Abstract
Mobile genetic elements, such as plasmids, phages, and transposons, are important sources for evolution of novel functions. In this study, we performed a large-scale screening of metagenomic phage libraries for their ability to suppress temperature-sensitivity in Salmonella enterica serovar Typhimurium strain LT2 mutants to examine how phage DNA could confer evolutionary novelty to bacteria. We identified an insert encoding 23 amino acids from a phage that when fused with a bacterial DNA-binding repressor protein (LacI) resulted in the formation of a chimeric protein that localized to the outer membrane. This relocalization of the chimeric protein resulted in increased membrane vesicle formation and an associated suppression of the temperature sensitivity of the bacterium. Both the host LacI protein and the extracellular 23-amino acid stretch are necessary for the generation of the novel phenotype. Furthermore, mutational analysis of the chimeric protein showed that although the native repressor function of the LacI protein is maintained in this chimeric structure, it is not necessary for the new function. Thus, our study demonstrates how a gene fusion between foreign DNA and bacterial DNA can generate novelty without compromising the native function of a given gene.
Keywords: gene fusion, phage, evolution of new function, bacteria, lacI
Introduction
How new genes emerge and evolve is a long-standing central question in evolutionary biology, and it is of importance in the contexts of evolution of diversity (Blount et al. 2012; Chen et al. 2013; Farr et al. 2017), adaptive radiations (Wood and Erwin 2018; Zhang et al. 2018), and speciation (Jenkin 1933; Bridges 1935). Different approaches have been used to investigate the basis of how new genes are formed from pre-existing genes, with some early experimental work done by Sturtevant (1925) and later by Muller (1936). During the last two decades, comparative genomics and phylogenetic analysis have given us a better understanding of these mechanisms (Altschmied et al. 2002; Xing et al. 2006; Zhou et al. 2008; Dubruille et al. 2012; Carvalho et al. 2015), and although several models (e.g., duplication–divergence, de novo, gene fusion/fission) have been proposed for evolution of new genes, experimental studies of the origin of novelty are more limited (Nasvall et al. 2012; Digianantonio and Hecht 2016; Farr et al. 2017; Knopp et al. 2019).
One mechanism for creating new genes is the gene fusion model that involves reorganization of existing genes, such that fragments of different genes fuse together resulting in novel functionality. Comparative genomics have provided support for the origin of new genes by fusion. Among these, one class includes fusion of entire domains from different proteins that results in the novel protein performing functions of both the domains. Examples of evolution of new genes by this mechanism include the origin of tRNA synthesases (Berthonneau and Mirande 2000; Eswarappa et al. 2018) and fatty acid chain desaturases (McCarthy and Hardie 1984). This mechanism for the formation of new genes was also observed during experimental evolution studies in Pseudomonas fluorescens, where fusion of different domains of a desaturase and a di-guanylate cyclase resulted in the generation of an adaptive phenotype (Farr et al. 2017). The second class among the gene fusion models includes nonspecific chimeric formation where different fragments of different genes fuse together. Origins of adh-jingwei and adh-twain genes in Drosophila species are examples of this category (Long and Langley 1993; Jones et al. 2005).
A second mechanism of origin of new genes, especially observed in but not limited to bacteria, is from extracellular mobile elements that includes phage DNA and conjugative elements (transposons and plasmids) (Treangen and Rocha 2011; Wiedenbeck and Cohan 2011; Blount et al. 2012; Jerlstrom Hultqvist et al. 2018). These mobile elements often result in immediate innovative changes in a one-step genetic event and hence are an important source of generating novelty. Examples of evolution of novel genes by contribution of these mobile elements include the evolution of metabolic pathways (Pal et al. 2005; Homma et al. 2007), diversification of cell-envelope surface structures, synthesis of lipopolysaccharides, and novel regulatory interactions (Nakamura et al. 2004).
We describe here an experimental example of an origin of a new gene where both of the above-mentioned mechanisms interplay. Our experiments show how phage DNA when fused with an existing bacterial gene results in novel functionality. More specifically, a chimeric gene is formed by addition of a 169-bp fragment of foreign DNA to a truncated lacI gene. When translated into a protein, due to an internal stop codon, this 169-bp region adds only 23 amino acids to the C-terminal of the truncated LacI protein. When expressed, the chimeric protein can suppress temperature sensitivity in a mutant of Salmonella enterica serovar Typhimurium strain LT2 (designated S. Typhimurium throughout the text) at nonpermissive temperatures. The gene fusion results in relocalization of the chimeric LacI protein to the outer membrane, which results in an increase in membrane vesicle formation and suppression of the temperature-sensitive phenotype. Furthermore, the native repressor functions (i.e., DNA binding and inducer response) of the LacI protein are maintained in the chimeric protein, even though they are not needed for the novel function.
Results
Screening and Rescue of Temperature-Sensitive S. Typhimurium Mutants at Nonpermissive Temperatures
Phage metagenomic libraries were prepared to identify the different mechanisms by which mobile genetic elements can contribute to the rescue of temperature-sensitive S. Typhimurium mutants at nonpermissive temperatures. These libraries were prepared by ligating DNA extracted from different environmental phage samples into the cloning vector pCA24N –gfp (Kitagawa et al. 2005), and they have previously been described in detail (Jerlstrom Hultqvist et al. 2018). The temperature-sensitive S. Typhimurium mutants were generated by chemical mutagenesis, as previously described (Schmid et al. 1989). We further characterized these temperature-sensitive mutants on the basis of the range of nonpermissive temperatures where growth was not observed (37, 40, and 43 °C). Fifty of these temperature-sensitive mutants were screened with five metagenomic phage libraries (Supplementary Table 1a from Jerlstrom Hultqvist et al. [2018]) and were selected at appropriate nonpermissive temperatures. Using this experimental design, suppression of temperature sensitivity was observed for four different temperature-sensitive mutants (supplementary table 1, Supplementary Material online). In three cases (DA27906, DA27913, DA27926), suppression of temperature sensitivity was the result of transfer of cognate genes from another organism, which was confirmed by identifying the causal mutation for the temperature-sensitive phenotype in these strains (supplementary table 1, Supplementary Material online). In the fourth case (DA27987), rescue for growth at nonpermissive temperature was observed as a result of a serendipitous truncation of the last 80 bp of the lacI gene on the cloning vector and the subsequent fusion with a phage-derived 169-bp DNA fragment (fig. 1 and supplementary table 1, Supplementary Material online). This was most likely the outcome of the nonspecific cutting of the lacI gene on the cloning vector by the restriction enzyme that was used for generation of the libraries (Jerlstrom Hultqvist et al. 2018). The temperature sensitivity in this mutant was observed at 37 °C and higher, but the chimeric LacI protein suppressed the temperature sensitivity only at 37 °C.
Fig. 1.
Chimeric LacI protein that allows growth of temperature-sensitive mutant at nonpermissive temperatures. Formation of a chimeric protein as a result of fusion of a 169-bp insert to a truncated LacI protein (deletion of last 80 bp of the native lacI gene). The insertion results in addition of 23 amino acids (due to an internal stop codon) to the truncated LacI protein.
