Abstract
Variants in genes encoding synaptic adhesion proteins of the neuroligin family, most notably neuroligin-4, are a significant cause of autism spectrum disorders in humans. Although human neuroligin-4 is encoded by two genes, NLGN4X and NLGN4Y, that are localized on the X-specific and male-specific regions of the two sex chromosomes, the chromosomal localization and full genomic sequence of the mouse Nlgn4 gene remain elusive. Here, we analyzed the neuroligin-4 genes of numerous rodent species by direct sequencing and bioinformatics, generated complete drafts of multiple rodent neuroligin-4 genes, and examined their evolution. Surprisingly, we find that the murine Nlgn4 gene is localized to the pseudoautosomal region (PAR) of the sex chromosomes, different from its human orthologs. We show that the sequence differences between various neuroligin-4 proteins are restricted to hotspots in which rodent neuroligin-4 proteins contain short repetitive sequence insertions compared with neuroligin-4 proteins from other species, whereas all other protein sequences are highly conserved. Evolutionarily, these sequence insertions initiate in the clade eumuroidea of the infraorder myomorpha and are additionally associated with dramatic changes in noncoding sequences and gene size. Importantly, these changes are not exclusively restricted to neuroligin-4 genes but reflect major evolutionary changes that substantially altered or even deleted genes from the PARs of both sex chromosomes. Our results show that despite the fact that the PAR in rodents and the neuroligin-4 genes within the rodent PAR underwent massive evolutionary changes, neuroligin-4 proteins maintained a highly conserved core structure, consistent with a substantial evolutionary pressure preserving its physiological function.
Keywords: autism spectrum disorders, synaptogenesis, pseudoautosomal, NLGN4, neural cell-adhesion molecule, eumuroidea
Introduction
Eutherian genomes differ extensively in the number and sizes of their chromosomes, but share a common mode of sex determination based on two specialized chromosomes, X and Y (Graves 2016). Generally, the two sex chromosomes have different sizes and gene contents but always include in their telomeric regions a “pseudoautosomal” region (PAR). PARs contain a cluster of shared genes that recombine like autosomes and are referred to as pseudoautosomal genes. The PAR mediates proper pairing of X- and Y-chromosomes during male meiosis and is thought to facilitate sex chromosome separation. Based on current hypotheses of sex chromosome evolution, the X- and Y-chromosomes arose from autosomes, underwent a specialization, and, thus, lost their ability to recombine over their full length, leaving the PARs as the only recombining segments. The hypothesis of “addition and attrition” in sex chromosome evolution posits that new translocations of autosomal genes were necessary to keep the PAR intact in the face of constant “eroding” forces resulting from a lack of Y-chromosomal recombination (Wallis et al. 2008; Wilson and Makova 2009a; Graves 2016). Although genes localizing to the PAR are identical on the X- and Y-chromosomes, the precise border between PAR and X/Y-specific genes appears to vary between eutherians. As a result, a few genes in a sex chromosome-specific segment of the X-chromosome are neither subject to pseudoautosomal recombination nor to X-chromosomal inactivation. These X- and Y-homologs (“gametologs”) can be distinguished by sequence variations and/or differences in their expression pattern, and include in the human genome, for instance, the genes encoding neuroligin-4 and amelogenin (Salido et al. 1992; Iwase et al. 2003; Wilson and Makova 2009b; Bellott et al. 2014; Johansson et al. 2016; Raudsepp and Chowdhary 2016).
Comprehensive information on the human- and mouse-genome sequences has now been available for almost two decades, and most of the human and mouse genes have been chromosomally mapped and sequenced. Whereas the human PAR has been fully sequenced up to the telomers (Ross et al. 2005), the mouse PAR has evaded full sequencing owing to the presence of extensive stretches of repetitive elements that vary considerably even between closely related laboratory mouse strains (Harbers et al. 1986, 1990; Kipling et al. 1996). Accordingly, only the steroid sulfatase (Sts; Salido et al. 1996), N-acetylserotonin O-methyl-transferase (Asmt; Kasahara et al. 2010), and midline-1 genes (Mid1; Palmer et al. 1997) have so far been mapped to the mouse PAR based on fluorescence in situ-hybridization (FISH). Further complicating the functional analysis of the mouse PAR, the massive prevalence of repetitive sequences and the scarcity of relevant genome sequence data have largely prevented functional analyses of mouse homologs of human PAR and gametologous genes by gene targeting, so that the only relevant mouse models at present were derived from targeting of Mid1 (Lancioni et al. 2010) and gene-trapping of Nlgn4 (Jamain et al. 2008).
