Abstract
Background
Microsatellites or simple sequence repeats (SSRs) have become the most significant DNA marker technology used in genetic research. The availability of complete draft genomes for a number of Palmae species has made it possible to perform genome-wide analysis of SSRs in these species. Palm trees are tropical and subtropical plants with agricultural and economic importance due to the nutritional value of their fruit cultivars.
Objective
This is the first comprehensive study examining and comparing microsatellites in completely-sequenced draft genomes of Palmae species.
Methods
We identified and compared perfect SSRs with 1–6 bp nucleotide motifs to characterize microsatellites in Palmae species using PERF v0.2.5. We analyzed their relative abundance, relative density, and GC content in five palm species: Phoenix dactylifera, Cocos nucifera, Calamus simplicifolius, Elaeis oleifera, and Elaeis guineensis.
Results
A total of 118241, 328189, 450753, 176608, and 70694 SSRs were identified, respectively. The six repeat types were not evenly distributed across the five genomes. Mono- and dinucleotide SSRs were the most abundant, and GC content was highest in tri- and hexanucleotide SSRs.
Conclusion
We envisage that this analysis would further substantiate more in-depth computational, biochemical, and molecular studies on the roles SSRs may play in the genome organization of the palm species. The current study contributes a detailed characterization of simple sequence repeats in palm genomes.
Electronic supplementary material
The online version of this article (10.1007/s13258-020-00924-w) contains supplementary material, which is available to authorized users.
Keywords: Arecaceae, Palmae family, Microsatellite, SSR abundance, Molecular marker
Introduction
Plants in the palm family (Arecaceae or Palmae) are important economic crops that are widely cultivated in arid and semi-arid regions of North Africa, the Sahara, the Middle East, and eastward to the Indus Valley. Palmae is a distinct family of monocotyledon species with up to 2800 species currently known, which are distributed over 202 genera (Xiao et al. 2016). Palm plants are critical ecological and socioeconomic resources for many countries, including Saudi Arabia; they play important roles in food security, wood for building, ornamentals, and industrial materials (Barrow 1998; Aberlenc-Bertossi et al. 2014). The date palm (Phoenix dactylifera), coconut (Cocos nucifera), and African oil palm (Elaeis oleifera) are the most economically important fruit crops in the palm family. There are more than 3000 cultivars of date palm worldwide, of which 60 are considered to be important in the global market (Moussouni et al. 2017). Despite the increasing number of genomic studies on Palmae trees, little genome-wide characterization has been performed on these plants for the purposes of conservation and genetic assessment.
Assessment of genetic diversity is crucial for the conservation of palm cultivar germplasm. Estimates of the genetic diversity of palm plant germplasm have traditionally been based on morphological information (Elhoumaizi et al. 2002). However, morphological markers do not reliably provide accurate assessments because they are highly affected by environmental factors. Molecular markers are more informative at any developmental stage of the plant. Molecular breeding through marker-assisted selection would also expedite the genetic improvement of palm cultivars (Zhao et al. 2012). Microsatellites or simple sequence repeats (SSRs) are very useful markers for the analysis of plant diversity. In addition, SSR markers can be used for DNA fingerprinting to distinguish among closely-related palm cultivars. SSRs are tandem repeats of one to six base pairs per repeat unit, and are widely distributed in eukaryotic and prokaryotic genomes (Xu et al. 2016; Yang et al. 2003). Rapid expansions and contractions of these repeats due to replication slippage may make them useful for carrying out population genetics studies within a species (Huntley and Golding 2006).
The recent release of draft whole-genome sequences for several palm species provides an opportunity to carry out post-genomic analysis in order to identify and compare the distributions of SSRs across palm genomes. To date, draft genome sequences have been released for five species in the Palmae family: P. dactylifera, C. nucifera, Calamus simplicifolius, E. oleifera, and E. guineensis. This study aimed to screen these five genome sequences for microsatellites, detect SSR motifs, and analyze the frequency and distribution of SSRs.
Materials and methods
Genome sequences
Draft genome sequences for five palm tree species (P. dactylifera, C. nucifera, C. simplicifolius, E. oleifera, and E. guineensis) were selected for the analysis of SSR distributions at the genomic level. These genomes have been assembled at the scaffold level according to the genomic resources of the National Center for Biotechnology Information (NCBI). The genome sequences with accession numbers of GCA_000413155.1, GCA_003604295.1, GCA_900491605.1, GCA_000441515.1, and GCA_001672495.1 were downloaded in FASTA format from the Genomes FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
Genomic quality assessment
Completeness of the genome assemblies was assessed with Benchmarking Universal Single Copy Ortholog (BUSCO) v3.0.2 (Simão et al. 2015) with default settings. BUSCO genes are good candidates for evaluating genome completeness because from an evolutionary perspective, they are expected to be found in the tested genome (Simão et al. 2015; Waterhouse et al. 2017). The BUSCO tool analyzed each genome assembly state in terms of complete BUSCOs, complete and single-copy BUSCOs, complete and duplicated BUSCOs, fragmented BUSCO, and missing BUSCOs using a plant-specific database (embryophyta_odb9) that consisted of 1440 total BUSCO groups from 30 species.
Identification of microsatellites
Genome-wide SSR mining was performed by scanning each entire genome with the software PERF v0.2.5 (Avvaru et al. 2017). A number of criteria were adopted to identify perfect SSRs. Specifically, repeat sizes of 1 to 6 nucleotides long were searched, and minimum repeat numbers were restricted to 12 repeats for mononucleotides, 7 repeats for dinucleotides, 5 repeats for trinucleotides, and 4 repeats for tetra-, penta- and hexanucleotides, consistent with previous studies (Liu et al. 2017; Qi et al. 2018). The remaining parameters were set as default. Repeats with unit patterns being circular permutations and/or reverse complements were deemed as one type in this study (Jurka and Pethiyagoda 1995; Li et al. 2009). For instance, ACT contains ACT, TAC, CTA, TGA, ATG, and GAT in different reading frames or on the complementary strand. Different types of SSR repeats or motifs were compared in terms of relative frequency (the number of SSRs per Mb) and relative density (the total length of SSRs in bp per Mb). All graphical and statistical analyses were performed in the R programming environment (version 3.4.3) (R Core Team, 2017).