Both the Phage and Bacterial Parts of the Chimeric LacI Protein Are Required for the Novel Phenotype
To characterize the chimeric LacI protein, genetic constructs of different lengths and properties were designed. The 169-bp DNA fragment fused with the lacI gene, when read in-frame with the truncated lacI, had a stop codon positioned after 23 amino acids (fig. 1 and supplementary fig. 1, Supplementary Material online). Thus, we designed two gene constructs: the first with the truncated lacI gene along with the complete 169-bp insert and the second with the fused insert but only up to the stop codon. Both constructs resulted in growth of the temperature-sensitive mutant at 37 °C (figs. 2A and 3; supplementary fig. 1, Supplementary Material online) confirming that the fusion of only the 23-amino acid fragment was sufficient for the novel phenotype. We then designed a construct changing the sequence of this chimeric protein at the nucleotide level, but maintaining the sequence at the level of the amino acids (supplementary fig. 2 and supplementary table 2, Supplementary Material online). This recoded fragment also rescued growth of the temperature-sensitive mutant confirming that the translated protein product, rather than the transcribed RNA, caused the rescue (supplementary fig. 1, Supplementary Material online). Next, we investigated if the truncated lacI gene (without the last 80 bp) and the fused 169-bp insert (in the same reading frame as present in the fused protein and with a start codon in place) each individually by themselves were sufficient for the rescue of the temperature-sensitive mutant. No rescue was observed for either of these constructs, confirming that both parts of the chimeric LacI protein are required for the observed new function (fig. 2A and supplementary fig. 1, Supplementary Material online). We also designed a construct such that the truncated lacI gene and the phage DNA insert were both expressed from the same fragment, but were not fused together. This was obtained by inserting a stop codon after the truncated lacI gene, which was followed by inserting a ribosome binding site and a start codon upstream of the phage DNA insert. The reading frame of the phage DNA insert was kept the same as what was observed in the chimeric LacI protein. No rescue was observed for this construct either (fig. 2A and supplementary fig. 1B, Supplementary Material online). To investigate the minimum length of the insert needed for the chimeric protein to suppress temperature sensitivity, we removed three, five, or seven amino acids, respectively, from the C-terminal end of the phage insert and fused these shortened fragments to the truncated LacI. None of these constructs was able to rescue the temperature-sensitive mutant (supplementary fig. 1D, Supplementary Material online). Furthermore, we also did not find significant homology of this 23-amino acid insert to sequences in the NCBI database (supplementary table 3, Supplementary Material online).
Fig. 2.

Different constructs tested for their ability to rescue the temperature-sensitive phenotype at 37 °C. Ability to rescue is denoted by ✓ and inability to rescue is denoted by ✗. (A) Both components of chimeric LacI protein are essential for the novel phenotype. (B) Specificity of LacI protein and to the 23-amino acid insert sequence for the novel phenotype.
Fig. 3.
Recloning of the fused LacI protein under the control of an inducible promoter. The full-length chimeric LacI protein (construct 1) and the chimeric LacI protein till the internal stop codon (construct 2) were inserted in front of an IPTG-inducible promoter and checked for the ability to suppress temperature sensitivity. All growth rate values are normalized to growth rate of the temperature-sensitive mutant with the original plasmid containing the chimeric protein at 37 °C. Error bars represent SD.
The Novel Phenotype Is Specific to the LacI Protein and to the 23-Amino Acid Insert Sequence
To investigate whether any unspecific protein when fused to the 23-amino acid insert would result in the rescue of the temperature-sensitive mutant, we constructed another chimeric protein fusing together a LacI-like DNA-binding protein, FruR to the 23-amino acid insert. First, we inserted the 23-amino acid insert at the C-terminal of the complete FruR protein, and second, we deleted 26 amino acids at the C-terminal of FruR protein (to mimic the observed truncation in LacI protein) and then fused the 23 amino acids to this truncated FruR (fig. 2B). None of these constructs could rescue the temperature-sensitive phenotype, suggesting that the new function of the chimeric protein requires some characteristics of the LacI protein (supplementary fig. 1, Supplementary Material online). To investigate the specificity of the 23-amino acid insert sequence, we generated a library where randomized sequences (∼106 unique inserts) of length 23 amino acids were fused to the C-terminus of the truncated LacI protein (fig. 2B). The temperature-sensitive mutant could not be rescued when transformed with this library, suggesting that there is specificity at the level of the insert sequence as well.
The Native Function of LacI Protein Is Maintained in the Chimeric Protein, But It Is Not Required for the Novel Phenotype
To investigate if the chimeric LacI protein was suppressing temperature sensitivity by differentially regulating genes as compared with the native LacI, we first investigated if the chimeric LacI protein still functioned as a DNA-binding protein. To test this, we used a S. Typhimurium strain carrying the LacO operator sequence upstream of a gene coding for a yellow fluorescent protein (YFP). The LacO operator sequence is bound by LacI protein, which consequently then inhibits transcription of downstream genes. Thus, in the presence of native LacI, the cells will not produce YFP. We then inserted the native LacI and the chimeric LacI, separately, on the pBAD plasmid vector under the control of paraBAD (a native promoter that regulates expression of arabinose metabolism genes). When induced using 0.1% L-arabinose, we observed a decrease in the fluorescent signal for both the chimeric LacI protein and the native LacI protein. This implies that the chimeric LacI protein still performs its native function as a DNA-binding repressor (supplementary fig. 3, Supplementary Material online). Furthermore, we also investigated if this repression was removed in the presence of IPTG, which is a synthetic analog of lactose and removes repression by interacting directly with the LacI protein. We observed derepression for both the native LacI protein and the chimeric LacI protein in the presence of 100 µM IPTG (supplementary fig. 3, Supplementary Material online). These results demonstrate that the chimeric LacI protein maintained both of its native functions, that is, interaction with the DNA and with the inducer molecule. Next, we investigated if the DNA-binding function of LacI was contributing to the novel phenotype. To test this, we made two previously described point mutations (Ala10Thr and Ser21Phe) in the DNA-binding domain of LacI protein that have been shown to abolish the DNA-binding ability of the LacI repressor (Betz and Fall 1988). Using the same construct as described earlier (LacO upstream of the fluorescent protein), we observed that both these mutations also result in loss of DNA-binding function in the chimeric LacI protein (supplementary fig. 3, Supplementary Material online), however, these variants were still able to suppress the temperature sensitivity of the mutant. These results demonstrate that although the DNA-binding ability of the chimeric LacI protein is maintained, it is not needed for the novel phenotype. To further determine the characteristics of the chimeric LacI, we also investigated if expression of the chimeric LacI protein resulted in a fitness cost in the wild-type strain, as compared with the growth rate observed when only the native LacI protein is expressed. Strains where the native LacI and the chimeric LacI were inserted on the chromosome were used for this experiment. The rationale behind using these strains was to specifically measure the physiological costs of the chimeric LacI protein by itself, without the cost that is associated with the plasmid vector. We did not observe any fitness cost for the expression of the chimeric LacI protein as measured by exponential growth rate (supplementary fig. 4, Supplementary Material online).