The present study was triggered by our futile attempts to identify the gene for mouse neuroligin-4 (Nlgn4) and its chromosomal location in publicly available data sets. Neuroligin-4 belongs to the highly conserved neuroligin family of neural cell-adhesion proteins that are localized to the postsynaptic side of interneuronal synapses, interact transsynaptically with presynaptic neurexins, and play a key role in the functional maturation and validation of synapses (Krueger-Burg et al. 2017; Südhof 2017). In mice, neuroligins exhibit a synapse-type-specific localization and function, so that Nlgn1 is primarily present in excitatory glutamatergic synapses (Song et al. 1999; Chubykin et al. 2007; Poulopoulos et al. 2009), Nlgn2 in inhibitory and cholinergic synapses (Graf et al. 2004; Varoqueaux et al. 2004), Nlgn3 in subsets of both excitatory and inhibitory synapses (Budreck and Scheiffele 2007; Tabuchi et al. 2007), and Nlgn4 in glycinergic and GABAergic synapses (Hoon et al. 2011; Hammer et al. 2015). Changes in the human NLGN1 (OMIM 600568), NLGN3 (OMIM 300336), and NLGN4X (OMIM 300427) genes were repeatedly associated with nonsyndromic monogenic forms of autism spectrum disorders (ASDs) and other neuropsychiatric diseases (Jamain et al. 2003; Avdjieva-Tzavella et al. 2012; Steinberg et al. 2012; Xu et al. 2014; Bourgeron 2015; Kilaru et al. 2016; Nakanishi et al. 2017; Parente et al. 2017). As neuroligins determine the formation and composition of synapses, their genetic perturbation causes functional synaptic defects and consequent changes in the functionality of neuronal networks (Südhof 2008). Supporting the notion of a causal involvement of NLGN gene variants in the etiology of ASDs, corresponding genetically modified mouse models with altered Nlgn3 and Nlgn4 genes exhibit complex synaptic defects and ASD-related behavioral deficits, such as repetitive behavior, restricted interests, and reduced social interaction (Tabuchi et al. 2007; Jamain et al. 2008; Rothwell et al. 2014; Nakanishi et al. 2017). In this context, however, Nlgn4/NLGN4 poses a puzzling problem. NLGN4 variants are the most frequent ASD-linked neuroligin variants, with more than 50 mutations—mostly causing a loss of function—identified in individuals with ASD so far (Südhof 2008; Zhang et al. 2009; Kleijer et al. 2014). Strikingly, though, and unlike those of neuroligin-1 to -3, the sequences of mouse and human neuroligin-4 have diverged substantially during evolution (Bolliger et al. 2008; Jamain et al. 2008). Furthermore, the mouse draft genome sequence contains no neuroligin-4 gene, confounding an assessment of its evolutionary history. Consequently, the relevance of data on mouse Nlgn4, for instance, obtained with a gene-trap Nlgn4 knock-out mouse (Jamain et al. 2008; Hammer et al. 2015), for human NLGN4 and associated ASD cases, have been questioned based on the argument that the apparent lack of evolutionary conservation indicates that mouse Nlgn4 is of subordinate functional relevance.
In the present study, we defined the sequence, evolution, and chromosomal location of the mouse Nlgn4 gene. We show that mouse Nlgn4 maps to the PAR and has been subject to the accumulation of increased GC content and repetitive sequences. Furthermore, we found that most rodent neuroligin-4 proteins are virtually identical to their human homolog, that excessive sequence variations are only evident in the clade eumuroidea, which includes the mouse and hamster families, and that at the same time, additional dramatic evolutionary changes occurred in the organization and gene content of the eumuroidea PARs. Our data allow us to draft a map of the laboratory mouse PAR and its gene content, and show that a selected set of genes, including Nlgn4, withstood a massive “erosion” of the eumuroidea PARs, indicating that they must have been under substantial evolutionary pressure and hence of functional relevance.
Results
Chromosomal Localization of the Mouse Neuroligin-4 Gene
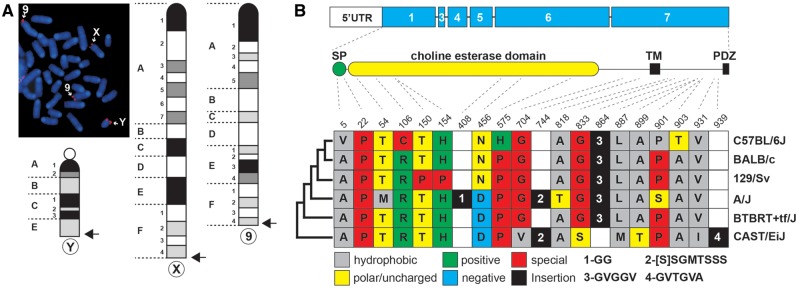
Despite the emergence of next-generation sequencing technologies and vast amounts of mouse-genome sequences becoming available over the last decade, the mouse Nlgn4 gene is still absent from the current mouse-genome build. As the Nlgn4 gene clearly exists in mice (Jamain et al. 2008; “Nlgn4*,” Bolliger et al. 2008), it must be present on a yet unsequenced area of the mouse genome. To address the enigma of the mouse Nlgn4 gene, we first examined its chromosomal localization. We performed FISH using a probe that was derived from a phage clone containing part of the mouse Nlgn4* gene (Bolliger et al. 2008). Fluorescence signals were detected in the telomeric regions of the X- and Y-chromosomes and of chromosome 9 (fig. 1A), reminiscent of FISH-staining results for Asmt (Kasahara et al. 2010). Thus, the murine Nlgn4 gene, is localized in the PAR in contrast to its human homologs, NLGN4X and NLGN4Y.