Results
Assessing the completeness of the genome assemblies
We adopted the BUSCO plant lineage dataset, which consisted of 1440 single-copy orthologs for the Embryophyta lineage, to assess the completeness of each of the five genome assemblies. The C. nucifera genome assembly had the highest BUSCO scores among those surveyed (Fig. 1), with 1311 (91%) complete BUSCOs (1200 complete single-copy and 111 complete duplicated BUSCOs); 3.80% of sequences were fragmented (54 BUSCOs) and 5.20% were considered missing (75 BUSCOs). The BUSCO scores of C. nucifera, P. dactylifera, and C. simplicifolius genome assemblies were comparable, and higher than the two palm assemblies from genus Elaeis (E. oleifera and E. guineensis). However, the E. guineensis genome assembly showed low BUSCO scores relative to all four of the other assemblies (Fig. 1). In the E. guineensis genome assembly, only 60 (4.20%) complete BUSCOs were identified (54 complete single-copy and 6 complete duplicated BUSCOs).
Fig. 1.
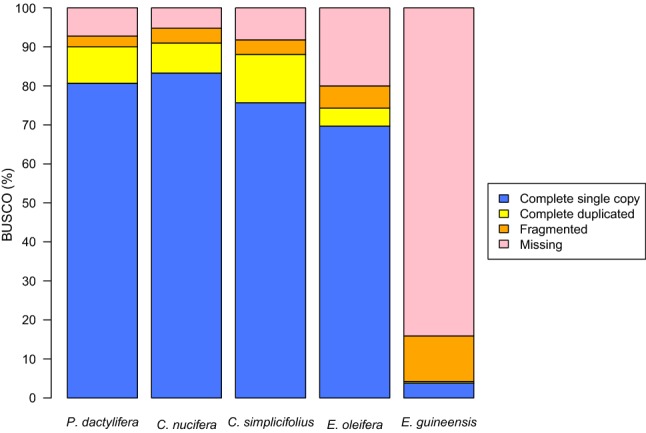
Genome assembly evaluation. The BUSCO embryophyta_odb9 dataset, including 1440 BUSCOs, was used to assess the five genome assemblies
Identification and characterization of microsatellites in palm genomes
In the five palm genomes (P. dactylifera, C. nucifera, C. simplicifolius, E. oleifera, and E. guineensis), a total of 118241, 328189, 450753, 176608 and 70694 perfect SSRs were identified (Files S1–S5) with frequencies ranging from 125.90 to 229.88 SSR/Mb, respectively (Table 1). About 0.40, 0.36, 0.44, 0.23, and 0.26% of the genome was occupied by perfect SSRs, respectively. The relative densities ranged 2297.25–4373.04 SSR/Mb, while the mean lengths of SSRs were approximately 19 bp for C. nucifera and C. simplicifolius, and approximately 18 bp for P. dactylifera, E. oleifera, and E. guineensis.
Table 1.
Overview of the five palm genomes
| Parameter | P. dactylifera | C. nucifera | C. simplicifolius | E. oleifera | E. guineensis |
|---|---|---|---|---|---|
| Common name | Date palm | Coconut palm | Rattan palm | American oil palm | African oil palm |
| Genome size (Mb) | 556.48 | 1839.17 | 1960.81 | 1402.73 | 499.03 |
| GC content (%) | 35.86 | 31.79 | 39.65 | 28.12 | 32.10 |
| Number of SSRs | 118241 | 328189 | 450753 | 176608 | 70694 |
| Total length of SSRs (bp) | 2233944 | 6522297 | 8574690 | 3222415 | 1279910 |
| Average length (bp) | 18.89 | 19.87 | 19.02 | 18.25 | 18.11 |
| Frequency (SSR/Mb) | 212.48 | 178.44 | 229.88 | 125.90 | 141.66 |
| Density (bp/Mb) | 4014.41 | 3546.32 | 4373.04 | 2297.25 | 2564.80 |
| Genome SSRs content (%) | 0.40 | 0.36 | 0.44 | 0.23 | 0.26 |
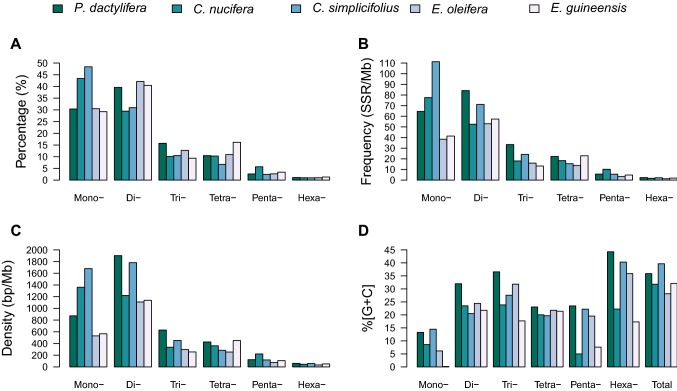
The number, length, relative frequency, relative density, and percentage of the six types of SSRs are shown in Table 2. The percentage, relative frequencies, and densities of different SSR types were found to vary greatly between the five palm genomes (Fig. 2). Dinucleotide SSRs were the most frequent type in P. dactylifera, E. oleifera, and E. guineensis, with the highest frequencies of 84.15, 53.01, and 57.37 SSR/Mb, accounting for 39.61, 42.11, and 40.50% of SSRs in these genomes, respectively (Fig. 2a, b). Mononucleotide SSRs were the most abundant type in C. nucifera and C. simplicifolius, with the highest frequencies of 77.53 and 111.28 SSR/Mb, occupying about 43.45 and 48.41% of all SSRs in those genomes, respectively. Mononucleotide SSRs were also the second most frequent in P. dactylifera, E. oleifera, and E. guineensis, while dinucleotide SSRs were the second most abundant type in C. nucifera and C. simplicifolius. Tri- and tetranucleotide SSRs were more frequent than pentanucleotide SSRs in all five genomes. Hexanucleotide SSRs were the least abundant across all five genomes, with a frequency of below 2.38 SSR/Mb, and accounted for only 1.12, 1.00, 0.98, 1.01, and 0.07% of all SSRs in these genomes, respectively (Fig. 2b). Dinucleotide SSRs were found to have the highest densities, ranging from 1109.95 to 1901.36 bp/Mb in P. dactylifera, C. simplicifolius, E. oleifera, and E. guineensis, whereas mononucleotide SSRs had the highest density (1360.92 bp/Mb) in C. nucifera (Fig. 2c).
Table 2.