Four Different Mutations Contribute to the Temperature-Sensitive Phenotype
To understand the mechanism of action of the chimeric LacI protein, we first had to identify the underlying mutations in the temperature-sensitive mutant and understand how they generated temperature sensitivity. Whole-genome sequencing of the temperature-sensitive mutant led to the identification of 48 mutations as compared with the parental S. Typhimurium. To identify the causal mutations, we transduced the temperature-sensitive mutant with a P22 lysate that was generated using the wild-type S. Typhimurium with the rationale that transductants selected for growth at nonpermissive temperatures would carry the corrected wild-type allele for the gene that is needed for the temperature-sensitive phenotype. The transductants were selected at both 37 and 43 °C. Two different mutations were identified to contribute to the temperature-sensitive phenotype. The first one was a mutation in gene rfaC S151N that when corrected allowed the strain to grow at both 37 and 43 °C. The second mutation was in gene degP G207D that when corrected allowed the strain to grow at 37 °C but not at 43 °C. The rfaC gene encodes the enzyme heptosyl transferase that is involved in the synthesis of the lipid A component of the outer membrane, whereas gene degP codes for a protein that functions as a chaperone and a protease especially for outer membrane proteins OmpC, OmpF, OmpA, and LamB (Krojer et al. 2008). We reconstructed these two mutations in the wild-type S. Typhimurium, both individually and in combination. However, neither individually nor in combination did these mutations result in the temperature-sensitive phenotype at 37 °C (supplementary fig. 5, Supplementary Material online), and only the rfaC S151N mutation had a temperature-sensitive phenotype at 43 °C. Additionally, the chimeric LacI protein did not suppress the temperature sensitivity caused by the rfaC S151N mutation. This result implied that additional mutations were contributing to the temperature sensitivity at 37 °C. As both these mutations are known to affect outer membrane integrity and cause misfolding of outer membrane proteins (Coleman and Deshpande 1985; Krojer et al. 2008; Klein et al. 2014), we examined additional mutations that might exacerbate this affect resulting in complete inhibition of growth. Two other mutations were identified: a mutation in ompR gene (Q223H), which regulates expression of outer membrane proteins, proteases, and chaperones, and a mutation in aas gene (P72S), which is an acyltransferase involved in membrane phospholipid turnover. Strain reconstruction showed that the presence of these four mutations resulted in temperature sensitivity at 37 °C, and that the temperature sensitivity of this quadruple mutant could be rescued by the chimeric LacI protein.
Suppression of Temperature Sensitivity by the Chimeric Protein Is Not Mediated via General Stress Response nor by Increased Expression of Target Genes
To identify the mechanism for suppression of the temperature sensitivity, we performed a series of genetic experiments. First, we selected mutants of the temperature-sensitive strain (DA27987) that were able to grow at 37 °C. Whole-genome sequencing of these mutants resulted in identification of mutations in the heat-shock response genes dnaK and dnaJ (supplementary table 4, Supplementary Material online). Given that one of these mutations results in loss-of-function of protein DnaK, it is likely that these revertants have increased expression of genes involved in the heat-shock response (Liberek and Georgopoulos 1993), which could potentially be a mechanism that allows growth of the temperature-sensitive mutant. To test this hypothesis, we transformed the temperature-sensitive mutant (DA27987) with a plasmid from the ASKA collection (Kitagawa et al. 2005) encoding the rpoH gene under an IPTG-inducible promoter. The rpoH gene encodes the regulator of the heat-shock response genes in S. Typhimurium. Induction of rpoH from this plasmid was able to rescue the temperature sensitivity of the mutant, supporting our previous results that induction of the heat-shock regulon could suppress temperature sensitivity (supplementary fig. 6, Supplementary Material online). Secondly, we also observed that overexpression of the wild-type alleles of any of the four genes that in combination result in the temperature sensitivity corrects for the growth defect at 37 °C (supplementary fig. 6, Supplementary Material online). Overexpression was obtained by using plasmids from the ASKA collection (see Materials and Methods) containing the wild-type alleles for these genes under an IPTG-inducible promoter. Lastly, transformation of the temperature-sensitive mutant with plasmids from the ASKA library (see Materials and Methods) followed by selection at 37 °C showed that the rescue of the mutant was also possible by overexpression of the gene rpoE, which is a regulator induced under conditions of membrane-stress (supplementary fig. 6, Supplementary Material online).
Given these results, our first hypothesis regarding the mechanism of action of the chimeric LacI protein was that it was inducing the general stress responses in the cell, that is, the membrane-stress response and/or the heat-shock response. To investigate this hypothesis, mRNA expression levels of genes rpoE and degP (constituting the membrane-stress response) and of genes rpoH, dnaK, and dnaJ (constituting the heat-shock response) were analyzed (fig. 4). As the temperature-sensitive mutant cannot grow at 37 °C without the chimeric LacI protein, we only compared expression level changes between the wild-type S. Typhimurium expressing the chimeric LacI protein to a wild-type S. Typhimurium strain with an empty vector (expressing the native LacI protein). These cultures were grown at 44 °C to mimic, to some extent, the physiological state of the temperature-sensitive mutant. We did not observe any difference in expression levels for these genes in the presence of the chimeric LacI protein. Our second hypothesis of how the chimeric protein suppressed the temperature-sensitive mutant was that it increased the expression of the target genes of rfaC, ompR, degP, and aas. However, no changes in expression level were observed for any of these genes (fig. 4).
Fig. 4.
Chimeric LacI does not change expression levels of genes involved in general stress response, or of the target genes. qPCR data measuring expression levels for genes involved in general stress response or of the target genes, in the wild-type strain either with native LacI protein (black circles) or with chimeric LacI protein (gray circles). In each case, strains were grown at 44 °C. Values for each gene are normalized to values observed in wild-type strain expressing the native LacI protein and are plotted on a log2 scale. Error bars represent SD.
Additionally, we had initially observed that at 37 °C, the presence of NaCl (1%) in the growth media resulted in growth of a triple mutant consisting of three out of the four target mutations, that is, rfaC S151N degP G207D ompR Q223H with an atypical growth curve trajectory (fig. 5A). This atypical growth curve was also observed for the original temperature-sensitive mutant DA27987 in the presence of NaCl at 37 °C (supplementary fig. 7, Supplementary Material online). We reasoned that this atypical growth curve might be the result of the same physiological defect that causes temperature sensitivity at 37 °C in the absence of NaCl. This assumption was further strengthened by the observation that the defect is corrected for in the presence of the chimeric LacI protein (fig. 5A and supplementary fig. 7, Supplementary Material online). Thus, to examine any putative changes in the levels of protein that are caused due to the chimeric LacI protein, this triple mutant (rfaC S151N degP G207D ompR Q223H) was grown in the presence of NaCl either with an empty vector carrying the native LacI protein or in the presence of the chimeric LacI protein, and whole-cell proteomics was performed. Whole-cell proteomic analysis was also performed for the wild-type strain, in the presence of either the native LacI protein or the chimeric LacI protein. In neither of these cases, did we observe upregulation of the general stress response genes or of the target genes (fig. 5B and supplementary table 6, Supplementary Material online), confirming our earlier results that the chimeric protein is not alleviating temperature sensitivity by these mechanisms.
Fig. 5.
Growth of the triple mutant (rfaC S151N degP G207D ompR Q223H) at 37 °C in the presence of NaCl. (A) Growth curve defect observed in the triple mutant when grown in the presence of NaCl at 37 °C expressing the native LacI (black). The defect is corrected by expression of the chimeric LacI protein (gray). Six biological replicates were used in each case. (B) Protein level changes in the wild-type and triple mutant in the presence of native LacI protein (black circles) or chimeric LacI protein (gray circles). Cells are grown in NaCl-containing LB at 37 °C. All values are normalized to expression levels observed in wild-type strain containing the native LacI protein and are plotted on a log2 scale. Error bars represent SD.