Fig. 1.
Nlgn4 gene and Nlgn4 protein in mice. (A) A phage clone comprising parts of the Nlgn4 sequence (“Nlgn4*,” Bolliger et al. 2008) was used for fluorescence in situ-hybridization (FISH) to determine the physical localization of Nlgn4 in the mouse genome. FISH-positive signals were found in the pseudoautosomal region of both sex chromosomes as well as in a telomeric region of chromosome 9. Arrows indicate relative chromosomal locations of FISH signals. (B) Schematic overview illustrating the protein-coding region on the different exons (blue) as well as the relative position of the signal peptide (SP), cholinesterase-like domain, transmembrane domain (TM), and PDZ-binding motif (PDZ) within the NLGN4 protein. The protein sequence of five selected laboratory mouse strains and the wild-derived strain CAST/EiJ were aligned, and synonymous and nonsynonymous amino acid variations were highlighted using the one-letter amino acid code. Numerals represent varying amino acid insertions found in several strains. The relative evolutionary relationship of the different mouse strains is depicted on the left side of the colored boxes (not drawn to scale).
Complete Rodent Neuroligin-4 Gene Sequences Reveal a Common Pattern of Diversity
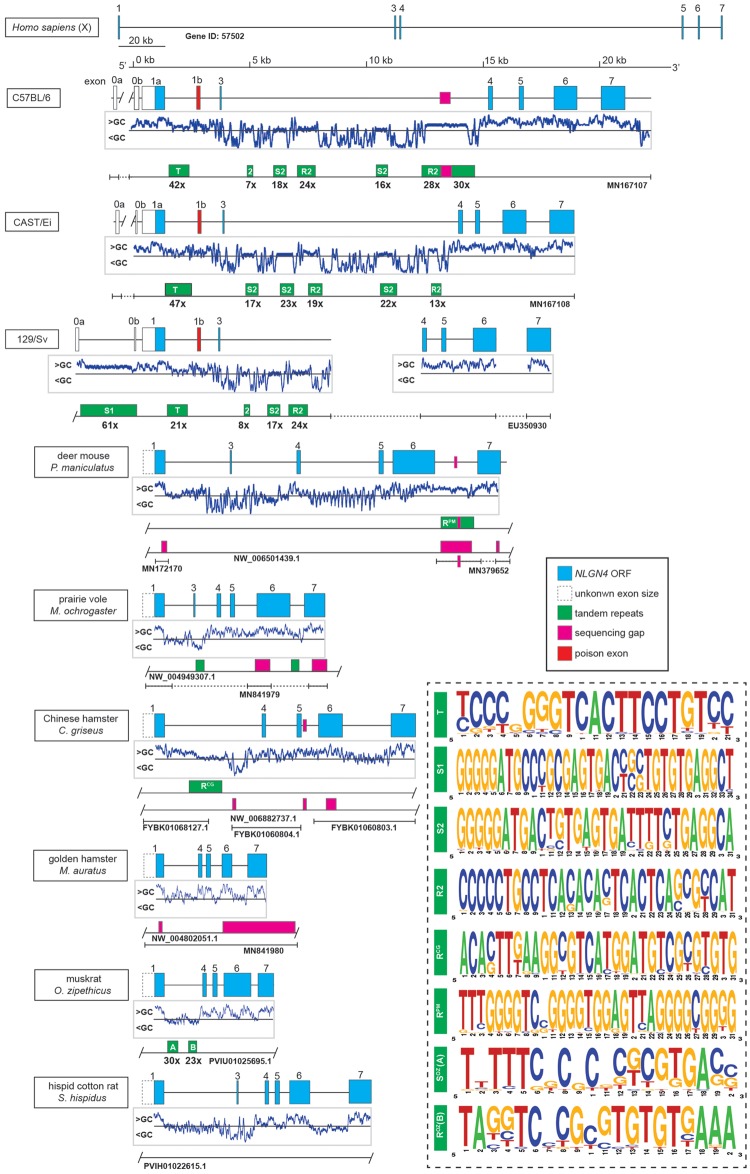
Based on the previously published but incomplete mouse Nlgn4 gene sequence (Nlgn4*) and limited cDNA sequence information (Bolliger et al. 2008; Jamain et al. 2008), we hypothesized that the lack of information on the mouse Nlgn4 gene could be partially due to GC-rich sequences and/or extensive stretches of repetitive elements. Therefore, we pursued a number of polymerase chain reaction (PCR) and sequencing strategies using isolated genomic DNA of selected laboratory mouse strains as a template to obtain a complete sequence of the mouse Nlgn4 gene. The obtained fragments were directly sequenced without further subcloning to avoid possible sequence rearrangements in bacterial hosts that may be caused by repetitive elements. After refining suitable PCR and sequencing strategies, we were able to fully sequence the mouse Nlgn4 gene in the wild-derived, inbred strain CAST/EiJ (Mus musculus castaneus), covering exons 1–7. Our coverage of the common lab strain C57BL/6J is >98% leaving only 400 bases of a highly repetitive tandem repeat unsequenced located in intron 3 (fig. 2). With a total of seven exons, Nlgn4 covers a genomic region of ∼24 kb (C57BL/6J) (fig. 2). Several introns contain large insertions of repetitive elements that share similar motifs (fig. 2 andsupplementary data sheet 1, Supplementary Material online). In addition to our previous reports (Bolliger et al. 2008; Jamain et al. 2008), we confirmed two 5′-UTR exons, which we named exon 0a and exon 0b, accordingly (fig. 2). Immediately, downstream of the ATG-bearing exon (now exon 1a), we identified an alternatively spliced “poison” exon (exon 1b) whose