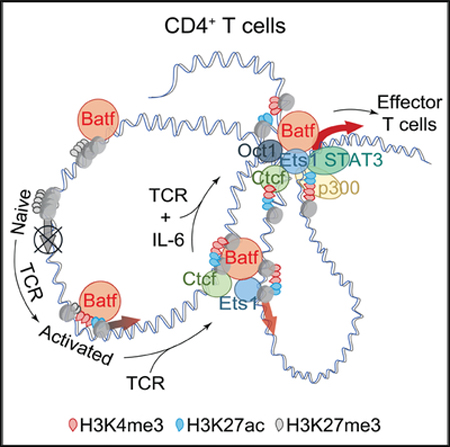
SUMMARY
The basic leucine zipper transcription factor activating transcription factor-like (Batf) contributes to transcriptional programming of multiple effector T cells and is required for T helper 17 (Th17) and T follicular helper (Tfh) cell development. Here, we examine mechanisms by which Batf initiates gene transcription in developing effector CD4 T cells. We find that, in addition to its pioneering function, Batf controls developmentally regulated recruitment of the architectural factor Ctcf to promote chromatin looping that is associated with lineage-specific gene transcription. The chromatin-organizing actions of Batf are largely dependent on Ets1, which appears to be indispensable for the Batf-dependent recruitment of Ctcf. Moreover, most of the Batf-dependent sites to which Ctcf is recruited lie outside of activating protein-1-interferon regulatory factor (Ap-1-Irf) composite elements (AICEs), indicating that direct involvement of Batf-Irf complexes is not required. These results identify a cooperative role for Batf, Ets1, and Ctcf in chromatin reorganization that underpins the transcriptional programming of effector T cells.
In Brief
Pham et al. uncover mechanisms by which Batf restructures the chromatin landscape during CD4+ effector T cell differentiation. Batf controls Ctcf recruitment to lineage-specifying gene loci in an Ets1-dependent manner to promote chromatin looping and lineage-specific gene transcription, thereby identifying a heretofore unknown cooperativity of these factors in effector T cell development.
Graphical abstract
INTRODUCTION
The immune response to diverse pathogens relies on the antigen-driven differentiation of naive CD4 T cells down different effector pathways, including T helper 1 (Th1), T helper 2 (Th2), T helper 17 (Th17), and T follicular helper (Tfh). Because effector CD4 T cells are derived from a common precursor, it is implicit that modulation of the epigenetic landscape of the naive T cell to activate or repress genes most suited for combating a particular pathogen underlies effector specification. Each of the helper T cell subtypes express a “master regulator” transcription factor (Tbet for Th1, Gata3 for Th2, and Rorγt for Th17) that, although essential for enforcing expression of lineage-specifying genes, does not appear to be the initiator of the primary epigenetic events that launch phenotype selection (Ciofani et al., 2012). A major study investigating Th17 development identified two cooperatively interacting T cell receptor (TCR)-induced factors: basic leucine zipper transcription factor activating transcription factor (ATF)-like (Batf) and interferon regulatory factor 4 (Irf4), which initiated chromatin remodeling before the recruitment of lineage-inducing transcription factors, including Rorgt at Th17specifying loci, and were thus deemed Th17 pioneer factors (Ciofani et al., 2012).
Pioneer transcription factors bind target sequences within nucleosomal DNA to initiate nucleosomal clearing thereby enabling recruitment of trans-factors involved in de novo gene expression during cell differentiation (Zaret and Carroll, 2011). Pioneers are diverse transcription factors that target distinct DNA sequences via zinc finger, basic helix-loop-helix, POU, and forkhead domains (Gifford and Meissner, 2012). A recent study confirmed the pioneering functions of proteins with these DNA binding domains and additionally identified the basic leucine zipper (bZip) factor Creb/Atf (Sherwood et al., 2014). The activating protein-1 (Ap-1) transcription factor family, of which Batf is a member, also possesses basic leucine zipper structures and can promote chromatin accessibility (Biddie et al., 2011). In Th17 cells, diminished chromatin accessibility and transcription factor recruitment at lineage-specifying loci in the absence of Batf has led to the designation of Batf as a pioneer factor (Ciofani et al., 2012). Despite the universal expression of Batf after TCR stimulation in CD4 T cells and its recent designation as a pioneer factor, how Batf is able to bind and modulate nucleosomal chromatin remains poorly understood.
DNA binding by Batf at Ap-1 consensus sequences requires dimerization with the Jun subfamily of Ap-1 factors (Murphy et al., 2013). This includes Jun (Echlin et al., 2000), JunB (Carr et al., 2017; Hasan et al., 2017; Schraml et al., 2009; Yamazaki et al., 2017), and JunD (Carr et al., 2017; Li et al., 2012).Heterodimers formed with JunB and JunD are the preferred binding partners in Th17 cells (Li et al., 2012), with JunB appearing to have a dominant role (Carr et al., 2017; Hasan et al., 2017; Yamazaki et al., 2017). The finding that Batf and Irf4 deficiencies phenocopied each other (Murphy et al., 2013), particularly in Th17 cells (Brustle et al., 2007; Schraml et al., 2009), led to the discovery that Batf-Jun heterodimers formed a trimeric complex with Irf4 or Irf8, which binds composite consensus motifs, referred to as activating protein-1-interferon regulatory factor (Ap-1-Irf) composite elements or AICEs (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012; Murphy et al., 2013). Despite their important role as targets for Batf and Irf binding at genes central to Th17 and Tfh development and function (Ise et al., 2011; Schraml et al., 2009), Irf-independent Batf binding at non-AICE elements is well documented (Ciofani et al., 2012; Li et al., 2012), although of unknown functional significance.
Enhancer activation by both pioneer and lineage-specifying transcription factors is an essential step in the development of the unique transcriptome of each CD4 T cell subset. The organization of these transcription-factor-bound regulatory elements is facilitated by the CCCTC-binding factor Ctcf. Originally described as an insulator, Ctcf is now also recognized as an architectural factor that organizes higher-order chromatin structure and looping interactions, facilitating enhancer-promoter interactions (Bonev and Cavalli, 2016; Ong and Corces, 2014). In naive T cells, interactions between Ctcf functions and Oct-1 mediate inter-chromosomal interactions between the Th2 cytokine locus and the Il17a/f locus, limiting the transcription of Il17a (Kim et al., 2014). Ctcf deficiency has been reported to diminish Ifng expression in Th1 cells (Ribeiro de Almeida et al., 2009; Sekimata et al., 2009) and increase Il17a expression in Th17 cells early (Kim et al., 2014), but not late (Ribeiro de Almeida et al., 2009). Although Ctcf is important in the spatial reorganization of regulatory elements in subset-specific loci, it is unclear what drives Ctcf recruitment to these specific genomic regions.
In this report, we investigated the function of Batf in regulating genomic targets to initiate the programming of gene expression during the transition from naive to effector CD4 T cell. We find that Batf binds closed chromatin and is required for induced chromatin accessibility, enabling the recruitment of additional factors to newly accessible sites. We define a previously undescribed function of Batf in mediating the recruitment of Ctcf to a subset of Ctcf binding sites that induce higher-order chromatin restructuring and the looping of distal regulatory elements at key Batf-dependent gene loci during effector T cell development. Further, we find that Batf contributes to increased expression of Ets1, which appears to be indispensable for the Batf-dependent recruitment of Ctcf. Our study highlights the requirement of a Batf-Ets1 axis in controlling both the remodeling of nucleosomal DNA and the ensuing reorganization of genomic architecture to alter the expression of lineage-specifying genes during effector T cell development.
RESULTS
Batf Pioneers Chromatin Accessibility in CD4 T Cells
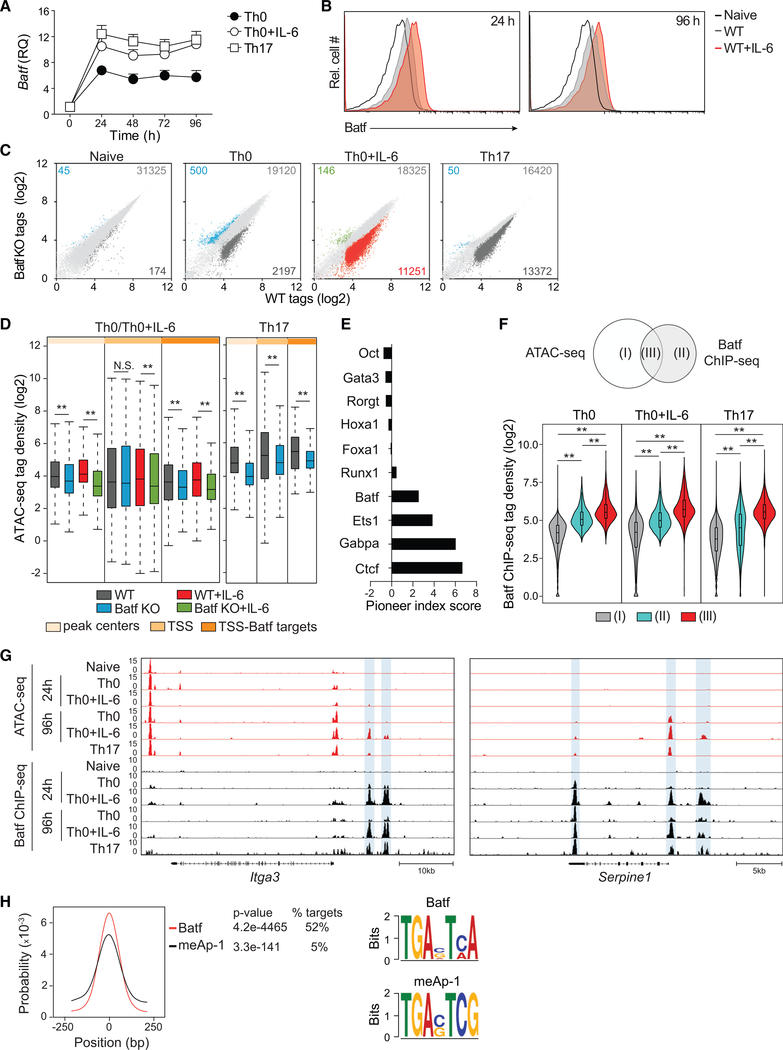
Batf, acting primarily as a heterodimeric complex with JunB (Dorsey et al., 1995; Murphy et al., 2013), has been ascribed pioneering functions in T cell development, although detailed studies of its function in initiating chromatin accessibility have not been reported. To address that, we first established a kinetic profile of Batf expression after TCR stimulation of naive CD4 T cells in vitro. Activation was performed without exogenous cytokine addition (Th0) or under conditions known to polarize effector development down Batf-dependent pathways: Tfh-like polarization (modeled by the addition of exogenous interleukin6 (IL-6); hereafter, referred to as “Th0+IL-6”) or Th17 polarization (Schraml et al., 2009). In accord with prior studies, Batf transcription was rapidly induced upon TCR stimulation (Figure 1A) (Ciofani et al., 2012), increasing approximately 5-fold within 24 h and maintained thereafter at a relatively stable level for at least 4 days. Batf expression was enhanced ~2-fold by the addition of IL-6 and, consistent with upregulated Batf transcription, Batf protein expression was also increased (Figure 1B). Based on the observed kinetics, time points between 24 and 96 h were selected to investigate Batf-driven chromatin remodeling.
Figure 1. Batf Is Required for Chromatin Accessibility in CD4 + T Cells.
(A) qRT-PCR analysis of Batf expression in naive WT CD4+CD62hi T cells activated for the indicated time points with anti-CD3 and anti-CD28 in the presence or absence of IL-6 or under Th17 culture conditions. Results are relative to naive cell expression (day 0).
(B) Naive WT CD4+CD44lowCD62Lhi T cells were activated with anti-CD3 and anti-CD28, with or without IL-6, for 24 h and 96 h. Cells were harvested and stained intracellularly for Batf.
(C and D) Differential genome-wide chromatin accessibility in WT versus Batf-deficient T cells. Nuclei isolated from naive, 24-h-activated CD4+ T cells ± IL-6, and 48-h Th17 cells from WT and Batf KO cells were subjected to ATAC-seq. Scatterplots of normalized tag density identify chromatin accessibility in WT compared with Batf KO cells (fold change > 2; false discovery rate [FDR] < 0.05) in naive, Th0, Th0+IL-6, and Th17 cells (C). Boxplots indicate normalized signal density ± 500 bp of peak centers (light orange), transcription start sites (TSSs; medium orange), and TSS of Batf target genes (dark orange) that compare WT and Batf KO cultured cells (D).
(E) Factors with pioneering characteristics are enriched in Th0+IL-6 cells. ATAC-seq data from 24-h IL-6-stimulated CD4+ T cells interrogated with the Protein Interaction Quantitation algorithm (PIQ) identified enriched motifs. Greater pioneer index scores correlate with greater chromatin opening.
(F–H) Venn diagram displaying overlapping regions of Batf ChIP-seq and ATAC-seq peaks (left). Violin plots show normalized tag density of Batf binding in naive, Th0 (24 h), Th0+IL-6 (24 h), and Th17 (48 h) cells centered ± 500 bp of unique and overlapping sites between Batf-occupied sites and accessible regions (F). ATAC-seq and Batf ChIP-seq tracks visualized with the Integrated Genome Browser (IGB) identify open chromatin (red) and Batf binding (black) in the Itga3 and Serpine1 genomic regions. Blue highlights indicate Batf binding in closed chromatin regions (G). Batf and meAP-1 motif-enrichment analysis of pooled Batf-bound closed chromatin from naive, 24-h activated CD4+ T cells ± IL-6 and 48-h Th17 cells (equivalent to cluster II in Figure 1F) (H).
Data are means ± SEM of three to five independent experiments with one mouse per experiment (A and B), or representative of two independent experiments with similar results (C–H). RQ, relative quantification; N.S., not significant. **p < 0.001 (Mann-Whitney test).