The Chimeric Protein Affects the Cell Membrane
To determine other potential mechanisms that could allow for growth of the temperature-sensitive mutant at 37 °C, we investigated whether the presence of divalent cations (Mg2+ or Ca2+) or the presence of noncationic osmolytes like sucrose in the growth media could result in growth of the mutant at nonpermissive temperatures. We observed that the presence of salts (100 mM for Mg2+, 10 and 100 mM for Ca2+, supplementary fig. 8, Supplementary Material online) resulted in growth of the mutant at nonpermissive temperatures, but growth was not observed in the presence of different concentrations of sucrose (0.5%, 2%, and 5%). It is thus likely that in presence of agents that stabilize the outer membrane growth of the mutant is observed. To further test a possible change in integrity of the outer membrane, we investigated susceptibility of the wild-type S. Typhimurium strain, expressing either the native LacI protein or the chimeric LacI protein, to antibiotics vancomycin, rifampicin, and tetracycline (table 1). S. Typhimurium is intrinsically resistant to vancomycin and rifampicin due to outer membrane permeability barrier (Sukupolvi et al. 1984). As a result mutants with altered outer membrane properties become susceptible to these antibiotics. We found that the wild-type strain became more susceptible to both vancomycin and rifampicin when the chimeric LacI protein was expressed, whereas no change in susceptibility was observed for tetracycline. This was observed irrespective of whether the chimeric LacI protein was being expressed from the chromosome (as a single copy) or from the plasmid. These results suggest that the chimeric LacI protein, either directly or indirectly, affects cell-membrane integrity and stability.
Table 1.
Membrane Integrity Was Analyzed by Measuring Susceptibility of Wild-Type S. Typhimurium to Vancomycin, Rifampicin, and Tetracycline (used as control).
| Strain | Antibiotic Used |
||
|---|---|---|---|
| Vancomycin | Rifampicin | Tetracycline | |
| Wild type (no plasmid) | 96 | 2 | 1 |
| Wild-type/empty plasmid | 96 | 2 | 1 |
| Wild-type/fused protein | 24 | 0.5 | 1 |
Note.—Absolute values obtained from E-tests are reported.
The Chimeric Protein Localizes in the Cellular Outer Membrane and Induces Outer Membrane Vesicle Formation
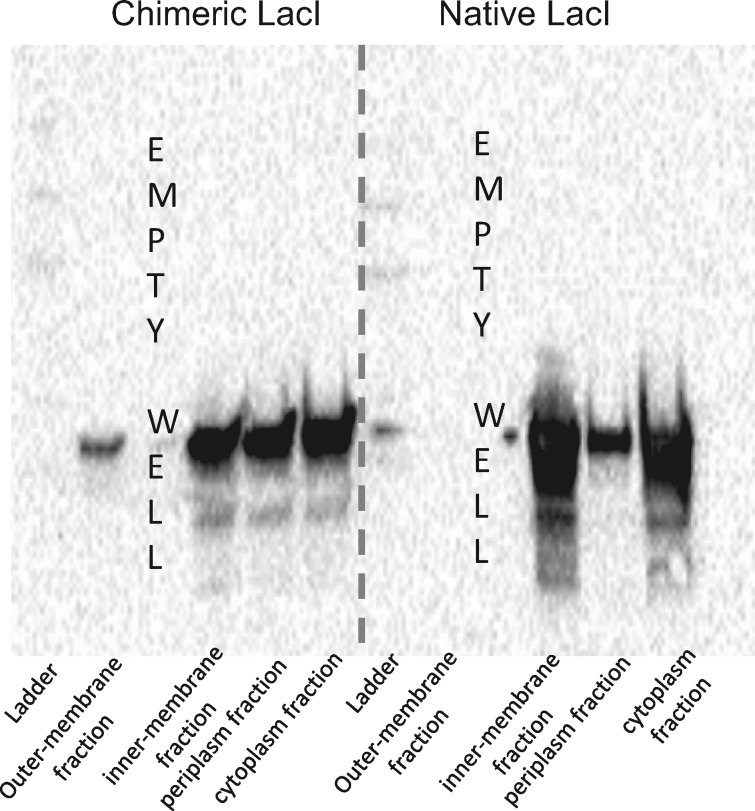
As the chimeric LacI protein affected membrane integrity, we investigated whether it was directly interacting with the membrane by determining the localization of the chimeric protein in the cell. To this end, FLAG tags were added to both the native LacI and the chimeric LacI, and it was confirmed that the FLAG-tagged chimeric LacI still suppressed temperature sensitivity. These localization experiments were performed in the wild-type strain expressing either the tagged native LacI protein or the tagged chimeric LacI protein. Different fractions of cell were then isolated (cytoplasmic, periplasm, inner membrane, and outer membrane fractions), and Western blot was performed on these isolated fractions using an anti-FLAG antibody. The native protein was found in the cytoplasm, periplasm, and inner membrane, whereas chimeric protein was found in all fractions, including the outer membrane (fig. 6).
Fig. 6.
Differential localization of chimeric LacI protein to the cell outer membrane. Western-blot analysis of different fractions of cells in the presence of either native protein or chimeric protein. Chimeric protein is differentially relocalized in the outer membrane fraction.
How could localization of the chimeric protein to the outer membrane suppress the temperature-sensitive phenotype? The mutations that conferred this phenotype are all involved in membrane biogenesis and conceivably a defect in biogenesis could result in accumulation of misfolded proteins and generation of the temperature-sensitive phenotype. This assumption was supported by the fact that we observed an upregulation of the CpxR, CpxA, CpxP, and DegP proteins in the triple mutant rfaC S151N degP G207D ompR Q223H (fig. 7A). The first three of these proteins are involved in sensing misfolded proteins in the inner membrane, periplasm, and the outer membrane (Raivio et al. 1999, 2013), whereas DegP acts as a protease and chaperone for misfolded outer membrane porins. Importantly, this upregulation was abrogated in the presence of the chimeric LacI protein (fig. 7A), suggesting that the chimeric LacI protein reduces the level of misfolded proteins. A well-known mechanism for expulsion of misfolded proteins (and other periplasmic content) from bacteria is via outer membrane vesicles (McBroom and Kuehn 2007). One possibility is therefore that the interaction of the chimeric protein with the outer membrane increases outer membrane vesicle formation, resulting in release of toxic misfolded proteins. We tested this hypothesis by measuring the amounts of outer membrane vesicles produced by the wild-type strain expressing either the chimeric LacI protein or the native LacI protein at 44 °C. As is seen in figure 7B, we observed a slight increase in the amount of outer membrane vesicles produced by the wild-type S. Typhimurium in the presence of the chimeric LacI protein as compared with the native LacI protein (P = 0.04). Furthermore, we also show that the temperature sensitivity of the mutant can be removed by independently inducing membrane vesicle formation, which was obtained by deleting the nlpI gene (supplementary fig. 9, Supplementary Material online). Deletion of this gene has been shown to increase membrane vesicle formation, without an upregulation of the rpoE regulon (McBroom and Kuehn 2007). This disconnect between induction of outer membrane vesicles and the upregulation of rpoE is important, as we have shown that upregulation of rpoE, by itself, also results in rescue of the temperature-sensitive phenotype. These results are compatible with the hypothesis that relocalization of the chimeric protein causes an increase in outer membrane vesiculation that, by removing the toxic misfolded proteins, ameliorates the temperature sensitivity.
Fig. 7.