integration in the nascent mRNA leads to a premature in-frame translational stop (supplementary data set 1, Supplementary Material online, page 6: 18A–C and 21A and B). The presence of exon 1b, however, is not an artifact, because quantitative PCR analyses with different primer combinations revealed that the levels of transcripts with or without exon 1b are comparable, likely contributing further to considerable lower expression levels of Nlgn4 protein in mice as compared with other neuroligin family members (Varoqueaux et al. 2006). Size and base variations of this poison exon were also present in some of the mouse strains sequenced by the mouse-genome projects judged by the conservation of suitable splice acceptor and donor sites (Lilue et al. 2018; Thybert et al. 2018) (sequence references in supplementary data sheet 4, Supplementary Material online).
Fig. 2.
Organization of the neuroligin-4 gene in different mouse strains and hamster species. Schematic overview of the Nlgn4 gene in three mouse strains (C57BL/6J, MN167107; CAST/EiJ, MN167108; 129/Sv, EU350930), six hamster species (see supplementary data sheet 2, Supplementary Material online) and the human NLGN4X gene. The genes are drawn to scale with the exception of human NLGN4X. Neuroligin-4 coding regions are depicted in blue, open boxes represent upstream exon/s encoding the 5′-UTRs (in laboratory strains), partially dashed boxes (in hamster species) represent undetermined exon 1 sizes due to yet unspecified splice acceptor sites. Below, the mouse and hamster genes profiles of the relative GC contents are depicted indicating relatively high GC content (GC>50%) primarily in and around all exons. Additionally, initial sequencing gaps (magenta) from public sequence sources as well as repeat clusters (green) are placed on a separate bar. Bars with T-shaped ends indicate that beyond this point no sequence information is available, whether from public sources or our own sequencing efforts for a given stretch of DNA. Bar ends with a slash indicate that the given DNA information from a public source is only partially displayed. As not all repeat stretches are consistent in nature, only tandem repeats are displayed with their repeat numbers (for a general analyses of all repeats, nonmotif, or tandem, cf. supplementary data sheet 1, Supplementary Material online). The consensus motif of different tandem repeats is presented as a weblogo in the accompanying dashed box indicating the relative frequency of each base. The identity and relative position of the repeats in all lab mouse strains were comparable, only the repeat count varied. All mouse repeats were calculated using information from C57BL/6J, except for the repeat S1, which was calculated from the 129/Sv sequence deposited previously (Bolliger et al. 2008). An overview of the sequence sources can be found in the supplementary text file 3, Supplementary Material online.
