Abstract
Background
There are about 2.4 hundred thousand new cases and 1.5 hundred thousand deaths of ovarian cancer (OC) annually in the world. Chronic inflammation is a risk factor for OC. C-X-C motif chemokine ligand 1 (CXCL1) defects may facilitate inflammation and transactivate EGFR in ovarian cancer, but the precise haplotypes associated with the potential diseases remained largely unknown. In this work, we characterized CXCL1 gene variations to elucidate their possible associations with OC.
Methods
We analyzed the CXCL1 gene for 300 OC patients with 400 healthy participants as controls. The statistical analyses and Hardy-Weinberg equilibrium tests of the patients and control populations were conducted using the SPSS software (version 19.0) and Plink (version 1.9).
Results
The variants rs11547681, rs201090116, rs199791199, rs181868085, rs4074 and rs1814092 within or near the CXCL1 gene were characterized. The genetic heterozygosity of rs11547681 and rs4074 was very high. Statistical analysis showed that the variant rs11547681 in the gene was closely associated with the risk of OC in the Chinese Han population, although this variant was not associated with FIGO stages or pathological grades of the patients.
Conclusions
Rs11547681 in CXCL1 gene was associated with the risk of OC in the Chinese Han population.
Keywords: Ovarian cancer, Chronic inflammation, Chemokines, CXCL1, 5’UTR
Introduction
Globally, there are about 2.4 hundred thousand new ovarian cancer (OC) cases and about 1.5 hundred thousand deaths caused by this disease annually [1]. OC is the 7th most common and 5th most lethal cancer among women worldwide [2]. The incidence rates of OC are highest in northern and eastern Europe, and lowest in Asia and Africa [1]. In the past decades, however, the morbidity has considerably decreased in many previously high incidence countries but, in contrast, is increasing in some low incidence countries, such as China [1, 3].
The 5-years survival rates of OC range from 30.3 to 44.1%, depending on the specific subtypes and stages at the time of diagnosis or treatment [4, 5]. Early detection and treatment of this disease can significantly increase the survival rate of OC, but, due to the lack of specific signs or symptoms at early stages, this disease is usually diagnosed only at late stages [6]. The recurrence rate of this disease is also very high, often leading to death of the patient, who might have achieved a clinical complete remission after primary therapy [5, 6].
So far, several genetic and non-genetic factors have been found to be associated with OC. Women with affected first-degree relatives usually have higher risks for OC [7], and women, who have relatives diagnosed with OC below 50 years old, have even higher OC risks [8]. Many gene mutations have been associated with sporadic OC patients, including BRCA1 and 2 [9], BRIP1 [10] and RAD51 [11]. Oral contraceptive use and tubal ligation seem to be protective factors [12, 13], whereas older menopausal ages, obesity, menopausal hormone therapy use, a history of endometriosis and smoking are all risk factors [14–16].
Chronic inflammation, such as those caused by asbestos or talc exposure, endometriosis or pelvic inflammatory diseases, has also been suggested to be a risk factor for OC [17, 18]. On the other side, anti-inflammatory medications, e.g., acetaminophen and low-dose of aspirin are protective factors for OC [19, 20]. Immunotherapies, including immune checkpoint blockade and cancer vaccines, also have many special roles in immune recognition and immune regulation of the OC cells [21].
Chemokines expressed in tumor or stromal cells may facilitate tumor angiogenesis and in the meantime suppress immune-mediated tumor elimination, and as a result are associated with cancers [22–24]. C-X-C motif chemokine ligand 1 (CXCL1) is a member of the CXC subfamily of chemokines and an oncogenic factor in many cancers [25, 26]. It can transactivate EGFR in OC through binding to the G-protein coupled receptor CXC receptor 2 (CXCR2) [27]. Abnormal expression of CXCL1 is associated with many tumors [28], but the associated haplotypes remained largely unknown.
In this work, we investigated variants in the CXCL1 gene for their associations with the risk of OC in the Chinese Han population. We found that variant rs11547681 was associated with ovarian cancer and demonstrated the 5’UTR for the functions of CXCL1.