To identify genomic sites targeted for Batf-dependent chromatin accessibility, assay for transposase-accessible chromatin sequencing (ATAC-seq) was performed on activated and naïve CD4 T cells from wild-type (WT) and Batf-deficient (Batf knockout [KO]) mice (Figure 1C). The chromatin landscape of naıїve, WT CD4 T cells was not significantly different from that of Batf KO-naive cells (Figure S1A). In contrast, approximately 10% of all accessible peaks in Th0 cells and greater than 30% in Tfh-like and Th17 cells were lost in Batf KO (Figure 1C). Calculated and normalized tag densities around peak centers and transcription start sites (TSSs), as well as TSSs of Batf-target genes previously identified (Ciofani et al., 2012), defined permissive sites genome-wide (Figure 1D). Of the remaining peaks, Batf deficiency resulted in significantly diminished chromatin accessibility particularly at TSSs, including those of Batf-target genes. Moreover, Tfh cells induced in vivo, which are dependent on Batf expression for optimal development and function (Betz et al., 2010), also exhibited significantly reduced chromatin accessibility in the absence of Batf (Figures S1B and S1C). Notably, in IL-6-activated T cells (Tfh-like and Th17 cells), relatively few peaks (<200) were induced in the absence of Batf (Figure 1C), suggesting that Batf has minimal repressive activities under conditions of enhanced Batf expression driven by IL-6. Together these studies indicate that a substantial fraction of accessible sites induced in naive T cells subsequent to TCR signaling is Batf dependent, particularly in the context of IL-6 co-signaling.
Protein interaction quantitation (PIQ) is an algorithm that integrates chromatin accessibility with genome-wide transcription factor (TF) motif discovery and identifies pioneer factors by assigning index scores based on motif-specific local increases in accessibility (Sherwood et al., 2014). TFs detected by PIQ from Th0+IL-6 cell ATAC-seq data, ranked by their index scores, identified Batf as a putative pioneer factor, along with a set of known pioneer factors that included Ets1, Gabpa, and Ctcf (Figure 1E).
To assess the pioneering function of Batf-containing complexes, regions of Batf recruitment were identified by chromatin immunoprecipitation sequencing (ChIP-seq) and correlated with chromatin accessibility in Th0, Th0+IL-6, and Th17 cells activated for 24 h (Figure 1F). Three types of sites could be identified: open chromatin not bound by Batf (I), Batf-bound closed chromatin (II), and Batf-bound open chromatin (III). Notably, although the magnitude of binding was significantly greater in open regions (cluster III), ~60% of the total sites of Batf recruitment occurred at closed chromatin (cluster II) (Table S1). Indeed, more than one-half of the 962 Batf-bound peaks residing in loci that encode Batf-regulated genes (Ciofani et al., 2012) were associated with closed chromatin at 24 h, as exemplified by Itga3 and Serpine1 (Figure 1G; Table S1). Moreover, Batf-bound closed chromatin was generally associated with increases in the repressive mark H3K27me3, which was diminished at open regions bound by Batf (Figure S1D). Similarly, the permissive histone modifications H3K4me3 and H3K27ac correlated strongly with Batf-bound open chromatin, whereas Batf-bound closed chromatin was largely devoid of these marks.
Notably, many inaccessible sites to which Batf was initially bound at 24 h became accessible by 96 h (Th0, 20%; Th0+IL-6, 69%) (Figure 1G), consistent with Batf’s participation in pioneering those sites. In addition, the amplitude of many ATAC-seq peaks was significantly greater in cells that received IL-6 signaling, consistent with a contribution of increased Batf expression to the probability that a given site will be remodeled. How Batf-containing complexes bind closed chromatin is unclear, but it has been reported that members of the Ap-1 family can recognize consensus motifs containing a CpG dinucleotide (meAP-1 TGAnTCG) in heterochromatin and can activate gene expression at silenced promoters (Gustems et al., 2014). Using motif analysis, we found enrichment of the meAP-1 element at Batf sites within inaccessible regions (cluster II) (Figure 1H), identifying a potential mechanism by which Batf associates with closed chromatin. Collectively, these data indicate that Batf-containing complexes bind nucleosomal chromatin and can, therefore, act as pioneer factors, perhaps in part by recognizing methylated CpG motifs within target DNA sequences.
Irf4 Augments the Pioneering Activity of Batf Despite Its Own Lack of Pioneering Activity
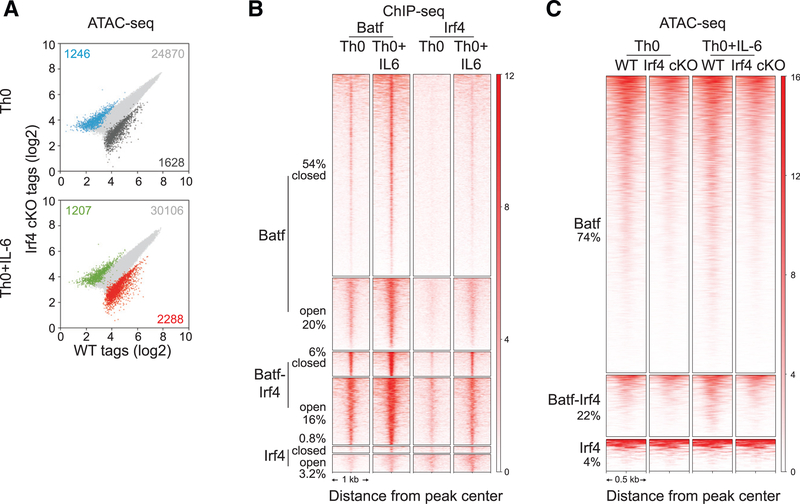
In CD4 T cells, Batf and Irf4 are induced within 4 h after TCR engagement (Yosef et al., 2013) and are cooperatively recruited to AICEs. Previous studies (Grusdat et al., 2014; Li et al., 2012; Murphy et al., 2013) have suggested that Batf-Irf complexes perform pioneer functions by creating accessibility for binding of subset-specifying TFs, such as RORγt in Th17 cells (Ciofani et al., 2012). To assess the contribution of Irf4 to the pioneering function of Batf-containing complexes, ATAC-seq was performed on WT or Irf4-deficient Th0 and Th0+IL-6 cells (Figure 2A). In contrast to Batf-deficient cells (Figure 1C), Irf4 deficiency had modest effect on genome-wide chromatin accessibility. In agreement with previous results (Grusdat et al., 2014; Li et al., 2012; Murphy et al., 2013), Irf4 ChIP-seq showed that most of the Irf4 binding sites (76%) were co-bound by Batf, whereas only 22% of Batf binding sites showed coordinate recruitment of Irf4 (Table S1; Figure 2B). Moreover, whereas most initial Batf binding was to closed chromatin—irrespective of co-binding of Irf4—Irf4 binding to closed chromatin was highly dependent on co-binding with Batf (Figure 2B; Table S1); ~90% of Irf4-bound sites in closed chromatin was associated with Batf co-binding. Although Irf4 was largely devoid of pioneering activity without Batf, ATAC-seq tag densities in Irf4-deficient cells at Batf and Batf-Irf4 sites were reduced compared with those of WT cells (Figure 2C), indicating a cooperative effect of Irf4. Thus, although Batf-containing complexes could modulate chromatin structure independent of Irf4, Irf4 functioned to augment that activity.
Figure 2. Irf4 Is Dispensable for Batf-Regulated Chromatin Accessibility in CD4+ T Cells.
(A) ATAC-seq analysis of 24-h activated WT andIrf4 conditional KO (cKO) Th0 and Th0+IL-6 cells. Scatterplots of normalized ATAC-seq tag density compare chromatin accessibility (fold change > 2; FDR < 0.05) between 24-h activated WT and Irf4 cKO Th0 and Th0+IL-6 cells.
(B) Irf4 ChIP-seq peaks from WT cells along with Batf ChIP-seq peaks (Figures 1F and 1G) integrated with chromatin accessibility (Figure 1F) identify unique Batf and Irf4 sites along with sites of Batf and Irf4 co-binding that are associated with closed and open chromatin regions. Heatmap density shows normalized Batf and Irf4 ChIP-seq signals in WT Th0 and Th0+IL-6 cells centered ± 1,000 bp of the indicated clusters. Percentages of Batf and Irf4 sites associated with closed and open chromatin are indicated.
(C) Heatmap density shows normalized ATAC-seq signals centered ± 500 bp of unique Batf and Irf4 sites along with co-bound Batf and Irf4 sites with percentages of sites compared between WT and Irf4-deficient cells in Th0 and Th0+IL-6 conditions.
Data are representative of two independent experiments with similar results
Batf-Induced Gene Expression Is Linked to Increased Chromatin Accessibility
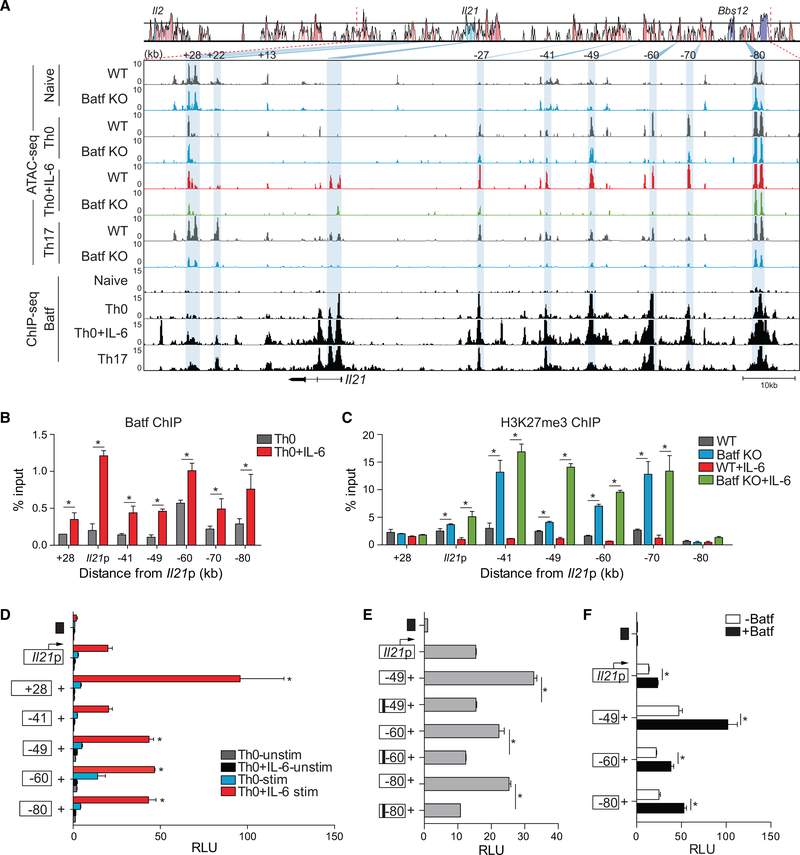
To perform a more detailed analysis of chromatin remodeling by Batf, we chose to focus on the Il21 locus because it is among the genes expressed in an IL-6- and Batf-dependent manner by multiple Th cell lineages (Ciofani et al., 2012; Kaplan et al., 2011; Schraml et al., 2009; Zhou et al., 2007) (Figure S2A). ATAC-seq data from WT and Batf KO naive and stimulated CD4 T cells (Figure 1C) were used to identify differentially accessible regions in the extended Il21 locus, which is closely linked to the Il2 locus (Figure 3A). This demonstrated that naive WT and Batf KO T cells have a similar chromatin landscape in this locus; there was limited or no chromatin accessibility, and it was flanked by potential boundary elements marked by activation-independent accessibility 80 kb upstream and 28 kb downstream of the transcriptional start site of Il21, the latter of which appears to insulate the Il21 locus from the Il2 locus (see below). At sites with limited accessibility across the locus in naive WT cells (Figure 3A), there were increased H3K27me3 marks (Figure S2B), indicating that the Il21 locus exists in a transcriptionally repressed state in naive CD4 T cells. Upon activation, these marks were markedly decreased (Figure S2B) and coincided with increased accessibility, particularly at several conserved non-coding sequences, including conserved non-coding sequences −49, −60, −70, and −80 kb (Figure 3A, blue highlights).This was in contrast to conserved non-coding sequence +27, in which accessibility was lost upon activation, except in WT Th17 cells. IL-6 co-signaling was required to induce accessibility within the Il21 promoter and first intron, also enhancing accessibility and recruitment by Batf locus-wide compared with TCR stimulation alone (Figures 3A and 3B). This correlated with further reduction in H3K27me3, albeit more modest than that induced by TCR alone (Figures 3A and S2B). Accordingly, accessibility of these elements was greatly diminished in the absence of Batf (Figure 3A) and was associated with increased H3K27me3 (Figure 3C). Similar findings for loci encoding other Batf-target genes, including Ahr, Il23r, Il17a/f, and Maf (Figure S3) further demonstrated that control of local chromatin accessibility by Batf is associated with modulation of gene transcription.
Figure 3. Altered Chromatin Accessibility at Genomic Regions Containing Batf Target Genes.
(A) Comparative chromatin accessibility and Batf recruitment to the mouse Il21 locus in WT and Batf KO naive, Th0, Th0+IL-6, and Th17 cells. ATAC-seq peaks (Figures 1C and 1D) and ChIP-seq peaks (Figures 1F and 1G) are shown aligned to a VISTA plot highlighting sequence conservation at key intergenic conserved non-coding sequence elements. Blue shading indicates diminished chromatin accessibility in Batf KO compared with WT cells.
(B and C) ChIP-qPCR analysis of Batf and H3K27me3 binding to Il21 genomic regions. Batf recruitment to the Il21 locus in WT Th0 (gray) and Th0+IL-6 (red) shown in (B) and H3K27me3 binding in WT Th0 (gray), Th0+IL-6 (red), Batf KO Th0 (blue), and Th0+IL-6 (green) in (C). Data are normalized to total input samples and IgG control.
(D) The mouse Il21 promoter was linked to a firefly luciferase reporter construct and selected Il21 conserved non-coding sequence elements were juxtaposed upstream of the promoter. Day-3 Th0 and Th0+IL-6 cells were transfected with an empty control vector (black box), the Il21 promoter construct alone, or constructs containing the Il21 promoter and the indicated conserved non-coding sequence and then rested overnight, restimulated with phorbol myristate acetate (PMA) and ionomycin for 5 h and assessed for luciferase expression.
(E and F) Day-3 WT Th0+IL-6 cells were transfected as in (D) with WT or mutated Batf consensus motif-containing conserved non-coding sequence (black bars) (E); or in the presence or absence of a Batf-expressing vector (F). Data are normalized to the empty vector control.
Data are means ± SEM of three to four independent experiments with one mouse per experiment (B–F) or representative of two independent experiments with similar results (A). RLUs, relative light units. *p < 0.05 (Student’s t test or one-way ANOVA, followed by Tukey’s test).