We identified numerous sequence differences in the Nlgn4 gene between C57BL/6J and CAST/EiJ strains. Some of these were due to base changes in the coding region, as had been shown for the Nlgn4 cDNAs of BALB/c and C57BL/6J mice (Bolliger et al. 2008). Additionally, insertions, deletions, and copy number variations of the tandem repeats obscured the overall sequence similarity between mouse strains. As single base variations in human NLGN4 genes were reported to cause ASD, we extended our analysis to the laboratory strains A/J and BTBRT+tf/J, which were both suggested to exhibit ASD-related behavioral phenotypes (Moy et al. 2007). We found a considerable number of nonsynonymous base changes that resulted in amino acid changes (fig. 1B). Some of the changes included insertions of amino acids, likely due to sequence duplications on the DNA level (fig. 1B andsupplementary fig. S1, Supplementary Material online). To assess the potential damaging character of single missense changes within the amino acid sequence, we superimposed these changes onto the human NLGN4X protein sequence (Polyphen-2, HumVar output; cf. supplementary data set 2, Supplementary Material online). However, the tested substitutions appeared to have no damaging impact. In contrast to numerous amino acid changes found in Nlgn4 (supplementary table S1, Supplementary Material online), the Nlgn3 protein sequences in C57BL/6J and CAST/EiJ mice differed only by a single amino acid from those present in the strains A/J (P11S) and BTBRT+tf/J (C637R) (accession numbers, see supplementary data sheet 2, Supplementary Material online).
Diversity and Phylogeny of NLGN4 Genes in Rodents and Primates
The low-sequence identity between human NLGN4X and mouse Nlgn4* proteins (59.7%; Bolliger et al. 2008; Jamain et al. 2008) prompted us to expand our Nlgn4 gene analysis to other available rodent genomes and to the NLGN4 genes of primates in order to assess whether this evolutionary divergence is a general feature of neuroligin-4 genes in different species. Typically, the neuroligin-4 coding sequence is distributed over six exons. Exon 1 encodes the translation start site and contains 472 bases from the start ATG to the downstream splice donor site. The alternatively spliced sequence A1 that is encoded by exon 2 commonly present in NLGN1 and NLGN3 is absent in NLGN4 and Nlgn4 genes. Nlgn4 exons 3–7 consist of 60 (i.e., splice site A2), 153, 186, 790, and 850 bases, respectively. Various species have predicted or validated additional exons preceding exon 1, including mice (Bolliger et al. 2008), which we here refer to as exon 0a and exon 0b (this report is the first one on the presence of exon 0b) (fig. 2). To date, about 20 genomes of the rodent lineage have been published in the NCBI database. In the current builds of these genomes, Nlgn4 is not fully sequenced and in some cases not even annotated. Based on sequence comparison, we fully annotated the NLGN4 genes in guinea pig, degu, darma-mole rat, chinchilla, naked mole, blind mole, 13-lined squirrel, and Chinese hamster, and we outlined the NLGN4 gene in marmots (fig. 3). Further, we sequenced missing parts of NLGN4 in the prairie vole, golden hamster, and deer mouse (fig. 2 andsupplementary data sheet 2, Supplementary Material online). Regarding their genomic organization—for example, individual exon size, total gene size, and neighboring genes—the genomic NLGN4 coding sequences of many rodents were almost identical to their human homologs (fig. 3). Strikingly, however, major differences in genome organization were found in the infraorder myomorpha, comprising the “mouse-like” families of hamsters and mice (figs. 2 and 3 and cf. supplementary fig. S2, Supplementary Material online, to appreciate rodent diversity). Here, we observed several key changes in the NLGN4 gene, specifically 1) fundamental changes in the presumptive signal peptide (size and amino acid sequence), 2) a collapse of the total gene size (from 113–338 to <20 kb), 3) a massive expansion of repeats within introns, which appear to be species-specific, and 4) expansion of the coding region, resulting in a calculated molecular weight of the translated product of up to 130 kDa (summarized in table 1). The differences within the hamster family (cricetidae) are particularly noteworthy. The prairie vole, for instance, displays a possibly critical change within the neuroligin-4 coding region that is anticipated to affect neuroligin/neurexin binding at the protein level (Araç et al. 2007). Indeed, PolyPhen-2 analysis of the amino acid change Y385H within the conserved region Q379 to D386 in human NLGN4X indicates a potentially damaging change (supplementary data set 2, Supplementary Material online). The golden hamster has the most deteriorated NLGN4 gene due to the loss of exon 3 and partial sequence deletions in exons 6 and 7, but the expression of the corresponding translational product of 636 amino acids needs to be demonstrated.
Fig. 3.