Materials and methods
Study population
A total of 300 sporadic OC cases and 400 normal controls (Table 1) were assembled for this study at the Department of Gynaecology and Obstetrics and Medical Examination Center of the Second Affiliated Hospital of Harbin Medical University, Harbin, China. From each participant, we obtained a written informed consent. This work has been reviewed and approved by the Ethics Committee of Harbin Medical University. We also confirmed that all experiments were performed in accordance with relevant guidelines and regulations and were consistent with the 1975 Declaration of Helsinki.
Table 1.
Clinical characteristics of study population
| Parameter | CRC | Control | F | t | P | 95%CI | |
|---|---|---|---|---|---|---|---|
| Up | Low | ||||||
| Sample (n) | 300 | 400 | – | – | – | – | – |
| Age (years) | 50.39 ± 12.21 | 49.68 ± 7.66 | 9.977 | 0.639 | 0.523 | −1.47926 | 2.90141 |
Data are shown as mean ± SD; between the two groups, there were no statistical differences of the age and gender composition
Medical histories of the enrolled participants were recorded in detail, and all the participants received physical examinations. The diagnostic criteria for sporadic OC patients were those of FIGO (Federation International of Gynecology and Obstetrics) and the patients had no history of other systemic abnormalities or previous tumor or familial history of tumor. The exclusion criteria for the control participants were any diseases or systemic abnormalities.
DNA analysis
We used standard protocols to extract the genomic DNA from the peripheral blood leukocytes as described previously [29]. The CXCL1 gene was amplified by PCR with the primers shown in Table 2. PCR products were sequenced using standard protocols [30, 31] for genotype analysis.
Table 2.
PCR primers used for CXCL1 gene sequence analysis
| Exon | Forward primer | Reverse primer | Size (bp) | Tm (°C) |
|---|---|---|---|---|
| Exon1 | GCGGGCTGCATCAGTGGA | CGGGACTTACATGACTTCGGT | 595 | 59.8 |
| Exon2 | CTGCTGCTCCTGCTCCTGGTA | GGAAGGGAATCTCGTGAGGC | 370 | 59.4 |
| Exon3 | AAACCGAAGTCATGTAAGTCC | CAATAATCCCAATTTCTAGTCC | 336 | 54.0 |
| Exon4a | TTAGAGGTCCCTGCCACA | ATTCCCCTGCCTTCACAA | 629 | 52.2 |
| Exon4b | TGCAACATGCCAGCCACT | ATAGCAAATTGAACACCC | 460 | 50.0 |
SNP genotyping and statistical analysis
The variations within or near the CXCL1 gene were determined for the 300 sporadic OC cases and 400 normal controls. The DNA regions were amplified and the PCR products were sequenced to determine the genotypes; two researchers conducted the measurements independently. Overall OC genetics correlation analysis was also conducted. As previously reported, the statistical analyses and Hardy–Weinberg equilibrium tests of the patients and control populations were conducted [29, 32–34].
Results
Clinical data
The clinical diagnosis of all the participants was confirmed by specialists in Department of Gynaecology and Obstetrics in the Second Affiliated Hospital of Harbin Medical University, Harbin, China. These OC patients had no history of other systemic abnormalities or previous tumor or familial history of tumor. All the OC patients (n = 300, female, average age was 50.39 years, the min and max age were 18 and 81 respectively) and normal controls (n = 400, female, the average age was 49.68, the min and max age were 31 and 67 respectively) were recruited specifically for this study. There were no statistical differences in age composition between the two groups (Table 1).