Number, length, frequency, and density of mono- to hexanucleotide repeats in palm genomes
| Repeat type | Parameter | P. dactylifera | C. nucifera | C. simplicifolius | E. oleifera | E. guineensis |
|---|---|---|---|---|---|---|
| Mono- | Number of SSRs | 35942 | 142586 | 218192 | 53939 | 20684 |
| Total length (bp) | 485527 | 2502957 | 3293664 | 743482 | 282341 | |
| Average length (bp) | 13.51 | 17.55 | 15.10 | 13.78 | 13.65 | |
| Frequency (SSR/Mb) | 64.59 | 77.53 | 111.28 | 38.45 | 41.45 | |
| Density (bp/Mb) | 872.50 | 1360.92 | 1679.75 | 530.03 | 565.78 | |
| Di- | Number of SSRs | 46830 | 96689 | 139482 | 74362 | 28630 |
| Total length (bp) | 1058068 | 2245158 | 3490730 | 1556956 | 567482 | |
| Average length (bp) | 22.59 | 23.22 | 25.03 | 20.94 | 19.82 | |
| Frequency (SSR/Mb) | 84.15 | 52.57 | 71.14 | 53.01 | 57.37 | |
| Density (bp/Mb) | 1901.36 | 1220.75 | 1780.25 | 1109.95 | 1137.17 | |
| Tri- | Number of SSRs | 18603 | 33229 | 47311 | 22446 | 6616 |
| Total length (bp) | 350310 | 619926 | 885591 | 415284 | 127803 | |
| Average length (bp) | 18.83 | 18.66 | 18.72 | 18.50 | 19.32 | |
| Frequency (SSR/Mb) | 33.43 | 18.07 | 24.13 | 16.00 | 13.26 | |
| Density (bp/Mb) | 629.51 | 337.07 | 451.65 | 296.06 | 256.10 | |
| Tetra- | Number of SSRs | 12396 | 33721 | 30357 | 19309 | 11425 |
| Total length (bp) | 236976 | 662892 | 556244 | 356136 | 224040 | |
| Average length (bp) | 19.12 | 19.66 | 18.32 | 18.44 | 19.61 | |
| Frequency (SSR/Mb) | 22.28 | 18.34 | 15.48 | 13.77 | 22.89 | |
| Density (bp/Mb) | 425.85 | 360.43 | 283.68 | 253.89 | 448.95 | |
| Penta- | Number of SSRs | 3149 | 18699 | 11011 | 4762 | 2402 |
| Total length (bp) | 68845 | 408660 | 233375 | 102875 | 52750 | |
| Average length (bp) | 21.86 | 21.86 | 21.20 | 21.60 | 21.96 | |
| Frequency (SSR/Mb) | 5.66 | 10.17 | 5.62 | 3.40 | 4.81 | |
| Density (bp/Mb) | 123.72 | 222.20 | 119.02 | 73.34 | 105.71 | |
| Hexa- | Number of SSRs | 1321 | 3265 | 4400 | 1790 | 937 |
| Total length (bp) | 34218 | 82704 | 115086 | 47682 | 25494 | |
| Average length (bp) | 25.90 | 25.33 | 26.16 | 26.64 | 27.21 | |
| Frequency (SSR/Mb) | 2.37 | 1.78 | 2.24 | 1.28 | 1.88 | |
| Density (bp/Mb) | 61.49 | 44.97 | 58.69 | 33.99 | 51.09 |
Fig. 2.
Comparison of percentage, frequency, density, and GC content of SSRs in palm genomes. Percentages were calculated according to the total number of each SSR type divided by the total number of SSRs. ABCD represent percentage, frequency, density, and GC content of SSRs, respectively
The GC content of different types of SSRs was investigated for the five genomes (Fig. 2d). Hexanucleotide SSRs had the highest GC content in P. dactylifera (44.24%), C. simplicifolius (40.28%), and E. oleifera (35.87%). Trinucleotide SSRs were found to have the highest GC content in C. nucifera (23.83%), while dinucleotide SSRs had the highest GC content in E. guineensis (21.76%). Mononucleotide SSRs were found to have the least GC content in P. dactylifera, C. simplicifolius, E. oleifera, and E. guineensis, at only 13.29, 14.50, 6.12, and 0.16%, respectively, while pentanucleotide SSRs had the least GC content in C. nucifera, at only 4.97%.
Abundance and repeat numbers for different microsatellite motifs
The microsatellites in palm genomes were determined to be relatively AT-rich. To gain insight into this characteristic, we analyzed SSR motif composition. The most abundant SSR motifs were found to vary with species. The degenerated number of repeat motifs was found to be 2, 4, and 10; these were identical between species for mono- to trinucleotide repeat types and were different for tetranucleotide, pentanucleotide, and hexanucleotide repeat types.
Mononucleotide repeats
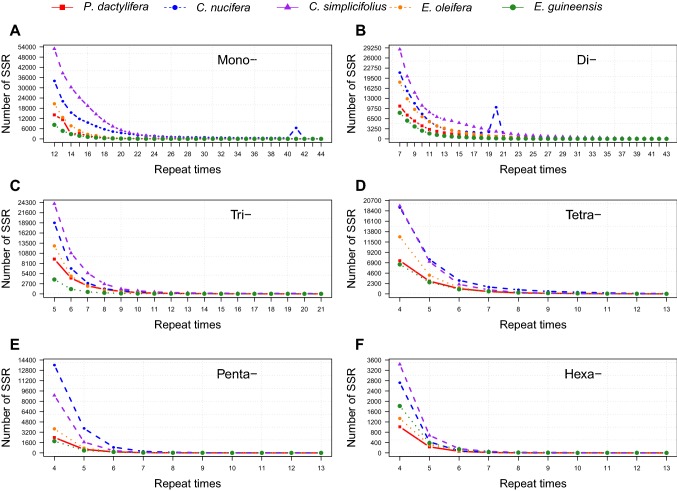
The predominant mononucleotide motif type was , with a total number of 31368, 131905, 188454, 50637 and 20651 SSRs in the five genomes, accounting for 87.27, 92.51, 86.37, 93.88, and 99.84% of all mononucleotide SSRs, respectively (Table 3). The frequencies and densities of were 36.10–96.11 SSR/Mb and 497.57–1436.24 bp/Mb, respectively, while the average lengths were 13.42–17.35 bp. The motif type was far less abundant than , accounting for only 0.16–13.63% of all mononucleotide SSRs in the five genomes. Mononucleotide repeats ranged from 12 to 115, 12 to 44, 12 to 175, 12 to 65, and 12 to 83 repeats in length, respectively. Repeat numbers between 12–22, 12–41, 12–36, 12–24, and 12–21 accounted for 99.18, 99.98, 99.71, 99.01, and 98.66% of the total number of mononucleotide SSRs in these genomes, respectively (Fig. 3a).
