Abstract
Parkinson’s disease (PD) is the second most prevalent neurological disorder and has been the focus of intense investigations to understand its etiology and progression, but it still lacks a cure. Modeling diseases of the central nervous system in vitro with human induced pluripotent stem cells (hiPSC) is still in its infancy but has the potential to expedite the discovery and validation of new treatments. Here, we discuss the interplay between genetic predispositions and midbrain neuronal impairments in people living with PD. We first summarize the prevalence of causal Parkinson’s genes and risk factors reported in 74 epidemiological and genomic studies. We then present a meta-analysis of 385 hiPSC-derived neuronal lines from 67 recent independent original research articles, which point towards specific impairments in neurons from Parkinson’s patients, within the context of genetic predispositions. Despite the heterogeneous nature of the disease, current iPSC models reveal converging molecular pathways underlying neurodegeneration in a range of familial and sporadic forms of Parkinson’s disease. Altogether, consolidating our understanding of robust cellular phenotypes across genetic cohorts of Parkinson’s patients may guide future personalized drug screens in preclinical research.
Subject terms: Induced pluripotent stem cells, Cellular neuroscience
Introduction
Parkinson’s Disease (PD) is a complex neurological disease, affecting approximately 2% of the population over 60 years of age. Since the first reports of PD correlation with the SNCA gene1–5, an increasing number of genomic predispositions has been identified as direct contributors or risk factors for the disease6–15. In the last decade, advances in cellular reprogramming and induced pluripotent stem cell (iPSC) technology have led to the development of patient-derived brain tissue engineering16–18. The culturing of live patient-derived neurons provides a unique opportunity to study the cellular mechanisms of genetically-linked diseases in vitro19,20. IPSC studies of brain disorders remain laborious and expensive, which limits the number of cell lines that can be studied in a single laboratory and may raise the issue of statistical power and reproducibility. The field is still in its infancy, and neuronal differentiation protocols are continuously improved upon, which makes technical harmonization between studies challenging21. Like any model, iPSC studies have their weaknesses, and current limitations are being addressed collectively by the research community22,23. Nevertheless, the urgent need for better treatments for brain disorders justifies the requirement for pioneering iPSC studies modeling those diseases in vitro now. Since 2011, more than 385 neuronal lines from PD patients (and control subjects) have been generated across 67 original independent studies24–92. Independent phenotypic characterization of these lines revealed known and novel impairments in a range of cellular functions associated with PD. Moving forward, it is essential to integrate and compare the results obtained from these studies to identify the most reliable disease neuronal phenotypes before expanding the work to large drug screens. The identification of robust phenotypes of neurons derived from Parkinson’s patients’ iPSCs may provide the basis for a new paradigm in preclinical drug development and accelerates the progression towards clinical trials.
To quantify the prevalence and penetrance of genes known to be associated with PD, we analyzed 50 epidemiological93–129 and 24 genomic studies8,10–14,130–148 and a genome-wide association studies (GWAS) database149. Within this context, we then examined recent findings from human induced pluripotent stem cell (hiPSC) models that shed light on the interplay between genetic predispositions and brain cell phenotypes in people living with PD. We present an analysis of 385 human iPSC-derived neuronal lines from 67 studies, which point towards specific impairments of dopaminergic neurons from people living with PD. In comparison with the prevalence of causal PD genes in epidemiological studies, our analysis underlines the current bias of PD iPSC studies towards a subset of familial genetic predispositions. We also summarize the methodological overlap and differences between these studies, which use a broad range of reprogramming methods. Finally, our meta-analysis highlights the possibility of converging molecular and cellular pathways underlying neurodegeneration in familial and sporadic PD with diverse genetic predispositions. In particular, we discuss the convergence of various genetic predispositions on the impairment of cellular pathways underlying metabolic function, synaptic communication, inflammation, and the recycling of damaged protein and organelles. The collective insights from independent iPSC disease studies, which are pioneering this blooming field of preclinical research, may guide us towards translating fundamental discoveries into effective treatments for patients.
The interplay between genomic predispositions and environmental factors leads to Parkinson’s
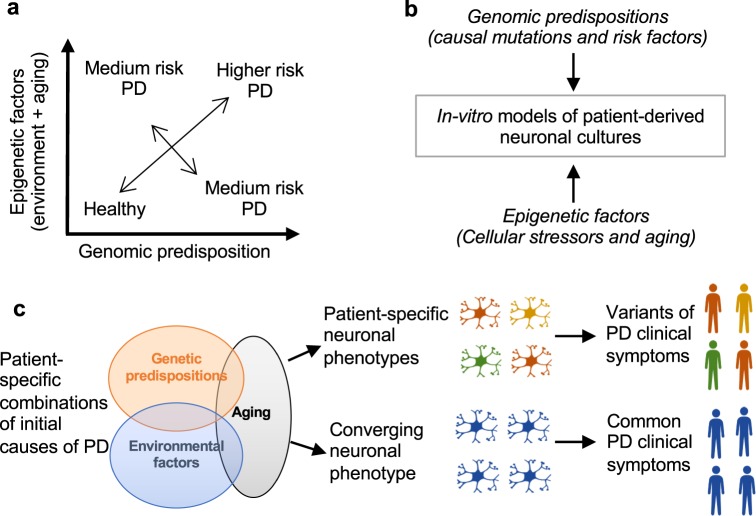
In the mid-1990s, the connection between PD and underlying genetic mutations was established4,5,150. It is now evident that varying degrees of the interplay between genomic predispositions and aging and cellular stressors impose a risk for disease151 (Fig. 1a). Previous studies have shown vascular insults to the brain, repeated head trauma, neuroleptic drugs, exposure to pesticides, and manganese toxicity increase the risks of developing symptoms of PD152–154. In addition, advancing age can also cause a cascade of stressors within the substantia nigra, which weakens the neurons and their ability to respond to further insults155,156. Ultimately, the uniqueness of the interactions between genes and the environment makes the development of a single treatment for PD difficult as they give rise to a spectrum of neuronal phenotypes that can be unique to individual patients (Fig. 1c). The development of a model with the ability to replicate the genomic and epigenetic aspects of the disease is crucial (Fig. 1b). As increasing evidence suggests that genetic mutations are key modulators of disease initiation and progression, the identification and understanding of the various genomic predispositions are required for the development of better-targeted treatments to slow the disease progression.
Fig. 1. A combinatorial spectrum of genetic risks, cellular stressors, and brain cell dysfunctions causes Parkinson’s disease.
a Graphical overview of PD risk associated with genomic predispositions and epigenetic factors. b Schematic overview of PD etiological trajectories in iPSC models. c Unique cellular phenotypes may cause PD symptoms in a subset of patients (individualized etiology) and the convergence of various initial causes into common cellular phenotypes may cause other symptoms (convergent etiology).
Learning from genetic analyses of PD case–control studies
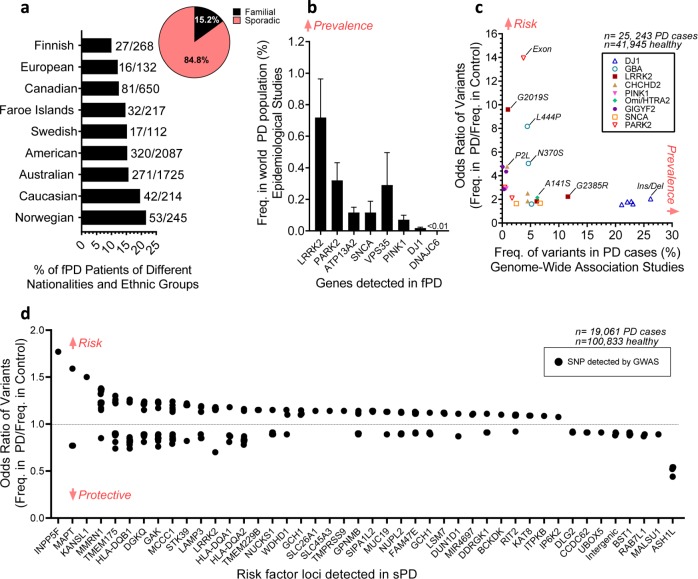
We analyzed the reports from 12 international studies94,157–167, totaling 5650 persons living with PD in North America, Europe, and Australia. We confirmed that globally only 15% of patients report a family history of PD symptoms, while the remaining 85% of the PD population are classified as sporadic PD (Fig. 2a). However, the distinction between genetic predispositions in familial and sporadic PD is blurry. No single-gene mutation in PD has a 100% penetrance. Instead, most likely, multiple genetic risk factors act in synergy to increase the chances of both familial and sporadic PD. Such genetic susceptibilities interplay with aging and environmental factors in both familial and sporadic PD.
Fig. 2. The genomics of Parkinson’s disease: prevalence and penetrance.
a In the world-wide population of people living with PD, ~85% of PD cases are sporadic (sPD) and the remaining are familial (fPD) (n = 5650 PD cases combined, refer to “Methods”). b Genetic mutations occur at low (< 1%) and varying frequencies (Freq.) in the PD world population (n=488 patients carrying mutation, 32,012 total PD cases used for analysis, refer to “Methods”). Data represented as the mean±SEM. c GWAS data suggests risk variants (OR>1.5) in fPD genes tend to be less prevalent in PD cases (n = 25,243 PD cases, 41,945 healthy, refer to “Methods”). d Single nucleotide polymorphisms (SNPs) in over 44 genomic regions show significant (p < 5 x 10−8) association to PD. Each point presents an independent SNP hit associated with PD.
Mounting evidence demonstrates the role of genetic predispositions in PD pathogenesis, and the prevalence and penetrance of each variant start to emerge. Typically, it appears that the most common PD gene variants found in the global population have the lowest penetrance. On the other hand, most penetrant mutations are rare, and most often associated with familial PD. Variants on at least 16 PARK genes, referred to as “causal” because they instigate a relatively high familial inheritance of PD or early-onset PD symptoms, have been identified and are highlighted in Table 15,119,132,135,147,168–178. To estimate the frequency of these mutations in the Parkinson’s population, we examined the published reports from 50 epidemiological studies. The high penetrance and hereditary nature of causal mutations can amplify their occurrence in specific ethnic groups (i.e., Japanese134, Chinese133, and Ashkenazi Jews11). However, globally our analysis shows that the presence of any known causal PD mutation is rare, occurring in less than 2% of the PD population (Fig. 2b). The two most common mutated genes associated with familial PD are LRRK2 and PRKN and are reported in 0.7% and 0.3% of all the people showing PD symptoms, respectively (Fig. 2b). However, the frequency of at least some risk variants (OR > 1.5), which we measured in a large genomic data set including 25,243 total PD cases (combination of NHGRI-EBI catalog and 24 independent genetic studies), appears higher than in the epidemiological studies, which would tend to focus on the variants with the highest penetrance. For example, LRRK2 G2385R is present in >10% of the PD patients and PRKN exon rearrangements are present in >3% of the patients (Fig. 2c). Our analysis also highlights that known mutations in familial PARK genes confer a broad range of risks to develop PD symptoms. PRKN(exon rearrangements), LRRK2(G2019S), GBA (L444P, N370S), and CHCHD2(P2L) appear to be the most penetrant mutations, increasing the chances of getting PD symptoms by up to a factor 14, 10, 8 and 5, respectively (Fig. 2c). However, these penetrant variants are relatively rare, occurring in ~4%, 2%, 5%, and 2% of the PD population, respectively (Fig. 2c). Overall, the data tends to confirm that the most prevalent variants are less penetrant (See DJ-1 variants for example in Fig. 2c). Recent GWAS allowed for the detection of common low penetrance mutations associated with PD. To date, over 44 novel risk loci have been identified and associated with sporadic PD9,14,141,179,180. Mutations within these risk loci can be disease modulating (OR > 1) or protective (OR < 1) for PD (Fig. 2d). Despite GWAS and genetic studies pointing to several novel molecular targets and pathways in sporadic PD, many of them remain unconfirmed. The presence of mutations on a single risk locus often poses a low risk for disease, yet mutations in multiple risk loci can collectively be disease modulating. The lack of a preponderant single definitive causative gene for most PD patients is a challenge for both the design of experimental models and the discovery of well-targeted treatments. Novel approaches for disease modeling in vitro are required to unravel the polygenic and complex mechanisms underlying sporadic disease successfully. Brain tissue engineering from patient-derived stem cells provides a unique opportunity to fulfill such need.
Table 1.