Conserved non-coding sequence elements tend to have regulatory functions, behaving as transcriptional enhancers, silencers, or locus-control regions (Fields et al., 2004; Hardison, 2000; Hatton et al., 2006; Lee et al., 2003; Zheng et al., 2010). To investigate the effect of Batf-dependent accessible regions on Il21 transcription, these elements were cloned upstream of the Il21 promoter and examined in a promoter/reporter assay (Figures 3D–3F). Although conserved non-coding sequences +28, −49, −60, and 80 significantly enhanced the transcriptional activity of the Il21 promoter (Figure 3D), conserved non-coding sequence-41 had minimal effect, despite robust Batf binding (Figures 3B and 3D). Mutation of Batf consensus sites within the conserved non-coding sequence elements impaired enhancer activity (Figure 3E), whereas overexpression of Batf augmented the activity (Figure 3F). Thus, recruitment of Batf to multiple elements in the Il21 locus appeared to enhance Il21 transcription in association with increased chromatin accessibility.
Batf Induces Increased Recruitment of the Chromatin Remodeling Factor Ctcf
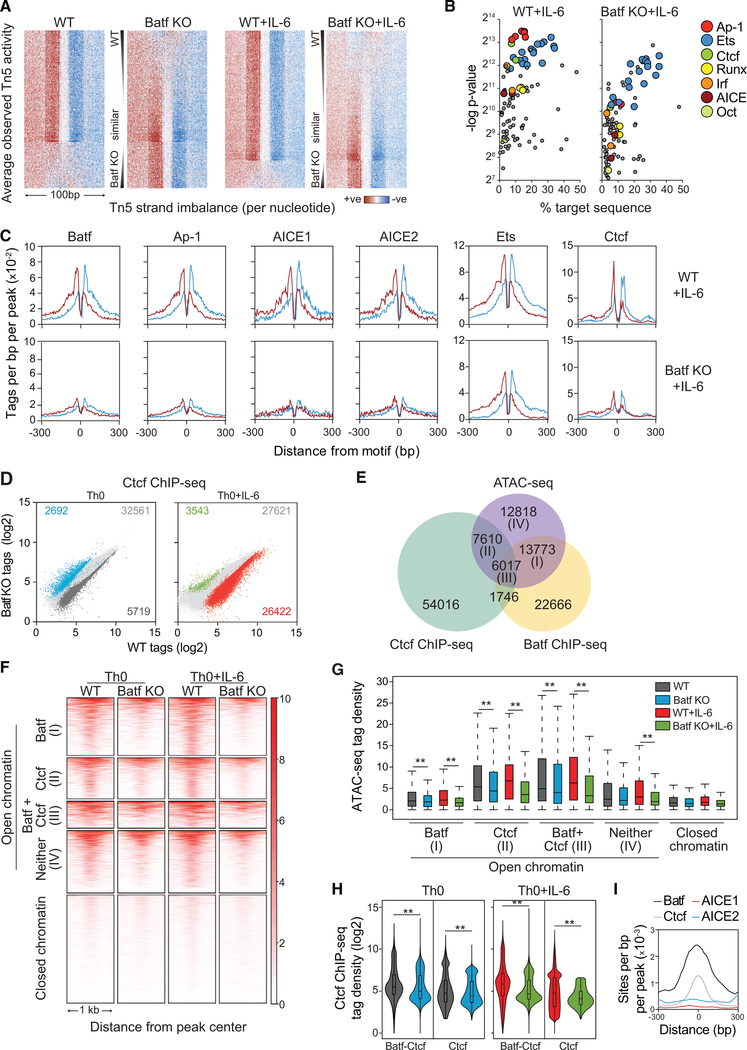
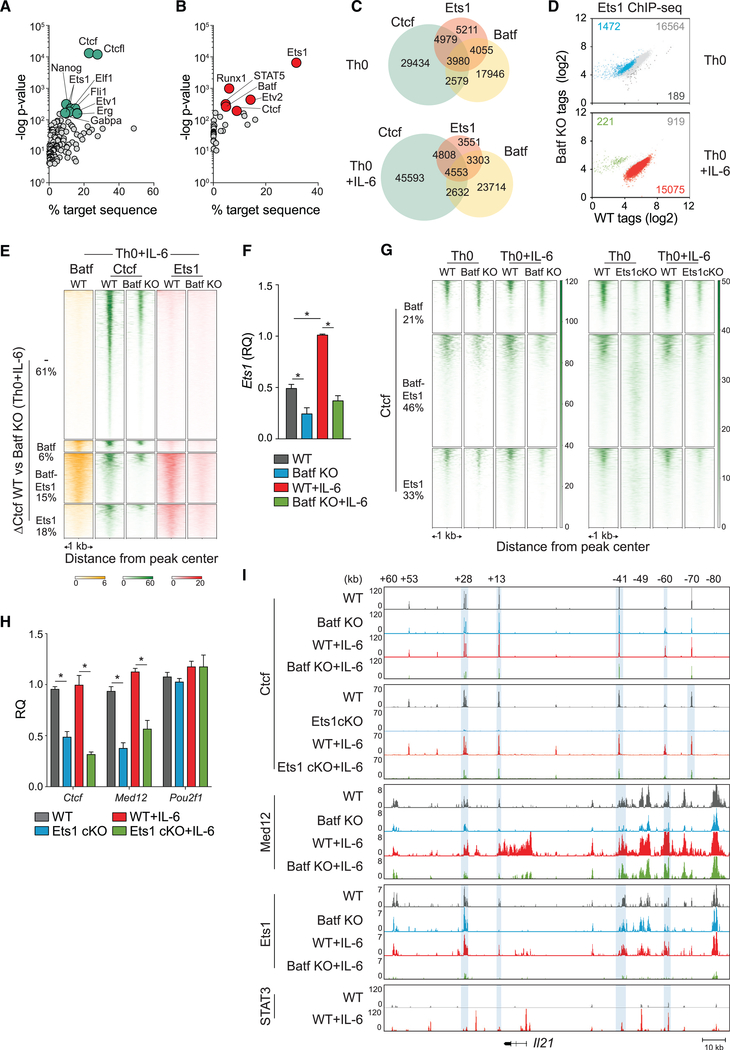
A hallmark of pioneer factors is their ability to alleviate chromatin constraints on the recruitment of transcription factors and histone-modifying complexes that regulate gene transcription (Sherwood et al., 2014; Zaret and Carroll, 2011). To identify factors whose recruitment might be linked to Batf, ATAC-seq data from WT and Batf KO T cells activated with and without IL-6 were analyzed by Wellington bootstrap footprinting, an algorithm that identifies the differential occurrence of transcription factor footprints in two accessible chromatin datasets (Piper et al., 2013, 2015) (Figures 4A–C and S4). Consistent with increased Batf-dependent chromatin accessibility, most transcription factor footprints identified in WT cells were lost in Batf KO cells (Figure 4A). Motif-enrichment analysis of identified footprints revealed that a subset of transcription factor motifs was significantly diminished in Batf KO cells (Figures 4B, 4C, and S4A–S4C). Not surprisingly, this included Ap-1 family members (e.g., Batf itself), Irf4, and associated AICE1 and AICE2 elements. Interestingly, however, Ets, Runx, Oct, and Ctcf consensus motifs were also identified. Particularly intriguing was the identification of Ctcf, a DNA binding factor that acts both as a chromatin insulator, defining boundary domains, and as a mediator of DNA looping, functioning to juxtapose distal regulatory elements with gene promoters to modulate transcription (Ong and Corces, 2014; Ren et al., 2017). Diminished Ctcf motifs in Batf-deficient cells suggested that Batf might facilitate Ctcf recruitment, thereby affecting chromatin remodeling and higher-order chromatin structure.
Figure 4. Batf-Mediated Chromatin Accessibility Is Required for Transcription Factor Recruitment in Activated CD4+ T Cells.
(A–C) ATAC-seq data from WT and Batf KO CD4+ T cells activated for 24 h ± IL-6 were subjected to Wellington footprinting analysis (Figures 1C and 1D).Heatmaps show differential footprints compared between WT and Batf KO cells; red indicates positive strand cuts (+ve) over negative strand cuts (ve) per nucleotide positions, and blue indicates negative strand cuts. Binding sites are sorted from top to bottom in order of decreased footprint occupancy score (A). Footprints from WT and Batf KO Th0+IL-6 cells (A) were subjected to motif-enrichment analysis and are represented by scatterplots (B). Histograms show specific transcription factor motif enrichment from footprinting analysis (A), with red indicating positive strand cuts and blue indicating negative strand cuts in WT and Batf KO Th0+IL-6 cells (C).
(D) Scatterplots show normalized Ctcf ChIP-seq tag density with numbers of differential peaks (fold change > 2; FDR < 0.05) from WT and Batf KO Th0 andTh0+IL-6 cells.
(E) Venn diagram displays shared binding of Batf and Ctcf to accessible chromatin regions identified by Batf and Ctcf ChIP-seq from WT Th0+IL-6 cells peaks andmerged ATAC-seq peaks from WT Th0 and Th0+IL-6 and Batf KO Th0 and Th0 +IL-6 cells using the HOMER function mergePeaks with parameter −d given.
(F and G) Heatmap density (F) and boxplots (G) show normalized ATAC-seq signals centered ± 500 bp of the indicated clusters (E).
(H and I) Violin plots of Ctcf occupancy ± 500 bp of overlapped Batf-Ctcf sites or unique Ctcf sites in 96-h WT and Batf KO Th0 and Th0+IL-6 (H). Enriched Batf, Ctcf, AICE1, and AICE2 motifs at genomic sites co-bound by Batf and Ctcf (I).
Data are representative of two independent experiments with similar results. **p < 0.001 (Mann-Whitney test)
In support of a role for Batf in recruiting Ctcf, Ctcf ChIP-seq revealed a significant reduction in genome-wide Ctcf-occupied sites in the absence of Batf (Figure 4D; Tables S2 and S3), particularly in T cells activated in the presence of IL-6. Integration of ATAC-seq peaks with Batf and Ctcf ChIP-seq peaks revealed substantial enrichment of co-binding of Ctcf and Batf in accessible chromatin (Figure 4E). Although this represented a minority of total Ctcf binding (~80% of total Ctcf was bound to sites in inaccessible chromatin), it accounted for ~44% of the total Ctcf-bound accessible sites (Figure 4E, region III). Similarly, although most Batf binding did not coincide with Ctcf recruitment, ~30% of Batf-bound sites in accessible chromatin did. This was in marked contrast to ~4% of Batf sites co-bound by Ctcf in closed chromatin. Thus, coordinate binding of Batf and Ctcf was enriched at accessible sites, suggesting that Batf acts to recruit Ctcf to sites with gene regulatory function.
Notably, a large fraction of the Batf-Ctcf co-bound sites was enriched for loci that contain genes expressed in the context of Th17 cell differentiation (Ciofani et al., 2012) (Tables S2 and S3). However, Batf also contributed significantly to Ctcf binding at sites at which Batf did not bind directly. Accordingly, although Batf deficiency resulted in reduced ATAC-seq signal densities at accessible sites co-bound by Batf and Ctcf in WT T cells (cluster III; Figures 4F and 4G), it also reduced signal intensities at sites that bound Ctcf independent of Batf (cluster II; Figures 4F and 4G). Consistent with these findings, in addition to reducing Ctcf binding at sites to which Batf was co-bound, deficiency of Batf, although not significantly altering the expression of Ctcf (Figure S4D), also significantly reduced Ctcf binding at a substantial number of sites to which it did not co-bind (Figure 4H). Thus, the increased recruitment of Ctcf induced by Batf appeared to be mediated both by direct interactions—that is, co-binding of Batf and Ctcf to the same sites in the genome—and indirect mechanisms that involved Batf-dependent regulation of the expression of other factors that promote Ctcf recruitment.
Although Irf4 was dispensable for a substantial portion of Batf-mediated chromatin accessibility (Figures 2A and 2C), we sought to determine whether Batf-dependent Ctcf recruitment required cooperative Irf4 binding. Motif analysis of Batf-bound or Ctcfbound sites showed enrichment of Batf and AICE motifs or Ctcf motifs, respectively, as expected (not shown). Interestingly, however, although Batf and Ctcf motifs were enriched at sites of Batf-Ctcf co-occupancy, AICE motifs were not (Figure 4I). Moreover, although Irf4 showed the expected co-recruitment to Batf-bound sites, there was limited recruitment to sites of Batf-Ctcf co-binding (Figure S4E). As predicted, given Irf4’s modest effect on genome-wide chromatin modulation (Figures 2A and 2C), Irf4 deficiency had no significant effect on chromatin accessibility at Ctcf sites (Figure S4F). Thus, co-binding of Batf and Ctcf occurs principally at sites of Batf recruitment at which there is no coordinate recruitment with Irf4, i.e., at non-AICE sites.
Batf Upregulates and Cooperates with Ets1 to Mediate Ctcf Recruitment
In view of our finding that Batf affected Ctcf binding at many sites independent of co-binding with Ctcf, we speculated that Batf might act indirectly by modulating the expression of factors that cooperate in regulating chromatin reorganization at these sites. We, therefore, examined candidates identified in our footprint analysis of Batf-dependent accessible chromatin—Ets, Runx, and Oct (Figures 4B, 4C, and S4A–S4C). To determine whether these factors, or others, might act cooperatively with, or downstream of, Batf to regulate Ctcf recruitment, motif analysis was performed on Ctcf-bound sites that were subject to control by Batf but that did not directly bind Batf (Figure 5A). Along with Ctcf and its related factor, Ctcf-like (Ctcfl or Boris)—which is poorly expressed in T cells—motifs corresponding to Nanog and several members of the Ets-domain family of transcription factors were found to be enriched. Although Nanog, Etv1, and Erg have little or no expression in T cells, Ets1 is expressed and to a greater degree than other Ets factors previously identified in Th17 cells (Ciofani et al., 2012). Ets1 is a putative pioneer factor (Bevington et al., 2016; Cauchy et al., 2016) (and see Figure 1E) and was, therefore, a potential co-contributor with Batf and/or other factors in promoting Ctcf binding.
Figure 5. Ets1 and Batf Are Required for Modulating Ctcf Occupancy in the Genome of Activated CD4+ T Cells.
(A) Motif enrichment analysis of Batf-dependent Ctcf peaks exclusive of Batf binding represented by scatterplot with cyan dots indicating top 10 motifs with lowest p value. Analyzed peaks were collected by integrating Ctcf ChIP-seq data from 96-h WT Th0 and Th0+IL-6 versus Batf KO Th0 and Th0+IL-6 with Batf ChIP-seq peaks (Figure 1F).
