hata AN, Lockerman EL, Kalsy A, et al. (2015). Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 5, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco AO, and Huang S (2015). Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: ‘What does not kill me strengthens me’. Br J Cancer 112, 1725–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, et al. (2016). Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7, 10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben A, Gittelman R, Gao J, Zhang J, Yusko EC, Wu CJ, Emerson R, Zhang J, Tipton C, Li J, et al. (2017). TCR Repertoire Intratumor Heterogeneity in Localized Lung Adenocarcinomas: An Association with Predicted Neoantigen Heterogeneity and Postsurgical Recurrence. Cancer Discov 7, 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, and Wolchok JD (2018). Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risom T, Langer EM, Chapman MP, Rantala J, Fields AJ, Boniface C, Alvarez MJ, Kendsersky ND, Pelz CR, Johnson-Camacho K, et al. (2018). Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat Commun 9, 3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson-Tessi M, Gillies RJ, Gatenby RA, and Anderson AR (2015). Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res 75, 1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K, et al. (2017). Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 170, 875–888 e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, et al. (2013). Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, Lund T, Tanic M, Reading JL, Joshi K, et al. (2019). Neoantigen-directed immune escape in lung cancer evolution. Nature 567, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, and Ignatiadis M (2019). Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res 79, 2798–2804. [DOI] [PubMed] [Google Scholar]
- Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magri A, Novara L, et al. (2019). Adaptive mutability of colorectal cancers in response to targeted therapies. Science 366, 1473–1480. [DOI] [PubMed] [Google Scholar]
- Rye IH, Trinh A, Saetersdal AB, Nebdal D, Lingjaerde OC, Almendro V, Polyak K, Borresen-Dale AL, Helland A, Markowetz F, and Russnes HG (2018). Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors. Molecular oncology 12, 1838–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. (2012). An immunosurveillance mechanism controls cancer cell ploidy. Science 337, 1678–1684. [DOI] [PubMed] [Google Scholar]
- Seth S, Li CY, Ho IL, Corti D, Loponte S, Sapio L, Del Poggetto E, Yen EY, Robinson FS, Peoples M, et al. (2019). Pre-existing Functional Heterogeneity of Tumorigenic Compartment as the Origin of Chemoresistance in Pancreatic Tumors. Cell Rep 26, 1518–1532 e1519. [DOI] [PubMed] [Google Scholar]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. (2017). Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. (2007). Molecular definition of breast tumor heterogeneity. Cancer Cell 11, 259–273. [DOI] [PubMed] [Google Scholar]
- Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, et al. (2016). Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, and Settleman J (2010). EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, et al. (2015). Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21, 827. [DOI] [PubMed] [Google Scholar]
- Spitzer MH, and Nolan GP (2016). Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacker SA, Williams SP, Karnezis T, Shayan R, Fox SB, and Achen MG (2014). Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat Rev Cancer 14, 159–172. [DOI] [PubMed] [Google Scholar]
- Storchova Z, and Pellman D (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5, 45–54. [DOI] [PubMed] [Google Scholar]
- Tang YC, and Amon A (2013). Gene copy-number alterations: a cost-benefit analysis. Cell 152, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane K, Eriksson H, Maaskola J, Hansson J, and Lundeberg J (2018). Spatially Resolved Transcriptomics Enables Dissection of Genetic Heterogeneity in Stage III Cutaneous Malignant Melanoma. Cancer Res 78, 5970–5979. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, and Vogelstein B (2015). Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Novelli MR, and Bodmer WF (1996). The mutation rate and cancer. Proc Natl Acad Sci U S A 93, 14800–14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, et al. (2017). Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Velde R, Yoon N, Marusyk V, Durmaz A, Dhawan A, Myroshnychenko D, LozanoPeral D, Desai B, Balynska O, Poleszhuk J, et al. (2019). Resistance to targeted therapies as a multifactorial, gradual adaptation to inhibitor specific selective pressures. bioRxiv, 504837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Bafna V, and Mischel PS (2019). Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 19, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, and Kinzler KW (2004). Cancer genes and the pathways they control. Nat Med 10, 789–799. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. (2012). Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz D, and Komarova NL (2005). Emergence and prevention of resistance against small molecule inhibitors. Seminars in cancer biology 15, 506–514. [DOI] [PubMed] [Google Scholar]
- Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, Levy R, Jimenez-Sanchez A, Trabish S, Lee JS, et al. (2019). UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 179, 219–235 e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. (2015). Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y (2016). Spatial Heterogeneity in the Tumor Microenvironment. Cold Spring Harb Perspect Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AW, McPherson A, Milne K, Kroeger DR, Hamilton PT, Miranda A, Funnell T, Little N, de Souza CPE, Laan S, et al. (2018). Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell 173, 1755–1769 e1722. [DOI] [PubMed] [Google Scholar]
- Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. (2014). Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang PL, and Gray NS (2009). Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, and Vijg J (2018). Somatic Mutagenesis in Mammals and Its Implications for Human Disease and Aging. Annu Rev Genet 52, 397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Magee BB, Magee PT, Holland BR, Rodrigues E, Holmes AR, Cannon RD, and Schmid J (2015). Selective Advantages of a Parasexual Cycle for the Yeast Candida albicans. Genetics 200, 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]