Summary of Parkinson’s genes first identified in familial and early-onset cases of PD and risk factors.
| Gene (Chromosomal position) | Gene product | Function | Inheritance | Mutations studied in iPSC studies | Reference (iPSC studies) |
|---|---|---|---|---|---|
| SNCA (4q21) | a-synuclein | Suppression of apoptosis, chaperone activity, antioxidation, neuronal differentiation, regulation of DA biosynthesis | AD | A53T; E46K; duplication; triplication | 28,30,33–35,41,42,49,55,60,66,73,80,83,84,89,90, |
| PRKN(6q25.2-q27) | Parkin | Mitochondrial quality control | AR | V324A; T240R; R275W; R42P; −1 bp del 255A; Exon 3–4 del; Exon 2 del; Exon 3/5 del; Exon 6/7 del; Exon 2/4 del; Exon 5/6 del; Exon5 del; Exon 3 del | 24,29,31,39,40,42,54,64,68,71,74,76,77,84,88,91, |
| PINK1 (1p35-p36) | PTEN-induced kinase 1 | Mitochondrial quality control | AR | G309D; V170G; Q456X | 31,32,52,62,70,71,85, |
| DJ-1 (1p36) | DJ-1 | Protection against oxidative stress | AR | E46D | 27 |
| LRRK2 (12q12) | Leucine rich repeat kinase 2 | Membrane trafficking, mitochondrial membrane maintenance | AD | G2019S; R1441C; R1441G; I2020T | 26,32,37,38,42–44,46–48,50,51,53,56,57,61,65,78,79,82,84,86,87, |
| ATP13A2 (1p36) | Cation-transporting ATPase 13A2 | Cation homeostasis, lysosomal function | AR | N/A | N/A |
| VPS35 (16q11.2) | Vacuolar protein sorting 35 | Recycling of membrane proteins between endosomes and the trans-Golgi network | AD | N/A | N/A |
| DNAJC6 (1p31.3) | DNAJ subfamily C member 6 | Clathrin mediated endocytosis | AR | N/A | N/A |
| FBXO7 (22q12–q13) | F-box protein 7 | Phosphorylation-dependent ubiquitination | AR | N/A | N/A |
| SYNJ1 (21q22.11) | Synaptojanin-1 | Regulation of synaptic vesicle endocytosis | AR | N/A | N/A |
| PLA2G6 (22q12–q13) | Phospholipase A2, group 6 | Phospholipid remodeling, mitochondrial function | AR | N/A | N/A |
| CHCHD2 (7p11.2) | Coiled-coil-helix-coiled-coil-helix domain 2 | Regulation of mitochondrial metabolism under oxygen stress | AD | N/A | N/A |
| GIGYF2 (2q36–q37) | GRB10 interacting GYF protein 2 | Negative regulation of cell growth | AD | N/A | N/A |
| Omi/HTRA2 (2p13) | High-temperature requirement A2 | Neuroprotection | AR | N/A | N/A |
| VPS13C (15q22.2) | Vacuolar protein sorting 13C | Maintenance of mitochondrial function | AR | N/A | N/A |
| EIF4G1 (3q27.1) | Eukaryotic translation initiation factor 4 gamma 1 | Regulation of mRNAs translation | AD | N/A | N/A |
| UCHL1 (4p13) | Ubiquitin C-terminal hydrolase L1 | Ubiquitin–proteasome system and neuronal survival | AD | N/A | N/A |
| GBA (1q21) | Glucosylceramidase Beta | Lysosomal function | RF | N370S; L444P; RecNil | 30,36,58,59,69,84, |
| MAPT (17q21.31) | Microtubule-associated protein Tau | Modulates the stability of axonal microtubules | RF | Haplotype H1/H2 | 25 |
N/A indicates mutations not studied in iPSC-PD studies.
AD autosomal dominant; AR autosomal recessive; RF risk factor; del deletion.
Studies of PD with patient-derived iPSCs
The discovery of iPSC technology181–183 has offered the capacity to generate live brain tissue from healthy subjects and patients for studying neurodegenerative diseases184. Directed reprogramming and neuronal differentiation of iPSCs allows the study of specific neuronal subtypes. Human-derived neurons offer a unique opportunity for modeling real cases of human genetic diseases in vitro. The ability to generate neurons both from PD patients and healthy control individuals allows the identification of early disease-linked phenotypes and provides a new paradigm for preclinical drug development and validation (Fig. 3a).
Fig. 3. Using brain cells generated from patient-derived iPSC to study PD in vitro.
Data from this figure was extracted and analyzed from 67 iPSC-PD studies, refer to “Methods”. a A schematic pipeline of in vitro disease modeling and preclinical drug screening with patient-derived brain cells. b The number of iPSC studies that used human neuronal lines with corresponding mutations on specific genes associated with PD (also refer to Table 1). Categories in bold and darker bars represent the total number of studies examining that gene. c The types of control and PD cell lines are displayed as the percentage of total cell lines. d The number of PD and control cell lines used in iPSC-PD studies. Data presented as the mean ± SEM. e Donor cell types and reprogramming methods used in hiPSC-PD studies. N/R indicates that details were not reported in these studies. f The diagram summarizes the different type of tissue culture trajectories used to differentiate cultures of iPSCs into midbrain neurons. Line thickness and percentages (in the “neurons” box) represent the proportion of studies in corresponding trajectories. The percentage displayed for each intermediate stage shows the proportion of studies that uses the corresponding cell type. EB embryoid bodies, NPCs neural progenitors g Neural induction duration indicates the number of days (average + range) required for the generation of terminal neural precursor cell types (last stage before neuronal maturation: NPCs, neurospheres, rosettes, or EZ spheres depending the stages that were skipped) from iPSC. h Neural maturation duration indicates the average number of weeks from terminal neural precursor cell type (NPCs or previous stage if NPC stage was skipped) to the neuronal cells used for phenotypic evaluation. i Small molecules and growth factors were used at various stages of midbrain dopaminergic neuronal differentiation. Data presented as the percentage of hiPSC-PD studies that report the corresponding factors in the tissue culture media composition. j The proportions (mean + SEM) of neurons (bIII-Tub/DAPI) and midbrain dopamine neurons (TH/DAPI) in cultures vary between differentiation protocols and trajectories. Each data point is the average percentage reported in a single study (n = 33 independent studies, refer to “Methods”). The first column labeled as “all” groups all the studies regardless of their differentiation trajectories. Relevant immunohistochemistry quantification was not reported in studies using neural differentiation trajectories A and D.
Since 2011, over 385 hiPSC lines have been generated from PD patients (n = 215) and controls (n = 170). Despite the large epidemiological preponderance of sporadic cases of PD (85% sporadic vs. 15% familial PD, Fig. 2a), only a few studies have modeled sporadic PD with iPSCs (Fig. 3b). Majority of iPSC-PD studies modeled LRRK2-G2019S, PRKN exon deletions, PINK1 Q456X, SNCA triplication, and GBA N370S (Fig. 3b). When combined all together, less than 20% of the iPSC disease lines were sporadic and 80% of the disease cell lines modeled single causal mutations occurring in familial PD (Fig. 3c). Extended application of iPSC technology beyond monogenic disease to multigenic sporadic disease is required to reveal cellular features relevant to the majority of people affected by PD. An increasing number of iPSC-based models of neurological and psychiatric disorders show that even complex brain disorders with limited heritability lead to neuronal phenotypes in vitro185–187. Despite this current bias in the literature towards familial PD, these pioneer studies are extremely valuable and, in the sections below, we will compare their methods and summarize the overlap and discrepancies of the phenotypes reported in PD neuronal lines.
How many patient neuronal lines should we use for iPSC models of PD?
This is a crucial question for which a consensus remains difficult to reach. Due to the cumbersome and costly nature of iPSC studies, a balanced trade-off between a large number of neuronal lines and the depth of the analysis is inevitable. For comparison, other kinds of PD patient-control studies vary quite substantially from GWAS generally including thousands of case–control subjects to postmortem brain tissue studies including cohorts of 11 ± 5 patients and 8 ± 6 controls, on average188. We report here that current iPSC-PD studies, on average, use five individuals per study (two control and three patient lines) and a maximum of 12 controls and 11 PD line. Fewer than 12% (8/67) of published studies used more than ten cell lines (Fig. 3d)37,42,56,70,79,84,92. Interestingly, it has been estimated that sample sizes of 10–30 individuals per hiPSC study may be required to achieve a statistical power of 80%, assuming that the cellular readouts variance is high and the disease effect is small (>0.7 relative heterogeneity, which is the ratio between within-group standard deviation and mean group difference)189. In this situation, most iPSC studies of PD may fall below the suggested sample size requirements. However, it is important to note that due to the novelty of iPSC models, predicting the statistical power of a study remains approximative, and the number of lines required highly depends on the variance of the cellular phenotypes obtained with a specific analytical readout. It may be possible to reduce the variance without increasing the number of cell lines. For example, the patients from which the biopsies are taken may be selected based on gender, age, ethnicity, social context (i.e., farmers exposed to pesticides or professional athletes exposed to repetitive trauma), genotypes, and clinical severity of the symptoms. Ultimately, the clinical and genotypical homogeneity of the subjects may reduce the variance of the cellular phenotypes and may justify using a smaller number of cell lines. This may explain the current bias in the literature towards iPSC models of familial cases of PD (Fig. 3b, c). Contrastingly, there is an assumption that high genetic heterogeneity in sporadic disease contributes to a range of cellular phenotypes and therefore requires a higher number of cell lines to obtain statistical power19,45,190. However, this assumption might be wrong if multigenic sporadic PD predispositions converge to similar clinical symptoms caused by common brain cell phenotypes. Despite the current limitation of a relatively low number of cell lines used in each study, and regardless whether familial or sporadic patients were included, significant PD phenotypes have been reported, thus demonstrating the value of this research model. However, to translate these results into clinical trials and maximize their chance of success, validation in a larger number of cell lines is desired. Studies including a high number of cell lines will benefit from recent advances in high-content technologies. However, unfortunately, performing in-depth analysis of 10–30 cell lines requires substantial resources that most laboratories do not have access to. The strategy taken in this review to combine the results from several independent studies may be a necessary compromise between in-depth analysis and a high number of cell lines. The optimization and harmonization of tissue culture methods may also be key to facilitate the identification of robust phenotypes across studies and will be discussed below. In addition, new methods decreasing the variance of neuronal phenotypes in iPSC models and decreasing the cost of the analyses are necessary and will also be discussed in the next sections.
Choosing the right control: healthy matched subjects or genetically edited isogenic lines?
Disease-linked cellular phenotypes are identified by comparing neuronal lines from patients and controls. To date, healthy subjects are the most commonly used control cell lines (Fig. 3c). However, differences in genetic background may give rise to variance in neuronal phenotypes that are unrelated to the disease. Some studies try to address this by using asymptomatic carriers, which are typically siblings or first-degree relatives of the patient27,28,33,35,57,62,69,70. Despite reducing genetic variability by around 50%, the limited availability of these controls hampers their use. In addition, asymptomatic carriers may express mild disease phenotypes57,69,70, which can increase the threshold for detecting the early disease phenotypes of progressive neurodegenerative disorders in symptomatic carriers. In order to reduce further genetic heterogeneity between control and disease lines, the development of genetic editing techniques such as TALEN, ZFN, and CRISPR/Cas9 has enabled the generation of isogenic lines191–194. Differing in only one single known mutant gene, the comparison of isogenic neurons allows the precise analysis of the role played by a specific mutation in disease modulation. Gene editing techniques have been particularly useful for studying monogenic forms of PD. Specifically, restorative isogenic lines of common genetic variants including LRRK2, SNCA, and GBA have highlighted complex roles of these mutations in protein aggregation, autophagy, and lysosomal dysfunction observed in PD49,55,58. In addition, PD mutations have been introduced into healthy subject or embryonic stem cell lines34,43,51,53,66. Having access to isogenic lines minimizes genetic background variability between patient and control lines, thereby reducing the threshold of detection for disease-related cellular phenotypes. However, considering the low penetrance of most familial PD genes, it may also be argued that the “genetic background” of a patient contributes to the cellular phenotype in a multigenic synergic way.
Generating relevant neuronal cell types for PD
The cellular reprogramming toolbox for researchers is rapidly expanding and includes a panoply of neuronal differentiation protocols to generate cells representing various brain regions21. PD is a debilitating motor system disorder resulting from the selective degeneration of midbrain dopamine (mDA) neurons located in the substantia nigra pars compacta. Protocols have been established to specifically generate dopaminergic neurons and brain cells with a midbrain molecular profile195,196.
More than 86% of published hiPSC-PD studies started with donor skin cells (fibroblasts and keratinocytes) and only 5% used blood cells (peripheral blood mononuclear cells, CD34+ cord blood cells or lymphoblastoid cells) (Fig. 3e). More than 70% of iPSCs were generated using integrating viral vectors (retroviral and lentiviral), 17% used non-integrating viral vectors (Sendai virus) and only 2% used viral-free approaches (episomal) (Fig. 3e). Several neuronal differentiation methods used across the 67 iPSC-PD studies were also analyzed in this review26,36,39,44,86,197–204. Different methods followed distinct neuronal differentiation trajectories (from iPSC to neurons), involving the generation of various intermediate cell types (Fig. 3f). The intermediate stages may include embryoid bodies (EB, stem cells in suspension), EZ sphere (aggregates of early neural stem cells in suspension), neural rosettes (early neural stem cells with radial arrangements), neurosphere (neural progenitors in suspension), and neural progenitor cells (NPC, neural precursors with bipolar morphology). The generation of cell types such as EBs, neural rosettes, and NPCs mimics various stages of in utero neurogenesis21. Few protocols also generated unconventional cell types such as EZ spheres202 or neurospheres205. Such alternatives aim to eliminate laborious differentiation steps involving EB formation and manual picking of rosettes. On average, the generation of precursor cell types such as EZ spheres, neurospheres, or NPCs can range from 11 to 45 days (Fig. 3g) and neural maturation can range between 2 and 28 weeks (Fig. 3h). Temporal manipulation of small molecules and growth factors is used to induce neural induction, selective brain region patterning, differentiation, and maturation (Fig. 3i). Diverse combinations of molecules and choice of neural reprogramming trajectory can increase the efficiency and specificity of mDA neuronal differentiation. Midbrain DA neurons can also be generated using only small molecules or with the aid of lentiviral vectors70,86,200. Varying differentiation protocols may influence the quality of cells. For example, neurons generated from floor-plate intermediates exhibited PD phenotypes, while cells generated on a stromal feeder layer from neural rosette intermediate did not31. Therefore, in some cases, phenotypic variability in iPSC-PD studies can be a consequence of varying neural differentiation protocols.