Table 3.
Number, length, frequency, and density of the most frequent SSR motifs for each SSR type in palm genomes
| Repeat motif type | Parameter | P. dactylifera | C. nucifera | C. simplicifolius | E. oleifera | E. guineensis |
|---|---|---|---|---|---|---|
| A | Number of SSRs | 31368 | 131905 | 188454 | 50637 | 20651 |
| Total length (bp) | 421010 | 2288571 | 2816200 | 697952 | 281897 | |
| Frequency (SSR/Mb) | 56.37 | 71.72 | 96.11 | 36.10 | 41.38 | |
| Density (bp/Mb) | 756.56 | 1244.35 | 1436.24 | 497.57 | 564.89 | |
| C | Number of SSRs | 4574 | 10681 | 29738 | 3302 | 33 |
| Total length (bp) | 64517 | 214386 | 477464 | 45530 | 444 | |
| Frequency (SSR/Mb) | 8.22 | 5.81 | 15.17 | 2.35 | 0.07 | |
| Density (bp/Mb) | 115.94 | 116.57 | 243.50 | 32.46 | 0.89 | |
| AT | Number of SSRs | 16405 | 49505 | 73131 | 36840 | 15913 |
| Total length (bp) | 384960 | 1194804 | 2064214 | 801040 | 321888 | |
| Frequency (SSR/Mb) | 29.48 | 26.92 | 37.30 | 26.26 | 31.89 | |
| Density (bp/Mb) | 691.78 | 649.64 | 1052.74 | 571.06 | 645.03 | |
| AG | Number of SSRs | 24498 | 37493 | 56621 | 27114 | 6043 |
| Total length (bp) | 548096 | 854432 | 1214612 | 549626 | 115996 | |
| Frequency (SSR/Mb) | 44.02 | 20.39 | 28.88 | 19.33 | 12.11 | |
| Density (bp/Mb) | 984.93 | 464.57 | 619.44 | 391.83 | 232.44 | |
| AC | Number of SSRs | 5716 | 9393 | 9444 | 10139 | 6583 |
| Total length (bp) | 121652 | 191274 | 207532 | 202016 | 128190 | |
| Frequency (SSR/Mb) | 10.27 | 5.11 | 4.82 | 7.23 | 13.19 | |
| Density (bp/Mb) | 218.609 | 104.00 | 105.84 | 144.02 | 256.88 | |
| AAT | Number of SSRs | 4702 | 14147 | 15355 | 6666 | 3073 |
| Total length (bp) | 100233 | 287169 | 324243 | 137616 | 66840 | |
| Frequency (SSR/Mb) | 8.45 | 7.69 | 7.83 | 4.75 | 6.16 | |
| Density (bp/Mb) | 180.12 | 156.14 | 165.36 | 98.11 | 133.94 | |
| AAG | Number of SSRs | 5349 | 10634 | 14152 | 7636 | 2179 |
| Total length (bp) | 97662 | 187221 | 245298 | 135789 | 37794 | |
| Frequency (SSR/Mb) | 9.61 | 5.78 | 7.22 | 5.44 | 4.37 | |
| Density (bp/Mb) | 175.50 | 101.80 | 125.10 | 96.80 | 75.74 | |
| AGG | Number of SSRs | 2977 | 2357 | 3512 | 2774 | 204 |
| Total length (bp) | 54216 | 41088 | 63447 | 48822 | 3342 | |
| Frequency (SSR/Mb) | 5.35 | 1.28 | 1.79 | 1.98 | 0.41 | |
| Density (bp/Mb) | 97.43 | 22.34 | 32.36 | 34.81 | 6.70 | |
| ATC | Number of SSRs | 1501 | 1846 | 7215 | 1301 | 623 |
| Total length (bp) | 25269 | 31590 | 130437 | 22332 | 10743 | |
| Frequency (SSR/Mb) | 2.70 | 1.00 | 3.68 | 0.93 | 1.25 | |
| Density (bp/Mb) | 45.41 | 17.18 | 66.52 | 15.92 | 21.53 | |
| ACAT | Number of SSRs | 2598 | 11404 | 4124 | 6140 | 6786 |
| Total length (bp) | 54624 | 260300 | 85240 | 123208 | 141664 | |
| Frequency (SSR/Mb) | 4.67 | 6.20 | 2.10 | 4.38 | 13.60 | |
| Density (bp/Mb) | 98.16 | 141.53 | 43.47 | 87.84 | 283.88 | |
| AAAT | Number of SSRs | 4217 | 11376 | 10768 | 5458 | 2406 |
| Total length (bp) | 79320 | 205164 | 192744 | 97060 | 43100 | |
| Frequency (SSR/Mb) | 7.58 | 6.19 | 5.49 | 3.89 | 1.72 | |
| Density (bp/Mb) | 142.54 | 111.55 | 98.30 | 69.19 | 30.73 | |
| AAAG | Number of SSRs | 2115 | 3419 | 7449 | 3011 | 695 |
| Total length (bp) | 39336 | 60516 | 134672 | 52956 | 12220 | |
| Frequency (SSR/Mb) | 3.80 | 1.86 | 3.80 | 2.15 | 1.39 | |