(B–E) Ets1 ChIP-seq peaks from 96-h WT and Batf KO Th0 and Th0+IL-6 subjected to motif-enrichment analysis identified Batf and Ctcf motifs represented by scatterplot with red dots indicating top motifs with the lowest p value (B). Ctcf, Batf, and Ets1 coincident and independent binding derived from Batf ChIP-seq (Figure 1F), Ctcf ChIP-seq (Figure 4D) and Ets1 ChIP-seq data from 96-h WT and Batf KO Th0 and Th0+IL-6 (as in B) (C). Scatterplots show normalized Ets1 ChIPseq tag density with numbers of differential peaks (fold change > 2; FDR < 0.05) compared between WT and Batf KO cells (D). Batf and Ets1 ChIP-seq peaks from WT and Batf KO cells integrated with enriched Ctcf peaks in WT compared with Batf KO (DCtcf WT versus Batf KO) in Th0+IL-6 cells identify Ctcf sites that are cobound with Ets1 and Batf or with Ets1 or Batf alone. Heatmap density shows normalized Batf, Ctcf, and Ets1 ChIP-seq signals compared between WT and Batf KO in Th0+IL-6 cells centered ± 1 kb of the indicated clusters. Percentages of ΔCtcf (WT versus Batf KO) sites associated with Batf and Ets1 sites are shown (E).
(F) Ets1 expression in WT and Batf KO Th0 and Th0+IL-6 cells 96-h after stimulation assessed by RT-PCR. Values were normalized to expression in WT+IL-6 cells to calculate relative quantification.
(G) Comparison of Batf- and Ets1-dependent Ctcf recruitment derived from Ctcf ChIP-seq peaks from Ets1cKO Th0 and Th0+IL-6 cells compared with WT and Batf KO ChIP-seq regions. Heatmap density shows normalized Ctcf ChIP-seq signals center ± 1 kb of clusters corresponding to Ctcf binding along with co-bound Ets1, Batf, or both. Percentages of Ctcf sites associated with Batf and Ets1 sites are shown.
(H) Naive WT and Ets1 cKO CD4+CD62hi T cells were activated with anti-CD3 and anti-CD28 ± IL-6 for 96 h; total RNA was extracted, and gene expression was assessed by RT-PCR. WT cells were used as controls to calculate relative quantification.
(I) Ctcf, Med 12, and Ets1 ChIP-seq data from WT, Batf KO, and Ets1 cKO Th0 and Th0+IL-6 and STAT3 ChIP-seq data from WT Th0 and Th0+IL-6 aligned to the extended Il21 locus. Blue shading highlights reduced binding in Batf KO and Ets cKO cells compared with WT cells. Data for STAT3 ChIP--seq are from GSE:65621.
Data are means ± SEM of three to five independent experiments with one individual mouse per experiment (F and H), or representative of two independent with similar results (A–E, G, and I). RQ, relative quantification. *p < 0.05 (one-way ANOVA followed by Tukey’s test).
To define possible sites of interaction between Ets1 with Ctcf, Batf, or both and to assess the effect of Batf deficiency on Ets1 binding genome wide, ChIP-seq for Ets1 was performed in WT and Batf-deficient Th0 and Th0+IL-6 cells (Figures 5B–5E and 5G). Motif analysis revealed that Batf and Ctcf motifs were highly enriched in Ets1-bound sites (Figure 5B). This suggested direct interactions of Ets1 with Batf and Ctcf and implied a role for Ets1 in promoting Ctcf genome-wide binding. Indeed, of the total sites of Ets1 binding in WT cells (Figure 5C), most (~53%) also bound Ctcf and a substantial fraction (~46%) co-bound Batf. Moreover, of the total sites of Batf-Ctcf co-binding genome wide, most (~62%) also bound Ets1. Thus, Ets1 showed substantial co-binding at sites of Ctcf recruitment, whether coordinately with, or independent of, Batf.
Because these findings suggested that Ets1 might be downstream of Batf in regulating Ctcf recruitment, we examined the effect of Batf expression on Ets1-binding genome wide and determined how Batf deficiency affected Batf-dependent and -independent sites of Ets1-Ctcf co-binding (Figures 5D and 5E). Interestingly, Batf deficiency had only a modest effect on global Ets1 recruitment in Th0 cells, whereas it had a profound effect in Th0+IL-6 cells (Figure 5D), even though Ets1 binding was comparable between WT Th0 and Th0+IL-6 cells (Figure S5A); 93% of genome-wide Ets1 binding—including Batf-dependent Ctcf sites co-bound by Ets1, Batf, or both (Figure 5E)—was reduced in Th0+IL-6 cells in the absence of Batf. Substantial reductions in chromatin accessibility (Figure 1C) with concomitant diminution of motif enrichment for Ets (Figures 4B and S4B) likely contributed to the difference in Ets1 recruitment observed in Batf KO Th0+IL-6 cells compared with Batf-deficient Th0 cells. In agreement with a recent report (Karwacz et al., 2017), Batf deficiency also significantly reduced Ets1 expression in both Th0 and Th0+IL-6 cells (Figure 5F). Collectively, these findings suggest that Batf affected Ets1 global genome binding at least in part via effects on Ets1 expression but also by creating permissive sites for Ets1 binding that together served to control Ctcf recruitment to Batf-dependent sites.
To examine this further, Ctcf ChIP-seq was performed on Ets1-deficient Th0 and Th0+IL-6 cells, and the effects on Ctcf binding were compared with those conferred by Batf deficiency (Figure 5G). Of the total sites bound by Ctcf plus Batf, Ctcf plus Ets1, or Ctcf plus Batf and Ets1 (Figure 5C), Ets1 deficiency significantly reduced Ctcf binding at sites at which Ets1 normally co-bound, irrespective of binding by Batf and also at sites to which it did not bind. Similarly, in agreement with our findings above (Figures 4D, 4G, and 4H), Batf deficiency significantly reduced Ctcf binding, both at sites where it co-bound with Ctcf and Ets1, Ctcf alone, or neither. Because IL-6 signaling phosphorylates STAT3 and induces its nuclear localization and DNA binding in concert with enhanced Batf expression (Durant et al., 2010), we also surveyed STAT3 binding. Analyses of STAT3-, Ctcf-, Batf-, and Ets1-bound regions in cells co-activated with IL-6 revealed that greater than 50% of Ctcf-BatfEts1- and Batf-Ets1-bound sites also bound STAT3, as it did to nearly all of the remaining sites of Ctcf and Ets1 binding. This suggests that STAT3 contributes to Ctcf recruitment in cooperation with Batf and Ets1 in IL-6-stimulated cells (Figure S5B). Thus, Batf appears to modulate the activation-dependent recruitment of Ctcf both by its binding to shared sites and by its induction of Ets1, which may act coordinately with, or independent of, Batf, and these effects appear to be critically modulated by IL-6-induced STAT3 co-binding.
The pattern of reduction in Ctcf binding consequent to Ets1 deficiency suggested that Ets1 might facilitate Ctcf recruitment via both direct interactions at sites of co-binding and other mechanisms. We, therefore, examined the effect of Ets1 deficiency on the expression of Ctcf and Med12, a central component of the mediator complex that can interact with Ctcf to modulate chromatin reorganization (Bonev and Cavalli, 2016) (Figure 5H). Ets1 deficiency significantly compromised the expression of both Ctcf and Med12, but had no significant effect on the expression of Pou2f1. The latter encodes Oct1, an established interaction partner of Ctcf in T cells (Kim et al., 2014). In accord with its marked effect on Ctcf and Med12 expression, Ets1 bound these gene loci (Figure S5C). We found that Ets1 deficiency also affected the expression of IL-6 target genes, including Batf (Figure S5D). Because optimal Ets1 expression is Batf dependent, this revealed an interdependent relationship of both factors. Collectively, our findings indicate that Ets1 modulates chromatin remodeling both by regulating the availability of Ctcf and Batf (and the mediator complex) and serves as a central co-factor with Batf to recruit Ctcf to sites across the genome.
To examine the interplay of Batf, Ets1, Ctcf, and Med12 with greater precision, we again interrogated the Il21 locus (Figure 5I). We found that Ctcf binding at conserved non-coding sequences +28, +13, −41, and −60 in the Il21 locus was impaired by Batf deficiency, particularly in IL-6-stimulated cells (Figures 5I and S5E). In accord with a critical role for Ets1 in the genome-wide recruitment of Ctcf, Ctcf binding across the Il21 locus was markedly reduced in Ets1-deficient cells stimulated with or without IL-6, indicating an indispensable role for Ets1 in the recruitment of Ctcf to this locus (Figures 5I and S5E). Consistent with the Batf-dependent Ets1 induction, recruitment to Ctcf-bound regions was also reduced in the absence of Batf, particularly when cells were stimulated with IL-6. Ets1 recruitment to Ctcf-bound sites was also significantly reduced in the absence of Batf, particularly in the presence of IL-6 signaling. Med12 binding was also substantially diminished at sites of Ctcf binding, as well as at the Il21 promoter and gene body (Figure 5I). Notably, although STAT3 binding to the Il21 locus downstream of IL-6 signaling localized to regions bound by Ctcf and Ets1, there was also prominent STAT3 binding at sites that bound Batf and Irf4 (i.e., AICE elements) but not Ctcf or Ets1 (Figure 5I), including the promoter. Analysis of additional Batf target genes, including Ahr, Il12rb2/Il23r, Il17a/f, and Maf, demonstrated a consistent theme of Batf-dependent co-occupancy of Ets1 and Med12 at Ctcf-bound sites (Figure S5F). Collectively, these data indicate that Batf, acting through and in cooperation with Ets1, is central to the recruitment of chromatin looping factors that likely underpin competency for gene expression during effector CD4 T cell development.
Batf Is Required for Ctcf-Mediated Chromatin Looping at the Il21 and Il17a/f Loci
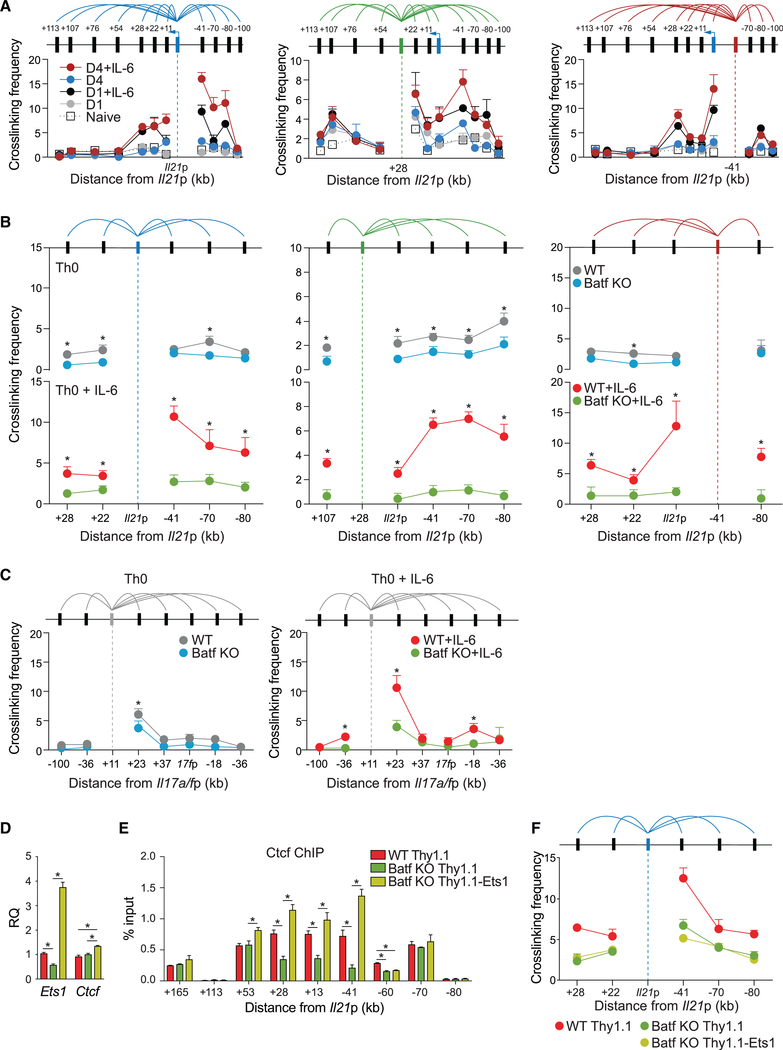
Ctcf contributes to chromatin looping between regulatory regions to modulate gene expression (Merkenschlager and Odom, 2013). In CD4 T cells, Ctcf has been reported to facilitate interactions between regulatory elements that control Ifng, Il4/ Il5/Il13, and Il17a/Il17f expression in Th1, Th2, and Th17 cells, respectively (Kim et al., 2014; Lee et al., 2003; Ribeiro de Almeida et al., 2009; Sekimata et al., 2009). Because we found that Batf induces enhanced Ctcf recruitment, whether directly or indirectly, we hypothesized that Batf is required for chromatin looping that favors transcriptional activity in Batf target genes. Batf-dependent looping at the Il21 locus was, therefore, interrogated with the chromosome conformation capture (3C) assay (Figure 6), incorporating the Il21 promoter and two of the most Batf-sensitive, Ctcf-bound conserved non-coding sequence elements (conserved non-coding sequences +28 and −41; Figure S5E) as anchor fragments.
Figure 6. Batf Is Required for Chromatin Looping at the Il21 Locus in Activated CD4+ T Cells.
(A–C) Chromosome conformation capture (3C) assay performed on 24-h and 96-h WT Th0 and Th0+IL-6 cells (A–C) and 96-h Batf KO cell (B and C) shows relative crosslinking frequencies between selected anchor fragments for the Il21 locus (promoter, +28 and −41; A and B) and Il17a/f locus (Il17a conserved non-coding sequence +11; C) and EcoRI fragments containing the indicated regions. Crosslinking frequencies were corrected for differences in ligation and PCR efficiency using reference DNA and normalized to control interaction frequencies with primer pairs within Gapdh locus.
(D–F) Naive WT and Batf KO cells were activated under Th0+IL-6 conditions. On day 2, cells were transduced with control retrovirus vector (Thy1.1) or retroviral vector expressing Ets1-Thy1.1. Thy1.1-positive cells sorted on day 4 were used to assess gene expression by RT-PCR (D), Ctcf binding by ChIP-qPCR (E), and chromatin looping between Il21 promoter and indicated regions by 3C assay (F).
Data are means ± SEM of three to five independent experiments with one individual mouse per experiment. *p < 0.05 (Student’s t test or one-way ANOVA followed by Tukey’s test).