SNP gene analyses
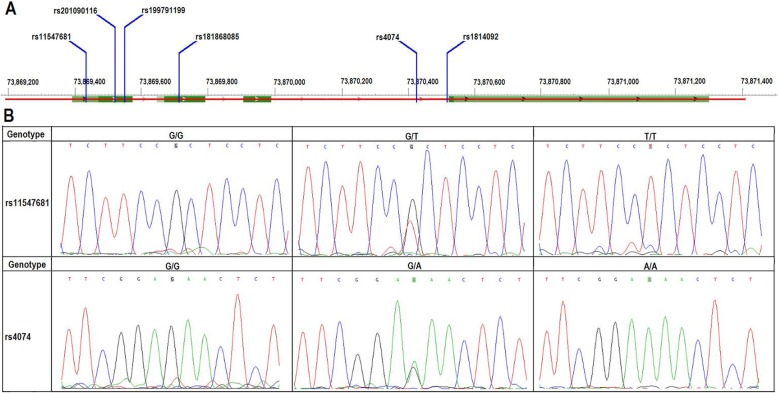
In order to test the hypothesis that germline common genetic variants in CXCL1 gene may be associated with the susceptibility to OC, we extracted the genomic DNA from the peripheral blood leukocytes of the participants and sequenced the CXCL1 gene to detect SNPs. We found six SNPs distributed on the gene, including rs11547681, rs201090116, rs199791199, rs181868085, rs4074 and rs1814092 (Fig. 1a). Analysis of these SNPs showed that the genetic heterozygosity of rs11547681 and rs4074 was very high (Fig. 1b), whereas that of rs201090116, rs199791199, rs181868085 and rs1814092 was very low and were excluded from further analysis.
Fig. 1.
Schematic diagrams and DNA sequence chromatogram of SNPs in CXCL1 gene. a: locations of rs11547681, rs201090116, rs199791199, rs181868085, rs4074 and rs1814092 within the CXCL1 gene; b: DNA sequence chromatogram of the three polymorphisms identified in the CXCL1 gene in all the population used for disease-association analyses
Polymorphism-disease association analyses
To test the hypothesized associations between CXCL1 variations and OC, we conducted analyses on the SNPs and found the variant rs11547681 within the 5’UTR of the gene was associated with the risk of OC in the Chinese Han population (Tables 3 and 4). Further, we analyzed the Hardy–Weinberg equilibrium test for the study population groups, and it was in line with equilibrium (Table 5).
Table 3.
The genotype and allele frequency of variations in 300 Chinese Han ovarian cancer patients and 400 normal controls
| SNP | Group | Genotype frequency (%) | Allele frequency (%) | ||||
|---|---|---|---|---|---|---|---|
| rs11547681 | Genotype | G/G | G/T | T/T | G | T | |
| OC | 300 | 185 (61.7) | 107 (35.7) | 8 (2.7) | 477 (79.5) | 123 (20.5) | |
| Controls | 400 | 294 (73.5) | 94 (23.5) | 12 (3.0) | 682 (85.3) | 118 (14.8) | |
| rs4074 | Genotype | G/G | G/A | A/A | G | A | |
| OC | 300 | 84 (28.0) | 153 (51.0) | 63 (21.0) | 321 (53.5) | 279 (46.5) | |
| Controls | 400 | 120 (30.0) | 207 (51.8) | 73 (18.3) | 447 (55.9) | 353 (44.1) | |
Note: OC Ovarian Cancer
Table 4.
rs11547681 variations within 5’UTR of the CXCL1 gene associated with risk of ovarian cancer in Chinese populations
| Variations | Type | Pearson Chi-square | Risk | |||||
|---|---|---|---|---|---|---|---|---|
| Value | Min count | df | Asymp. Sig. (2-sided) | Value | 95% CI-low | 95% CI-up | ||
| rs11547681 | Genotype | 12.412 | 8.57 | 2 | 0.002a | – | – | – |
| Allele | 7.954 | 103.29 | 1 | 0.005a | 0.671 | 0.508 | 0.886 | |
| rs4074 | Genotype | 0.921 | 58.29 | 2 | 0.631 | – | – | – |
| Allele | 0.781 | 270.86 | 1 | 0.377 | 0.909 | 0.735 | 1.124 | |
a: statistically significant
Table 5.