| Density (bp/Mb) | 70.69 | 32.90 | 68.68 | 37.75 | 24.49 | |
| AATT | Number of SSRs | 167 | 2166 | 1764 | 1007 | 279 |
| Total length (bp) | 2844 | 38332 | 32752 | 17628 | 4864 | |
| Frequency (SSR/Mb) | 0.30 | 1.18 | 0.90 | 0.72 | 0.56 | |
| Density (bp/Mb) | 5.11 | 20.84 | 16.70 | 12.57 | 9.75 | |
| AAAAT | Number of SSRs | 652 | 6646 | 3360 | 887 | 508 |
| Total length (bp) | 14160 | 143850 | 70780 | 18960 | 10740 | |
| Frequency (SSR/Mb) | 1.17 | 3.61 | 1.71 | 0.63 | 1.02 | |
| Density (bp/Mb) | 25.45 | 78.22 | 36.10 | 13.52 | 21.52 | |
| AAAAG | Number of SSRs | 846 | 1761 | 1228 | 1101 | 432 |
| Total length (bp) | 18895 | 37900 | 26140 | 23915 | 9365 | |
| Frequency (SSR/Mb) | 1.52 | 0.96 | 0.63 | 0.79 | 0.87 | |
| Density (bp/Mb) | 33.95 | 20.61 | 13.33 | 17.05 | 18.77 | |
| AATAT | Number of SSRs | 612 | 2662 | 946 | 889 | 761 |
| Total length (bp) | 13630 | 61295 | 20375 | 19400 | 17180 | |
| Frequency (SSR/Mb) | 1.10 | 1.45 | 0.48 | 0.63 | 1.53 | |
| Density (bp/Mb) | 24.49 | 33.33 | 10.39 | 13.83 | 34.43 | |
| AAATT | Number of SSRs | 35 | 6313 | 309 | 502 | 385 |
| Total length (bp) | 725 | 138175 | 6600 | 11295 | 8790 | |
| Frequency (SSR/Mb) | 0.06 | 3.43 | 0.16 | 0.36 | 0.77 | |
| Density (bp/Mb) | 1.30 | 75.13 | 3.37 | 8.05 | 17.61 | |
| AGAGGG | Number of SSRs | 189 | 78 | 107 | 198 | 7 |
| Total length (bp) | 4836 | 1920 | 2736 | 5184 | 174 | |
| Frequency (SSR/Mb) | 0.34 | 0.04 | 0.06 | 0.14 | 0.01 | |
| Density (bp/Mb) | 8.69 | 1.04 | 1.40 | 3.70 | 0.35 | |
| AAAAAT | Number of SSRs | 146 | 938 | 457 | 83 | 96 |
| Total length (bp) | 3738 | 23424 | 11382 | 2064 | 2382 | |
| Frequency (SSR/Mb) | 0.26 | 0.51 | 0.23 | 0.06 | 0.19 | |
| Density (bp/Mb) | 6.72 | 12.74 | 5.81 | 1.47 | 4.77 | |
| ACATAT | Number of SSRs | 87 | 360 | 421 | 303 | 426 |
| Total length (bp) | 2490 | 9678 | 11988 | 8736 | 12360 | |
| Frequency (SSR/Mb) | 0.16 | 0.20 | 0.22 | 0.22 | 0.85 | |
| Density (bp/Mb) | 4.48 | 5.26 | 6.11 | 6.23 | 24.77 | |
| AAAAAG | Number of SSRs | 109 | 239 | 209 | 120 | 67 |
| Total length (bp) | 2790 | 5976 | 5232 | 3096 | 1704 | |
| Frequency (SSR/Mb) | 0.20 | 0.13 | 0.11 | 0.09 | 0.13 | |
| Density (bp/Mb) | 5.01 | 3.25 | 2.67 | 2.21 | 3.42 |
Fig. 3.
Repeat counts for different SSR types in the palm genomes. ABCDEF represent mono-, di-, tri-, tetra-, penta-, and hexanucleotide SSR types, respectively
Dinucleotide repeats
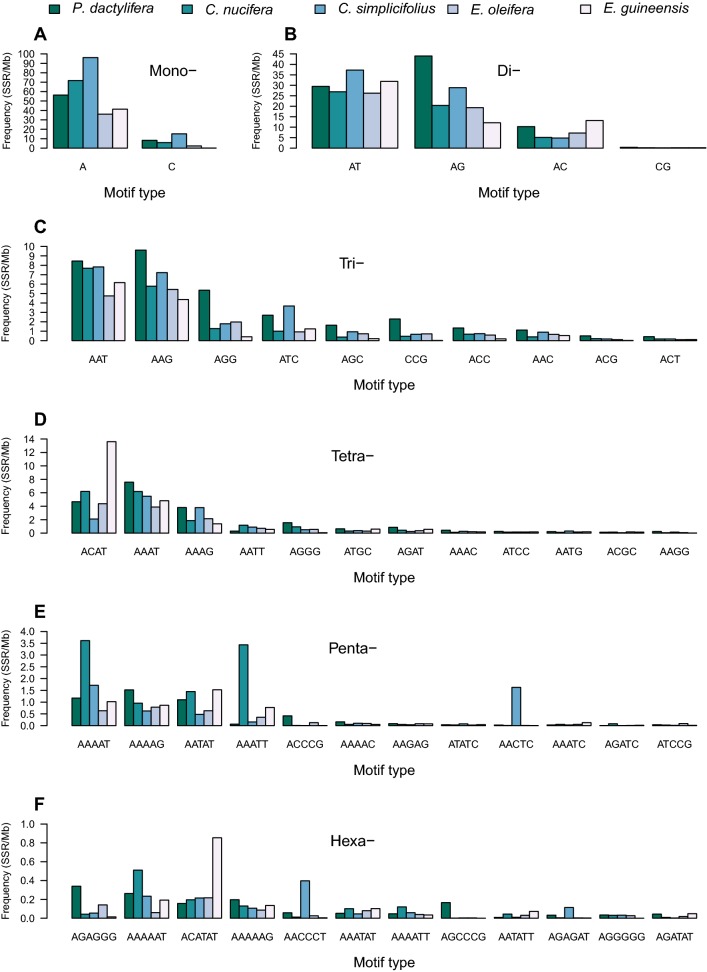
The motif type was the most predominant dinucleotide SSR in P. dactylifera, with a frequency of 44.02 SSR/Mb and occupying about 52.31% of all dinucleotide SSRs in this genome (Fig. 4b). The most frequent dinucleotide motif in C. nucifera, C. simplicifolius, E. oleifera, and E. guineensis was , with frequencies of 26.26–37.30 SSR/Mb and accounting for 51.20, 52.43, 49.54, and 55.58% of all dinucleotide SSRs in these genomes, respectively (Table 3). The motif was the least frequent dinucleotide SSR (0.15–0.38 SSR/Mb) in all of the five genomes. Dinucleotide repeats ranged from 7–86, 7–31, 7–85, 7–118, and 7–41 repeats in length, respectively. The most predominant repeat numbers ranged between 7–28, 7–24, 7–42, 7–25, and 7–21, which accounted for 99.16, 99.93, 99.56, 99.38, and 97.65% of all dinucleotide SSRs, respectively (Fig. 3b).
Fig. 4.