Increased expression of CpxARP and DegP proteins in the triple mutant (rfaC S151N degP G207D ompR Q223H) and increased production of outer membrane vesicle formation in the presence of chimeric LacI protein in the wild-type strain. (A) Protein expression levels of the CpxARP and DegP showing increased level of misfolded proteins in the triple mutant. All values are normalized to expression levels observed in wild-type strain containing the native LacI protein and are plotted on a log2 scale. Error bars represent SD. (B) Relative concentration of outer membrane vesicles produced by the wild-type strain at 44 °C in the presence of either native LacI or chimeric LacI. Asterisk indicates statistical significance at p = 0.05. Values are normalized to those observed for the wild-type strain containing the native LacI protein. Error bars represent SD.
Discussion
Plasmids and phages are important sources of raw material for the evolution of novel genes in microorganisms (Nakamura et al. 2004; Pal et al. 2005; Homma et al. 2007; Treangen and Rocha 2011; Wiedenbeck and Cohan 2011; Jerlstrom Hultqvist et al. 2018). They are not only sources of horizontal gene transfer and of genetic variation in the host but any recombination between these elements and host genes can result in evolutionary innovations. In this study, we observed how phage DNA could contribute to formation of novel genes by fusing with a bacterial gene. Our results show that a 23-amino acid fragment when fused with a truncated version of the host DNA-binding protein LacI results in the ability to suppress the temperature-sensitive phenotype of a mutant bacterium. To explain how this occurs, we also had to define the underlying cause of the temperature-sensitive phenotype. Our results show that a combination of mutations in four genes (rfaC, degP, ompR, and aas) generates the temperature sensitivity. A common denominator for these genes is that they encode proteins involved in membrane function: rfaC and aas encode enzymes involved in membrane biogenesis, ompR encodes a protein that is a transcriptional regulator of chaperones and outer membrane proteins, and degP encodes a protein that is both a chaperone and a protease for outer membrane proteins OmpC, OmpF, OmpA, and LamB. Loss-of-function mutations in rfaC result in an unstable outer membrane at 37 °C (Coleman and Deshpande 1985). Although the exact nature of the contribution of gene aas to the temperature-sensitive phenotype is unclear, given its role in phospholipid turnover, it is conceivable that mutations in this gene further affect the membrane integrity. Furthermore, instability in the outer membrane has been shown to result in accumulation of misfolded OMPs (Sen and Nikaido 1991; de Cock and Tommassen 1996; Bogdanov and Dowhan 1999; Bulieris et al. 2003) and this misfolding is expected to be exacerbated due to inactivation of the DegP protein. Finally, a mutation in gene ompR is also needed. We show that the observed mutation in ompR by itself results in differential regulation by the OmpR protein with a 6-fold increase in expression levels of ompF (supplementary fig. 10, Supplementary Material online). We also observed increased expression levels of the CpxR, CpxA, and CpxP proteins in the triple mutant at (rfaC S151N degP G207D ompR Q223H) 37 °C. These proteins are involved in sensing misfolded proteins in periplasm, inner membrane, and outer membrane (Raivio et al. 1999, 2013) indicating greater accumulation of misfolded proteins in the temperature-sensitive mutant. Additionally, overexpression of wild-type allele of degP on a plasmid suppresses temperature sensitivity in the quadruple mutant at 37 °C. Overall, these data are highly suggestive with the idea that the four mutations result in inhibition of growth because of accumulation of misfolded proteins.
How can the chimeric protein remove the growth inhibition caused by accumulation of misfolded proteins? Based on the data, we propose that the chimeric LacI protein suppresses the temperature sensitivity by localizing to the outer membrane where it induces the formation of outer membrane vesicles that in turn increases secretion of the toxic misfolded proteins. Outer membrane vesicles are known to be beneficial to bacterial cells due to their ability to remove toxic products, and they have been shown to provide benefits under various stress conditions, including nutrient limiting conditions (Li et al. 2016; Lin et al. 2017), presence of antibiotics (Kulkarni et al. 2015; Kim et al. 2018), and cold-stress (Frias et al. 2010). However, it is also possible that the observed increase in outer membrane vesicle formation in the presence of the chimeric LacI protein is only correlational, and that the chimeric LacI protein is suppressing temperature sensitivity via a different mechanism; for example, the association of the chimeric protein with the outer membrane might be stabilizing the outer membrane, and hence allowing growth of the mutant at nonpermissive temperatures.
Our results also demonstrate that despite the addition of 23-amino acid insert at the C-terminal of the truncated LacI protein, the chimeric LacI protein was still functional as a DNA-binding protein. Thus, even though the protein is relocalized and is interacting with the outer membrane, there is sufficient amount of the protein in the cytoplasm to still allow for it to function as a DNA-binding repressor and inducer-responsive (IPTG) regulatory protein.
Two still unanswered questions are as follows: 1) why the chimeric protein localizes to the outer membrane? and 2) how the chimeric protein stimulates formation of outer membrane vesicles? With regard to the former question, although an analysis of 40 amino acids at the C-terminal of the chimeric protein does not show any predicted transmembrane region (supplementary table 5, Supplementary Material online, prediction done using the TMHMM tool v .2.0), one possibility is that the 23-amino acid stretch acts as a recognition signal allowing translocation of the chimeric protein from the inner membrane to the outer membrane. With regard to the second question, there are different possibilities. Outer membrane vesicles are formed by bulging and pinching off the outer membrane, thus, any protein that affects the fluidity and bending of the outer membrane could affect the rate of formation of outer membrane vesicles. As the chimeric LacI protein is associated with the outer membrane, it can possibly affect the cross-linking of different components of the outer membrane changing fluidity or the bending of the outer membrane, hence effecting outer membrane vesicle formation.
Finally, from a broader evolutionary perspective, our results show how a new function—modulation of outer membrane vesicle formation—can emerge from fusion of a short piece of phage DNA with a native host gene. The chimeric protein can modulate a process that is present across all domains of life and it can provide the host with an advantage under several different growth conditions. The new function is not dependent on the original function (DNA binding) of the LacI protein, and conversely, the fusion event does not influence the native functions of LacI. Thus, the chimera has acquired a new function while maintaining the native function, highlighting how gene fusion can generate novel functions from existing genes without a necessary trade-off.
Materials and Methods
Screening of Temperature-Sensitive Mutants of S. Typhimurium with Metagenomic DNA Libraries
The temperature-sensitive S. Typhimurium mutants used in this study were generated by chemical mutagenesis and have been described previously (Schmid et al. 1989). Each mutant was further characterized based on whether it could grow at temperatures of 37, 40, and 43 °C. The preparation of environmental metagenomic libraries has been described in (Jerlstrom Hultqvist et al. 2018). Briefly, different environmental samples were first enriched for bacteriophage DNA by filtering the samples through a 0.22-µm filter, treating the filtrate with FeCl3 (1 mg Fe/l final concentration), and collecting the flocculating viral particles by filtering with polycarbonate membrane filters. These samples were then concentrated using Amicon UltraCel-30k and were then DNAase treated. After this, these samples were used for DNA extraction. The extracted DNA was then amplified using illustra Ready-To-Go GenomiPhi v3 or HY DNA amplification kit (GE Healthcare) as per the manufacturers protocol, digested with enzyme StuI, and ligated to a high-copy number plasmid pCA24N, –gfp. Each temperature-sensitive mutant was transformed with five different environmental libraries by electroporation (Supplementary Table 1a in Jerlstrom Hultqvist et al. [2018]). The transformants were recovered in lysogeny broth (LB) containing 100 µM of IPTG for a period of 1 h, after which the transformants were plated on LB agar plate containing 15 µg/ml chloramphenicol and 100 µM IPTG. The plates were incubated at appropriate nonpermissive temperatures for a maximum of 2 weeks.