A survey of the locus using the promoter as anchor revealed that chromatin looping in the Il21 locus was absent in naive CD4 T cells and minimal early after TCR stimulation alone (day 1; Figure 6A). Consistent with a previous report (Park et al., 2016), only with the addition of IL-6 signaling did we observe significant increases in chromatin looping, which was greatest on day 4 and was internally consistent with each of the anchor fragments (Figure 6A). Because of its accessibility in naive T cells (Figure 3A) and its recruitment of Ctcf (Figure 5I), the intergenic +28-kb element would appear to insulate the Il21 locus from the adjacent Il2 locus. As predicted, Batf-deficient T cells showed significantly impaired looping—particularly in the presence of IL-6—establishing the importance of Batf in modulating chromatin structure to support chromatin looping (Figure 6B). Similar Batf-dependent chromatin looping was observed at the activation-dependent conserved non-coding sequence +11-accessible region of the Il17a/f locus (Figure 6C). In view of the significant loss of Ctcf at looping nodes in Batf- and Ets1-deficient T cells and the similar loss of mediator and Ets1 binding to the same sites in Batf-deficient cells (Figure 5I), a critical role for Ets1 in Batf-induced chromatin looping was indicated. Indeed, although ectopic expression of Ets1 in Batf-deficient Th0+IL6 cells only modestly enhanced Ctcf expression (Figure 6D), it restored Ctcf binding at regions +28, +13, and −41 of the Il21 locus, demonstrating that Batf-induced Ets1 functions to recruit Ctcf to those sites. This was in contrast to conserved non-coding sequence 60, where Ctcf recruitment was dependent on cooperative binding of Batf and Ets1, even in the presence of abundant expression of Ctcf (Figure 6E). However, despite recovery of Ctcf binding at +28, +13, and −41 of the Il21 locus in Ets1-transduced Batf KO Th0+IL-6 cells, chromatin looping between Batf-dependent conserved non-coding sequences and the Il21 promoter was not restored to WT levels (Figures 6F), underscoring an essential function for Batf in directing chromatin organization at select loci.
Batf Is Required for Ctcf-Mediated Chromatin Looping Genome-wide in CD4 T Cells
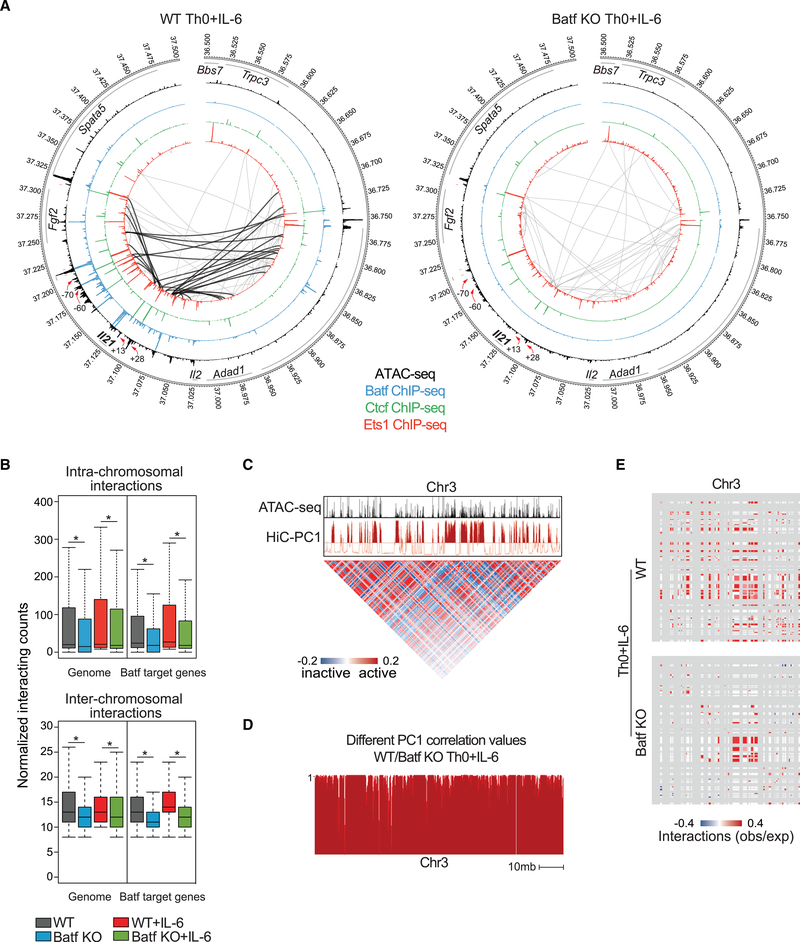
Our locus-level studies indicated that Batf had a major role in Ctcf-dependent chromatin looping as a mechanism to control Batf-dependent gene expression. To extend those findings genome-wide, we used Hi-C to assess more-extended chromatin interactions. Naive and activated WT and Batf-deficient CD4 T cells were processed according to the Hi-C protocol, and data were analyzed with the HOMER Hi-C analysis package to identify significant interactions and create contact maps (Lin et al., 2012). Looping interactions identified in the Il21-Il2 genome by Hi-C (Figure 7A) mirrored those found with 3C (Figure 6), validating the genome-wide approach. Consistent with substantial repressive histone marks at the Il21 locus in naive CD4 T cells (Figure S2B), minimal looping was observed in either WT or Batf-deficient cells (Figures S6A and 7A). In contrast, looping interactions were greatly diminished in activated Batf-deficient cells, particularly at conserved non-coding sequence +28 and conserved non-coding sequence +13 in which there was coordinate binding of Batf, Ets1, Ctcf, and Med12 in WT cells (Figures 5I and 7A). More than five Batf-dependent interactions were observed at conserved non-coding sequences +28 and +13, suggesting that these two conserved non-coding sequences act as key interaction nodes for Ctcf-mediated regulation of Il21 gene expression.
Figure 7. Batf Is Required for Global Chromatin Interaction in Activated CD4+ T Cells.
(A) Circos plots of open chromatin, transcription factor occupancy, and intra-chromosomal interactions in the extended Il21 locus in WT (left) and Batf KO (right) Th0+IL-6 cells; 24-h WT and Batf KO Th0+IL-6 ATAC-seq data are displayed in the largest inner rings (black), and Batf (blue), Ctcf (green), and Ets1 (red) occupancy from 96-h WT and Batf KO Th0+IL-6 ChIP-seq data are shown in sequentially smaller inner rings. Significant (p < 0.005, resolution 10 kb) intra-chromosomal interactions identified by Hi-C analysis of 96-h WT and Batf KO Th0+IL-6 are shown by gray connecting lines. Black lines (left) indicate interactions that are lost in Batf KO cells. Red arrows indicate reduced Batf, Ets1, and Ctcf binding in Batf KO cells compared with WT cells.
(B) Boxplots show the normalized intra- (top, 10 kb resolution) or inter- (bottom, 100 kb resolution) acting frequencies of chromosomal connections across the entire genome or regions containing Batf target genes in 96-h WT and Batf KO Th0 and Th0+IL-6 cells.
(C) Principal-component 1 (PC1) from Hi-C data identified permissive chromatin across chromosome 3 (chr3; red track) aligned with ATAC-seq data (A). Matrix illustrates Pearson’s correlation coefficient of intra-chromosomal interactions in chromosome 3 normalized to observed interaction frequency to expected interaction frequency at 100 kb resolution. Blue, lower than expected (inactive); red, higher than expected (active).
(D) Principal-component analysis of Hi-C data shows inter experimental correlation (different correlation values) calculated by comparing the two interaction profiles at each locus with PC1 values, defined at 50-kb intervals of chr3 for WT and Batf KO Th0+IL-6 cells. The different correlation value is high (close to 1) if the locus is likely to interact with similar regions in both samples and is low if the locus interacts with different regions.
(E) Enrichment of interactions at accessible chromatin (50 kb resolution) in WT (top) and Batf KO (bottom) Th0+IL-6 cells using Structured Interaction Matrix Analysis (SIMA). Colors (red/blue) indicate association calculated as the log ratio of observed frequency to expected frequency (obs/exp).
Data are representative of two independent experiments with similar results. *p < 0.05 (Mann-Whitney test).
Similarly, intra-chromosomal interactions within 1 mb of genomic regions containing the Batf target genes Ahr, Il23r, Il17a/f, and Maf revealed that the extent of chromatin looping was significantly reduced in Batf-deficient cells (Figure S6B)— consistent with impaired Ctcf recruitment to those loci (Figures 5I and S5F)—whereas intra-chromosomal interactions of Batfindependent genes in WT and Batf-deficient Th0+IL-6 cells were comparable (Figure S7). Genome-wide calculated and normalized interaction reads demonstrated significant impairment of intra- and inter-chromosomal interactions in Batf-deficient cells, whether assessed globally or at genomic regions containing Batf target genes (Figure 7B). Principal-component analysis of the normalized interaction matrix for chromosome 3, which contains the Il21-Il2 gene locus, segregated regions into active (positive principal-component 1 [PC1] values) and inactive (negative PC1 values) chromatin, and consistent with the correlation between chromatin accessibility and Batf-mediated Ctcf binding (Figures 4F and 4G), chromatin looping was tightly associated with open chromatin (Figure 7C). Moreover, the compartment structure of chromosome 3, defined by PC1 value and presented as different correlation values, becomes less permissive in Batf-deficient cells compared with WT cells under Th0+IL-6 conditions (Figure 7D). Using Structured Interaction Matrix Analysis (SIMA) to further correlate regions of open chromatin with interacting domains, we found that associations of accessible regions in chromosome 3 with intra- and interdo-main interactions were substantially reduced in Batf-deficient T cells compared with WT cells (Figure 7E), demonstrating a requirement for Batf in broadly directing chromatin interactions. Taken together, our results establish that Batf promotes local and long-range chromosomal interactions through direct and indirect control of the expression and recruitment of Ctcf to key regulatory nodes.
DISCUSSION
Batf is central to the development and function of several effector T cell subsets. In this study, we have identified mechanisms by which Batf acts to restructure the chromatin landscape to enable new gene transcription during the programming of effector T cell differentiation. Although Batf has previously been referred to as a pioneer factor, through a more detailed analysis of its function in chromatin remodeling, we have established that Batf binds to many sites of nucleosomal chromatin genome-wide before nucleosomal clearing to facilitate additional transcription factor recruitment at these sites. Notably, we found that a key trans-factor, Ctcf, is recruited in a Batf-dependent manner to both Batf-bound and -unbound sites, functioning to promote chromatin looping that reorganizes Batf target genes during effector T cell development. We further find that Batf promotes Ets1 expression, with which it cooperates to induce Ctcf expression and binding to control the formation of higher-order chromatin architecture. In view of the TCR-induced rapid induction of Batf in naive CD4 T cells and the amplification of its expression by IL-6 signaling, our findings provide insight into Batf’s non-redundant role in IL-6-dependent effector T lineage specification. Finally, we establish the importance of non-AICE Batf target sequences as sites for Ctcf recruitment and chromatin remodeling.
Our findings and those of others (Bevington et al., 2016) indicate that TCR signaling in naive T cells results in a remarkable increase in genome-wide chromatin accessibility. This early checkpoint is common to the differentiation of all effector T cells and implies that similar mechanisms may prepare the enhancer landscape for cytokine-induced lineage-specific transcription factor recruitment. The requirement for Batf in Th17 and Tfh cell development (Ise et al., 2011; Schraml et al., 2009) and the Batf-dependent induction of chromatin accessibility in Th17 cells has been attributed to the pioneering function of Batf (Ciofani et al., 2012), which is reinforced herein; the rapid induction of Batf by TCR signaling and its ability to bind closed chromatin that is opened contingent upon its expression strongly implicate Batf as a pioneer factor. A similar function of Batf has been described for “Tr1” cells (Karwacz et al., 2017), and given the requirement for Batf in the optimal development Th2 and Th9 cells (Bao et al., 2016; Betz et al., 2010; Jabeen et al., 2013), it likely participates in the induction of chromatin accessibility in these subsets as well. An outlier would appear to be Th1 cells, in which the production of interferon-γ (IFN-γ) is unperturbed by deficiency of Batf despite its normal expression by those cells (Ise et al., 2011; Schraml et al., 2009). However, because Th1 cells also express substantial amounts of Batf3, a functionally equivalent homolog of Batf (Murphy et al., 2013), it remains to be determined whether there are deficits in genome-wide chromatin accessibility and Th1 development in the absence of both Batf and Batf3, or whether Ap-1 factors other than Batf may contribute to chromatin remodeling in Th1 cells. In any case, Batf appears to have a broad role in the induction of chromatin accessibility that is a prerequisite for altered gene expression during effector T cell development.
Defining features of pioneer factors are their ability to bind nucleosomal DNA and initiate nucleosome clearing to enable binding of other transcription factors to assemble enhanceosomes. We find that Batf binds thousands of sites in closed chromatin and is indispensable for nucleosome clearance at many. It is unclear how Batf achieves that. Remarkably, few studies have examined Ap-1 family member binding in the context of chromatin accessibility. Although molecular modeling suggests that Fos-Jun binding to nucleosomal DNA may be sterically hindered(He etal., 2013),findings herein establish that Batf binds nucleosomal DNA; nucleosome remodeling at many sites bound by Batf does not occur in Batf-deficient T cells, and Batf binding precedes nucleosomal clearingatmanysites.IthasbeenreportedthatAp-1heterodimers can directlybindBAF60a (Ito etal.,2001),a component of the SWI/ SNF (BAF) nucleosome-remodeling complex that mediates nucleosome clearing and repositioning. Because Batf lacks a transactivation domain and preferentially binds JunB or JunD in T cells (Murphy et al., 2013) and because JunB, but not JunD, binds BAF60a, only by partnering with JunB might Batf directly effect nucleosomal clearing. In this regard, it is notable that JunB—like Batf—is required for optimal Th17 development (Carr et al., 2017;Hasanetal.,2017;Yamazakietal.,2017),although its contribution to the pioneering function of Batf is undefined. Further studies to dissect the possible roles of Batf and Jun proteins in creating accessible chromatin should be informative.
Through the interrogation of Batf-dependent accessible sites genome wide by motif analysis, two interrelated discoveries were made: Batf regulated the DNA binding and looping function of the chromatin architecture factor Ctcf, and Batf regulated the expression and recruitment of Ets1, which, in turn, regulated the expression and recruitment of Ctcf and the Med12 component of the mediator complex that is required for gene transcription. The Batf-and Ets1-induced expression and recruitment of Ctcf were functionally important because both intra-and inter-chromosomal interactions were profoundly impaired by Batf deficiency, particularly in T cells activated by concurrent TCR and IL-6 signaling. Although Ap-1-dependent chromatin looping has been previously described at locus- and gene-specific levels, Ctcf was not examined (Chavanas et al., 2008; Qiao et al., 2015). Our findings link early Batf expression during effector T cell differentiation to direct and indirect mechanisms that control Ctcf-mediated chromatin remodeling.