Harmonizing rigorous protocols across studies
Regardless of the differentiation method, cell cultures can be widely heterogeneous. On average, iPSC-PD studies, which use midbrain patterning protocols, yield 64% neurons and 27% dopaminergic neurons of the total cells (Fig. 3j). Proportions of neurons (bIII-Tub+) and dopaminergic neurons (TH+) vary vastly between studies even when using identical differentiation techniques (Fig. 3j). Quantitative analyses conducted at discordant timepoints along the maturation process may further contribute to differences in reported proportions of cell type between studies. In addition, arbitrary thresholds of immunocytochemical staining may alter the quantification of cell type numbers reported. Some studies exclusively focus on the analysis of dopaminergic neurons, but many others consistently report the presence of astrocytes, neural stem cells and multiple midbrain neuronal subtypes (i.e., glutaminergic and GABAergic) in their cultures41,74. Mixed cultures of human neurons and glia are more accurate in representing the neurophysiology of the human brain in vivo but add some variance in the model, which needs to be taken into consideration during experimental planning. For disease modeling, inconsistencies in both study design and analyses between laboratories can hinder the reproducibility of the results. The artificial nature of reprogramming protocols makes it difficult to agree on the superiority of a single protocol. As the field progresses, the gap between the quality of in vitro and in vivo brain tissue will tighten. The methods generating brain tissue as close as possible to their in vivo counterpart will become gold-standard methodologies, which will help to harmonize culture models between laboratories and may help expedite the clinical translation of the most robust and reproducible results. However, in some cases, for example large-scale studies requiring fast and cheap protocols, a trade-off between brain tissue quality and ease of the methods may be necessary.
An alternative and complementary strategy is to use a top-to-bottom approach, which consists of identifying homogenous cell types of interest in mixed cultures before comparative analysis206. The identification of desired cell types can be achieved with molecular or functional analyses. The selection or enrichment of specific cells, for example electrophysiologically mature dopaminergic neurons, can be achieved during the cellular reprograming process or right before the final comparative analysis. Fluorescence-activated cell sorting (FACS) can facilitate the removal of non-neural cell types to focus the analysis on a pure population of mDA neurons68,69. However, homogeneity based on single markers may be insufficient. Recent large-scale single-cell transcriptomics revealed the complexity of multiple cell types in neuronal culture as well as brain in vivo207. We and others also reported that batch or tissue culture protocol variations, such as passage number or basal media, can generate a mixture of neurons at various electrophysiological states of maturity206,208. To ensure an unbiased comparison restricted to functionally mature neurons, a thorough analysis of neuronal properties using electrophysiological techniques is necessary. The recent development of Patch-Seq technology, which combines single-cell RNA-sequencing (RNA-seq) and patch clamping, enables electrophysiological, transcriptomic, and morphological profiling of single neurons206,209,210. The multimodal characterization of cellular subtypes can be used to eliminate functional immaturity bias in iPSC-based disease modeling206,210.
How environmental factors and aging can be recapitulated in vitro
An obvious limitation of in vitro models is the lack of environmental context. The influence of nongenetic factors is not recapitulated in the basal phenotype of patient-derived neurons. For example, the influence of head trauma of a boxer with sporadic PD will not be recapitulated by default in reprogrammed neurons. An alternative would be to transplant the patient-derived neurons in animals and simulate the trauma on the animal. Similarly, influence of decades of aging of the human brain is difficult to reproduce in vitro in a few months within the boundaries of feasible experimental design. Brains in a dish will always be an imperfect experimental model. However, many tricks can be used to recapitulate the environmental and aging stress in vitro. Table 2 summarizes a list of reagents that have already been used in iPSC neuronal culture to mimic oxidative stress, proteostatic stress, mitochondrial stress, synaptic stress, ER stress, inflammation, and cellular aging. An interesting example is progerin, a truncated form of lamin A associated with premature aging. Increasing the expression of progerin in iPSC neurons can recapitulate at least some aspect of cellular aging in vitro71. Human iPSC-derived dopamine neurons overexpressing progerin displayed specific phenotypes such as neuromelanin accumulation. In addition, PD patient-derived neurons revealed disease-related phenotypes that required both genetic susceptibility and induced-aging in vitro71.
Table 2.
Cellular stressors used in hiPSC-PD models.
| Type of stressor | Reagent | What is it? | Mechanism of action | Environmental and biological relevance |
|---|---|---|---|---|
| Proteostatic | MG132 | Peptide-aldehyde (proteasome inhibitor) | Initiates apoptosis and reduces degradation of ubiquitin-conjugated proteins by inhibiting 20S proteasome activity | * |
| Cycloheximide | Naturally occurring fungicide | Inhibits global transcriptional function | * | |
| Lactacystin | Proteasomal inhibitors | Inhibits ubiquitin–proteasome proteolysis pathway | * | |
| Epoxomicin | Naturally occurring proteasome inhibitor | Inhibits protein degradation by inhibiting chymotrypsin-like proteasome | Used to induce PD-like symptoms in animal models | |
| Leupeptin | Naturally occurring protease inhibitor | Inhibits lysosomal proteolysis by inhibiting cysteine protease | Used to induce aging associated degeneration of neural processes in animal models | |
| Ammonium Chloride | Weak Base | Inhibits lysosomal proteolysis by altering pH of lysosomes | Component of fertilizers | |
| Bafilomycin | Toxic macrolide antibiotic | Inhibits autophagy by preventing maturation of autophagic vacuoles. Targets vacuolar type H+-ATPase causes lysosome neutralization and inhibition of autophagosome and lysosome fusion. | * | |
| Chloroquine | Prototype anti-malarial drug | Inhibits autophagy by increasing lysosomal pH and preventing fusion of autophagosomes and lysosomes. | * | |
| Rapamycin | Macrolide compound | Increases autophagy by inducing autophagosome formation via inhibition of mTOR | * | |
| Mitochondrial | FCCP/CCCP | Protonophore (respiratory chain uncoupler) | Reduce cell respiratory capacity by inhibiting mitochondrial cell volume and membrane potential | * |
| Rotenone | Insecticide | Inhibits mitochondria respiration and ATP generation via inhibition of mitochondrial complex I | Insecticide associated with PD epidemiology | |
| Valinomycin | Antibiotic/Ionophore | Induced loss of mitochondrial membrane potential and mitochondrial translocation via facilitating potassium transfer across lipid layers | * | |
| Oligomycin | Antibiotic | Blocks ATP synthesis via inhibition of ATP synthase preventing oxidative phosphorylation of ADP | * | |
| Antimycin A | Antibiotic | Inhibits mitochondrial electron transport by inhibition of complex III | * | |
| Paraquat | Herbicide | Increases ROS | Herbicide associated with PD epidemiology | |
| MPTP | Impure by-product of recreational drug design | Induces cell death due to ATP depletion via inhibition of mitochondrial complex I | Causes Parkinsonism in users | |
| Maneb | Fungicide | Alters cellular redox reactions by catalysing DA oxidation and chelation of metals | Fungicide used to generate PD in animal models | |
| Concanamycin A | Antibiotic | Inhibits mitophagy by preventing acidification of organelles via inhibition of H+-ATPase | * | |
| Synaptic | KCl | Metal halide salt | Induces dopamine release by increasing calcium response | KCl found in some foods and used as a salt substitute |
| Glutamate | Neurotransmitter | Activates extra-synaptic and synaptic excitatory glutamatergic receptors via AMPA, NMDA Kainite receptors, and alterations in intracellular calcium | Main component of food flavourer monosodium glutamate (MSG) | |
| Caffeine | CNS stimulant | RyR agonist that increases the calcium release from the calcium-sensitive internal stores | Widely consumed psychoactive drug present in many foods | |
| Colchicine | Microtubule depolarizing agent | Inhibits microtubule polymerization by binding to tubulin | Promote neurite retraction in vitro | |
| Oxidative | Hydrogen Peroxide | Strong oxidizer | Directly increase oxidation and ROS | Induce oxidative stress linked to aging |
| No antioxidants | Removing antioxidants from neuronal culture media | Reduce the prevention of the oxidation of molecules | Antioxidants in food | |
| 6-OHDA | Synthetic compound (enters neurons via neurotransmitter uptake transporters) | Produce ROS specifically in dopaminergic and noradrenergic neurons; inhibition of mitochondrial complexes I and IV | Selective destruction of DAn used to generate PD in animal models | |
| Dopamine | Neurotransmitter | Induced dopamine toxicity by increased dopamine oxidation by monoamine oxidases (MAO) in cytoplasm | Mimic chronic use of Levodopa | |
| Cadmium | Transition metals | Induce oxidative stress by depleting cells’ antioxidants | Heavy metals present in contaminated air, water. soil and food | |
| Exogenous a-synuclein | Major component of Lewy bodies | Increase basal ROS levels | Mimic effect of a-synuclein accumulation in vitro | |
| ER | A23187 | Calcium ionophore | Induced ER stress by increasing cytosolic calcium concentrations | * |
| Brefeldin A | Fungal metabolite | Inhibits intracellular protein transport by preventing association of COP-I coat to the Golgi membrane | * | |
| DTT | Reducing agent | Induces ER stress by preventing inhibition of disulfide bonds | * | |
| Age-inducing | Progerin | Truncated version of the lamin A protein | Increases the frequency of unrepaired double-strand breaks in DNA | Mimics aging processes |
| Inflammation | IL-17 | Hematopoietic growth factor (cytokine) | Induces inflammation through IL-17–IL-17R signaling and activation of NFkB | Naturally occurring inflammatory cytokine |
These compounds were used to stress cells to explore mechanisms of PD not to cause PD in models.
An asterisk indicates a synthetic compound not naturally occurring or that does not have environmental or biological relevance.
Human iPSC studies of PD highlight converging molecular and cellular pathways across genetic subgroups
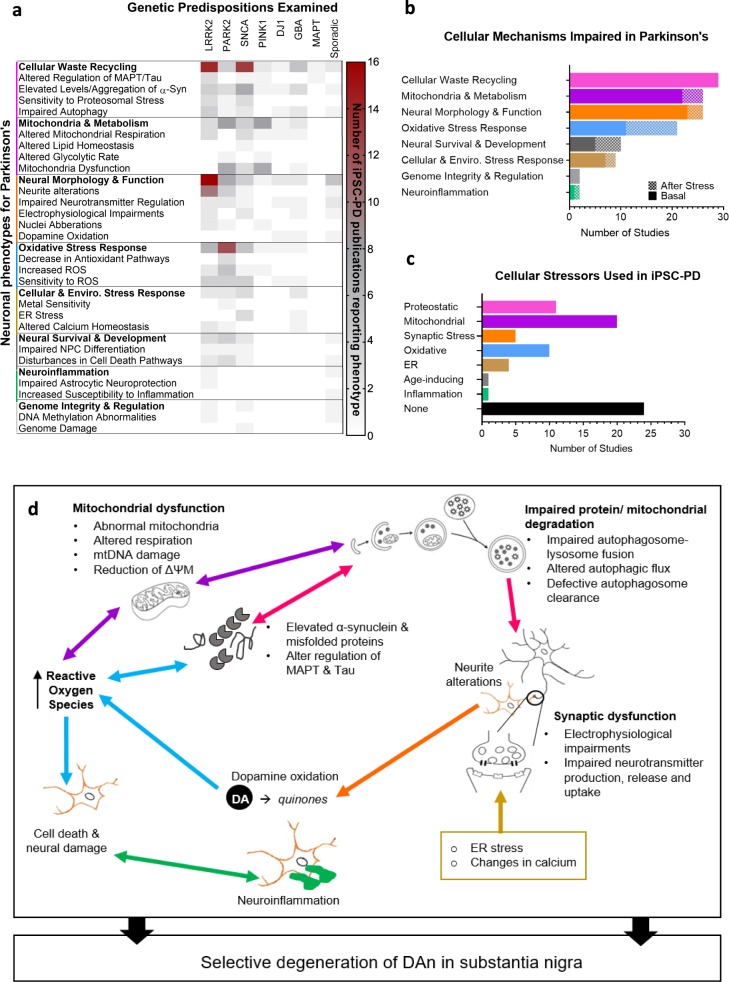
Our analysis of 385 iPSC-derived cell lines from 67 published studies reveals that many PD neuronal phenotypes are shared between genetically heterogeneous familial and sporadic patients (Fig. 4a). Notably, impairments in mechanisms involved in cellular waste recycling, mitochondrial function, neuronal morphology and physiology, and sensitivity to reactive oxygen species (ROS) are most common across patient lines with varying genetic predispositions (Fig. 4b). The studies measured cellular phenotypes that occurred either spontaneously or in response to chemicals mimicking cellular aging and stress (Table 2 and Fig. 4a, c). It is important to note that the frequency of reported phenotypes in our meta-analysis may be biased because only few studies (~19%, 13/67) reported negative results (absence of phenotypes)31,32,36,37,40,45,48,52,59,64,74,76,86. In addition, most cell lines were not systematically phenotyped without prior hypothesis and thus, there is likely to be an ascertain bias in these phenotypes. Less hypothesis-driven multimodal or omics analysis will help to address such bias41,72,76–80,87,90. Phenotypes caused by genomic predispositions allude to crosstalk and impairments in multiple pathways that act collectively to mediate selective degeneration of dopaminergic neurons in the substantia nigra (Fig. 4d) and will be discussed in detail below.