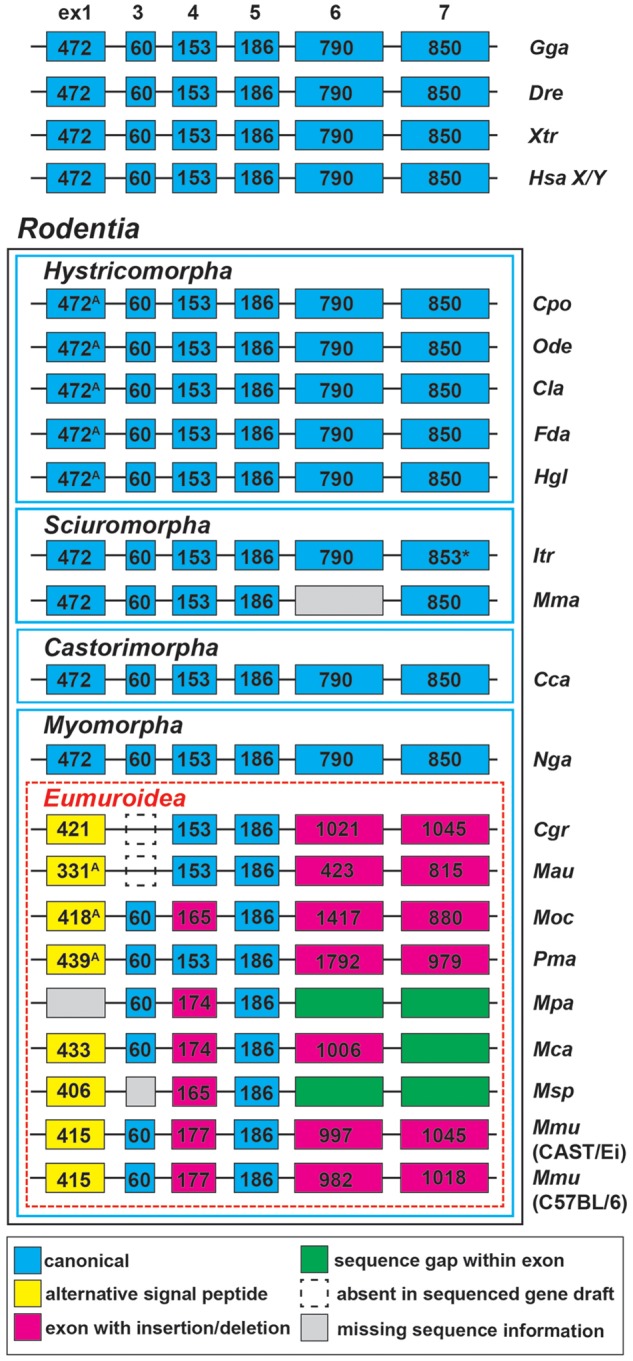
Variations of NLGN4 exon sizes in selected vertebrates and rodents. NCBI deposited DNA sequence information of 18 rodent species and four selected vertebrate species including human (Homo sapiens, Hsa; NLGN4X Gene ID: 57502, NLGN4Y: 22829), chicken (Gallus gallus, Gga; 428006), zebrafish (Danio rerio, Dre; LOC108709161/LOC108707974), and clawed frog (Xenopus tropicalis, Xtr; NLGN4a: 561122, NLGN4b: 100147967) were used for comparison with available rodent NLGN4 coding sequences. Notably, the selected vertebrate species as well as guinea pig (Cavia porcellus, Cpo; 100714631/106025955), degu (Octodon degus, Ode; 101591647), chinchilla (Chinchilla lanigera, Cla; 102015543), darma-mole rat (Fukomys darmarensis, Fda; 104848054), naked mole (Heterocephalus glaber, Hgl; 101715794), 13-lined squirrel (Ictidomys tridecemlineatus, Itr; 101974873), American beaver (Castor canadensis, Cca; LOC109703453, LOC109679926, LOC109674687), and the upper-galilee mole (Nannospalax galili, Nga; 103725635) display identical exon sizes. Within the clade eumuroidea (i.e., the rodent infraorder myomorpha excluding the family spalacidae; to which Nga belongs), the neuroligin-4 coding sequence encoded by exon 1 consistently decreases whereas the sizes of exons 6 and 7 increase. NLGN4 exon 3 is absent in both, the golden hamster (Mesocricetus auratus, Mau; 101837222) and Chinese hamster (Cricetulus griseus, Cgr; NW_006882737.1) gene sequences; however, it is still present in the prairie vole (Microtus ochrogaster, Moc; 102001548) and deer mouse (Peromyscus maniculatus, Pma; 102911426). Within the genus Mus size variations are present in nearly all exons despite exons 3 and 5 (Mus pahari, Mpa: NW_018393795.1; Mus caroli, Mca: NW_018389566.1; Mus spretus, Msp: LVXV01024335.1). *, insertion of an additional single base triplet; A, possibility of alternative start codons. Labeling of exon boxes reflects respective base count. Blue frames separate species of different rodent infraorders, a dashed red frame outlines the clade eumuroidea. An extended version with a more refined clustering of additional families within the clade eumuroidea can be found in supplementary figure S2, Supplementary Material online.