The most frequent SSR motif types in palm genomes. ABCDEF represent mono-, di-, tri-, tetra-, penta-, and hexanucleotide SSR types, respectively
Trinucleotide repeats
was the most frequent trinucleotide motif in P. dactylifera and E. oleifera, with frequencies of 9.61 and 5.44 SSR/Mb and accounting for 28.75 and 34.02% of all trinucleotide SSRs in these two genomes, respectively (Fig. 4c). The repeat was the most abundant motif in C. nucifera, C. simplicifolius, and E. guineensis, with frequencies of 7.69, 7.83, and 6.16 SSR/Mb and comprising about 42.57, 32.46, and 46.45% of all trinucleotide SSRs in these genomes, respectively. The and were also more frequent than other trinucleotide motifs, together accounting for 12.50–24.07% of all trinucleotide SSRs in the five palm genomes. and motifs were the least frequent trinucleotide SSRs in P. dactylifera, C. nucifera, C. simplicifolius, and E. oleifera, whereas and were the least abundant motifs in E. guineensis. Trinucleotide repeat counts ranged from 5–97, 5–21, 5–32, 5–59, and 5–42, respectively. Repeat numbers between 5–12, 5–20, 5–17, 5–12, and 5–9 accounted for 97.84, 99.94, 99.30, 97.45, and 91.84% of all trinucleotide SSRs in these genomes, respectively (Fig. 3c).
Tetranucleotide repeats
The motif was the most abundant tetranucleotide repeat in P. dactylifera and C. simplicifolius, with frequencies of 7.58 and 5.49 SSR/Mb and occupying about 34.02 and 35.47% of all tetranucleotide SSRs in these two genomes, respectively (Table 3). The most frequent tetranucleotide motif in E. oleifera and E. guineensis was , accounting for 31.80 and 59.40% of all tetranucleotide SSRs in those genomes, respectively. and repeats were the predominant tetranucleotide motifs in C. nucifera, with almost identical frequencies of approximately 6.20 SSR/Mb, together comprising about 67.55% of all tetranucleotide SSRs in the genome (Fig. 4d). and motifs were relatively frequent repeats in all five genomes. Tetranucleotide repeat counts ranged from 4–96, 4–15, 4–35, 4–31, and 4–33, respectively. Repeat numbers between 4–9, 4–12, 4–9, 4–9, and 4–10 accounted for 98.61, 99.60, 99.45, 99.01, and 98.62% of all tetranucleotide SSRs in these five genomes, respectively (Fig. 3d).
Pentanucleotide repeats
The motif was the most frequent pentanucleotide repeat in P. dactylifera and E. oleifera, followed by and motifs. These three motif types together accounted for 67.01 and 60.42% of all pentanucleotide SSRs in those two genomes, respectively (Table 3). The most abundant motif in C. nucifera was , which accounted for 35.54% of all pentanucleotide SSRs; meanwhile, was the predominant pentanucleotide motif in E. guineensis, accounting for 31.68% of all pentanucleotide SSRs in that genome. The motif was also relatively frequent in E. guineensis, being the second most abundant type. and motifs were the most abundant types in C. simplicifolius with similar frequencies of approximately 1.7 SSR/Mb, together accounting for 59.48% of the total number of pentanucleotide SSRs in this genome (Fig. 4e). Pentanucleotide repeat counts ranged from 4–73, 4–12, 4–10, 4–24, and 4–16, respectively. Repeat numbers between 4–6, 4–7, 4–6, 4–6, and 4–6 accounted for 97.90, 99.22, 99.23, 98.15, and 97.34% of all pentanucleotide SSRs in the five palm genomes, respectively (Fig. 3e).
Hexanucleotide repeats
Hexanucleotide motifs were found to have far lower frequency and density compared to other microsatellite repeat types. and motifs were the most abundant hexanucleotide types in P. dactylifera, together accounting for 25.36% of all hexanucleotide SSRs (Fig. 4f). The most abundant motifs in C. nucifera were and , together accounting for 39.76% of all hexanucleotide SSRs, while and were the predominant hexanucleotide motifs in C. simplicifolius, together accounting for 28.07% of all hexanucleotide SSRs in the genome. The motif was found to be the most frequent type in E. oleifera and E. guineensis, with frequencies of below 0.86 SSR/Mb. Hexanucleotide repeat lengths ranged from 4–10, 4–11, 4–21, 4–17, and 4–20, respectively. Repeat numbers between 4–5, 4–5, 4–6, 4–5, and 4–5 accounted for 94.02, 96.45, 97.18, 91.68, and 90.08% of all hexanucleotide SSRs in the five palm genomes, respectively (Fig. 3f).
Discussion
The availability of genomic sequences for several palm species provides the opportunity to elucidate and compare the distributions of microsatellites across these genomes. In a previous study, genomic microsatellite loci were screened for two Palmae species (P. dactylifera and E. oleifera) (Xiao et al. 2016). To the best of our knowledge, the present study is the first comprehensive report on the identification of microsatellites with 1–6 bp nucleotide motifs in five Palmae species: P. dactylifera, C. nucifera, C. simplicifolius, E. oleifera, and E. guineensis. Consistent search parameters were used to perform the same analysis for all five palm genomes. Computational approaches were utilized to elucidate and compare the relative frequency, relative density, and GC content of SSRs in these species. Perfect microsatellites were found to comprise 0.23–0.44% of the five palm genomes. The percentages of SSRs in species within the same genus (E. oleifera and E. guineensis) were comparable, and lower than in the other three palm genomes. This variation in the percentage of genome SSR content may arise from differences in computational methods used for SSR identification, the relative completeness of different genome assemblies as obviously observed in the genome assembly of E. guineensis, or real variation in microsatellite content among these species (Sharma et al. 2007).
The six types of SSRs were not equally represented in all five palm genomes. In general, mono- and dinucleotide repeats were found to prevail. More precisely, mononucleotide SSRs were the most frequent repeat type in C. nucifera and C. simplicifolius, consistent with previous findings in monocots and dicots (Sonah et al. 2011) and similar to what has been found for eukaryotic genomes overall (Sharma et al. 2007; Qi et al. 2015). Dinucleotide SSR repeats were the most abundant type in P. dactylifera, E. oleifera, and E. guineensis, which is consistent with prior findings for dicotyledons (Kumpatla and Mukhopadhyay 2005). Tri- and tetranucleotide SSR types were found to have very similar frequencies in the five palm genomes. Hexanucleotide repeats were the least frequent SSR type in all five species, which is similar to what has been seen in previous studies (Subramanian et al. 2002; Liu et al. 2017; Manee et al. 2019).
Previously, microsatellite abundances were found to be similar in species of the same genus (Shi et al. 2014). Here, only E. oleifera and E. guineensis are classified into the same genus, and these species did not have similar profiles overall. Interestingly, the overall frequency and density of SSRs were about the same in P. dactylifera and C. simplicifolius, suggesting potential similarity in the genomic structures of these two palm species. This is further supported by these genomes having similar abundances of SSRs by type, with the exception of mononucleotide SSRs.