Analogous to the cooperation of Batf-Jun complexes with Irf4 at AICEs in T cells (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012), members of the Ets family of transcription factors bind cooperatively with Irf4 or Irf8 at Ets-Irf composite elements (EICEs) to control fate decisions in B cells, macrophages, and dendritic cells (Glasmacher et al., 2012). Ets factors have been thought to be less important in T cells because of their more restricted expression, although Th17 cells have been shown to express Ets1 (Moisan et al., 2007). Indeed, Ets1 has been reported to repress Th17 differentiation through an indirect mechanism involving the modulation of Il2 expression (Moisan et al., 2007). Nevertheless, we found that expression of Ets1 was significantly enhanced by Batf and had a direct role in Ctcf and Med12 expression by T cells. Moreover, coordinate binding of Ets1 to sites of Batf-dependent Ctcf binding was substantially greater than Batf itself. Accordingly, Batf-dependent recruitment of Ctcf was markedly impaired by Ets1 deficiency. This strongly implicates Ets1 as an important factor mediating Batf-dependent chromatin remodeling. Because Batf is not expressed by naive T cells, this places Batf downstream of TCR signals and upstream of Ets1 in a transcription factor cascade that controls chromatin looping at a number of important gene loci that are developmentally regulated during effector T cell differentiation.
Despite the importance of cooperativity between Batf-Jun heterodimers with Irf4 at AICEs, we found that Irf4, although clearly important for the induction of chromatin accessibility at many sites, was dispensable at most sites at which Batf-dependent recruitment of Ctcf was found. Although it is unknown whether Irf factors contribute to the pioneering functions of Batf-Jun heterodimers or Ets family members, Ets1 binds nucleosomal DNA during early T cell development in the thymus (Cauchy et al., 2016) and, as a member of the Ets family of transcription factors, has pioneering activity (Sherwood et al., 2014). Moreover, Ets1 and Ap-1 proteins can physically interact (Bassuk and Leiden, 1995), raising the possibility that Ets1 binding could cooperate with Batf at sites at which both bind to create accessible sites for Ctcf recruitment. Although Batf-dependent Ctcf recruitment to AICEs and EICEs was evident, this represented a minority of sites to which Ctcf recruitment was controlled by Batf. This suggests that both Batf and Ets1 likely act with factors other than Irf4 to recruit Ctcf and will require further study. Moreover, we found that most Batf-dependent Ctcf binding was not associated with co-binding of either Batf or Ets1, indicating that binding of Ctcf to those sites may have higher affinity for Ctcf and result from increased Ctcf expression alone, may be dependent on other factors whose expression and/or function is downstream of Batf, or both. Studies to characterize these sites in more detail will be needed.
The requirement for Batf and Ets1 in the induced binding of Ctcf genome-wide is remarkable given that Ctcf is itself a pioneer factor (Ong and Corces, 2014; Sherwood et al., 2014). In fact, most sites bound by Ctcf were already occupied by Ctcf in naive T cells before TCR signaling-induced activation—and were thus Batf independent—and resided in closed chromatin even after effector T cell differentiation. This suggests a possible hierarchy of Ctcf functions, where the pre-existing sites of occupancy may well be enriched for insulators and long-range topological interactions that are deposited earlier in T cell development, whereas sites of Batf- or Ets1-dependent Ctcf binding induced during effector T cell development may be enriched for enhancers that participate in shorter-range chromatin looping that regulates de novo gene transcription. The observed decrement in looping to TSSs in Batf-deficient cells is consistent with that possibility. This may reflect differential binding affinities of Ctcf consensus sequences to which Ctcf can bind in closed chromatin versus those to which it binds only after nucleosomal clearing.
In view of Batf’s enhanced expression by STAT3 downstream of IL-6 signaling in developing Th17 and Tfh cells, and its critical role for both Rorgt and Bcl6 expression in those lineages (Ise et al., 2011; Schraml et al., 2009), it will be important to understand how STAT3 is integrated with other transcription factors that control early Batf expression—and with Batf itself at downstream target genes. Unlike Batf, STAT3 does not possess pioneering functions (Vahedi et al., 2012, 2013), yet it participates in the assembly of lineage-specific enhanceosomes by recruiting p300 (Durant et al., 2010; Vahedi et al., 2012). STAT3 recruitment to IL-6-responsive genes has been found to be BRG1 dependent (Ni and Bremner, 2007), suggesting a mechanism whereby Batf and/or Ets1 recruitment of the SWI/SNF complex could promote STAT3 binding to sites pioneered by Batf or Ets1. Additionally, and not mutually exclusively, STAT3 has been reported to bind the C-terminal region of Jun to mediate IL-6-induced transcription (Zhang et al., 1999). In preliminary studies, we found that sites of chromatin accessibility identified in the extended Batf locus were already present in naive CD4 T cells and are enriched for Ap-1, nuclear factor kB (NF-kB) and STAT3 motifs that would appear poised for rapid response downstream of TCR and IL-6 signaling without requirement for pioneering activity (D.P., C.T.W., R.D.H., unpublished data). Future investigations examining the interplay of Ap-1 complexes in addition to Batf-Jun, Ets1, the BAF complex, and STATs will allow a better understanding of how expression of Batf is controlled and how Batf controls the chromatin landscape in developing effector T cells via Ctcf-mediated chromatin reorganization.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Casey Weaver (cweaver@uabmc.edu). Mice and plasmids generated in this study are available upon request via a material transfer agreements (MTA).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J, Batf−/− (Batf KO) (Schraml et al., 2009), Irf4fl/fl (Irf4 cKO) (Klein et al., 2006), and CD4-Cre mice were purchased from The Jackson Laboratory (Bar Harbor, ME USA). Ets1fl/fl CD4-Cre+ (Ets1 cKO) were previously described (Zook et al., 2016). 6–8 week old male and female mice (C57BL/6J background) were used in all experiments. Littermates were used as wild-type (WT) control mice. Mice were maintained under specific pathogen-free conditions. All experiments were performed with the approval of the University of Alabama Institutional Animal Care and Use Committee.
Cell culture conditions
Naive CD4+CD62Lhi T cells were isolated from spleen and lymph nodes using a MACS isolation system (Miltenyi Biotec) or sorted by flow cytometry for CD4+CD44lowCD62Lhigh naive cell population. Naive CD4+ T cells were activated with plate-bound anti-CD3 (2 ug/ml, 145–2C11) and soluble anti-CD28 (37.51, 0.5 ug/ml) for Th0 with additional cytokines and antibodies to generate Th17 cells (20 ng/ml IL-6; 5 ng/ml human TGF-β; 10 ng/ml IL-1 β, 10 ug/ml anti-IL-4, 11B11; and 10 ug/ml anti-IFN-γ, XMG). 20 ng/ml IL-6 was added to Th0 in some culture conditions (Th0+IL-6). Cells were expanded after 3 days without additional cytokines (Th0, Th0+IL-6) or half concentration (Th17) of the original cytokines in fresh medium. Recombinant cytokines and antibodies are from BD Biosciences or R&D.
METHOD DETAILS
Listeria monocytogenes (L.m.) infection
6–8 weeks WT and Batf KO mice were infected with 5×107 cfu/ml L.m by intravenous injection (i.v.). Mice were sacrificed after 48 h of infection, and splenocytes were isolated and processed for further analysis such as cell staining and ATAC-seq analysis.
Cell sorting and FACS
Cells were surfaced-stained with fluorochrome conjugated antibodies for 30 min at 4°C, washed with 2% BSA/PBS and fixed with 2% formaldehyde for 10 min at room and sorted using a BD FACS Aria. Sorted cells were used for total RNA isolation to assess gene expression by RT-PCR.
Gene expression by RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen Life Technologies) and reversed transcribed to make cDNA using iScript cDNA Synthesis Kit according to the manufacturer’s instructions (Bio-Rad). Gene expression was assessed by RT-PCR and normalized to housekeeping gene expression (β2-microglobulin, β2 m). The relative gene expression was calculated by the change-in-threshold (-ΔΔCT) method.
Constructs and luciferase assay
The Il21 promoter and conserved noncoding sequences were PCR amplified using listed primers and cloned into pGL3 luciferase plasmid. Plasmids containing Batf mutated sites were constructed using QuikChange XL Site-Directed Mutagenesis Kit (Agilent). The Batf-expression vector was previously described (Jabeen et al., 2013). For T cell transfection experiments, naive CD4+ T cells were activated with 2 ug/ml anti-CD3 and 0.5 ug/ml anti-CD28 with or without 20 ng/ml IL-6. After 72 h, cells were transfected with the luciferase plasmids and control plasmid using Nucleofector Kits for Mouse T Cells (Lonza). Cells were rested overnight, restimulated with PMA and ionomycin for 5 h, and followed by assessment of luciferase expression with the dual luciferase system (Promega).
Retroviral transduction experiments
Ets1 (Origene, cat no. MR207015) cDNA was digested and sub-cloned into the MSCV-Thy1.1 bicistronic retroviral plasmid and viral stocks were prepared as described (Jabeen et al., 2013). CD4+ T cells were transduced on day 2 of activation with titered supernatants containing control or Ets1-expressing retrovirus by centrifugation at 2000 rpm at 25°C for 1h in the presence of 8 ug/ml polybrene. Virus-containing supernatant was removed, and cells were rested overnight, expanded on day 3 with addition of fresh media, and analyzed on day 4.
Chromosome conformation capture assay (3C)
3C assay was performed as described (Hagège et al., 2007; Naumova et al., 2012) with some modifications. 107 cells were cross-linked with 2% formaldehyde for 10 min at room temperature, quenched with 0.125 M Glycine for 5 min, and lysed with ice-cold lysis buffer. Nuclei were resuspended in restriction enzyme buffer (CutSmart) containing 0.095% SDS and incubated for 10 min at 65°C. 1% Triton X-100 was added, mixed, and cross-linked DNA was digested overnight with 800 U EcoRI (NEB) containing 1 mM ATP at 37°C. Enzyme was inactivated by addition of 1.6% SDS for 20 min at 65C follow by the addition of 1% Triton X-100. DNA fragments were ligated with 4000 U T4 ligase (NEB) overnight at 16C. Crosslinked DNA were reversed by incubation with 400 ug proteinase K overnight at 65C. Additional proteinase K was added and incubated for 2 h at 65C follow by incubation with 300 ug RNase for 30 min at 37°C. Ligated DNA were purified by phenol-chloroform extraction and ethanol precipitation. Ligation products were quantified by RT-PCR. BAC clones were used to generate reference DNA to correct for differences in ligation and PCR efficiency. Equimolar amounts of two BAC clones spanning the mouse Il21 locus (RP23–290D8 and RP23–128L14) and BAC spanning the mouse Gapdh locus (RP23–410F11) from CHORI were mixed, digested with EcoRI, phenol chloroform extracted and ethanol precipitated, and ligated at a DNA concentration of 300 ng/ul. Relative crosslinking frequencies between the analyzed pairs were calculated as described (Hagège et al., 2007). Crosslinking frequencies were corrected for differences in ligation and PCR efficiency using reference DNA generated from BAC clones and were normalized to control interaction frequencies using primer pairs within Gapdh locus.
Hi-C
HiC assay was performed similarly as 3C assay with some modifications. Naive WT and Batf-deficient CD4 T cells isolated by FACS sorting for CD4+CD44lowCD62Lhigh cell population were analyzed directly or after stimulation with anti-CD3, anti-CD28 and IL-6 for 96 h. 5×106 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, quenched with 0.125 M Glycine for 5 min, and lysed with ice-cold lysis buffer. Nuclei were resuspended in restriction enzyme buffer (CutSmart, NEB) containing 0.095% SDS and incubated for 10 min at 65°C. 1% Triton X-100 was added, mixed, and cross-linked DNA was digested overnight with 800 U HindIII (NEB) containing 1mM ATP at 37C. DNA ends were filled in and marked with biotin by incubating digested DNA with 1.5 ul of each1.5 ul 10 mM dATP, 10 mM dGTP, 10 mM dTTP; 37.5 ul 0.4 mM biotin-14-dCTP, and 50 U large (Klenow) fragment (NEB) for 45 min at 37°C. Enzyme was inactivated by addition of 1.6% SDS for 25 min at 65C while shaking at 225 rpm. DNA fragments were ligated with 4000 U T4 ligase (NEB) overnight at 16°C. Crosslinked DNA were reversed by incubation with 400 ug proteinase K overnight at 65C. Additional proteinase K was added and incubated for 2 h at 65°C follow by incubation with 300 ug RNase for 30 min at 37°C. Ligated DNA were purified by phenol-chloroform extraction and ethanol precipitation. DNA concentration was measured using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Marked and ligated DNA was checked for efficiency by amplifying a Gapdh region and purifying PCR product using MinElute kit (QIAGEN). Purified DNA was digested with NheI (NEB) enzyme for 2 h at 37°C and run on 2% agarose gel for quality control. Successful marked and ligated DNA generated 3 bands at 300 bp, 130 bp, and 170 bp after restriction enzyme digestion.
Biotin from unligated ends was removed by incubating ligated DNA with 10 mM dATP, 10 mM dGTP and T4 DNA polymerase in NEB2.1 buffer at 12°C for 2 h. Reaction was stopped with 2 ul 0.5 M EDTA pH8. DNA was purified by phenol-chloroform extraction and ethanol precipitation. Ligated DNA was sheared to a size 100–500 bp using Bioruptor (Diagenode). Marked, ligated, and sonicated DNA was used for end-repair and dA-tailing reactions using NEBNext DNA Library Prep Master Mix Set for Illumina (NEB). Biotinylated DNA was captured using magnetic streptavidin beads (Invitrogen), used for adaptor ligation reaction, and PCR amplification using NEBNext DNA Library Prep Master Mix Set for Illumina and NEBNext Multiplex Oligos for Illumina (Index Primers Set, NEB). Hi-C libraries were sequenced with paired-end 50 bp using Illumina HiSeq 2500.