Table 1.
Characteristics of the NLGN4 Gene, cDNA, and Protein in Human and Rodents.
| Species | Gene | cDNA |
Protein |
|||
|---|---|---|---|---|---|---|
| Size (kb) | GC (%) | GC3 (%) | AA | pI | MW (kDa) | |
| Homo sapiens (X chr.) | 259 | 52.9 | 66.5 | 836 | 5.65 | 94.0 |
| Homo sapiens (Y chr.) | 219 | 51.3 | 63.8 | 836 | 5.71 | 94.2 |
| Castor canadensis | >243a | 55.7 | 71.8 | 836 | 5.70 | 94.0 |
| Cavia porcellus | 274 | 55.5 | 74.2 | 836b | 5.77 | 94.1 |
| Chinchilla lanigera | 305 | 54.5 | 71.3 | 836b | 5.70 | 94.0 |
| Cricetulus griseus (E) | 11 | 73.3 | 98.2 | 888c | 6.22 | 95.1 |
| Fukomys darmarensis | 257 | 58.9 | 81.4 | 836b | 5.63 | 93.8 |
| Heterocephalus glaber | 277 | 56.5 | 76.3 | 836b | 5.71 | 93.9 |
| Ictidomys tridecemlineatus | 113 | 62.5 | 91.6 | 837 | 5.75 | 93.8 |
| Mesocricetus auratus (E) | 4.7 | 74.7 | 85.2 | 635c | 10.45 | 65.4 |
| Microtus ochrogaster (E) | 7.5 | 74.0 | 89.3 | 1,043 | 5.73 | 107.0 |
| Mus musculus (E) | 18 | 75.2 | 96.8 | 954 | 5.83 | 98.2 |
| Nannospalax galili | 266 | 52.5 | 65.6 | 836 | 5.82 | 94.1 |
| Octodon degus | 338 | 58.4 | 81.4 | 836b | 5.66 | 93.8 |
| Ondatra zibethicus (E) | 5.2 | 71.7 | 86.9 | 853c | 6.26 | 91.3 |
| Peromyscus maniculatus (E) | 15 | 70.2 | 87.4 | 1,202 | 5.62 | 129.3 |
| Sigmodon hispidus (E) | 9.3 | 71.0 | 94.5 | 876c | 6.14 | 94.0 |
note.—Total gene sizes are rounded to full kb starting from the protein coding ATG to the translational stop codon. GC content, frequency at any position in the cDNA; GC3 content, frequency of either G or C in the third position; AA, number of amino acids in each protein; pI, isoelectric point; MW, calculated molecular weight; E, species member of the clade eumuroidea.
Minimal estimated gene size due to a sequencing gap.
Alternative ATG might increase amino acid count to 841.
Absence of exon 3.
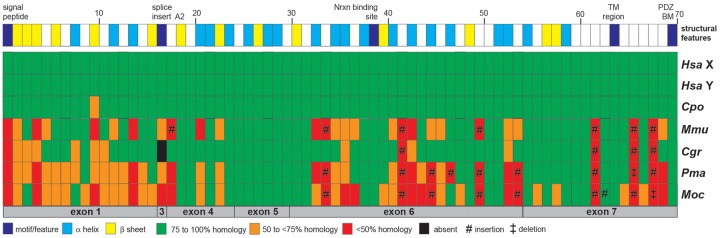
Based on the crystal structure of human NLGN4X (Leone et al. 2010, http://www.rcsb.org/pdb/protein/Q8N0W4, last accessed December 2019), we performed a sequence alignment with amino acid sequences of human NLGN4X/Y and guinea pig, mouse, prairie vole, deer mouse, and Chinese hamster NLGN4 in order to assess whether the observed insertions might affect the protein structure (fig. 4). The resulting alignment was split up into 70 segments, key features (signal peptide, splice site A2, neurexin-binding domain, transmembrane domain, and PDZ-binding motif) were assigned, and alpha helices and beta strands were inferred based on the human NLGN4X protein structure. The neuroligin-4 homologs retained highest homology within segments 19–32, encoded by exons 4 and 5, and the beginning of exon 6, at the neurexin-binding site (segments 38–40), within segments 55–61, and around the transmembrane region (segments 63–65). Low-sequence homology and sequence insertions were consistently observed in identical segments throughout all hamster and mouse species as compared with the human and guinea pig sequences.
Fig. 4.