Fig. 4. Phenotypic insights from iPSC studies of Parkinson’s disease.
a A heatmap representation of neuronal phenotypes reported in genetically heterogeneous PD lines examined in 67 hiPSC studies. Categories in bold represent a sum of all sub-phenotypes. Reported absences of phenotypes are not represented in the figure. b Summary of the impairments in cellular mechanisms which were reported in iPSC-PD studies. Data represented as total number of studies reporting impairment with or without induced stress (“after stress” or “basal”, respectively). c Types of artificial cellular stressors used across hiPSC-PD studies (refer to Table 2). Data presented as the number of studies that used stressor to induce or investigate PD. d Schematic representation of crosstalk between cellular mechanisms involved in PD pathogenesis.
Genetic predispositions reduce mitochondrial respiration in patient-derived neurons
Epidemiological studies outlined in Fig. 2 have pointed to several genes involved in mitochondrial function and recycling (e.g., CHCHD2, PRKN, PLA2G6, VSP13C, PINK1, LRRK2; see also Table 1). An increase in dysfunctional mitochondria was identified in 26 independent hiPSC-PD studies using LRRK2, PRKN, PINK1, GBA, and SNCA patient-derived neuronal lines24,27,31,32,37–39,52,55,57,59,61,62,64,67,68,70,71,73,74,77,85,89,90. Neurons derived from PD patients displayed an increase in mitochondrial copy number62 and a greater proportion of abnormal mitochondria31,39. Abnormal mitochondria exhibited either signs of enlargement31,71, shortening32, thinning64, irregular cristae structures and network59,74, or fragmentation24,70. Mitochondria in the proximal axon of neurons from individuals carrying the LRRK2 R1441C mutation were 20% shorter than individuals with LRRK2-G2019S mutations32. An increase in bidirectional mobility of mitochondria was also reported in LRRK2 lines but not in PINK132. Overall, cellular respiration and metabolism were altered in neuronal lines derived from LRRK2, PRKN, SNCA, PINK1, DJ-1, and GBA patients27,37,55,58,59,61,67,70,73,83,85. Basal mitochondrial respiration is consistently compromised across PD lines, exhibited as reductions in maximal oxygen consumption rate, diminished ADP and ATP levels and NAD+/NADH redox states27,55,58,59,61,67,73,85. Impaired mitochondrial respiration was also associated with an increase in mitochondrial ROS and lysosomal hyperactivity67. The glycolytic rate of patient-derived neurons was higher in PINK170 and PRKN lines77. This was not observed in the transcriptomic analysis of SNCA lines90. An increase of passive proton leakage from the inner membrane was reported in PINK1 mutant neurons but not in LRRK232. In addition, the degradation and translocation of mitochondrial membrane proteins such as PARKIN and MIRO1 were impaired in LRRK2 and PRKN neurons38,62,73.
Overall neuronal metabolism may be impaired by endogenous dysfunction of mitochondria25,31,37–39,52,55,57,59,61,64,67,68,70,73,74,77,85,89,90, or indirect external stressors24,32,62,71,90, or by abnormal recycling of damaged mitochondria38,39,52,68. For example, some patient neurons displayed signs of impaired mitochondrial quality control related to aberrant mitophagy38,52,68, mitochondrial DNA damage37,57 and consistent decreases in mitochondrial membrane potential24,38,67,83,85.
Genetic predispositions that reduce the constant supply of energy to neurons ultimately disrupt the functioning of brain circuits. Mitochondrial impairments can confer increased neuronal vulnerability to oxidative, nitro-oxidative stress, and metal ions24,32,39,55,58,67,68,74. The high metabolic demand of substantia nigra neurons might explain their selective degeneration211.
Impaired oxidative stress-buffering capacity increases neuronal susceptibility to cell death in PD
Impairments in multiple cellular mechanisms increase oxidative burden in patient-derived neurons with mutations in LRRK2, PRKN, PINK1, GBA, DJ-1, and SNCA (20 studies)24,27–29,32,33,38,40,42,46,48,53,55,58,68,77,88,89. Elevated basal ROS levels reported in patient-derived neurons increase cell susceptibility to cell death such that patient lines treated with oxidative and mitochondrial stressors (i.e., H2O2, rotenone and paraquat) exhibit significantly higher levels of the cell death marker caspase-3 compared with healthy controls27–29,33,39,46,68,77,88. Notably, antioxidant buffering in patient lines is reduced, marked by a low level of antioxidant enzymes and the downregulation of antioxidant pathways such as NRF2 and target gene NQO129,39,77. In addition, ten studies, which did not report oxidative stress phenotypes in basal conditions, reported and increase vulnerability when oxidative stress was induced with chemicals24,31,32,40,42,48,53,55,58,89. This suggests that PD genetic predispositions could be compensated by upregulating endogenous antioxidant pathways or providing antioxidant supplements. Several clinical trials testing antioxidants treatments are ongoing. However, unfortunately, so far, clinical trials failed to show that antioxidants such as MitoQ significantly slow down the progression of symptoms212. Antioxidants may need to be combined with other treatments for more positive outcomes.
Genetic predispositions increase the probability of protein aggregation in PD neurons
Alpha-synuclein and Tau protein accumulation constitute the most common phenotype reported in 28 studies investigating LRRK2, PRKN, PINK1, GBA, SNCA, and MAPT lines25–31,34–36,39,41,43,46,48–51,53,55,56,58,60,65,67,69,73,83. The upregulation MAPT (microtubule-associated protein tau) transcripts were associated with protein aggregation, which impaired axonal mitochondria movement and induced neurite aberrations in patient neurons25,53,213. The accumulation of α-synuclein and Tau has also been noted in the varicosities of contorted and fragmented axons of patients41. Elevated expression of SNCA mRNA was associated with the dysregulation of oxidative stress, protein aggregation, and cell death regulatory genes, proposed to induce or increase selective sensitivity of mDA neurons to these factors28,29. Alpha-synuclein protein levels were also elevated in patient-derived neurons27–30,34–36,39,41,46,50,51,55,56,65,69,73,83. Notably, the level of soluble27 and monomeric28,46 conformations of α-synuclein were increased. Increased events of α-synuclein oligomerization73, elevated levels of insoluble oligomers27,31,55 and the formation of inclusion bodies65 were also reported. Neurons derived from SNCA mutant iPSCs55 displayed an increase in the phosphorylation of α-synuclein at residue S129; this was not detected in LRRK251 derived neurons. Alpha-synuclein oligomers mediated neurotoxicity through metal redox reactions and increased the formation of fibril aggregates, forming Lewy bodies33. Nevertheless, neurotoxicity mediated by protein aggregation can be a consequence of impairments in various contributing cellular mechanisms including inflammation, mitochondrial function, autophagy and stress, suggesting a role in late PD pathogenesis36,39,44.
Dysregulation of autophagy and protein ubiquitination contributes to neurodegeneration
Across 15 hiPSC studies, evidence of impaired protein degradation in LRRK2, PRKN, GBA, DJ-1, SNCA, and sporadic PD lines has been reported26,27,29,36,45,48–50,53,56,58,63,69,75. Unbiased pathway analysis of cysteine-modified proteins revealed an enrichment of proteins involved in the ubiquitination and removal of unfolded protein76. Accumulation of autophagosomes and increased expression of key autophagy regulators (i.e., beclin1, p62) were also consistently noted in PD neurons, suggestive of an upregulation of autophagy initiation26,36,48,49,56,74. Increased protein levels of the autophagy marker LC3B-II in LRRK2 I2020T were associated with increased markers of protein oxidation48. However, reductions in microtubule-associated protein light chain 3 (LC3) and lysosomal marker colocalization reflective of unsuccessful autolysosome formation and maturation implied disturbances in autophagy progression45,56. Increasing genetic, epidemiological, and clinical studies draw biochemical and cellular links between PD and lysosomal storage disorders214,215. Mutations in lysosomal genes such as GBA, SMPD1, and ATP13A2 have been shown to impact the bidirectional feedback loop for processing and clearance of α-synuclein in PD124,146,216. Signs of enlarged lysosomal compartments paired with decreased lysosomal enzymatic activity suggested that poor autophagy completion contributed to protein aggregation in PD27,36,47,58,69,92. Interestingly, lysosomal impairment in sporadic patient-derived neurons was observed at a later timepoint (180 days) compared with DJ-1 mutant neurons (70 days)27. Young-onset iPSC models of PD revealed cycloheximide treatment specifically slowed the degradation rate of α-synuclein92. The dysregulation of ubiquitin genes also increased the susceptibility of patient neurons to proteasome stress29. Dysregulated autophagy and ubiquitin–proteasome mechanisms were associated with an increase in extracellular α-synuclein in patient’s dopamine neurons36,58. In addition, patient-derived neurons exhibited poor ability to degrade mutant LRRK2 protein compared with wildtype50. LRRK2 (G2019S) mutant protein displayed enhanced-binding to the lysosomal membrane and prevented the assembly of chaperone-mediated autophagy50. The inhibitory effect resulted in decreased degradation of long-lived proteins, which may result in unwanted protein–protein interactions mediating neurodegeneration50. PD genetic predispositions favoring either aberrant protein degradation or accumulation in patient-derived neurons were associated with increased vulnerability to proteasomal and oxidative stresses, and contributed to accelerated neurodegeneration27,29,48,53,58.
Patient-derived neurons exhibit morphological signs of neurodegeneration
Fifteen hiPSC-PD studies investigating LRRK2, PRKN, SNCA, and sporadic patient-derived lines reported alterations in neurite process morphology26,42,45,49,51,53,54,56,60,65,67,71,76,82,88. Consistently, studies reported reductions in neurite length, branching, and network complexity42,45,49,51,53,54,56,60,65,67,71,76,82,88. However, incidences of hyperbranching26 and lack of neurite alterations have also been reported37. In addition, irregular presynaptic membranous inclusions and dystrophic synaptic structures were reported in a neuronal model of synucleinopathy73. The exact mechanism underlying neurite alterations in PD neurons remains unclear and can be attributed either to dysfunction in neurite morphogenesis or increased neurite deterioration42. Elevation in autophagic flux, in response to α-synuclein and Tau accumulation, was associated with the reduction of neurite structure and outgrowth in patient lines49,53. Induction of the initiation of autophagy or inhibition of its completion also greatly exacerbated the neurite alterations in patient dopamine neurons26,56. Microtubule instability induced by the depolymerizing agent colchicine and loss of PRKN function also resulted in neural aberrations, though not specifically in TH+ cells54. Both autophagy and microtubule polymerization play key roles in neurite injury, growth and axonal formation, and impairments in either of these mechanisms can mediate the neurite alterations observed in PD neurons217,218.
Synaptic dysfunction increases dopamine oxidation in PD leading to neurodegenerative processes
Twelve human iPSC studies examining LRRK2, SNCA, GBA, and sporadic PD reported signs of impaired synaptic function27,31,40–42,47–49,56,60,69,71,91. Dysregulation of genes involved in synaptogenesis, synaptic vesicle mechanisms, synaptic transmission, and regulation of neurotransmitter release have been identified in SNCA, LRRK2, and PRKN patient-derived lines41,42,49,92. LRRK2 I2020T mutation decreased the spontaneous calcium-dependent release of dopamine48 but not PRKN mutant40. The loss of PARKIN in patient midbrain neurons has been shown to downregulate dopamine reuptake machinery40. Downregulation of synaptic proteins involved in synaptic vesicle endocytosis was associated with an increase of cytosolic dopamine47. Elevated levels of cytosolic dopamine mediate dopamine toxicity and contribute to neural damage and degeneration27,31,40,69,75. Excessive oxidation of dopamine by variants of monoamine oxidases in the cytoplasm increases the formation of ROS and toxic quinones, contributing to neurodegeneration40,69. Disruption of calcium homeostasis and endoplasmic reticular (ER) stress may mediate excessive neurotransmitter release. RNA-seq analyses also revealed an enrichment of genes associated with ER stress in PD patient-derived neurons80,90. More than 50% of generated patient lines overexpressing α-synuclein displayed signs of impaired electrophysiological activity, most notably demonstrated by reduced action potentials upon stimulation49. Neurons derived from SNCA, LRRK2, GBA, and sporadic patients also exhibited delayed or the absence of firing synchronicity, a decrease in the number of active channels and firing rates, as well as reduced spontaneous activity41–43,49,56,69. It has been suggested that, in vivo, neurons may go successively through hypo- and hyperactive phases, possibly as a homeostatic response to the cytotoxic effect of synaptic dysfunction219. Such phases remain to be described with human iPSC-derived neurons in vitro, but multiple genetic predispositions appear to directly or indirectly impair synaptic function, which contributes to a vicious neurodegenerative cascade.
Neuroinflammation exacerbates neurodegeneration in sporadic PD
Midbrain neurons derived from sporadic patients showed increased susceptibility to the effects of adaptive immune cells72. Sporadic patient neuronal lines co-cultured with T-lymphocytes exhibited substantial signs of cell death mediated by IL-17–IL-17R signaling and activation of NFkB72. Similarly, IL-17 treatment resulted in increased neuronal death72. Inflammation in the central nervous system and periphery are key hallmarks of PD220. Increasing evidence implicates the role of microglia in neuronal loss, though the underlying mechanisms remain to be determined221,222. RNA-seq analysis of astrocytes derived from LRRK2-G2019S iPSCs highlighted dysregulation in genes involved in the extracellular matrix, which may reduce the neuroprotective capacity of astrocytes in PD78. Investigating the role of neuroinflammation in patient-derived microglia may also contribute to the understanding of the selective vulnerability of mDA neurons in sporadic and late-onset PD223.