Hardy-Weinberg equilibrium test for the study population groups
| SNPs | Genotype | H-W equilibrium Testing | |||
|---|---|---|---|---|---|
| Homo/Hetero/Homozygote | O (HET) | E (HET) | P | ||
| rs11547681 | OC | 8/107/185 | 0.3567 | 0.3259 | 0.1541 |
| Controls | 12/94/294 | 0.2350 | 0.2515 | 0.2292 | |
| rs4074 | OC | 63/153/84 | 0.5100 | 0.4975 | 0.7281 |
| Controls | 73/207/120 | 05175 | 0.4931 | 0.3618 | |
Note: OC Ovarian Cancer
The genotype frequencies in the two groups were also analyzed by three genetic models (trend, dominant and recessive), and we found that the rs11547681 was associated with the risk of OC in trend and dominant models (Table 6). For the variant rs4074, we did not find statistical significance between the OC and control groups (chi-square tests; trend, dominant and recessive models). We compared the genotype frequency of the rs11547681 and rs4074 in the two groups and the data from the HapMap HCB population, but did not find the genotype frequency data of rs11547681 in the HapMap HCB population.
Table 6.
SNP rs11547681 within CXCL1 gene associated with the risk of ovarian cancer
| SNPs | Value | Trend model | Dominant model | Recessive model |
|---|---|---|---|---|
| rs11547681 | ChisQ | 8.0140 | 11.1100 | 0.0686 |
| P | 0.0046a | 0.0009a | 0.7933 | |
| rs4074 | ChisQ | 0.8121 | 0.3321 | 0.8282 |
| P | 0.3675 | 0.5644 | 0.3628 |
a: statistically significant
Clinical features comparative analysis
We also compared the clinical characteristics between the wild type, heterozygous variant and homozygous variant groups of the OC patients, but did not find statistically significant differences between the three groups in FIGO stages and pathological grades (Table 7).
Table 7.
Comparative analysis of clinical features between wild type, heterozygous variation and homozygous variation groups
| Clinical Index | Wild Type | heterozygous variation | homozygous variation | Chi-Square Test |
|---|---|---|---|---|
| TNM Stage (I/II/III/IV) | 59/32/92/2 | 30/23/54/1 | 2/1/5/0 | P = 0.956 |
| Pathological Grades(H/M/L/Non) | 101/15/36/33 | 54/7/31/15 | 5/0/3/0 | P = 0.375 |
H Pathological high Grade; M Pathological moderately Grade; L Pathological low Grade; Non No pathological grade
Discussion
In this study, we found that the SNP rs11547681 within the 5’UTR of CXCL1 gene was associated with OC. The microenvironment of tumors is an important factor in modulating cancer development, especially in organs that communicates with the outside environment [35, 36]. The cells surrounding those tumors usually release some factors, such as growth or inflammatory factors, which may regulate inflammation or progression of tumors [37, 38].
The human chemokines have strong activities on tumor cells, especially in cross talk of tumor cells and their host microenvironment [39]. CXCL1 is one member of the chemokines, which is a proinflammatory mediator in many inflammatory diseases. CXCL1 promotes and exacerbates growth and progression of many tumors [40]. By activating CXCR2, CXCL1 is associated with cancer cell growth and proliferation, tumor angiogenesis and metastasis [41, 42]. In tumor therapies, CXCL1 is also responsible for resistance of the cancer cells to several chemotherapeutic drugs [43].
In the OC cells, over-expression of the CXCL1 factor promotes the abilities of cellular proliferation and invasion in vitro [27, 44]. Progesterone and calcitriol can inhibit ovarian and endometrial cancer cell growth by attenuating the functions of CXCL1; if the expression of CXCL1 is reduced, the inhibitory effect of the two agents is also abrogated [45]. Conversely, when the expression of CXCL1 is increased, the activation of metastasis promoting gene p65 is also increased in OC cells [45]. The serum CXCL1 may be a novel tumor marker for OC diagnosis [46]. In this study, we found that the SNP rs11547681 in the CXCL1 gene was associated with OC. This finding provides novel insights into the special roles of CXCL1 factor for the pathogenesis, diagnosis and therapies of ovarian cancer.