Hi-C analysis
Raw paired-end sequencing data were mapped to mm9 genome and processed using HiC-Pro pipeline (Servant et al., 2015). Significant intra- (resolution 10kb, p < 0.005) and inter-chromosomal (resolution 100kb, p < 0.005) interactions, principle component analysis, and feature enrichment analysis were performed using findHiCInteractionsByChr.pl, analyzeHiC, runHiCpca, and annota-teInteractions perl scripts, respectively, that are part of the Hypergeometric Optimization of Motif Enrichment (HOMER) suite of tools for Motif Discovery and next-generation sequencing analysis (Heinz et al., 2010). Structured Interaction Matrix Analysis (SIMA), a feature of HOMER, was used to evaluate the compartmental interaction associated with chromatin accessibility. Intra-chromosomal interactions and other sequencing data were visualized using Circos (Krzywinski et al., 2009). The normalized intra- and inter-chromosomal interacting counts were used to generate boxplots (indicate interquartile range with whiskers ± 1.5 times) using R. Batf target genes were determined from WT and Batf KO Th17 RNA-seq dataset (accession GSE: 40918) (Ciofani et al., 2012) and used for downstream analysis.
Chromatin Immunoprecipitation (ChIP) and ChIP-seq
Naive T cells were isolated from spleen and lymph nodes of WT or Batf KO mice either by magnetic sorting (Miltenyi Biotec 130–106643) or by flow cytometry (gated on CD4+CD44lowCD62Lhigh population) prior to activation for ChIP-seq and ChIP assay, respectively. Cells (10–20×106) were cross-linked for 10 min with 1% formaldehyde, quenched with 0.125 M Glycine for 5 min and washed with PBS. Cell pellets were resuspended in lysis buffer (5 mM Pipes, 85 mM KCl, 0.5% NP-40 with protease inhibitor), homogenized by running cells through an 18-gauge needle for 10 times, and pelleted by centrifuging at 2,000 rpm at 4C for 5 min. Nuclei were resuspended in RIPA buffer (1X PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor) and sonicated to a size range of 200–500 bp using Bioruptor. Cells were pelleted, and RIPA buffer was added to supernatant to 1 ml. Antibodies (rabbit anti-Batf [Schraml et al., 2009], rabbit anti-Ctcf. (Active Motif, 61311), rabbit anti-Med12 (Bethyl, A300–774A), rabbit anti-Ets1 (Santa Cruz, sc-350 X), rabbit anti-Irf4 (Santa Cruz, sc-28696 X), rabbit anti-H3K4me3 (Millipore, 07–473), rabbit anti-H3K27ac (Abcam, ab4729), rabbit anti-H3K27me3 (Millipore, 07–449) and normal rabbit IgG (Millipore, 12–370)) were coupled to Dynabeads M-280 Sheep anti-rabbit IgG (Thermo Fisher Scientific) at 4C with rotation overnight then washed with PBS/BSA solution. Antibody-bound beads was added to sonicated chromatin and incubated at 4C with rotation overnight. The immunocomplexes were washed with 5 times LiCl buffer (100 mM Trish pH 7.5, 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate), once with TE buffer, resuspended in 200 ul elution buffer (1% SDS and 0.1 M NaHCO3), and incubated at 65C for 1 h. Supernatants were collected, incubated at 65C overnight, and immunoprecipitated DNA were purified using MinElute columns (QIAGEN). Reversed cross-links DNA were analyzed by RT-PCR. ChIP data were analyzed using percent input method 100 × 2 (Adjusted input - Ct (IP)) and adjusted to IgG control background.
For ChIP-seq assay, immunoprecipitated DNA was used to prepare library using NEBNext Ultra II DNA library prep kit and NEB-Next Multiplex Oligos for Illumina (NEB). ChIP-seq libraries were sequenced with single-end 50 bp using Illumina HiSeq 2500.
ChIP-seq analysis
Raw sequencing data were mapped to mm9 genome using Bowtie1 (Langmead et al., 2009) with options -m 1 k 1. ChIP-seq peaks were called using MACS2 (Zhang et al., 2008) with p value cut off at 1×105. The total input DNA was used as the control for peak-calling. Differential peak calling was performed using DESeq2 with fold change greater than 2 and fdr less than 0.05. Tag counts were normalized to 10 million tags and calculated using annotatePeaks function of HOMER (Heinz et al., 2010). Unique and overlapping peaks were determined using mergePeaks function of HOMER. The log2 transformed normalized read data were used to generate boxplots (indicate interquartile range with whiskers ± 1.5 times) and violin plots (boxplot with its probability density) using R. Heat-maps were visualized using deepTools (Ramírez et al., 2016). Motif analysis was performed using findMotifsGenome function of HOMER. ChIP-seq peaks were annotated to the nearest gene in mm9 genome using annotatePeaks function of HOMER. Gene ontology analysis of associated peaks was performed using annotatePeaks function of HOMER with option -go. ChIP-seq tracks were normalized to 10 million tags and viewed using the Integrated Genome Brower (IGB) (Nicol et al., 2009). Batf target genes from WT and Batf KO Th17 RNA-seq dataset (accession GSE: 40918) (Ciofani et al., 2012) were used for downstream analysis. H3K4me3, H3K27me3, and H3K27ac ChIP-seq for Th0 and Th17 cells were obtained from GSE: 102317, GSE: 57500, GSE: 14254, and GSE: 90788 (Cheung et al., 2017; Li et al., 2017; Wei et al., 2009; Zhang et al., 2014). STAT3 ChIP-seq for Th0 and Th0+IL-6 was obtained from GSE: 65621 (Hirahara et al., 2015).
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
ATAC-seq was performed as described (Buenrostro et al., 2013). Naive WT and Batf KO CD4+T cells isolated by FACS sorting (CD4+CD44lowCD62Lhigh) were stimulated with anti-CD3, anti-CD28 with or without IL-6 for 24 h or under Th17 conditions for 48 h. 5×104 cells were harvested, washed with 1X PBS, and nuclei were isolated. Transposition reaction was carried using Nextera DNA Library Preparation Kit (Illumina, 15028212), and PCR amplification was performed to generate libraries. ATAC-seq libraries were sequenced with paired-end 50 bp using Illumina HiSeq 2500.
ATAC-seq analysis
Raw paired-end sequencing data were mapped to mm9 genome using Bowtie2 (Langmead et al., 2009) with option -X 2000. Accessible regions were determined using MACS2 with option -g mm–nomodel–nolambda–broad. Differential peak calling was performed using DESeq2 with fold change greater than 2 and fdr less than 0.05. Tag counts were normalized to 10 million tags and calculated using annotatePeaks function of HOMER (Heinz et al., 2010), and heatmaps were visualized using deepTools (Ramírez et al., 2016). Unique and overlapping peaks were determined using mergePeaks function of HOMER. The log2 transformed normalized read data were used to generate boxplots and violin plots using R. Motif analysis was performed using findMotifsGenome function of HOMER. ATAC-seq peaks were annotated to the nearest gene in mm9 genome using annotatePeaks function of HOMER. Gene ontology analysis of associated peaks was performed using annotatePeaks function of HOMER with option -go. Footprinting detection were performed using pyDNase algorithm with fdr value cut-off at 0.05 (Piper et al., 2013, 2015). In short, the tagmentation activity of transposase Tn5, used in ATAC-seq, is high in accessible regions and low in inaccessible regions. Thus, protein-DNA binding sites, or footprints, are detected by exploring for a depletion of Tn5 cuts compared to cuts in the surrounding regions, presumably accessible regions. Since DNA fragments generated by Tn5 are likely predominantly from within accessible regions, and incorporate with footprints, upstream ends (aligned as positive sequence tag) and downstream ends (aligned as negative sequence tag) cut by Tn5 will have concentrated signals to the left and right, respectively, from the accessible regions. The footprint occupancy score is reduced as the DNA fragments are extended away from the accessible regions. Differential footprints between two ATAC-seq datasets were calculated using the Wellington-bootstrap algorithm with minimum p value to be considered significant for fdr calculation set at 20. ATAC-seq tracks were normalized to 10 million tags and viewed using IGB (Nicol et al., 2009). Batf target genes from WT and Batf KO Th17 RNA-seq dataset (accession GSE: 40918) (Ciofani et al., 2012) were used for downstream analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Information on experimental replicates, statistical details, and n numbers can be found in the figure legends. Data are mean ± SEM. Statistical analyses were calculated using Prism (GraphPad Software) or R. We determined significance using the Mann-Whitney test for unpaired data, two-tailed Student’s t test for paired data, or a one-way ANOVA followed by Tukey’s test for multiple comparisons where appropriate. p values < 0.05 is considered as statistical significance.
DATA AND CODE AVAILABILITY
ATAC-seq, ChIP-seq, and Hi-C data generated during this study have been deposited in the NCBI Gene Expression Omnibus with the accession GSE: 123209.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD4-APC/Cyanine7 (clone GK1.5) | Biolegend | Cat# 100414; RRID:AB_312699 |
| Anti-Mouse CD25-APC (clone 3C7) | Biolegend | Cat#101910; RRID:AB_2280288 |
| Anti-Mouse CD62L-PE/Cy7 (clone MEL-14) | Biolegend | Cat# 104417; RRID:AB_313102 |
| Anti-Mouse CD44-FITC (clone IM7) | Biolegend | Cat#103006; RRID:AB_312957 |
| Biotin Anti-Mouse CXCR5 (clone 2G8) | BD Biosciences | Cat#551960; RRID:AB_394301 |
| Anti-Mouse PD-1-PE ((clone 29F.1A12) | Biolegend | Cat#135205; RRID:AB_1877232 |
| Anti-Mouse CD69-PerCP/Cy5.5 (clone H1.2F3) | Biolegend | Cat#104522; RRID:AB_2260065 |
| Anti-Mouse ICOS-FITC (clone 7E.17G9) | eBioscience | Cat#11–9942-82; RRID:AB_11218290 |
| Streptavidin eFluor450 | eBioscience | Cat# 48–4317-82; RRID:AB_10359737 |
| Anti-Mouse Batf-PE (clone 9B5A13) | Biolegend | Cat#654804; RRID:AB_2563517 |
| Anti-Mouse CD3 (clone 145–2C11) | BD Biosciences | Cat#557306; RRID:AB_396632 |
| Anti-Mouse CD28 (clone 37.51) | BD Biosciences | Cat#553295; RRID:AB_394764 |
| Anti-Mouse IFN-g (clone XMG1.2) | This paper | N/A, generated in house |
| Anti-Mouse IL-4 (clone 11B11) | This paper | N/A, generated in house |
| Rabbit Anti-Mouse Irf4 | Santa Cruz | Cat#sc-28696X; RRID:AB_2127141 |
| Rabbit Anti-Mouse Ets1 | Santa Cruz | Cat#sc-350X; RRID:AB_2100688 |
| Normal rabbit IgG | Millipore | Cat#12–370; RRID:AB_145841 |
| Rabbit Anti-Mouse H3K27me3 | Millipore | Cat#07–449; RRID:AB_310624 |
| Rabbit Anti-Mouse H3K4me3 | Millipore | Cat#07–473; RRID:AB_1977252 |
| Rabbit Anti-Mouse H3K27ac | Abcam | Cat#ab4729; RRID:AB_2118291 |
| Rabbit Anti-Mouse Batf | Schraml et al., 2009 | N/A, generated in house |
| Rabbit Anti-Mouse Med12 | Bethyl Laboratory | Cat#A300–774A; RRID:AB_669756 |
| Rabbit Anti-Mouse Ctcf. | Active Motif | Cat#61311; RRID:AB_2614975 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Human IL-2 Protein | R&D Systems | Cat# 2G2-IL-G1G |
| Recombinant Mouse IL-6 Protein | R&D Systems | Cat#4G6-ML-G2S |
| Recombinant Human TGF-β Protein | R&D Systems | Cat#24G-B |
| Recombinant Mouse IL-1β Protein | R&D Systems | Cat#4G1-ML-G2S |
| Phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat#P18S-1MG |
| lonomycin | EMD Bioscience | Cat#4D7–9S2 |
| Dynabeads M-280 Sheep Anti-Rabbit IgG | ThermoFisher Scientific | Cat#112G3D |
| Dynabeads® MyOne Streptavidin C1 | ThermoFisher Scientific | Cat#6SGG1 |
| EcoRI-HF | NEB | Cat#R31G1M |
| HindIII-HF | NEB | Cat#R31G4M |
| NheI-HF | NEB | Cat#R3131L |
| T4 DNA Ligase | NEB | Cat#G2G2M |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | Cat#G21GM |
| Deoxynucleotide (dNTP) Solution Set | NEB | Cat# NG446S |
| Biotin-14-dCTP | ThermoFisher Scientific | Cat# 19S18G18 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | Cat#172S27S |
| SsoAdvanced Universal Probes Supermix | Bio-Rad | Cat#1725285 |
| iScript cDNA Synthesis Kit | Biorad | Cat# 17G8891 |
| Streptavidin | BDBiosciences | Cat#SS7S98 |
| FBS | HyClone | Cat#SH30109.03 |
| RPMI-1640 | Corning | Cat#10–040-CM |
| Penicillin/Streptomycin | Corning | Cat#30–002-Cl |
| 2-Mercaptoethanol | Sigma | Cat# M3148 |
| Non-Essential Amino Acids (MEM) | Corning | Cat#25025-Cl |
| L-Glutamine | Corning | Cat#25–005-Cl |
| HEPES | Corning | Cat#25–060-Cl |
| Sodium Pyruvate | Corning | Cat#25–000-Cl |
| Trizol | ThermoFisher | Cat# 15596018 |
| RNaseA | ThermoFisher | Cat# EN0531 |
| Critical Commercial Assays | ||
| CD4+CD62L+ T Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130–106-643 |
| QuikChange XL Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200517 |
| Nucleofector Kits for Mouse T Cells | Lonza | Cat#VPA-1006 |
| Dual-Luciferase Reporter Assay System | Promega | Cat#E1910 |
| NEBNext DNA Library Prep Master Mix Set for Illumina | NEB | Cat# E6040S |
| NEBNext Ultra II DNA Library Prep Kit for Illumina | NEB | Cat#E7645S |
| Nextera DNA Library Prep Kit (24 samples) | llumina | Cat# FC-121–1030 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Cat# Q32851 |
| MinElute PCR Purification Kit | QIAGEN | Cat#28004 |
| LS Columns | Miltenyi Biotec | Cat# 130–042-401 |
| MS Columns | Miltenyi Biotec | Cat# 130–042-201 |
| LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit | ThermoFisher Scientific | Cat# L34976 |
| Deposited Data | ||
| ATAC-seq | GEO database | GSE: 123209 |
| ChIP-seq | GEO database | GSE: 123209 |
| Hi-C seq | GEO database | GSE: 123209 |
| Experimental Models: Primary cells | ||
| Mouse: primary T lymphocytes | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: Batf KO | The Jackson Laboratory | Stock No: 013758 |
| Mouse: Irf4fl/fl | The Jackson Laboratory | Stock No: 009380 |
| Mouse: Ets1fl/fl | Zook et al., 2016 | N/A |
| Mouse: CD4-Cre | The Jackson Laboratory | Stock No: 022071 |
| Recombinant DNA | ||
| Mouse BAC DNA Il21 | 3ACPAC Resources Center | Cat#RP23–290D8 |
| Mouse BAC DNA Il21 | 3ACPAC Resources Center | Cat#RP23–128L14 |
| Mouse BAC DNA Gapdh | 3ACPAC Resources Center | Cat#RP23–410F11 |
| MSCV-IRES-Thy1.1 | Jabeen et al., 2013 | N/A |
| MSCV-IRES-Thy1.1 Batf | Jabeen et al., 2013 | N/A |
| pCMV6-Ets1 | Origene | Cat#MR207015 |
| MSCV-IRES-Thy1.1 Ets1 | This paper | N/A, generated in house |
| pRL-TK | Promega | Cat#E2231 |
| pGL3-Il2p | This paper | N/A, generated in house |
| pGL3-Il2p -35 | This paper | N/A, generated in house |
| pGL3-Il2p -47 | This paper | N/A, generated in house |
| pGL3-Il2p -64 | This paper | N/A, generated in house |
| pGL3-Il2p -77 | This paper | N/A, generated in house |
| pGL3-Il2p -84 | This paper | N/A, generated in house |
| pGL3-Il21p | This paper | N/A, generated in house |
| pGL3-Il21p -77 | This paper | N/A, generated in house |
| pGL3-Il21p -84 | This paper | N/A, generated in house |
| pGL3-Il21p -27 | This paper | N/A, generated in house |
| pGL3-Il21p -41 | This paper | N/A, generated in house |
| pGL3-Il21p -49 | This paper | N/A, generated in house |
| pGL3-Il21p -60 | This paper | N/A, generated in house |
| pGL3-Il21p -70 | This paper | N/A, generated in house |
| pGL3-Il21p -80 | This paper | N/A, generated in house |
| pGL3-Il2p -Δ47 | This paper | N/A, generated in house |
| pGL3-Il21p -Δ49 | This paper | N/A, generated in house |
| pGL3-Il21p -Δ60 | This paper | N/A, generated in house |
| pGL3-Il21p -Δ80 | This paper | N/A, generated in house |
| Sequence-Based Reagents | ||
| Primers for cloning luciferase reporter vectors, see Table S4 | This paper | N/A |
| Primers for mutating luciferase reporter vectors, see Table S4 | This paper | N/A |
| Primers for ChIP qPCR, see Table S4 | This paper | N/A |
| Primers for 3C, see Table S4 | This paper | N/A |
| Primer for gene expression RT-PCR, see Table S4 | This paper | N/A |
| Primers for ATAC-seq | Buenrostro et al., 2013 | N/A |
| NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) | NEB | Cat# E7335S |
| NEBNext Multiplex Oligos for Illumina (Index Primers Set 2) | NEB | Cat#E7500S |
| Software and Algorithms | ||
| Bowtie2 (v2.2.5) | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Bowtiel (1.2.0) | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.2.0/ |
| MACS2 (v2.1.0) | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| HOMER (v4.8) | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| HiC-Pro (2.7.0) | Servant et al., 2015 | https://github.com/nservant/HiC-Pro |
| IGB (9.0.0) | Nicol et al., 2009 | https://wiki.transvar.org/display/igbman/Home |
| deepTools (v2.0) | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/latest/ |
| GraphPad Prism (v7.0) | N/A | https://www.graphpad.com/ |
| pyDNase (v0.2.3) |
Piper et al., 2013 Piper et al., 2015 |
https://pythonhosted.org/pyDNase/index.html |
| Circos | Krzywinski et al., 2009 | http://circos.ca/ |
| TrimGalore (v0.4.1) | N/A | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Picard (v2.6.0) | N/A | http://broadinstitute.github.io/picard/ |
| R(v3.2.1) | N/A | https://www.r-project.org/ |
| FlowJo (v10) | FlowJo, LLC | https://www.flowio.com/ |
Highlights.