Genetic predispositions reducing differentiation yield of mDA neurons
In vitro neural development was impaired in neural lines derived from patients carrying LRRK2, PRKN, SNCA, and sporadic mutations43,49,74,93. In four independent studies, the differentiation potential of neural progenitor cells derived from patients was significantly reduced, demonstrated by low yields of neurons in comparison with control lines43,49,74,81,94. A recent review presented the idea that PD is attributed to significant neurodevelopmental defects, which may increase the susceptibility for disease onset224. If confirmed, identifying genetic predispositions that contribute to early developmental defects in iPSC-PD may assist the development of novel PD therapies. However, these phenotypes may appear in conflict with other studies53,55,76 capable of generating functional neurons from cell lines with similar mutations. The differences could be due to varying protocols, which may be more or less stressful for the cells.
Epigenomic alterations linked with PD in patient-derived neurons
The ability to capture unique epigenomic alterations associated with PD remains an important challenge. Reprogramming fibroblasts to iPSCs may erase age-associated225 and naive epigenetic signatures which could contribute to sporadic PD pathophysiology226. However, an epigenetic phenotype was reported in iPSC-derived PD patient neurons79,89. Neuronal lines derived from LRRK2 and sporadic patients exhibited epigenomic alterations when compared with healthy controls79. Hypermethylation was prominent in gene regulatory regions associated with the downregulation of transcription factors FOXA1, NR3C1, HNF4A, and FOSL279. Interestingly, LRRK2 mutant and sporadic PD patient neurons shared similar methylation patterns, which were absent in the original donor fibroblasts79. A spontaneous increase in the number of DNA strand breaks and genomic damage89 in PD patient-derived neurons could indirectly impact genomic regulation.
Do iPSC-PD models exhibit similar neural phenotypes to those observed in rodent models?
The use of rodent models to simulate human pathologies associated with PD has been extensively reviewed227–230. PD pathogenesis is mostly induced in animal models with 6-OHDA, rotenone, MPTP, paraquat, and amphetamine231. These models induce substantia nigra dopaminergic neuron death through inhibition of mitochondrial function and increasing production of ROS229. Rats and mice systemically exposed to rotenone and paraquat exhibited signs of Lewy body formation232,233. Knockout and transgenic rodent models are designed to investigate the consequential pathological impacts of known PD genetic mutations. Genetic mouse models have provided a platform for the functional studies of proteins associated with PD such as PARKIN, LRRK2, DJ-1, and α-synuclein234,235. However, the human genetic background of idiopathic PD cannot be fully recapitulated with animal models.
iPSC-PD patient-derived neurons displayed a reduction in striatal dopamine release, a reduction in neurite complexity, an increased in tau phosphorylation, impairments of dopamine neurotransmission, autophagy, and mitochondrial abnormalities (Fig. 4a, b). These impairments were also observed in transgenic mice PD models236–238. Such correlation between models is interesting, even though some of the hypothesis-driven studies in iPSC neurons may have been biased by previous animal studies. IPSC models can also shed light on cellular phenotypes absent in animal models. For example, dopamine oxidation was present in PD iPSC-derived neurons but was not observed in neurons derived from DJ-1 KO mice iPSC27. Any model has its advantages and limitations and animal and patient-derived neuronal models complement each other. Animal models provide the organism context that is lacking in tissue culture models, whereas patient-derived neuronal models more accurately represent human genetics, which may in turn increase chances of translational success.
Omics analysis of patient-derived PD neurons
To date 10/67 iPSC-PD studies analyzed have used proteomic, transcriptomic, or epigenomic profiling to phenotype PD patient-derived neurons41,72,76–80,87,90. Omics analyses may be less biased and data-driven as opposed to purely hypothesis-driven239. Data from omics studies can also help to describe biological relationships between complex intertwined cellular pathways and identify relevant druggable molecular pathways.
Genes associated with clinical PD phenotypes such as abnormal nervous physiology phenotypes, abnormal motor capabilities, coordination, and movement phenotypes were dysregulated in iPSC neurons derived from LRRK2-G2019S patients87. RNA-seq and proteomics analysis also revealed a heavy dysregulation of genes and proteins associated with mitochondrial function, protein ubiquitination, unfolded protein response, ER/calcium regulation, and oxidative stress in PD patient-derived neurons in comparison with healthy controls41,69,72,77,78,80,84,87,90. In addition, the epigenome in PD-derived lines was described as uniquely aberrant compared with healthy controls79. Together, omics studies highlighted that the proteomic, transcriptomic, and epigenomic profiles of PD patient-derived neurons exhibited disease-specific alterations, which can also correlate with other neurological diseases41. Identifying genes that are consistently dysregulated in the same direction in PD genetic lines across independent studies will help to confirm the key common pathways involved in PD pathogenesis. Much work remains to be done in this field. New human molecular insights into PD pathogenesis will help form a stronger foundation for therapeutic development, and may also benefit other neurological diseases.
Will findings from PD iPSC models translate to human clinical trials?
One of the most exciting applications of patient-derived iPSC models of PD is to validate pharmacological treatments before clinical trials. The field is still at the stage of improving human brain tissue engineering, and many different protocols are being tested and developed. However, the need for progress in clinical translation for brain disorders is extremely high, and there is no time to wait for brain tissue models to be perfect. Pioneering iPSC studies pave the road to success and identify limitations which help the community to reach a consensus on the minimal requirements to model brain disorders in vitro most accurately. It seems essential to improve the efficiency of reprogramming and differentiation protocols while trying to make those models as physiological and realistic as possible208,240. Some concerns are raised that in vitro neuronal development, maturation and function might be too artificial, suggesting that the model may overlook some of the critical processes that occur in vivo. Nevertheless, some defects observed in iPSC-derived neurons have already been confirmed in human postmortem brain tissues39,43,241–243. Although this is very encouraging, it is unclear whether significant in vitro phenotypes that cannot be confirmed in postmortem brain tissue should be disregarded. Most postmortem brains also have technical limitations and may represent later stages of the disease, whereas iPSC models may represent earlier stages, preceding neurodegeneration.
Given the apprehensions that in vitro studies may be too artificial, human iPSC-derived neural progenitors may be transplanted into animal brains244–247. Besides ethical barriers, xenografts also raise the possibility that the healthy host tissue compensate for the impairment of the transplanted cells. Yet, if the phenotypes observed in vitro are recapitulated in vivo, pharmacological treatments could be assessed in a systemic environment, with much more realistic dosage and administration methods.
Concluding remarks
PD is increasingly described as a spectrum disorder, with patients experiencing a multitude of motor and non-motor symptoms in a unique way. Similarly, PD genetic predispositions are broad. Hundreds of gene variants increase the risk of PD, but no genetic mutation causes a complete penetrance. We estimated the prevalence of each of the most penetrant mutations (increasing risks by >5 times) to occur in <5% of the global PD population. Despite the broad representation of genetic predispositions in PD patients, our review of current studies using iPSC-derived brain cells demonstrates commonality in cellular impairment susceptibilities. Independent reports highlight a recurring theme around dysfunction of mitochondria, proteasomal mechanisms, synapses, inflammation, and oxidative stress regulation. Constitutive and simultaneous dysregulation of multiple pathways can become overbearing, resulting in accelerated neurodegeneration. As a result, there may be an exaggerated emphasis on pathways which may represent later stages of pathogenesis. Future work will further establish the interdependence of these cellular functions and will help to isolate the initial cause from the downstream consequence of a cellular phenotype. It is also possible that current studies are biased towards studying already known or arbitrarily chosen phenotypes, and the integration of multi-omics analysis will help address this issue. Further optimization of the brain tissue engineering methods will also reduce the threshold for detecting disease-related phenotypes from tissue culture artefacts, facilitating the identification of early cellular phenotypes. Future studies also will have to include the analysis of larger pools of patients including sporadic PD with genetic predispositions more representative of the epidemiology.
The rapid expansion of iPSC disease modeling studies of PD is exciting. Altogether, the current work reviewed here suggests that a neuroprotective therapy, which will stop the neurodegeneration in people living with PD, will most likely require to target multiple pathways at once. The prospect of investigating the impact of multigenic predispositions on brain cell functions will provide information on key modulators of neurodegeneration in PD, and preclinical data for more personalized medicine.
Methods
Meta-analysis of epidemiological PD studies
Data from Fig. 2a was obtained from epidemiological studies sourced from PubMed and Google Scholar search engines using keywords “Parkinson’s disease”, “prevalence”, “incidence”, and “epidemiology”. Twelve international studies94,157–167 including a total of 5650 persons living with PD were used for the analysis. Datasets were each obtained from single independent studies (Finnish, European, Canadian, Fareo Islands, Swedish, Australian, and Norwegian populations) or combined from two independent studies (American and Caucasian populations). We estimated the proportion of fPD patients in the world population with a formula (% fPD = fPD population/total PD population).
Data from Fig. 2b was obtained from epidemiological studies were identified through Google Scholar search utilizing keywords “Parkinson”, “genetic”, “population”, “familial”, and the relevant gene names (i.e. LRRK2, PRKN/PARK2, ATP13A2, SNCA, VPS35, PINK1, DJ-1, and DNAJC6). A total of 50 epidemiological studies93–129 were used for the analysis (total n = 488 patients carrying various mutations and 32,012 PD cases). The frequency of mutation (PD population with mutation/total PD population) was averaged across studies examining the same gene. Only studies containing sample sizes of >100 subjects were included in the analysis.
Meta-analysis of genetic and GWAS studies
A meta-analysis of genetic and GWAS studies is displayed in Fig. 2c. Raw GWAS data were extracted 24 independent genetic studies8,10–14,130–148 (identified through Google scholar search with gene name and keywords “Parkinson’s”, “variant”, and “odds ratio”) combined with GWAS database (NHGRI-EBI Catalog, accessed 14th Jan 2019, and filtered for “Parkinson’s” as phenotype trait149). The collated data set consisted of 25,243 PD cases and 41,945 healthy controls. The frequency (%) of each variant was measured in 25,243 total PD cases. The odds ratio (OR) was calculated by dividing the frequency of the variant among patients by the frequency of the variant in healthy controls. Only variants with OR greater than 1.5 were plotted against the frequency.
Data extraction from PDgene GWAS database
GWAS data were extracted from PDgene database141 was accessed on 18th Jan 2019 (n = 19,061 PD cases and healthy controls). Datasets for the genes listed under “Top Results” were filtered for p < 5 × 10–8 and OR values were plotted on Fig. 2d.
Literature search of and meta-analysis of iPSC-PD studies
Original research papers were identified through Google Scholar search utilizing keywords “Parkinson”, “iPSC”, “induced pluripotency stem cell”, “patient-derived”, and “models”. Papers published between 2011 and 2020 which included phenotypic and/multimodal analysis of the patient-derived PD lines were selected for analysis (n = 6724–92). The meta-analyses of these studies were displayed in Fig. 3b–j.
In Fig. 3j, we meta-analyzed immunocytochemistry quantifications from 33 independent iPSC-PD studies. Studies reporting TUJ1 and TUBBIII were used for calculating the percentages of neurons, both label the bIII-Tubulin protein, and the names are used interchangeably. The percentage of neurons (bIII-Tub/DAPI) was reported by 23 studies (including two of them not reporting trajectories). Percentage of TH neurons (TH+/DAPI) is averaged for 32 studies. However, 12 of these 32 studies did not directly report the percentage of TH+/DAPI but instead reported TH+/TUJ1 and TUJ1+/DAPI. In this instance, for consistency in our analysis, we estimated the proportion of TH+/DAPI with a formula (TH/DAPI = TUJ1/DAPI × TH/TUJ1).
Supplementary information
Acknowledgements
The authors wish to thank Prof. Christopher Proud, Prof Stuart Brierley, Dr Tim Sargeant, A/Prof. Ville Petteri Makinen, Dr Mark van den Hurk, and the other members of the Bardy lab for the feedback. This work was generously supported by The Parkinson’s South Australia Foundation, Perpetual Impact Philanthropy, the Brain Foundation, Flinders Foundation, Rebecca L. Cooper Foundation, Ian Potter Foundation, Angelique and Michael Boileau Corporate Philanthropy, MRFF Australian Government and Department of Health (CB), and The Australian Government Research Training Program Scholarship (JT).
Author contributions
J.T. performed the meta-analysis of all iPSC studies and genomics studies. H.A. performed the meta-analysis of epidemiological studies. C.B. designed the study and supervised J.T. and H.A. J.T., H.A., and C.B. prepared the figures. C.B. and J.T. wrote the paper. All authors have approved the submitted version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41531-020-0110-8.