Many chemokines exist as monomers or dimers in vivo. They function by binding to tissue glycosaminoglycans (GAGs) heparan sulfate (HS), chondroitin sulfate (CS) or dermatan sulfate (DS) [47, 48]. GAGs bind to a diversity of protein classes [49], so in order to interact with GAGs, the sequence of chemokine must determine the selectivity, affinity and geometry [50]. CXCL1 belong to the CXC chemokines subset, characterized by the N-terminal‘ELR’motifs [51]. The amino acid residues located within the N terminal loop and C terminal helix of CXCL1 factor mediate HS binding, and the participation of other residues may result in a very different binding geometry for CXCL1 [50]. The SNP rs11547681 associated with OC is located within the 5’UTR of CXCL1 gene. The 5’UTR and 3’UTR sequences regulate expression of genes [52, 53]. The 5’UTR sequences of gene bind with miRNAs and may be involved in gene expression, protein translation or disease pathogenesis [54]. In previous studies, we reported SNPs within the 5’UTR or 3’UTR sequences that are associated with diseases [31, 32, 55]. The results of this work further emphasized the important roles of 5’UTR sequences for CXCL1 factor functions.
In conclusion, we demonstrated the associations of CXCL1 variants rs11547681 with the risk of ovarian cancer in the Chinese Han population and updated our understanding on 5’UTR for CXCL1 functions, providing new information on the pathogenesis of cancers especially ovarian cancer.
Acknowledgements
The authors thank the patients and their families for their cooperation and participation in this study.
Financial disclosures
There are no financial disclosures from any authors.
Informed consent
Informed consent was obtained from all individual participants.
Authors’ contributions
Conceptualization: FF L, SL L; methodology: FF L, SL L, M G, C X; software: FF L, SL L, C X; formal analysis: FF L, SL L, M G, C X; investigation: M G, C X, YZ C, QW S, Y Y, YH H; resources: M G; data curation: FF L, SL L, M G, C X; writing: FF L, SL L; funding acquisition: FF L, SL L. The author (s) read and approved the final manuscript.
Funding
This work was supported by grants from Health and Family Planning Commission of Heilongjiang province Foundation (2017–077), Postdoctoral Foundaction of Heilongjiang Province and grants of National Natural Science Foundation of China (NSFC81271786, 81030029, 81671980, 81871623), Heilongjiang Innovation Research Foundation for College Students (201810226070). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used in the present study are available from the corresponding authors with reasonable requests.
Ethics approval and consent to participate
Ethics Committee of Harbin Medical University.
Consent for publication
Not applicable.
Competing interests
All the authors have declared that no competing interests exist.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Man Guo and Chao Xu contributed equally to this work.
Contributor Information
Fei-Feng Li, Email: Lifeifeng@hrbmu.edu.cn.
Shu-Lin Liu, Email: slliu@hrbmu.edu.cn.
References
- 1.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loomis D, Huang W, Chen G. The International Agency for Research on Cancer (IARC) evaluation of the carcinogenicity of outdoor air pollution: focus on China. Chin J Cancer. 2014;33(4):189–196. doi: 10.5732/cjc.014.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karim-Kos HE, de Vries E, Soerjomataram I, Lemmens V, Siesling S, Coebergh JW. Recent trends of cancer in Europe: a combined approach of incidence, survival and mortality for 17 cancer sites since the 1990s. Eur J Cancer. 2008;44(10):1345–1389. doi: 10.1016/j.ejca.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 5.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H, Ardanaz E, et al. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE--5-a population-based study. Lancet Oncol. 2014;15(1):23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 6.Ottevanger PB. Ovarian cancer stem cells more questions than answers. Semin Cancer Biol. 2017;44:67–71. doi: 10.1016/j.semcancer.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA. A systematic review and meta-analysis of family history and risk of ovarian cancer. Br J Obstet Gynaecol. 1998;105(5):493–499. doi: 10.1111/j.1471-0528.1998.tb10148.x. [DOI] [PubMed] [Google Scholar]