Identification of Causal Genes Underlying the Temperature-Sensitive Phenotype
The mutations causing temperature sensitivity were identified by performing P22 transduction assays. P22 lysate was first generated on a wild-type S. Typhimurium strain by mixing 1 ml of P22 stock (1×106 pfu/ml) with 200 µl of an overnight grown culture of bacteria. The lysis was allowed to take place overnight at 37 °C. The next day 200 µl of chloroform was added to the mixture to kill surviving cells. This was centrifuged and the supernatant was used as the lysate to transduce the temperature-sensitive mutant. For transduction, 2 µl of the lysate was mixed with 100 µl of overnight grown culture of the mutant, and incubated with shaking at 30 °C for 2 h. The mixture was then plated on LB agar plates and was incubated at appropriate nonpermissive temperatures, for 24 h. Transductants and the parental temperature-sensitive mutants were both whole-genome sequenced to identify the mutation causing temperature sensitivity. Whole-genome sequencing was performed by generating the libraries using the NexteraXT kit. Samples were dual-indexed, pooled together, and run on Illumina’s Miseq using the Miseq reagent kit v2 (2×250 cycle). Analysis of the fastq files obtained from Miseq sequencing was performed using CLC genomics Workbench version 8. SNP calling and structural rearrangements were both assessed using this tool. Target genes were then identified by looking at the corrected mutations in the transductants as compared with the original temperature-sensitive strain. Previously described λ-red recombineering technique was employed to insert candidate mutations (Datsenko and Wanner 2000; Warsi et al. 2018).
Identification of Different Mechanisms Underlying the Removal of Temperature Sensitivity
To investigate the different routes for suppressing temperature sensitivity, we first selected mutants of the temperature-sensitive strain that were able to grow at 37 °C. Selection was performed by plating five independent 1 ml overnight grown cultures (∼109 cells) of the temperature-sensitive strain and incubating the plates at 37 °C for 24 h. The temperature-sensitive strain and three mutants were whole-genome sequenced to identify the compensatory mutations. Secondly, we transformed the temperature-sensitive strain with the ASKA plasmid library, which is a collection of individual ORFs from Escherichia coli cloned onto a plasmid and placed under an IPTG-inducible promoter (Kitagawa et al. 2005). This allowed us to identify genes, which when overexpressed, could suppress the temperature sensitivity.
Generating Different Constructs of the Chimeric LacI/FruR Protein and Library Construction
To identify the features of the chimeric LacI/FruR protein that were necessary for rescuing the temperature-sensitive strain, different genetic constructs were generated. In each case, suitable cloning primers were used, such that each primer consisted of sequences for recognition by restriction enzymes (XbaI and KpnI) at the 5′-end followed by sequence required for amplification of the different fragments of the chimeric lacI gene. After PCR amplification of the desired construct, the PCR products were purified, treated with DpnI (to remove plasmid template), digested using 2 µl of each restriction enzyme for 1 h, repurified, and used for ligation. The vector used in ligation was a modified version of the pCA24N –gfp plasmid that consisted of addition of the XbaI and KpnI restriction sites in the original cloning site on the plasmid. Besides the restriction digestion, the vector was also dephosphorylated for a period of 30 min using the Fast AP (Antartic phosphatase) enzyme. Ligations were carried out overnight at 16 °C, after which these were purified and transformed in E. coli cells by electroporation.
The library consisting of randomized 23-amino acid sequences inserted at the C-terminal of a truncated LacI protein was also generated in the same manner as above, however, using specific reverse primers for PCR amplification. These reverse primers consisted of three specific regions that included sequences for recognition by restriction enzymes (KpnI) at the 5′-end, followed by a region consisting of a stop codon and 69 randomized bases (N), which was then followed by sequences required for amplification from suitable position in lacI gene. Four independent PCR reactions were carried out and mixed together before proceeding for ligation. Four independent ligations were carried out, were then mixed together, and then used for transformations in wild-type E. coli strain. Controls were also carried out using ligation reaction consisting of only the digested vector or digested product that allowed the calculation of the overall diversity of the library, which was in the range of ∼2×106.
In each of the above cases, the clone products were then isolated and transformed into an S. Typhimurium strain consisting of the deletion of the DNA-restriction system, but having a functional DNA-modification system. The transformants were selected overnight on chloramphenicol plates (15 µg/ml), after which these were used for isolation of the cloned products and in subsequent transformation of the quadruple temperature-sensitive mutant.
Measurement of Expression Level Changes Using qPCR and Proteomics
To investigate the changes in gene expression levels qPCR analysis was performed. For each strain, overnight cultures were diluted 1:100 in 10 ml of salt-free LB media and were grown at 44 °C to an optical density (OD600) value of 0.2. 1 ml of cells were then mixed with 1 ml of RNA protect reagent, vortexed, and kept on ice for 5 min. These were centrifuged and the pelleted cells were then used for RNA extraction. RNA extraction was performed using the RNeasy Mini Kit (Qiagen) as per the manufacturer’s protocol. The extracted RNA was then treated with DNase using the Turbo DNA-free kit (Ambion) as per the manufacturer’s protocol, after which the RNA was run on a 1% agarose gel for visual inspection. About 500 ng of RNA (quantified using the Qubit RNA BR assay kit) was used for cDNA preparation using the High Capacity Reverse Transcription Kit (Applied Biosystems). PerfecTa Sybr Green SuperMix (Quanta Biosciences) was used to perform the RT-qPCRs. The transcript abundance measured for the different genes was normalized to the geometrical mean of the levels obtained for house-keeping genes cysG and hcaT. Three biological replicates and three technical replicates were used in each case. The averages mentioned are based on biological replicates. Error bars represent SD. We only considered differences of at least 2-fold to be biologically significant.
Whole-cell proteomics was performed to identify the regulatory changes induced in the bacteria by either the chimeric LacI protein or the native LacI protein. Overnight grown cultures of the triple mutant consisting of three out of the four target mutations (rfaC S151N degP G207D ompR Q223H) and of the wild-type strain, carrying either the empty vector or the chimeric lacI gene, were diluted 1:100 in 10 ml of LB media (which contains 1% of NaCl) and were grown at 37 °C to an optical density (OD600) value of 0.2. The cells were centrifuged and then washed with ice-cold Phosphate buffered saline three times. The cell pellet was then stored at −80 °C and was then analyzed by the Proteomics Core facility at University of Gothenburg. Briefly, the samples were first lysed and the total protein concentration in the lysates was measured. An aliquot of each sample was reduced, alkylated, and then digested with trypsin. The digested peptides were then chemically labeled with a tandem mass tag reagent. The tandem mass tag-based relative quantification was performed largely as described previously (Jerlstrom Hultqvist et al. 2018), with the following modifications: the sample was prefractionated via a basic-pH reversed-phase HPLC into eight fractions and each fraction was analyzed using the 90-min LC-MS method. In total, more than 24,000 of peptides were identified at the false discovery rate of 1%, making up 2741 Salmonella LT2 protein quantified across the samples (supplementary table 6, Supplementary Material online). Error bars represent SD. We only considered differences of at least 2-fold to be biologically significant.