References
- 1.Venda LL, Cragg SJ, Buchman VL, Wade-Martins R. α-Synuclein and dopamine at the crossroads of Parkinson's disease. Trends Neurosci. 2010;33:559–568. doi: 10.1016/j.tins.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussbaum RL. The identification of alpha-synuclein as the first Parkinson disease. Gene. J. Parkinsons Dis. 2017;7:S43–S49. doi: 10.3233/JPD-179003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanassiadou A, et al. Genetic analysis of families with Parkinson disease that carry the Ala53Thr mutation in the gene encoding alpha-synuclein. Am. J. Hum. Genet. 1999;65:555–558. doi: 10.1086/302486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz E, et al. Identification of Spanish familial Parkinson's disease and screening for the Ala53Thr mutation of the alpha-synuclein gene in early onset patients. Neurosci. Lett. 1997;235:57–60. doi: 10.1016/S0304-3940(97)00710-6. [DOI] [PubMed] [Google Scholar]
- 5.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Wood-Kaczmar A, Gandhi S, Wood NW. Understanding the molecular causes of Parkinson’s disease. Trends Mol. Med. 2006;12:521–528. doi: 10.1016/j.molmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Escott-Price V, et al. Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann. Neurol. 2015;77:582–591. doi: 10.1002/ana.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simón-Sánchez, J. et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet.41, 1308–1312 (2009). [DOI] [PMC free article] [PubMed]
- 9.Chang D, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;7:e1002142. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards TL, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, et al. Genome-wide association study identifies candidate genes for Parkinson's disease in an Ashkenazi Jewish population. BMC Med. Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankratz N, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2009;124:593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saad M, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population. Hum. Mol. Genet. 2011;20:615–627. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- 14.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 15.Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 16.Parr CJC, Yamanaka S, Saito H. An update on stem cell biology and engineering for brain development. Mol. Psychiatry. 2017;22:808–819. doi: 10.1038/mp.2017.66. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Dolmetsch R, Geschwind, Daniel H. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soldner F, Jaenisch R. Stem cells, genome editing, and the path to translational medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soldner F, Jaenisch R. iPSC disease modeling. Science. 2012;338:1155. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- 21.Mertens J, Marchetto MC, Bardy C, Gage FH. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 2016;17:424–437. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haston KM, Finkbeiner S. Clinical trials in a dish: the potential of pluripotent stem cells to develop therapies for neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 2016;56:489–510. doi: 10.1146/annurev-pharmtox-010715-103548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aboud AA, et al. PARK2 patient neuroprogenitors show increased mitochondrial sensitivity to copper. Neurobiol. Dis. 2015;73:204–212. doi: 10.1016/j.nbd.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beevers JE, et al. MAPT genetic variation and neuronal maturity alter isoform expression affecting axonal transport in iPSC-derived dopamine neurons. Stem Cell Rep. 2017;9:587–599. doi: 10.1016/j.stemcr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borgs L, et al. Dopaminergic neurons differentiating from LRRK2 G2019S induced pluripotent stem cells show early neuritic branching defects. Sci. Rep. 2016;6:33377. doi: 10.1038/srep33377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burbulla LF, et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science. 2017;357:1255. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byers B, et al. SNCA triplication Parkinson's patient's iPSC-derived DA neurons accumulate [alpha]-synuclein and are susceptible to oxidative stress. PLoS ONE. 2011;6:e26159. doi: 10.1371/journal.pone.0026159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang K-H, et al. Impairment of proteasome and anti-oxidative pathways in the induced pluripotent stem cell model for sporadic Parkinson's disease. Parkinsonism Relat. Disord. 2016;24:81–88. doi: 10.1016/j.parkreldis.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Chung CY, et al. Identification and rescue of α-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung SunY, et al. Parkin and PINK1 patient iPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and α-synuclein accumulation. Stem Cell Rep. 2016;7:664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper O, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deas E, et al. Alpha-synuclein oligomers interact with metal ions to induce oxidative stress and neuronal death in Parkinson's disease. Antioxid. Redox Signal. 2016;24:376–391. doi: 10.1089/ars.2015.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dettmer U, et al. Parkinson-causing α-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat. Commun. 2015;6:7314. doi: 10.1038/ncomms8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devine MJ, et al. Parkinson's disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat. Commun. 2011;2:440. doi: 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandes HugoJR, et al. ER stress and autophagic perturbations lead to elevated extracellular α-synuclein in GBA-N370S parkinson's iPSC-derived dopamine neurons. Stem Cell Rep. 2016;6:342–356. doi: 10.1016/j.stemcr.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howlett EH, et al. LRRK2 G2019S-induced mitochondrial DNA damage is LRRK2 kinase dependent and inhibition restores mtDNA integrity in Parkinson's disease. Hum. Mol. Genet. 2017;26:4340–4351. doi: 10.1093/hmg/ddx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh C-H, et al. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell. 2016;19:709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imaizumi Y, et al. Mitochondrial dysfunction associated with increased oxidative stress and α-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, et al. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat. Commun. 2012;3:668. doi: 10.1038/ncomms1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouroupi G, et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc. Natl Acad. Sci. USA. 2017;114:E3679–E3688. doi: 10.1073/pnas.1617259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L, et al. Molecular features underlying neurodegeneration identified through in vitro modeling of genetically diverse Parkinson’s disease patients. Cell Rep. 2016;15:2411–2426. doi: 10.1016/j.celrep.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Liu G-H, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López de Maturana R, et al. Mutations in LRRK2 impair NF-κB pathway in iPSC-derived neurons. J. Neuroinflammation. 2016;13:295. doi: 10.1186/s12974-016-0761-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marrone L, et al. Generation of iPSCs carrying a common LRRK2 risk allele for in vitro modeling of idiopathic Parkinson's disease. PLoS ONE. 2018;13:e0192497. doi: 10.1371/journal.pone.0192497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen HaN, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen M, Krainc D. LRRK2 phosphorylation of auxilin mediates synaptic defects in dopaminergic neurons from patients with Parkinson's disease. Proc. Natl Acad. Sci. USA. 2018;115:5576–5581. doi: 10.1073/pnas.1717590115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohta E, et al. I2020T mutant LRRK2 iPSC-derived neurons in the Sagamihara family exhibit increased Tau phosphorylation through the AKT/GSK-3β signaling pathway. Hum. Mol. Genet. 2015;24:4879–4900. doi: 10.1093/hmg/ddv212. [DOI] [PubMed] [Google Scholar]
- 49.Oliveira LMA, et al. Elevated α-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson's patient-derived induced pluripotent stem cells. Cell Death Dis. 2015;6:e1994. doi: 10.1038/cddis.2015.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orenstein SJ, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 2013;16:394. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qing X, et al. CRISPR/Cas9 and piggyBac-mediated footprint-free LRRK2-G2019S knock-in reveals neuronal complexity phenotypes and α-Synuclein modulation in dopaminergic neurons. Stem Cell Res. 2017;24:44–50. doi: 10.1016/j.scr.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Rakovic A, et al. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J. Biol. Chem. 2013;288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinhardt P, et al. Genetic correction of a LRRK2 mutation in human iPSCs links Parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Ren Y, et al. Parkin mutations reduce the complexity of neuronal processes in iPSC‐derived human neurons. STEM CELLS. 2014;33:68–78. doi: 10.1002/stem.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryan ScottD, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sánchez‐Danés A, et al. Disease‐specific phenotypes in dopamine neurons from human iPS‐based models of genetic and sporadic Parkinson's disease. EMBO Mol. Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanders LH, et al. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson's disease patients: reversal by gene correction. Neurobiol. Dis. 2014;62:381–386. doi: 10.1016/j.nbd.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schöndorf DC, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat. Commun. 2014;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- 59.Schöndorf DC, et al. The NAD + precursor nicotinamide riboside rescues mitochondrial defects and neuronal loss in iPSC and fly models of Parkinson’s disease. Cell Rep. 2018;23:2976–2988. doi: 10.1016/j.celrep.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Schwab AndrewJ, Ebert, Allison D. Neurite aggregation and calcium dysfunction in iPSC-derived sensory neurons with Parkinson’s disease-related LRRK2 G2019S mutation. Stem Cell Rep. 2015;5:1039–1052. doi: 10.1016/j.stemcr.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwab AJ, et al. Decreased sirtuin deacetylase activity in LRRK2 G2019S iPSC-derived dopaminergic neurons. Stem Cell Rep. 2017;9:1839–1852. doi: 10.1016/j.stemcr.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seibler P, et al. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J. Neurosci. 2011;31:5970. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaltouki A, Hsieh CH, Kim MJ, Wang X. Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson's models. Acta neuropathologica. 2018;136:607–620. doi: 10.1007/s00401-018-1873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiba-Fukushima K, et al. Evidence that phosphorylated ubiquitin signaling is involved in the etiology of Parkinson's disease. Hum. Mol. Genet. 2017;26:3172–3185. doi: 10.1093/hmg/ddx201. [DOI] [PubMed] [Google Scholar]
- 65.Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J. Neurosci. 2014;34:418–433. doi: 10.1523/JNEUROSCI.2712-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su Y-C, Qi X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum. Mol. Genet. 2013;22:4545–4561. doi: 10.1093/hmg/ddt301. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki S, et al. Efficient induction of dopaminergic neuron differentiation from induced pluripotent stem cells reveals impaired mitophagy in PARK2 neurons. Biochem. Biophys. Res. Commun. 2017;483:88–93. doi: 10.1016/j.bbrc.2016.12.188. [DOI] [PubMed] [Google Scholar]
- 69.Woodard ChrisM, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014;9:1173–1182. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azkona G, et al. LRRK2 expression is deregulated in fibroblasts and neurons from Parkinson patients with mutations in PINK1. Mol. Neurobiol. 2018;55:506–516. doi: 10.1007/s12035-016-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sommer A, et al. Th17 lymphocytes induce neuronal cell death in a human iPSC-based model of Parkinson’s disease. Cell Stem Cell. 2018;23:123–131.e126. doi: 10.1016/j.stem.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 73.Prots I, et al. alpha-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc. Natl Acad. Sci. USA. 2018;115:7813–7818. doi: 10.1073/pnas.1713129115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaltouki A, et al. Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Rep. 2015;4:847–859. doi: 10.1016/j.stemcr.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aflaki E, et al. A new glucocerebrosidase chaperone reduces alpha-synuclein and glycolipid levels in iPSC-derived dopaminergic neurons from patients with Gaucher disease and Parkinsonism. J. Neurosci. 2016;36:7441–7452. doi: 10.1523/JNEUROSCI.0636-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bogetofte H, et al. Perturbations in RhoA signalling cause altered migration and impaired neuritogenesis in human iPSC-derived neural cells with PARK2 mutation. Neurobiol. Dis. 2019;132:104581. doi: 10.1016/j.nbd.2019.104581. [DOI] [PubMed] [Google Scholar]
- 77.Bogetofte H, et al. PARK2 mutation causes metabolic disturbances and impaired survival of human iPSC-derived neurons. Front. Cell. Neurosci. 2019;13:297. doi: 10.3389/fncel.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Booth HDE, et al. RNA sequencing reveals MMP2 and TGFB1 downregulation in LRRK2 G2019S Parkinson's iPSC-derived astrocytes. Neurobiol. Dis. 2019;129:56–66. doi: 10.1016/j.nbd.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 79.Fernández-Santiago R, et al. Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson's disease patients. EMBO Mol. Med. 2015;7:1529–1546. doi: 10.15252/emmm.201505439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heman-Ackah SM, et al. Alpha-synuclein induces the unfolded protein response in Parkinson’s disease SNCA triplication iPSC-derived neurons. Hum. Mol. Genet. 2017;26:4441–4450. doi: 10.1093/hmg/ddx331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iannielli A, et al. Pharmacological inhibition of necroptosis protects from dopaminergic neuronal cell death in Parkinson’s disease models. Cell Rep. 2018;22:2066–2079. doi: 10.1016/j.celrep.2018.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korecka JA, et al. Neurite collapse and altered ER Ca2+ control in human Parkinson disease patient iPSC-derived neurons with LRRK2 G2019S mutation. Stem Cell Rep. 2019;12:29–41. doi: 10.1016/j.stemcr.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ludtmann MHR, et al. alpha-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson's disease. Nat. Commun. 2018;9:2293. doi: 10.1038/s41467-018-04422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Momcilovic O, et al. Derivation, characterization, and neural differentiation of integration-free induced pluripotent stem cell lines from Parkinson's disease patients carrying SNCA, LRRK2, PARK2, and GBA mutations. PloS ONE. 2016;11:e0154890–e0154890. doi: 10.1371/journal.pone.0154890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morais VA, et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 86.Reinhardt P, et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLOS ONE. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandor C, et al. Transcriptomic profiling of purified patient-derived dopamine neurons identifies convergent perturbations and therapeutics for Parkinson's disease. Hum. Mol. Genet. 2017;26:552–566. doi: 10.1093/hmg/ddw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tabata Y, et al. T-type calcium channels determine the vulnerability of dopaminergic neurons to mitochondrial stress in familial Parkinson disease. Stem Cell Rep. 2018;11:1171–1184. doi: 10.1016/j.stemcr.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vasquez V, et al. Chromatin-bound oxidized α-synuclein causes strand breaks in neuronal genomes in in vitro models of Parkinson's disease. J. Alzheimers Dis. 2017;60:S133–S150. doi: 10.3233/JAD-170342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zambon F, et al. Cellular α-synuclein pathology is associated with bioenergetic dysfunction in Parkinson’s iPSC-derived dopamine neurons. Hum. Mol. Genet. 2019;28:2001–2013. doi: 10.1093/hmg/ddz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhong P, Hu Z, Jiang H, Yan Z, Feng J. Dopamine induces oscillatory activities in human midbrain neurons with Parkin mutations. Cell Rep. 2017;19:1033–1044. doi: 10.1016/j.celrep.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laperle AH, et al. iPSC modeling of young-onset Parkinson's disease reveals a molecular signature of disease and novel therapeutic candidates. Nat. Med. 2020;26:289–299. doi: 10.1038/s41591-019-0739-1. [DOI] [PubMed] [Google Scholar]
- 93.Ahn TB, et al. alpha-Synuclein gene duplication is present in sporadic Parkinson disease. Neurology. 2008;70:43–49. doi: 10.1212/01.wnl.0000271080.53272.c7. [DOI] [PubMed] [Google Scholar]