- 8.Jervis S, Song H, Lee A, Dicks E, Tyrer J, Harrington P, Easton DF, Jacobs IJ, Pharoah PP, Antoniou AC. Ovarian cancer familial relative risks by tumour subtypes and by known ovarian cancer genetic susceptibility variants. J Med Genet. 2014;51(2):108–113. doi: 10.1136/jmedgenet-2013-102015. [DOI] [PubMed] [Google Scholar]
- 9.Alsop K, Fereday S, Meldrum C, DeFazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C et al: BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian ovarian Cancer study group. J Clin Oncol 2012, 30(21):2654–2663. [DOI] [PMC free article] [PubMed]
- 10.Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107(11). 10.1093/jnci/djv214.. [DOI] [PMC free article] [PubMed]
- 11.Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, Edlund CK, Conti D, Harrington P, Fraser L, et al. Contribution of Germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian Cancer in the population. J Clin Oncol. 2015;33(26):2901–2907. doi: 10.1200/JCO.2015.61.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beral V, Doll R, Hermon C, Peto R, Reeves G. Ovarian cancer and oral contraceptives: collaborative reanalysis of data from 45 epidemiological studies including 23,257 women with ovarian cancer and 87,303 controls. Lancet. 2008;371(9609):303–314. doi: 10.1016/S0140-6736(08)60167-1. [DOI] [PubMed] [Google Scholar]
- 13.Sieh W, Salvador S, McGuire V, Weber RP, Terry KL, Rossing MA, Risch H, Wu AH, Webb PM, Moysich K, et al. Tubal ligation and risk of ovarian cancer subtypes: a pooled analysis of case-control studies. Int J Epidemiol. 2013;42(2):579–589. doi: 10.1093/ije/dyt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wentzensen N, Poole EM, Trabert B, White E, Arslan AA, Patel AV, Setiawan VW, Visvanathan K, Weiderpass E, Adami HO, et al. Ovarian Cancer risk factors by histologic subtype: an analysis from the ovarian Cancer cohort consortium. J Clin Oncol. 2016;34(24):2888–2898. doi: 10.1200/JCO.2016.66.8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beral V, Gaitskell K, Hermon C, Moser K, Reeves G, Peto R. Menopausal hormone use and ovarian cancer risk: individual participant meta-analysis of 52 epidemiological studies. Lancet. 2015;385(9980):1835–1842. doi: 10.1016/S0140-6736(14)61687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, et al. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol. 2013;31(20):2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ness RB, Cottreau C. Possible role of ovarian epithelial inflammation in ovarian cancer. J Natl Cancer Inst. 1999;91(17):1459–1467. doi: 10.1093/jnci/91.17.1459. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen CB, Kjaer SK, Albieri V, Bandera EV, Doherty JA, Hogdall E, Webb PM, Jordan SJ, Rossing MA, Wicklund KG, et al. Pelvic inflammatory disease and the risk of ovarian Cancer and borderline ovarian tumors: a pooled analysis of 13 case-control studies. Am J Epidemiol. 2017;185(1):8–20. doi: 10.1093/aje/kww161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonovas S, Filioussi K, Sitaras NM. Paracetamol use and risk of ovarian cancer: a meta-analysis. Br J Clin Pharmacol. 2006;62(1):113–121. doi: 10.1111/j.1365-2125.2005.02526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baandrup L, Kjaer SK, Olsen JH, Dehlendorff C, Friis S. Low-dose aspirin use and the risk of ovarian cancer in Denmark. Ann Oncol. 2015;26(4):787–792. doi: 10.1093/annonc/mdu578. [DOI] [PubMed] [Google Scholar]
- 21.Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol 2017. 28(suppl_8):viii1–7. [DOI] [PMC free article] [PubMed]