Membrane Integrity Analysis, Outer Membrane Isolation, and Western-Blot Analysis
To investigate the effect of the identified mutations on membrane integrity, the temperature-sensitive mutant was grown at 37 °C in no-salt LB media either with or without divalent cations (Mg2+ and Ca2+), or with a noncationic osmolyte (sucrose). The salts (MgSO4 and CaCl2) were used at final concentrations of 10 and 100 mM, whereas sucrose was used at concentrations of 0.5%, 2%, and 5%. Susceptibility to large antibiotics rifampicin and vancomycin was also measured to determine if the chimeric LacI protein was affecting membrane integrity. Susceptibility to antibiotic tetracycline was used as a control in these assays. This was done for the wild-type S. Typhimurium strain in the presence of either the native LacI or the chimeric LacI.
To investigate the localization of the chimeric LacI protein, FLAG tags (DYKDHDGDYKDHDIDYKDDDDKL) were attached at the N-terminal of both the chimeric LacI and the native LacI protein. The fused constructs were expressed from the IPTG-inducible high-copy number pCA24N plasmid. We first confirmed that attaching the FLAG tag to the chimeric LacI protein did not affect its rescuing ability of the temperature-sensitive mutant at the nonpermissive temperature. After this, different fractions of the cell were isolated using a differential precipitation protocol. For each strain, cultures were grown in 50 ml of no-salt LB containing 100 µM IPTG and 15 µg/ml chloramphenicol to an optical density (OD600) of 0.5. Cells were harvested by centrifuging at 4149 x g for 10 min. The supernatant was discarded and the pellet was resuspended in 1 ml of 100 mM Tris–HCl pH 8.0, 20% sucrose solution. Resuspended cells were incubated on ice for 10 min and then centrifuged at 2400 x g for 10 min. The pellet was resuspended in 1 ml of 100 mM Tris–HCl pH 8.0, 20% sucrose, 10 mM EDTA solution. Lysozyme was added to a final concentration of 100 µg/ml and incubated on ice for 20 min, after which MgSO4 was added to a final concentration of 20 mM. This solution was then spun down on a 100-µl cushion of 50% sucrose solution for 10 min at 3500 x g. The supernatant contains the periplasm and was stored separately. The pellet was resuspended in 100 mM Tris–HCl pH 8.0, 20% sucrose, 10 mM EDTA solution. This was followed by addition of DNaseI and RNaseA to a final concentration of 5 µg/ml each. The cells were then disrupted by five freeze-thaw cycles using dry ice-ethanol mixture to freeze the solution and then placing it at 37 °C water bath to thaw. After this, the lysate was incubated on ice for 2 h, followed by centrifugation at 16,200 x g for 35 min. The supernatant contains cytoplasm fraction of the cell and was stored separately. The pellet was resuspended in 500 µl of 20 mM sodium phosphate buffer, pH = 7. The resuspension was centrifuged at 16,200 x g for 25 min. The pellet was resuspended in 1 ml 0.5% sarcosyl in 20 mM sodium phosphate buffer, pH = 7 and was incubated at room-temperature for 30 min. This selectively precipitates the outer membrane. The solution was centrifuged at 16,200 x g for 35 min. The supernatant contains inner membrane fraction and was stored separately. The pellet containing the outer membrane was then washed twice by resuspending it in 1 ml of 0.5% sarcosyl in 20 mM sodium phosphate buffer, pH = 7 and centrifuging at 16,200 x g for 30 min. The pellet was then resuspended in 1 ml of Lamelli sample buffer. 7.5 µl of sample is run on the ready to use Mini-PROTEAN TGX gels (BioRAD: Cat No. 456-1095). Western blot was performed by first transferring the samples from the gel to a nitrocellulose membrane using a Trans-Blot Turbo Transfer Pack (BioRAD: Cat No. 1704158). The membrane was blocked using 5% BSA solution prepared in Tris-buffered saline containing 1% Tween-20 (TBST) for 2 h. It was then incubated with the antiFLAG antibody solution (prepared in 5% BSA solution prepared in TBST) for 1 h. The antiFLAG antibody is conjugated to the horseradish peroxidase enzyme that allows for chemiluminescent detection of tagged bands. After the incubation, the membrane was washed thrice with TBST solution for 15 min each. Bands were visualized using the ECL western-blotting chemiluminescent reagents (Amersham RPN 2109) that contain the substrate for the peroxidase enzyme.
Quantitative and Qualitative Assessment of Formation of Outer Membrane Vesicles and Rescue of Temperature Sensitivity in the ΔnlpI Mutant
Outer membrane vesicles were isolated from 50 ml of overnight grown cultures in no-salt LB at 44 °C. The cultures were centrifuged at 3,800 x g for 2 h, after which the supernatant was filtered twice using a 0.22-µM polyethersulfone filter. About 20 µl of filtrate was spotted on LB agar plates to confirm that the supernatant was free of bacterial cells. The supernatant was centrifuged at 38,000 x g for 2 h, which allows pelleting of the outer membrane vesicles. The vesicles were resuspended in 200 µl PBS. The concentration of the outer membrane vesicles was obtained by nanoparticle tracking analysis performed using Nanosight LM10. Each sample was diluted 1:100 for the analysis. Three biological replicates were used in each case. To determine if knocking out the nlpI gene (which increases vesicle formation) can remove temperature sensitivity of the temperature-sensitive mutant, a ΔnlpI knock out mutant was constructed using λ-red recombineering technique (Datsenko and Wanner 2000). Growth was then compared between the temperature-sensitive mutant, and the temperature-sensitive mutant with the nlpI gene deleted using a BioscreenC analyzer at OD600. Three replicates were used for each strain.
Strain Construction for Identification of the Native Function of the Chimeric LacI Protein and Measurement of Fitness Cost of Expressing the Chimeric LacI Protein
To identify whether or not the chimeric LacI protein had retained its native DNA-binding function, a S. Typhimurium strain was used that consisted of a LacO operator sequence present upstream of a syfp gene, which codes for the yellow flourescent protein (YFP). The LacO sequence is the DNA-binding sequence for the LacI protein, which when bound to it represses the transcription of downstream genes. We inserted the chimeric lacI gene and the native lacI gene, both on a plasmid vector (pBAD) and on the chromosome; in each case, under the control of the paraBAD (promoter of araBAD operon). The insertion on the chromosome was performed using previously described (Datsenko and Wanner 2000; Warsi et al. 2018) λ-red recombineering technique and involved replacing the araBAD genes with the gene of interest. In each case, induction from paraBAD was done using an L-arabinose concentration of 0.1%.
To measure fitness costs associated with expression of the chimeric LacI protein, strains with the native lacI gene and the chimeric lacI gene inserted on the chromosomes were used. Exponential growth rates were measured in Tryptone broth containing 0.05% L-arabinose using a BioscreenC analyzer at OD600, with measurements taken every 4 min. KaleidaGraph software was used to calculate the maximum exponential growth rate from the OD600 data, using the OD values between 0.02 and 0.08. For each strain, overnight cultures were diluted 1:1,000, which was then used for measurement of exponential growth rate. Four biological replicates were used in each case. Relative exponential growth rate was calculated as the ratio between the exponential growth rate of the wild-type strain expressing the chimeric LacI protein and that of the wild-type strain expressing the native LacI protein. Error bars represent SD.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors greatly acknowledge the critical reading of the article by members of Dan Andersson group. They also like to thank the editor and two reviewers for their constructive and useful comments. Quantitative proteomic analysis was performed at the Proteomics Core Facility of Sahlgrenska Academy, University of Gothenburg by Egor Vorontsov and Carina Sihlbom. The Proteomics Core Facility is grateful to the Inga-Britt and Arne Lundbergs Forskningsstiftlese for the donation of the Orbitrap Fusion Tribrid MS instrument. The authors are also grateful to Prof. Anca Segall (San Diego State University) for providing the temperature-sensitive strains. This work was supported by grants from the Swedish Research Council and the Knut and Alice Wallenberg foundation to D.I.A.