Within each type of SSR, microsatellite motifs were found to vary greatly for each of the five palm genomes. Among mononucleotide repeats, the most abundant motif was , accounting for 86.37–99.84% of the total number of mononucleotide SSRs. This observation is consistent with previous results from Volvariella volvacea, Agaricus bisporus, and Coprinus cinereus (Wang et al. 2014). Of dinucleotide SSRs, the motif was the most frequent in all examined genomes except for P. dactylifera, and this trend was similar in dicots (Sonah et al. 2011), pineapple (Fang et al. 2016), cucumber (Cavagnaro et al. 2010), and sweet orange (Biswas et al. 2014). The most abundant dinucleotide repeat in P. dactylifera was , which is consistent with previous findings in Brachypodium distachyon (Sonah et al. 2011), wheat (Deng et al. 2016), and garden asparagus (Li et al. 2016). Among trinucleotide SSRs, the motif was the most predominant in C. nucifera, C. simplicifolius, and E. guineensis, and consistent with reports from garden asparagus (Li et al. 2016), cucumber (Cavagnaro et al. 2010), pineapple (Fang et al. 2016), and Medicago truncatula and Populus trichocarpa (Sonah et al. 2011). The was the dominant trinucleotide motif in P. dactylifera and E. oleifera, similar to previous reports in Arabidopsis thaliana (Sonah et al. 2011) and Brassica species (Shi et al. 2013). The AT-rich motifs (AAAT), (AAAG), , , , , , , , and were the most abundant tetra-, penta- and hexanucleotide SSRs in the five palm genomes. Overall, the overrepresentation of motifs in palm genomes can be explained by the fact that strand separation is easier for AT-rich than for GC-rich sequences, raising the possibility of slipped strand mispairing (Zhao et al. 2011). A previous study revealed that the , , , and motifs also predominated in Brassica species (Shi et al. 2013).
GC content varies greatly among different genomes because of different selective constraints. It is important to identify the driving force behind GC content diversity in order to understand genome evolution across species. The overall GC content of eukaryotic genomes does not vary widely (Šmarda and Bureš 2012). However, in plants, grass genomes are known to have high GC content compared to other angiosperm families (Barow and Meister 2002; Šmarda and Bureš 2012). This study found the five palm genomes analyzed had lower GC content (28.12–39.65%) than do grasses (43.57–46.90%) (Singh et al. 2016), a number of Poaceae species (Deng et al. 2016), five monocots (43.57–46.14%), and two green algae (55.70 and 63.45%) (Zhao et al. 2014). In addition, GC content was not evenly distributed in three of the species, the exceptions being C. nucifera and E. guineensis ( 32%). Variation in GC content within each SSR type was also observed across the five genomes, with the exception of tetranucleotide SSRs. Tri- and hexanucleotide SSRs were generally found to have the highest GC contents. The results also suggested that motifs are the most predominant in each genome, consistent with findings in previous reports (Sharma et al. 2007; Shi et al. 2013; Li et al. 2016). This can be interpreted as confirming high AT content in the majority of the analyzed SSRs.
SSRs make up a significant proportion of the eukaryotic genomes and are highly polymorphic, surpassing coding gene sequences in both respects (Katti et al. 2001). The high mutation rates of SSRs make them highly informative and useful for a wide range of applications such as evolutionary research, population genotyping, and marker-assisted breeding. Recent studies have utilized genome-wide approaches for the development of SSR markers in plants (Shi et al. 2014; Deng et al. 2016; Kumari et al. 2019). Perhaps the main advantage of this strategy is to produce a large number of SSR markers distributed evenly throughout the genome. The construction of a Palmae SSR database for the scientific community would evidently have a significant impact on genetic studies in those species.
Comparative analysis of SSRs in these five palm genomes will provide a better understanding of the nature of these important sequences and will facilitate research on the role of SSRs in genome organization. Such knowledge will serve many useful purposes, including, among many others, the isolation and development of abundant markers for genetic and evolutionary studies mentioned above. In particular, elucidating the most frequent repeats in palm genomes provides an essential starting point for the library-based selection of markers that will be informative in distinguishing populations and cultivars within a species, or even for cross-species applications. This further provides an important foundation for characterizing genetic diversity in palm germplasm and for performing selection on valuable or undesired attributes while also maintaining and/or improving diversity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Amer S. Alharthi at the National Center for Robotics Technologies and Intelligent Systems, King Abdulaziz City for Science and Technology, for his technical support.
Author contributions
MMM and MBA conceived and designed the experiments; MMM and BMA carried out the experiments; MMM and BMA analyzed the data; MMM wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was funded by the Life Science and Environment Research Institute (Grant 37-1271) and the Center of Excellence for Genomics (Grant 20-0078), King Abdulaziz City for Science and Technology, Saudi Arabia.