- 22.Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2(12):1125–1131. doi: 10.1158/2326-6066.CIR-14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 24.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354(6):610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 25.Kuo PL, Shen KH, Hung SH, Hsu YL. CXCL1/GROalpha increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-kappaB/HDAC1 epigenetic regulation. Carcinogenesis. 2012;33(12):2477–2487. doi: 10.1093/carcin/bgs299. [DOI] [PubMed] [Google Scholar]
- 26.Peng W, Ye Y, Rabinovich BA, Liu C, Lou Y, Zhang M, Whittington M, Yang Y, Overwijk WW, Lizee G, et al. Transduction of tumor-specific T cells with CXCR2 chemokine receptor improves migration to tumor and antitumor immune responses. Clin Cancer Res. 2010;16(22):5458–5468. doi: 10.1158/1078-0432.CCR-10-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolitho C, Hahn MA, Baxter RC, Marsh DJ. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr Relat Cancer. 2010;17(4):929–940. doi: 10.1677/ERC-10-0107. [DOI] [PubMed] [Google Scholar]
- 28.Lee CH, Syu SH, Liu KJ, Chu PY, Yang WC, Lin P, Shieh WY. Interleukin-1 beta transactivates epidermal growth factor receptor via the CXCL1-CXCR2 axis in oral cancer. Oncotarget. 2015;6(36):38866–38880. doi: 10.18632/oncotarget.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng X, Zhou J, Li FF, Yan P, Zhao EY, Hao L, Yu KJ, Liu SL. Characterization of nodal/TGF-lefty signaling pathway gene variants for possible roles in congenital heart diseases. PLoS One. 2014;9(8):e104535. doi: 10.1371/journal.pone.0104535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li FFWX, Zhu M, Xiao H, Yang Q, et al. Identification of a location at chromosome 19p in a big Chinese family with Charcot-Marie-tooth disease. J Mol Biomark Diagn. 2013;4:144. doi: 10.4172/2155-9929.1000144. [DOI] [Google Scholar]
- 31.Zhang Q, Zhou J, Lei H, Zhu CY, Li FF, Zheng D, Liu SL. RBPJ polymorphisms associated with cerebral infarction diseases in Chinese Han population: a clinical trial/experimental study (CONSORT compliant) Medicine (Baltimore) 2018;97(31):e11420. doi: 10.1097/MD.0000000000011420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li FF, Zhou J, Zhao DD, Yan P, Li X, Han Y, Li XS, Wang GY, Yu KJ, Liu SL. Characterization of SMAD3 gene variants for possible roles in ventricular Septal defects and other congenital heart diseases. PLoS One. 2015;10(6):e0131542. doi: 10.1371/journal.pone.0131542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li FF, Zhu XD, Yan P, Jin MH, Yue H, Zhang Q, Fu J, Liu SL: Characterization of variations in IL23A and IL23R genes: possible roles in multiple sclerosis and other neuroinflammatory demyelinating diseases. Aging (Albany NY) 2016. [DOI] [PMC free article] [PubMed]
- 34.Li X, Shi S, Li FF, Cheng R, Han Y, Diao LW, Zhang Q, Zhi JX, Liu SL. Characterization of soluble N-ethylmaleimide-sensitive factor attachment protein receptor gene STX18 variations for possible roles in congenital heart diseases. Gene. 2017;598:79–83. doi: 10.1016/j.gene.2016.10.043. [DOI] [PubMed] [Google Scholar]
- 35.Itoh H, Kadomatsu T, Tanoue H, Yugami M, Miyata K, Endo M, Morinaga J, Kobayashi E, Miyamoto T, Kurahashi R, et al. TET2-dependent IL-6 induction mediated by the tumor microenvironment promotes tumor metastasis in osteosarcoma. Oncogene. 2018;37(22):2903–2920. doi: 10.1038/s41388-018-0160-0. [DOI] [PubMed] [Google Scholar]
- 36.Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY, Kuo PL. Interaction between tumor-associated dendritic cells and colon cancer cells contributes to tumor progression via CXCL1. Int J Mol Sci. 2018;19(8). 10.3390/ijms19082427. [DOI] [PMC free article] [PubMed]