Author Contributions
O.W. and D.I.A. designed the experiments. O.W., M.K., S.S., and J.J.H. carried out the experiments. O.W. and D.I.A. wrote the article with input from all coauthors.
References
- Altschmied J, Delfgaauw J, Wilde B, Duschl J, Bouneau L, Volff JN, Schartl M.. 2002. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161(1):259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthonneau E, Mirande M.. 2000. A gene fusion event in the evolution of aminoacyl-tRNA synthetases. FEBS Lett. 470(3):300–304. [DOI] [PubMed] [Google Scholar]
- Betz JL, Fall MZ.. 1988. Effects of dominant-negative lac repressor mutations on operator specificity and protein stability. Gene 67(2):147–158. [DOI] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE.. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489(7417):513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W.. 1999. Lipid-assisted protein folding. J Biol Chem. 274(52):36827–36830. [DOI] [PubMed] [Google Scholar]
- Bridges CB. 1935. Salivary chromosome maps. J Hered. 26(2):60–64. [Google Scholar]
- Bulieris PV, Behrens S, Holst O, Kleinschmidt JH.. 2003. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 278(11):9092–9099. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Vicoso B, Russo CA, Swenor B, Clark AG.. 2015. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 112(40):12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M.. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet. 14(9):645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman WG Jr, Deshpande KS.. 1985. New cysE-pyrE-linked rfa mutation in Escherichia coli K-12 that results in a heptoseless lipopolysaccharide. J Bacteriol. 161(3):1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL.. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 97(12):6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cock H, Tommassen J.. 1996. Lipopolysaccharides and divalent cations are involved in the formation of an assembly-competent intermediate of outer-membrane protein PhoE of E. coli. EMBO J. 15(20):5567–5573. [PMC free article] [PubMed] [Google Scholar]
- Digianantonio KM, Hecht MH.. 2016. A protein constructed de novo enables cell growth by altering gene regulation. Proc Natl Acad Sci U S A. 113(9):2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R, Marais GA, Loppin B.. 2012. Repeated evolution of testis-specific new genes: the case of telomere-capping genes in Drosophila. Int J Evol Biol. 2012:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarappa SM, Potdar AA, Sahoo S, Sankar S, Fox PL.. 2018. Metabolic origin of the fused aminoacyl-tRNA synthetase, glutamyl-prolyl-tRNA synthetase. J Biol Chem. 293(49):19148–19156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr AD, Remigi P, Rainey PB.. 2017. Adaptive evolution by spontaneous domain fusion and protein relocalization. Nat Ecol Evol. 1(10):1562–1568. [DOI] [PubMed] [Google Scholar]
- Frias A, Manresa A, de Oliveira E, Lopez-Iglesias C, Mercade E.. 2010. Membrane vesicles: a common feature in the extracellular matter of cold-adapted Antarctic bacteria. Microb Ecol. 59(3):476–486. [DOI] [PubMed] [Google Scholar]
- Homma K, Fukuchi S, Nakamura Y, Gojobori T, Nishikawa K.. 2007. Gene cluster analysis method identifies horizontally transferred genes with high reliability and indicates that they provide the main mechanism of operon gain in 8 species of gamma-proteobacteria. Mol Biol Evol. 24(3):805–813. [DOI] [PubMed] [Google Scholar]
- Jenkin TJ. 1933. Interspecific and intergeneric hybrids in herbage grasses. J Genet. 28:205–264. [Google Scholar]
- Jerlstrom Hultqvist J, Warsi O, Soderholm A, Knopp M, Eckhard U, Vorontsov E, Selmer M, Andersson DI.. 2018. A bacteriophage enzyme induces bacterial metabolic perturbation that confers a novel promiscuous function. Nat Ecol Evol. 2:1321–1330. [DOI] [PubMed] [Google Scholar]
- Jones CD, Custer AW, Begun DJ.. 2005. Origin and evolution of a chimeric fusion gene in Drosophila subobscura, D. madeirensis and D. guanche. Genetics 170(1):207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, Lazarte JMS, Kim J, Seo JS, Kim JH, et al. 2018. Outer membrane vesicles from beta-lactam-resistant Escherichia coli enable the survival of beta-lactam-susceptible E. coli in the presence of beta-lactam antibiotics. Sci Rep. 8(1):5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H.. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12(5):291–299. [DOI] [PubMed] [Google Scholar]
- Klein G, Kobylak N, Lindner B, Stupak A, Raina S.. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem. 289(21):14829–14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp M, Gudmundsdottir JS, Nilsson T, Konig F, Warsi O, Rajer F, Adelroth P, Andersson DI.. 2019. De novo emergence of peptides that confer antibiotic resistance. MBio. 10(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T.. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453(7197):885–890. [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Nagaraj R, Jagannadham MV.. 2015. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol Res. 181:1–7. [DOI] [PubMed] [Google Scholar]
- Li J, Azam F, Zhang S.. 2016. Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ Microbiol. 18(11):3850–3866. [DOI] [PubMed] [Google Scholar]
- Liberek K, Georgopoulos C.. 1993. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci U S A. 90(23):11019–11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X.. 2017. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 8(1):14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Langley CH.. 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260(5104):91–95. [DOI] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ.. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 63(2):545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy AD, Hardie DG.. 1984. Fatty acid synthase – an example of protein evolution by gene fusion. Trends Biochem Sci. 9(2):60–63. [Google Scholar]
- Muller HJ. 1936. Bar duplication. Science 83(2161):528–530. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T.. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 36(7):760–766. [DOI] [PubMed] [Google Scholar]
- Nasvall J, Sun L, Roth JR, Andersson DI.. 2012. Real-time evolution of new genes by innovation, amplification, and divergence. Science 338(6105):384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ.. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 37:1372–1375. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Leblanc SK, Price NL.. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol. 195(12):2755–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Popkin DL, Silhavy TJ.. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 181(17):5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MB, Kapur N, Isaacson DR, Lindroos P, Sharpe C.. 1989. Genetic analysis of temperature-sensitive lethal mutants of Salmonella typhimurium. Genetics 123(4):625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen K, Nikaido H.. 1991. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 173(2):926–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1925. The effects of unequal crossing over at the bar locus in Drosophila. Genetics 10(2):117–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi S, Vaara M, Helander IM, Viljanen P, Makela PH.. 1984. New Salmonella typhimurium mutants with altered outer membrane permeability. J Bacteriol. 159(2):704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Rocha EP.. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7(1):e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsi OM, Andersson DI, Dykhuizen DE.. 2018. Different adaptive strategies in E. coli populations evolving under macronutrient limitation and metal ion limitation. BMC Evol Biol. 18(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM.. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 35(5):957–976. [DOI] [PubMed] [Google Scholar]
- Wood R, Erwin DH.. 2018. Innovation not recovery: dynamic redox promotes metazoan radiations. Biol Rev. 93(2):863–873. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA.. 2006. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci U S A. 103(47):17608–17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yoshizawa S, Sun Y, Huang Y, Chu X, Gonzalez JM, Pinhassi J, Luo H.. 2019. Repeated evolutionary transitions of flavobacteria from marine to non-marine habitats. Environ Microbiol.21(2):648–666. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, Li X, Ding Y, Yang S, Wang W.. 2008. On the origin of new genes in Drosophila. Genome Res. 18(9):1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.