- 94.Bentley SR, et al. Pipeline to gene discovery—analysing familial Parkinsonism in the Queensland Parkinson's Project. Parkinsonism Relat. Disord. 2018;49:34–41. doi: 10.1016/j.parkreldis.2017.12.033. [DOI] [PubMed] [Google Scholar]
- 95.Blanckenberg J, Ntsapi C, Carr JA, Bardien S. EIF4G1 R1205H and VPS35 D620N mutations are rare in Parkinson's disease from South Africa. Neurobiol. Aging. 2014;35:445.e441–445.e443. doi: 10.1016/j.neurobiolaging.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 96.Brueggemann N, et al. Re: Alpha-synuclein gene duplication is present in sporadic Parkinson disease. Neurology. 2008;71:1294. doi: 10.1212/01.wnl.0000338439.00992.c7. [DOI] [PubMed] [Google Scholar]
- 97.Camacho JLG, et al. High frequency of Parkin exon rearrangements in Mexican-mestizo patients with early-onset Parkinson's disease. Mov. Disord. 2012;27:1047–1051. doi: 10.1002/mds.25030. [DOI] [PubMed] [Google Scholar]
- 98.Chen Y, et al. VPS35 Asp620Asn and EIF4G1 Arg1205His mutations are rare in Parkinson disease from Southwest China. Neurobiol. Aging. 2013;34:1709.e1707–1709.e1708. doi: 10.1016/j.neurobiolaging.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 99.Djarmati A, et al. ATP13A2 variants in early-onset Parkinson's disease patients and controls. Mov. Disord. 2009;24:2104–2111. doi: 10.1002/mds.22728. [DOI] [PubMed] [Google Scholar]
- 100.Djarmati A, et al. Detection of Parkin(PARK2) and DJ1(PARK7) mutations in early-onset Parkinson disease: Parkinmutation frequency depends on ethnic origin of patients. Hum. Mutat. 2004;23:525–525. doi: 10.1002/humu.9240. [DOI] [PubMed] [Google Scholar]
- 101.Erer S, et al. Mutation analysis of the PARKIN, PINK1, DJ1, and SNCA genes in Turkish early-onset Parkinson's patients and genotype-phenotype correlations. Clin. Neurol. Neurosurg. 2016;148:147–153. doi: 10.1016/j.clineuro.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 102.Fiala O, et al. Parkin (PARK 2) mutations are rare in Czech patients with early-onset Parkinson's disease. PLoS ONE. 2014;9:e107585. doi: 10.1371/journal.pone.0107585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo JF, et al. VPS35 gene variants are not associated with Parkinson's disease in the mainland Chinese population. Parkinsonism Relat. Disord. 2012;18:983–985. doi: 10.1016/j.parkreldis.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 104.Healy DG, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hedrich K, et al. DJ-1 (PARK7) mutations are less frequent than Parkin (PARK2) mutations in early-onset Parkinson disease. Neurology. 2004;62:389–394. doi: 10.1212/01.WNL.0000113022.51739.88. [DOI] [PubMed] [Google Scholar]
- 106.Khan NL, et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson's disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 107.Klein C, et al. PINK1, Parkin, and DJ-1 mutations in Italian patients with early-onset parkinsonism. Eur. J. Hum. Genet. 2005;13:1086–1093. doi: 10.1038/sj.ejhg.5201455. [DOI] [PubMed] [Google Scholar]
- 108.Kumar KR, et al. Frequency of the D620N mutation in VPS35 in Parkinson disease. Arch. Neurol. 2012;69:1360–1364. doi: 10.1001/archneurol.2011.3367. [DOI] [PubMed] [Google Scholar]
- 109.Longo GS, et al. Alpha-synuclein A53T mutation is not frequent on a sample of Brazilian Parkinson's disease patients. Arq. Neuropsiquiatr. 2015;73:506–509. doi: 10.1590/0004-282X20150032. [DOI] [PubMed] [Google Scholar]
- 110.Macedo MG, et al. Genotypic and phenotypic characteristics of Dutch patients with early onset Parkinson's disease. Mov. Disord. 2009;24:196–203. doi: 10.1002/mds.22287. [DOI] [PubMed] [Google Scholar]
- 111.Mata IF, et al. Lrrk2 pathogenic substitutions in Parkinson's disease. Neurogenetics. 2005;6:171–177. doi: 10.1007/s10048-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 112.Mellick GD, et al. Screening PARK genes for mutations in early-onset Parkinson's disease patients from Queensland, Australia. Parkinsonism Relat. Disord. 2009;15:105–109. doi: 10.1016/j.parkreldis.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 113.Moura KC, et al. Exon dosage variations in Brazilian patients with Parkinson's disease: analysis of SNCA, PARKIN, PINK1 and DJ-1 genes. Dis. Markers. 2012;32:173–178. doi: 10.1155/2012/218642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nichols WC, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 115.Sharma M, et al. A multi-centre clinico-genetic analysis of the VPS35 gene in Parkinson disease indicates reduced penetrance for disease-associated variants. J. Med Genet. 2012;49:721–726. doi: 10.1136/jmedgenet-2012-101155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sheerin UM, et al. Screening for VPS35 mutations in Parkinson's disease. Neurobiol. Aging. 2012;33:838 e831–835. doi: 10.1016/j.neurobiolaging.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sudhaman S, et al. VPS35 and EIF4G1 mutations are rare in Parkinson's disease among Indians. Neurobiol. Aging. 2013;34:2442.e2441–2442.e2443. doi: 10.1016/j.neurobiolaging.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 118.Valente EM, et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 2004;56:336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 119.Vilarino-Guell C, et al. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu YR, et al. Genetic analysis of Parkin in early onset Parkinson's disease (PD): novel intron 9 g> a single nucleotide polymorphism and risk of Taiwanese PD. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:229–234. doi: 10.1002/ajmg.b.30977. [DOI] [PubMed] [Google Scholar]
- 121.Zimprich A, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann. Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 123.Alcalay RN, et al. Frequency of known mutations in early-onset Parkinson disease: implication for genetic counseling: the consortium on risk for early onset Parkinson disease study. Arch. Neurol. 2010;67:1116–1122. doi: 10.1001/archneurol.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alcalay RN, et al. SMPD1 mutations, activity, and alpha-synuclein accumulation in Parkinson's disease. Mov. Disord. 2019;34:526–535. doi: 10.1002/mds.27642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anwarullah, et al. Absence of SNCA polymorphisms in Pakistani Parkinsons disease patients. Jpma. J. Pak. Med. Assoc. 2017;67:1512–1516. [PubMed] [Google Scholar]
- 126.Hattori N, et al. Molecular genetic analysis of a novel Parkin gene in Japanese families with autosomal recessive juvenile parkinsonism: evidence for variable homozygous deletions in the Parkin gene in affected individuals. Ann. Neurol. 1998;44:935–941. doi: 10.1002/ana.410440612. [DOI] [PubMed] [Google Scholar]
- 127.Johnson J, et al. SNCA multiplication is not a common cause of Parkinson disease or dementia with Lewy bodies. Neurology. 2004;63:554–556. doi: 10.1212/01.WNL.0000133401.09043.44. [DOI] [PubMed] [Google Scholar]
- 128.Lucking CB, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl. J. Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 129.Rogaeva E, et al. Analysis of the PINK1 gene in a large cohort of cases with Parkinson disease. Arch. Neurol. 2004;61:1898–1904. doi: 10.1001/archneur.61.12.1898. [DOI] [PubMed] [Google Scholar]
- 130.Simón-Sánchez J, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur. J. Hum. Genet. 2011;19:655. doi: 10.1038/ejhg.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Marco EV, et al. DJ-1 is a Parkinson's disease susceptibility gene in southern Italy. Clin. Genet. 2010;77:183–188. doi: 10.1111/j.1399-0004.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 132.Deng H, Wu Y, Jankovic J. The EIF4G1 gene and Parkinson's disease. Acta Neurologica Scandinavica. 2015;132:73–78. doi: 10.1111/ane.12397. [DOI] [PubMed] [Google Scholar]
- 133.Foo JN, Liu J, Tan E-K. CHCHD2 and Parkinson's disease. Lancet Neurol. 2015;14:681–682. doi: 10.1016/S1474-4422(15)00098-8. [DOI] [PubMed] [Google Scholar]
- 134.Funayama M, et al. Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian population. Neuroreport. 2007;18:273–275. doi: 10.1097/WNR.0b013e32801254b6. [DOI] [PubMed] [Google Scholar]
- 135.Funayama M, et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson's disease: a genome-wide linkage and sequencing study. Lancet Neurol. 2015;14:274–282. doi: 10.1016/S1474-4422(14)70266-2. [DOI] [PubMed] [Google Scholar]
- 136.Gómez-Garre P, et al. Systematic mutational analysis of FBXO7 in a Parkinson's disease population from southern Spain. Neurobiol. Aging. 2014;35:727.e725–727.e727. doi: 10.1016/j.neurobiolaging.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 137.Huttenlocher J, et al. Heterozygote carriers for CNVs in PARK2 are at increased risk of Parkinson's disease. Hum. Mol. Genet. 2015;24:5637–5643. doi: 10.1093/hmg/ddv277. [DOI] [PubMed] [Google Scholar]
- 138.Kay DM, et al. Heterozygous parkin point mutations are as common in control subjects as in Parkinson's patients. Ann. Neurol. 2007;61:47–54. doi: 10.1002/ana.21039. [DOI] [PubMed] [Google Scholar]
- 139.Krüger R, et al. A large-scale genetic association study to evaluate the contribution of Omi/HtrA2 (PARK13) to Parkinson's disease. Neurobiol. aging. 2011;32:548.e549–548.e545.548E518. doi: 10.1016/j.neurobiolaging.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maraganore DM, et al. High-resolution whole-genome association study of Parkinson disease. Am. J. Hum. Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nalls MA, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 2014;46:989. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pihlstrøm L, et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson's disease. Neurobiol. Aging. 2013;34:1708.e1707–1708.e1713. doi: 10.1016/j.neurobiolaging.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 143.Ran C, et al. Strong association between glucocerebrosidase mutations and Parkinson's disease in Sweden. Neurobiol. Aging. 2016;45:212.e215–212.e211. doi: 10.1016/j.neurobiolaging.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ross OA, et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson's disease. Ann. Neurol. 2008;64:88–92. doi: 10.1002/ana.21405. [DOI] [PubMed] [Google Scholar]
- 145.Semenova EV, et al. Analysis of PARK2 gene exon rearrangements in Russian patients with sporadic Parkinson's disease. Mov. Disord. 2012;27:139–143. doi: 10.1002/mds.23901. [DOI] [PubMed] [Google Scholar]
- 146.Sidransky E, Lopez G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012;11:986–998. doi: 10.1016/S1474-4422(12)70190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Y, et al. The contribution of GIGYF2 to Parkinson's disease: a meta-analysis. Neurological Sci. : Off. J. Ital. Neurological Soc. Ital. Soc. Clin. Neurophysiol. 2015;36:2073–2079. doi: 10.1007/s10072-015-2316-9. [DOI] [PubMed] [Google Scholar]
- 148.Puschmann A, et al. Heterozygous PINK1 p.G411S increases risk of Parkinson's disease via a dominant-negative mechanism. Brain. 2017;140:98–117. doi: 10.1093/brain/aww261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Scott WK, et al. Genetic complexity and Parkinson's disease. Deane Laboratory Parkinson Disease Research Group. Science. 1997;277:387–388. doi: 10.1126/science.277.5324.387. [DOI] [PubMed] [Google Scholar]
- 151.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu. Rev. Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 152.Adler CH. Differential diagnosis of Parkinson's disease. Med Clin. North Am. 1999;83:349–367. doi: 10.1016/S0025-7125(05)70108-5. [DOI] [PubMed] [Google Scholar]
- 153.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 154.Brown TP, Rumsby PC, Capleton AC, Rushton L, Levy LS. Pesticides and Parkinson's disease-is there a link? Environ. Health Perspect. 2006;114:156–164. doi: 10.1289/ehp.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reeve A, Simcox E, Turnbull D. Ageing and Parkinson's disease: why is advancing age the biggest risk factor? Ageing Res Rev. 2014;14:19–30. doi: 10.1016/j.arr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Levy G. The relationship of Parkinson disease with aging. Arch. Neurol. 2007;64:1242–1246. doi: 10.1001/archneur.64.9.1242. [DOI] [PubMed] [Google Scholar]
- 157.Autere JM, Moilanen JS, Myllyla VV, Majamaa K. Familial aggregation of Parkinson's disease in a Finnish population. J. Neurol. Neurosurg. psychiatry. 2000;69:107–109. doi: 10.1136/jnnp.69.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bonifati V, Fabrizio E, Vanacore N, De Mari M, Meco G. Familial Parkinson's disease: a clinical genetic analysis. Can. J. Neurol Sci. 1995;22:272–279. doi: 10.1017/S0317167100039469. [DOI] [PubMed] [Google Scholar]
- 159.Elbaz A, et al. Familial aggregation of Parkinson's disease: a population-based case-control study in Europe. EUROPARKINSON Study Group. Neurology. 1999;52:1876–1882. doi: 10.1212/WNL.52.9.1876. [DOI] [PubMed] [Google Scholar]
- 160.Kurz M, Alves G, Aarsland D, Larsen JP. Familial Parkinson's disease: a community-based study. Eur. J. Neurol. 2003;10:159–163. doi: 10.1046/j.1468-1331.2003.00532.x. [DOI] [PubMed] [Google Scholar]
- 161.Linder J, Stenlund H, Forsgren L. Incidence of Parkinson's disease and parkinsonism in northern Sweden: a population-based study. Mov. Disord. 2010;25:341–348. doi: 10.1002/mds.22987. [DOI] [PubMed] [Google Scholar]
- 162.McDonnell SK, et al. Complex segregation analysis of Parkinson's disease: the Mayo Clinic Family Study. Ann. Neurol. 2006;59:788–795. doi: 10.1002/ana.20844. [DOI] [PubMed] [Google Scholar]
- 163.Payami H, Larsen K, Bernard S, Nutt J. Increased risk of Parkinson's disease in parents and siblings of patients. Ann. Neurol. 1994;36:659–661. doi: 10.1002/ana.410360417. [DOI] [PubMed] [Google Scholar]
- 164.Petersen MS, Bech S, Nosova E, Aasly J, Farrer MJ. Familial aggregation of Parkinson's disease in the Faroe Islands. Mov. Disord. 2015;30:538–544. doi: 10.1002/mds.26132. [DOI] [PubMed] [Google Scholar]
- 165.Shino MY, et al. Familial aggregation of Parkinson's disease in a multiethnic community-based case-control study. Mov. Disord. Off. J. Mov. Disord. Soc. 2010;25:2587–2594. doi: 10.1002/mds.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Uitti RJ, et al. "Familial Parkinson's disease"-a case-control study of families. Can. J. Neurol. Sci. 1997;24:127–132. doi: 10.1017/S0317167100021454. [DOI] [PubMed] [Google Scholar]
- 167.Olgiati S, et al. DNAJC6 mutations associated with early-onset Parkinson's disease. Ann. Neurol. 2016;79:244–256. doi: 10.1002/ana.24553. [DOI] [PubMed] [Google Scholar]
- 168.Yamamura Y. The long journey to the discovery of PARK2: the 50th anniversary of Japanese Society of Neuropathology. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2010;30:495–500. doi: 10.1111/j.1440-1789.2010.01144.x. [DOI] [PubMed] [Google Scholar]
- 169.Funayama M, et al. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 170.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset Parkinsonism. Science. 2003;299:256. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 171.Gandhi S, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 172.Di Fonzo A, et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68:1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 173.Edvardson S, et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE. 2012;7:e36458. doi: 10.1371/journal.pone.0036458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Burchell VS, et al. The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 2013;16:1257. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Quadri M, et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum. Mutat. 2013;34:1208–1215. doi: 10.1002/humu.22373. [DOI] [PubMed] [Google Scholar]
- 176.Lu C-S, et al. PLA2G6 mutations in PARK14-linked young-onset parkinsonism and sporadic Parkinson's disease. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012;159B:183–191. doi: 10.1002/ajmg.b.32012. [DOI] [PubMed] [Google Scholar]
- 177.Strauss KM, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum. Mol. Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 178.Schormair B, et al. Diagnostic exome sequencing in early-onset Parkinson's disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson's disease. Clin. Genet. 2018;93:603–612. doi: 10.1111/cge.13124. [DOI] [PubMed] [Google Scholar]
- 179.Keller MF, et al. Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson's disease. Hum. Mol. Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.International Parkinson's Disease Genomics ConsortiumWellcome Trust Case Control Consortium 2 A two-stage meta-analysis identifies several new loci for Parkinson's disease. PLoS Genet. 2011;7:e1002142–e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 182.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 183.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 184.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum. Mol. Genet. 2011;20:R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bardy, C., Greenberg, Z., Perry, S. W. & Licinio, J. in Personalized Psychiatry (ed Baune, B. T.) 127–146 (Academic Press, 2020).