- 37.Danelli L, Frossi B, Pucillo CE. Mast cell/MDSC a liaison immunosuppressive for tumor microenvironment. Oncoimmunology. 2015;4(4):e1001232. doi: 10.1080/2162402X.2014.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, Valle LD, Trillo-Tinoco J, Maj T, Zou W, et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances Cancer therapies. Cancer Immunol Res. 2015;3(11):1236–1247. doi: 10.1158/2326-6066.CIR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Yue J, Jiang Z, Zhou R, Xie R, Xu Y, Wu S. CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma. Cell Death Dis. 2017;8(5):e2790. doi: 10.1038/cddis.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Planaguma A, Domenech T, Pont M, Calama E, Garcia-Gonzalez V, Lopez R, Auli M, Lopez M, Fonquerna S, Ramos I, et al. Combined anti CXC receptors 1 and 2 therapy is a promising anti-inflammatory treatment for respiratory diseases by reducing neutrophil migration and activation. Pulm Pharmacol Ther. 2015;34:37–45. doi: 10.1016/j.pupt.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Divella R, Daniele A, DEL R, Simone M, Naglieri E, Savino E, Abbate I, Gadaleta CD, Ranieri G. Circulating levels of VEGF and CXCL1 are predictive of metastatic Organotropismin in patients with colorectal Cancer. Anticancer Res. 2017;37(9):4867–4871. doi: 10.21873/anticanres.11895. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Liu J, Jiang Q, Deng J, Xu F, Chen X, Cheng F, Zhang Y, Yao Y, Xia Z, et al. Human adipose-derived Mesenchymal stem cell-secreted CXCL1 and CXCL8 facilitate breast tumor growth by promoting angiogenesis. Stem Cells. 2017;35(9):2060–2070. doi: 10.1002/stem.2643. [DOI] [PubMed] [Google Scholar]
- 43.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150(1):165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Huang N, Wang Q, Yang ZJ, Li L. Effect of CXCL1 gene expression on the biological function of epithelial ovarian carcinoma cells in vitro. Zhonghua Zhong Liu Za Zhi. 2013;35(2):109–113. doi: 10.3760/cma.j.issn.0253-3766.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Kavandi L, Collier MA, Nguyen H, Syed V. Progesterone and calcitriol attenuate inflammatory cytokines CXCL1 and CXCL2 in ovarian and endometrial cancer cells. J Cell Biochem. 2012;113(10):3143–3152. doi: 10.1002/jcb.24191. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Li D, Zhang W, Tang B, Li QQ, Li L. Evaluation of proteomics-identified CCL18 and CXCL1 as circulating tumor markers for differential diagnosis between ovarian carcinomas and benign pelvic masses. Int J Biol Markers. 2011;26(4):262–73. . [DOI] [PubMed]
- 47.Rajarathnam K, Sepuru KM, Joseph PRB, Sawant KV, Brown AJ. Glycosaminoglycan interactions fine-tune chemokine-mediated neutrophil trafficking: structural insights and molecular mechanisms. J Histochem Cytochem. 2018;66(4):229–239. doi: 10.1369/0022155417739864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stone MJ, Hayward JA, Huang C, Z EH, Sanchez J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int J Mol Sci. 2017;18(2). 10.3390/ijms18020342. [DOI] [PMC free article] [PubMed]
- 49.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des. 2008;72(6):455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 50.Sepuru KM, Rajarathnam K. Structural basis of chemokine interactions with Heparan sulfate, chondroitin sulfate, and Dermatan sulfate. J Biol Chem. 2019. [DOI] [PMC free article] [PubMed]
- 51.Rajarathnam K, Schnoor M, Richardson RM, Rajagopal S. How do chemokines navigate neutrophils to the target site: dissecting the structural mechanisms and signaling pathways. Cell Signal. 2019;54:69–80. doi: 10.1016/j.cellsig.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lou F, Ma HN, Xu L, Chen M, Zhu YB. Two polymorphisms of CD44 3'UTR weaken the binding of miRNAs and associate with naso-pharyngeal carcinoma in a Chinese population. Eur Rev Med Pharmacol Sci. 2014;18(17):2444–2452. [PubMed] [Google Scholar]
- 53.Fujimuro T, Matsui T, Nitanda Y, Matta T, Sakumura Y, Saito M, Kohno K, Nakahata Y, Bessho Y. Hes7 3'UTR is required for somite segmentation function. Sci Rep. 2014;4:6462. doi: 10.1038/srep06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Li FF, Han Y, Shi S, Li X, Zhu XD, Zhou J, Shao QL, Li XQ, Liu SL. Characterization of transcriptional repressor gene MSX1 variations for possible associations with congenital heart diseases. PLoS One. 2015;10(11):e0142666. doi: 10.1371/journal.pone.0142666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the present study are available from the corresponding authors with reasonable requests.