Batf binds nucleosomal DNA and mediates clearing in developing effector T cells
Batf modulates Ctcf recruitment to open chromatin to control looping in CD4 T cells
Batf and Ets1 cooperate to increase Ctcf expression and recruitment to DNA
Batf and Ets1 enhance Ctcf DNA binding in a largely Irf4-independent manner
ACKNOWLEDGMENTS
The authors are grateful to members of the Weaver laboratory for helpful comments and suggestions. We thank B. Dale, A. Smith, and H. Turner for technical assistance; and M. Kaplan (Indiana University) and A. Weinmann (University of Alabama at Birmingham [UAB]) for the provision of retroviruses. We acknowledge the UAB Epitope Recognition and Immunoreagent Core Facility (NIH P30 AR048311) for antibody preparations and the CFAR Basic and Translational Sciences Core Facility (NIH P30 AI27667) for fluorescence-activated cell sorting (FACS) sorting. This work was supported by NIH R01 DK115172 to R.D.H. and C.T.W. and T32 AI007051–37 to D.P., NIH F30DK098911 to C.M., and UAB Institutional Funds to C.T.W.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.09.064.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bao K, Carr T, Wu J, Barclay W, Jin J, Ciofani M, and Reinhardt RL (2016). BATF modulates the Th2 locus control region and regulates CD4+ T cell fate during antihelminth immunity. J. Immunol 197, 4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, and Leiden JM (1995). A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity 3, 223–237. [DOI] [PubMed] [Google Scholar]
- Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, and Taparowsky EJ (2010). Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med 207, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, Gilding LN, Ott S, Bonifer C, and Cockerill PN (2016). Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J 35, 515–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung M-H, Trump S, Lightman SL, et al. (2011). Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, and Cavalli G (2016). Organization and function of the 3D genome. Nat. Rev. Genet 17, 661–678. [DOI] [PubMed] [Google Scholar]
- Brὒstle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, and Lohoff M (2007). The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol 8, 958–966. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr TM, Wheaton JD, Houtz GM, and Ciofani M (2017). JunB promotes Th17 cell identity and restrains alternative CD4+ T-cell programs during inflammation. Nat. Commun 8, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchy P, Maqbool MA, Zacarias-Cabeza J, Vanhille L, Koch F, Fenouil R, Gut M, Gut I, Santana MA, Griffon A, et al. (2016). Dynamic recruitment of Ets1 to both nucleosome-occupied and -depleted enhancer regions mediates a transcriptional program switch during early T-cell differentiation. Nucleic Acids Res. 44, 3567–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Adoue V, Mechin M-C., Ying S, Dong S, Duplan H, Charveron M, Takahara H, Serre G, and Simon M (2008). Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylargi-nine deiminase 3 gene in differentiated keratinocyte. PLoS ONE 3, e3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, Shen T, Lee J-Y, Ren C, Chen C-H, et al. (2017). Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation. Mol. Cell 65, 1068–1080.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey MJ, Tae HJ, Sollenberger KG, Mascarenhas NT, Johansen LM, and Taparowsky EJ (1995). B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 11, 2255–2265. [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun H-W, Kanno Y, Powrie F, and O’Shea JJ (2010). Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echlin DR, Tae HJ, Mitin N, and Taparowsky EJ (2000). B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene 19, 1752–1763. [DOI] [PubMed] [Google Scholar]
- Fields PE, Lee GR, Kim ST, Bartsevich VV, and Flavell RA (2004). Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 21, 865–876. [DOI] [PubMed] [Google Scholar]
- Gifford CA, and Meissner A (2012). Epigenetic obstacles encountered by transcription factors: reprogramming against all odds. Curr. Opin. Genet. Dev 22, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. (2012). A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusdat M, McIlwain DR, Xu HC, Pozdeev VI, Knievel J, Crome SQ, Robert-Tissot C, Dress RJ, Pandyra AA, Speiser DE, et al. (2014). IRF4 and BATF are critical for CD8+ T-cell function following infection with LCMV.Cell Death Differ. 21, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustems M, Woellmer A, Rothbauer U, Eck SH, Wieland T, Lutter D, and Hammerschmidt W (2014). c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 42, 3059–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, and Forne T (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc 2, 1722–1733. [DOI] [PubMed] [Google Scholar]
- Hardison RC (2000). Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 16, 369–372. [DOI] [PubMed] [Google Scholar]
- Hasan Z, Koizumi S-I, Sasaki D, Yamada H, Arakaki N, Fujihara Y, Okitsu S, Shirahata H, and Ishikawa H (2017). JunB is essential for IL-23dependent pathogenicity of Th17 cells. Nat. Commun 8, 15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, and Weaver CT (2006). A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729. [DOI] [PubMed] [Google Scholar]
- He X, Chatterjee R, John S, Bravo H, Sathyanarayana BK, Biddie SC, FitzGerald PC, Stamatoyannopoulos JA, Hager GL, and Vinson C (2013). Contribution of nucleosome binding preferences and co-occurring DNA sequences to transcription factor binding. BMC Genomics 14, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineagedetermining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K, Onodera A, Villarino AV, Bonelli M, Sciume G, Laurence A, Sun H-W, Brooks SR, Vahedi G, Shih H-Y, et al. (2015). Asymmetric action of STAT Transcription factors drives transcriptional outputs and cytokine specificity. Immunity 42, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, and Murphy KM (2011). The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol 12, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, and Iba H (2001). Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem 276, 2852–2857. [DOI] [PubMed] [Google Scholar]
- Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, Walsh D, Olson MR, Kim MH, Tepper RS, et al. (2013). Th9 cell development requires a BATF-regulated transcriptional network. J. Clin. Invest 123, 4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH, Glosson NL, Stritesky GL, Yeh N, Kinzfogl J, Rohrabaugh SL, Goswami R, Pham D, Levy DE, Brutkiewicz RR, et al. (2011). STAT3-dependent IL-21 production from T helper cells regulates hematopoietic progenitor cell homeostasis. Blood 117, 6198–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I,Chen X, Watters A, Carriero N, Awasthi A, et al. (2017). Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat. Immunol 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LK, Esplugues E, Zorca CE, Parisi F, Kluger Y, Kim TH, Galjart NJ, and Flavell RA (2014). Oct-1 regulates IL-17 expression by directing interchromosomal associations in conjunction with CTCF in T cells. Mol. Cell 54, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, and Dalla-Favera R (2006). Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol 7, 773–782. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, and Marra MA (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome.Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Griffin TJ, and Flavell RA (2003). Regulation of the Th2 cytokine locus by a locus control region. Immunity 19, 145–153. [DOI] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, and Leonard WJ (2012). BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Mitra S, Spolski R, Oh J, Liao W, Tang Z, Mo F, Li X, West EE, Gromer D, et al. (2017). STAT5-mediated chromatin interactions in superenhancers activate IL-2 highly inducible genes: Functional dissection of the Il2ra gene locus. Proc. Natl. Acad. Sci. USA 114, 12111–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, and Murre C (2012). Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol 13, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, and Odom DT (2013). CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285–1297. [DOI] [PubMed] [Google Scholar]
- Moisan J, Grenningloh R, Bettelli E, Oukka M, and Ho I-C (2007). Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med 204, 2825–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TL, Tussiwand R, and Murphy KM (2013). Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol 13, 499–509. [DOI] [PubMed] [Google Scholar]
- Naumova N, Smith EM, Zhan Y, and Dekker J (2012). Analysis of longrange chromatin interactions using Chromosome Conformation Capture. Methods 58, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, and Bremner R (2007). Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J. Immunol 178, 345–351. [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr., Raja A, and Loraine AE (2009). The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, and Corces VG (2014). CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet 15, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Choi Y, Song M-J, Park K, Lee J-J, and Kim H-P (2016). Dynamic long-range chromatin interaction controls expression of IL-21 in CD4+ T cells. J. Immunol 196, 4378–4389. [DOI] [PubMed] [Google Scholar]
- Piper J, Elze MC, Cauchy P, Cockerill PN, Bonifer C, and Ott S (2013). Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res 41, e201–e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, and Ott S (2015). Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics 16, 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Shiue C-N, Zhu J, Zhuang T, Jonsson P, Wright APH, Zhao C, and Dahlman-Wright K (2015). AP-1-mediated chromatin looping regulates ZEB2 transcription: new insights into TNFa-induced epithelial-mesenchymal transition in triple-negative breast cancer. Oncotarget 6, 7804–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Gru€ning B., Bhardwaj V, Kilpert F, Richter AS, Heyne S, Du€ndar F., and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, Larson DR, and Zhao K (2017). CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell 67, 1049– 1058.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Heath H, Krpic S, Dingjan GM, van Hamburg JP, Bergen I, van de Nobelen S, Sleutels F, Grosveld F, Galjart N, and Hendriks RW (2009). Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J. Immunol 182, 999–1010. [DOI] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee W-L, Smith WA-E, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. (2009). The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, and Wilson CB (2009). CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity 31, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert J-P, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, and Gifford DK (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol 32, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun H-W, Sartorelli V, Kanno Y, and O’Shea JJ (2012). STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, C Poholek A, Hand TW, Laurence A, Kanno Y, O’Shea JJ, and Hirahara K (2013). Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev 252, 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Tanaka Y, Araki H, Kohda A, Sanematsu F, Arasaki T, Duan X, Miura F, Katagiri T, Shindo R, et al. (2017). The AP-1 transcription factor JunB is required for Th17 cell differentiation. Sci. Rep 7, 17402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. (2013). Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Carroll JS (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wrzeszczynska MH, Horvath CM, and Darnell JE Jr. (1999). Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol 19, 7138–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Modelbased analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kinkel S, Maksimovic J, Bandala-Sanchez E, Tanzer MC, Naselli G, Zhang J-G, Zhan Y, Lew AM, Silke J, et al. (2014). The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood 124, 737–749. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, and Rudensky AY (2010). Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, and Littman DR (2007). IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol 8, 967–974. [DOI] [PubMed] [Google Scholar]
- Zook EC, Ramirez K, Guo X, van der Voort G, Sigvardsson M, Svensson EC, Fu Y-X, and Kee BL (2016). The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J. Exp. Med 213, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq, ChIP-seq, and Hi-C data generated during this study have been deposited in the NCBI Gene Expression Omnibus with the accession GSE: 123209.