- 186.Burkhardt MF, et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol. Cell Neurosci. 2013;56:355–364. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sutherland GT, et al. A cross-study transcriptional analysis of Parkinson's disease. PLOS ONE. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hoekstra SD, Stringer S, Heine VM, Posthuma D. Genetically-informed patient selection for iPSC studies of complex diseases may aid in reducing cellular heterogeneity. Front. Cell. Neurosci. 2017;11:164–164. doi: 10.3389/fncel.2017.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Byrne, S. M., Mali, P. & Church, G. M. in Methods in Enzymology, Vol. 546 (eds Doudna J. A. & Sontheimer E. J.) 119–138 (Academic Press, 2014).
- 192.Li M, Suzuki K, Kim NY, Liu G-H, Izpisua Belmonte JC. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J. Biol. Chem. 2014;289:4594–4599. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ding Q, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang G, et al. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat. Protoc. 2016;12:88. doi: 10.1038/nprot.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.La Manno G, et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580.e519. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Arenas E, Denham M, Villaescusa JC. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- 197.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Perrier AL, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2004;101:12543. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sanchez-Danes A, et al. Efficient generation of A9 midbrain dopaminergic neurons by lentiviral delivery of LMX1A in human embryonic stem cells and induced pluripotent stem cells. Hum. gene Ther. 2012;23:56–69. doi: 10.1089/hum.2011.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Swistowski A, et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ebert AD, et al. EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res. 2013;10:417–427. doi: 10.1016/j.scr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Matsumoto T, et al. Functional neurons generated from T cell-derived induced pluripotent stem cells for neurological disease modeling. Stem Cell Rep. 2016;6:422–435. doi: 10.1016/j.stemcr.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang XQ, Zhang SC. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 2010;584:355–366. doi: 10.1007/978-1-60761-369-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhou S, et al. Neurosphere based differentiation of human iPSC improves astrocyte differentiation. Stem Cells Int. 2016;2016:15. doi: 10.1155/2016/4937689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bardy C, et al. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatry. 2016;21:1573–1588. doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.van den Hurk M, Bardy C. Single-cell multimodal transcriptomics to study neuronal diversity in human stem cell-derived brain tissue and organoid models. J. Neurosci. Methods. 2019;325:108350. doi: 10.1016/j.jneumeth.2019.108350. [DOI] [PubMed] [Google Scholar]
- 208.Bardy C, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl Acad. Sci. 2015;112:E2725. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cadwell CR, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 2015;34:199. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.van den Hurk, M., Erwin, J. A., Yeo, G. W., Gage, F. H. & Bardy, C. Patch-Seq protocol to analyze the electrophysiology, morphology and transcriptome of whole single neurons derived from human pluripotent stem cells. Front. Mol. Neurosci.11, 261 (2018). [DOI] [PMC free article] [PubMed]
- 211.Pacelli C, et al. Elevated mitochondrial bioenergetics and axonal arborization size are key contributors to the vulnerability of dopamine neurons. Curr. Biol. 2015;25:2349–2360. doi: 10.1016/j.cub.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 212.Snow BJ, et al. A double-blind, placebo-controlled study to assess the mitochondria-targeted antioxidant MitoQ as a disease-modifying therapy in Parkinson's disease. Mov. Disord. 2010;25:1670–1674. doi: 10.1002/mds.23148. [DOI] [PubMed] [Google Scholar]
- 213.Kawakami F, et al. LRRK2 phosphorylates tubulin-associated Tau but not the free molecule: LRRK2-mediated regulation of the Tau-tubulin association and neurite outgrowth. PLOS ONE. 2012;7:e30834. doi: 10.1371/journal.pone.0030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Shachar T, et al. Lysosomal storage disorders and Parkinson's disease: Gaucher disease and beyond. Mov. Disord. 2011;26:1593–1604. doi: 10.1002/mds.23774. [DOI] [PubMed] [Google Scholar]
- 215.Klein AD, Mazzulli JR. Is Parkinson's disease a lysosomal disorder? Brain. 2018;141:2255–2262. doi: 10.1093/brain/awy147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dehay B, et al. Lysosomal dysfunction in Parkinson disease: ATP13A2 gets into the groove. Autophagy. 2012;8:1389–1391. doi: 10.4161/auto.21011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sainath R, Gallo G. Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 2015;359:267–278. doi: 10.1007/s00441-014-1955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Winans AM, Collins SR, Meyer T. Waves of actin and microtubule polymerization drive microtubule-based transport and neurite growth before single axon formation. eLife. 2016;5:e12387. doi: 10.7554/eLife.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Michel PatrickP, Hirsch, Etienne C, Hunot S. Understanding dopaminergic cell death pathways in Parkinson disease. Neuron. 2016;90:675–691. doi: 10.1016/j.neuron.2016.03.038. [DOI] [PubMed] [Google Scholar]
- 220.Doorn KJ, et al. Microglial phenotypes and toll-like receptor 2 in the substantia nigra and hippocampus of incidental Lewy body disease cases and Parkinson's disease patients. Acta neuropathologica Commun. 2014;2:90. doi: 10.1186/s40478-014-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Phani S, Loike JD, Przedborski S. Neurodegeneration and inflammation in Parkinson's disease. Parkinsonism Relat. Disord. 2012;18:S207–S209. doi: 10.1016/S1353-8020(11)70064-5. [DOI] [PubMed] [Google Scholar]
- 222.McGeer PL, McGeer EG. Inflammation and neurodegeneration in Parkinson's disease. Parkinsonism Relat. Disord. 2004;10:S3–S7. doi: 10.1016/j.parkreldis.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 223.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schwamborn JCIsParkinson's. Disease a neurodevelopmental disorder and will brain organoids help us to understand it? Stem Cells Dev. 2018;27:968–975. doi: 10.1089/scd.2017.0289. [DOI] [PubMed] [Google Scholar]
- 225.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.de Boni, L. & Wüllner, U. Epigenetic analysis in human neurons: considerations for disease modeling in PD. Front Neurosci.13, 276 (2019). [DOI] [PMC free article] [PubMed]
- 227.Magen, I. & Chesselet, M.-F. in Progress in Brain Research, Vol. 184 (eds Anders Björklund & M. Angela Cenci) 53–87 (Elsevier, 2010).
- 228.Potashkin JA, Blume SR, Runkle NK. Limitations of animal models of Parkinson's disease. Parkinson#39;s. Dis. 2011;2011:7. doi: 10.4061/2011/658083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Terzioglu M, Galter D. Parkinson’s disease: genetic versus toxin-induced rodent models. FEBS J. 2008;275:1384–1391. doi: 10.1111/j.1742-4658.2008.06302.x. [DOI] [PubMed] [Google Scholar]
- 230.Koprich JB, Kalia LV, Brotchie JM. Animal models of alpha-synucleinopathy for Parkinson disease drug development. Nat. Rev. Neurosci. 2017;18:515–529. doi: 10.1038/nrn.2017.75. [DOI] [PubMed] [Google Scholar]
- 231.Jagmag SA, Tripathi N, Shukla SD, Maiti S, Khurana S. Evaluation of models of Parkinson's disease. Front. Neurosci. 2016;9:503–503. doi: 10.3389/fnins.2015.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cicchetti F, et al. Systemic exposure to paraquat and maneb models early Parkinson's disease in young adult rats. Neurobiol. Dis. 2005;20:360–371. doi: 10.1016/j.nbd.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 233.Sherer TB, et al. Mechanism of toxicity in rotenone models of Parkinson's disease. J. Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson's disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lim KL, Ng CH. Genetic models of Parkinson disease. Biochim Biophys. Acta. 2009;1792:604–615. doi: 10.1016/j.bbadis.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 236.Ramonet D, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLOS ONE. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li Y, et al. Mutant LRRK2R1441G BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gubellini P, Picconi B, Di Filippo M, Calabresi P. Downstream mechanisms triggered by mitochondrial dysfunction in the basal ganglia: from experimental models to neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1802:151–161. doi: 10.1016/j.bbadis.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 239.Robert C. Microarray analysis of gene expression during early development: a cautionary overview. Reproduction. 2010;140:787–801. doi: 10.1530/REP-10-0191. [DOI] [PubMed] [Google Scholar]
- 240.Brennand KJ, et al. Creating patient-specific neural cells for the in vitro study of brain disorders. Stem Cell Rep. 2015;5:933–945. doi: 10.1016/j.stemcr.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Toulorge D, Schapira AH, Hajj R. Molecular changes in the postmortem parkinsonian brain. J. neurochemistry. 2016;139(Suppl 1):27–58. doi: 10.1111/jnc.13696. [DOI] [PubMed] [Google Scholar]
- 242.Hartmann A. Postmortem studies in Parkinson's disease. Dialogues Clin. Neurosci. 2004;6:281–293. doi: 10.31887/DCNS.2004.6.3/ahartmann. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Chailangkarn T, et al. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536:338–343. doi: 10.1038/nature19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Emborg ME, et al. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013;3:646–650. doi: 10.1016/j.celrep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kikuchi T, et al. Survival of human induced pluripotent stem cell-derived midbrain dopaminergic neurons in the brain of a primate model of Parkinson's disease. J. Parkinsons Dis. 2011;1:395–412. doi: 10.3233/JPD-2011-11070. [DOI] [PubMed] [Google Scholar]
- 246.Kikuchi T, et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson's disease model. Nature. 2017;548:592–596. doi: 10.1038/nature23664. [DOI] [PubMed] [Google Scholar]
- 247.Steinbeck JA, et al. Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson's disease model. Nat. Biotechnol. 2015;33:204–209. doi: 10.1038/nbt.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.