Compliance with ethical standards
Conflict of interest
The author declare there are no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aberlenc-Bertossi F, Castillo K, Tranchant-Dubreuil C, Chérif E, Ballardini M, Abdoulkader S, Gros-Balthazard M, Chabrillange N, Santoni S, Mercuri A, et al. In silico mining of microsatellites in coding sequences of the date palm (arecaceae) genome, characterization, and transferability. Appl Plant Sci. 2014;2(1):1300058. doi: 10.3732/apps.1300058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvaru AK, Sowpati DT, Mishra RK. PERF: an exhaustive algorithm for ultra-fast and efficient identification of microsatellites from large DNA sequences. Bioinformatics. 2017;27:573. doi: 10.1093/bioinformatics/btx721. [DOI] [PubMed] [Google Scholar]
- Barow M, Meister A. Lack of correlation between at frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry J Int Soc Anal Cytol. 2002;47(1):1–7. doi: 10.1002/cyto.10030. [DOI] [PubMed] [Google Scholar]
- Barrow SC. A monograph of phoenix l. (palmae: Coryphoideae) Kew Bull. 1998;20:513–575. doi: 10.2307/4110478. [DOI] [Google Scholar]
- Biswas MK, Xu Q, Mayer C, Deng X. Genome wide characterization of short tandem repeat markers in sweet orange (Citrus sinensis) PLoS One. 2014;9(8):e104182. doi: 10.1371/journal.pone.0104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus l.) BMC Genom. 2010;11(1):569. doi: 10.1186/1471-2164-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P, Wang M, Feng K, Cui L, Tong W, Song W, Nie X. Genome-wide characterization of microsatellites in triticeae species: abundance, distribution and evolution. Sci Rep. 2016;6:32224. doi: 10.1038/srep32224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhoumaizi MA, Saaidi M, Oihabi A, Cilas C. Phenotypic diversity of date-palm cultivars (Phoenix dactylifera l.) from morocco. Genet Resour Crop Evol. 2002;49(5):483–490. doi: 10.1023/A:1020968513494. [DOI] [Google Scholar]
- Fang J, Miao C, Chen R, Ming R. Genome-wide comparative analysis of microsatellites in pineapple. Trop Plant Biol. 2016;9(3):117–135. doi: 10.1007/s12042-016-9163-6. [DOI] [Google Scholar]
- Huntley MA, Golding GB. Selection and slippage creating serine homopolymers. Mol Biol Evol. 2006;23(11):2017–2025. doi: 10.1093/molbev/msl073. [DOI] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40(2):120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 2001;18(7):1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- Kumari R, Wankhede DP, Bajpai A, Maurya A, Prasad K, Gautam D, Rangan P, Latha M, John KJ, Bhat KV, et al. Genome wide identification and characterization of microsatellite markers in black pepper (piper nigrum): a valuable resource for boosting genomics applications. PLoS One. 2019;14(12):e0226002. doi: 10.1371/journal.pone.0226002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpatla SP, Mukhopadhyay S. Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome. 2005;48(6):985–998. doi: 10.1139/g05-060. [DOI] [PubMed] [Google Scholar]
- Li CY, Liu L, Yang J, Li JB, Su Y, Zhang Y, Wang YY, Zhu YY. Genome-wide analysis of microsatellite sequence in seven filamentous fungi. Interdiscip Sci Comput Life Sci. 2009;1(2):141–150. doi: 10.1007/s12539-009-0014-5. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang G, Li X, Wang L, Yuan J, Deng C, Gao W. Genome-wide identification and validation of simple sequence repeats (SSRS) from Asparagus officinalis. Mol Cell Probes. 2016;30(3):153–160. doi: 10.1016/j.mcp.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Liu S, Hou W, Sun T, Xu Y, Li P, Yue B, Fan Z, Li J. Genome-wide mining and comparative analysis of microsatellites in three macaque species. Mol Genet Genom. 2017;292(3):537–550. doi: 10.1007/s00438-017-1289-1. [DOI] [PubMed] [Google Scholar]
- Manee MM, Algarni AT, Alharbi SN, Al-Shomrani BM, Ibrahim MA, Binghadir SA, Al-Fageeh MB. Genome-wide characterization and analysis of microsatellite sequences in camelid species. Mammal Res. 2019;20:1–15. [Google Scholar]
- Moussouni S, Pintaud JC, Vigouroux Y, Bouguedoura N. Diversity of algerian oases date palm (Phoenix dactylifera l., arecaceae): heterozygote excess and cryptic structure suggest farmer management had a major impact on diversity. PLoS One. 2017;12(4):e0175232. doi: 10.1371/journal.pone.0175232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi WH, Jiang XM, Du LM, Xiao GS, Hu TZ, Yue BS, Quan QM. Genome-wide survey and analysis of microsatellite sequences in Bovid species. PLoS One. 2015;10(7):e0133667. doi: 10.1371/journal.pone.0133667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi WH, Jiang XM, Yan CC, Zhang WQ, Xiao GS, Yue BS, Zhou CQ. Distribution patterns and variation analysis of simple sequence repeats in different genomic regions of bovid genomes. Sci Rep. 2018;8(1):14407. doi: 10.1038/s41598-018-32286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PC, Grover A, Kahl G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007;25(11):490–498. doi: 10.1016/j.tibtech.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Shi J, Huang S, Zhan J, Yu J, Wang X, Hua W, Liu S, Liu G, Wang H. Genome-wide microsatellite characterization and marker development in the sequenced brassica crop species. DNA Res. 2013;21(1):53–68. doi: 10.1093/dnares/dst040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Huang S, Zhan J, Yu J, Wang X, Hua W, Liu S, Liu G, Wang H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res Int J Rapid Publ Rep Genes Genomes. 2014;21(1):53–68. doi: 10.1093/dnares/dst040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. Busco: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Singh R, Ming R, Yu Q. Comparative analysis of GC content variations in plant genomes. Trop Plant Biol. 2016;9(3):136–149. doi: 10.1007/s12042-016-9165-4. [DOI] [Google Scholar]
- Šmarda P, Bureš P. Plant genome diversity. Berlin: Springer; 2012. The variation of base composition in plant genomes; pp. 209–235. [Google Scholar]
- Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR. Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One. 2011;6(6):e21298. doi: 10.1371/journal.pone.0021298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2002;4:R13–R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen M, Wang H, Wang JF, Bao D. Microsatellites in the genome of the Edible Mushroom, Volvariella volvacea. BioMed Res Int. 2014;2014:1–10. doi: 10.1155/2014/281912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. Busco applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2017;35(3):543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Xia W, Ma J, Mason AS, Fan H, Shi P, Lei X, Ma Z, Peng M. Genome-wide identification and transferability of microsatellite markers between palmae species. Front Plant Sci. 2016;7:1578. doi: 10.3389/fpls.2016.01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hu Z, Wang C, Zhang X, Li J, Yue B. Characterization of perfect microsatellite based on genome-wide and chromosome level in Rhesus monkey (Macaca mulatta) Gene. 2016;592(2):269–275. doi: 10.1016/j.gene.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang J, Chen L, Yu J, Dong J, Yao ZJ, Shen Y, Jin Q, Chen R. Identification and characterization of simple sequence repeats in the genomes of Shigella species. Gene. 2003;322:85–92. doi: 10.1016/j.gene.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Zhao X, Tan Z, Feng H, Yang R, Li M, Jiang J, Shen G, Yu R. Microsatellites in different Potyvirus genomes: survey and analysis. Gene. 2011;488(1–2):52–56. doi: 10.1016/j.gene.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Williams R, Prakash C, He G. Identification and characterization of gene-based ssr markers in date palm (Phoenix dactylifera l.) BMC Plant Biol. 2012;12(1):237. doi: 10.1186/1471-2229-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C. Genome-wide analysis of tandem repeats in plants and green algae. G3: Genes Genomes Genet. 2014;4(1):67–78. doi: 10.1534/g3.113.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.