Quality of structural differences among mouse and hamster NLGN4 proteins. The NLGN4 protein sequences of human NLGN4X/Y (Hsa X, XP_016885179.1; Hsa Y, XP_011529732.1), guinea pig (Cpo, inferred from its gene, Gene ID: 100714631), mouse (Mmu, CAST/EiJ, MH051930), prairie vole (Moc), Chinese hamster (Cgr), and deer mouse (Pma) (all three hamster NLGN4 protein sequences are included in supplementary text file S1, Supplementary Material online) were aligned and structural features assigned based on the crystal structure information of human NLGN4X (Leone et al. 2010). The alignment was split up into 70 segments. The segments reflected assigned structural features such as the signal peptide, alpha helices, beta strands, the transmembrane region (TM), and the PDZ-binding motif (PDZ BM), but also insertions and low homology regions. Individual segments may vary in size. A detailed annotation of the sequence alignment can be found in Text File S1. The color-code of the heat map reflects the degree of similarity of all species (including human NLGN4Y) relative to the reference sequence of human NLGN4X, which by default is represented entirely by green boxes. Guinea pig NLGN4 was chosen as an example outside the clade eumuroidea due to its high similarity to both human homologs. All segments are displayed relative to their position within the respective exon (gray boxes on the bottom of the heat map). Alignment was performed using MultAlin with default settings (“Identity-0-1”).
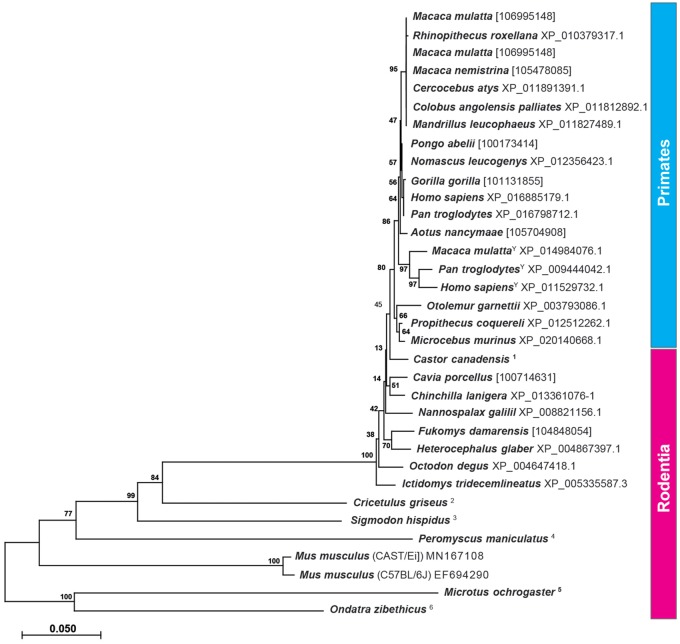
The strikingly high similarity of neuroligin-4 protein within most rodents (schematic overview: fig. 3; phylogenetic tree: fig. 5), with the notable exceptions in eumuroidea (fig. 5), raises the question as to whether gametologous isoforms that display the human X/Y divergence also exist in other species. Most genomes in the databases are from females with a few exceptions. We included in our calculation available information of NLGN4Y from primates (fig. 5) and found that it forms a separate branch at the base of the Old and New World monkeys, indicating that lemurs likely do not distinguish neuroligin-4 as a gametologous (X/Y) gene pair.
Fig. 5.
Phylogenetic analysis of NLGN4 in primates and rodents. The evolutionary history of neuroligin-4 in primates and rodents based on the translated protein sequences was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 1.50308039 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 34 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 1,444 positions in the final data set. Evolutionary analyses were conducted in MEGA X. Only protein sequences that included splice insert A2 have been used with the exception of Cricetulus griseus and Ondatra zibethicus in which splice site A2 (exon 3) is absent. In other species, which lacked available protein sequences including splice insert A2, exon 3 had been identified from the respective gene and translated into the final sequences. In these cases, the Gene ID is provided instead. For the primate species, Macaca mulatta, Pan troglodytes, and Homo sapiens, available NLGN4X and NLGN4Y sequences were included. Y, NLGN4Y protein sequence. For the following species, protein sequences were inferred from available genomic DNA: 1, LOC109703453, LOC109674687, and LOC109679926; 2, NW_006882737.1, FYBK01068127.1, FYBK01060804.1, FYBK01060803.1; 3, PVIH01022615.1; 4, NW_006501439.1, MN172170, MN379652; 5, NW_004949307.1, MN841979; 6, PVIU01025695.1.
The retention of gametologs seems to not merely be necessary for the maintenance of a comparable transcript level of functionally redundant copies of a former pseudoautosomal gene. Whereas the overall expression in males and females of total NLGN4X is not different (Werling et al. 2016), changes are found in a limited number of brain regions and nonneuronal tissues (supplementary data set 3, Supplementary Material online, based on RNAseq data from the Human Protein Atlas, Fagerberg et al. 2014).
Erosion of the PAR Region in the Infraorder Myomorpha
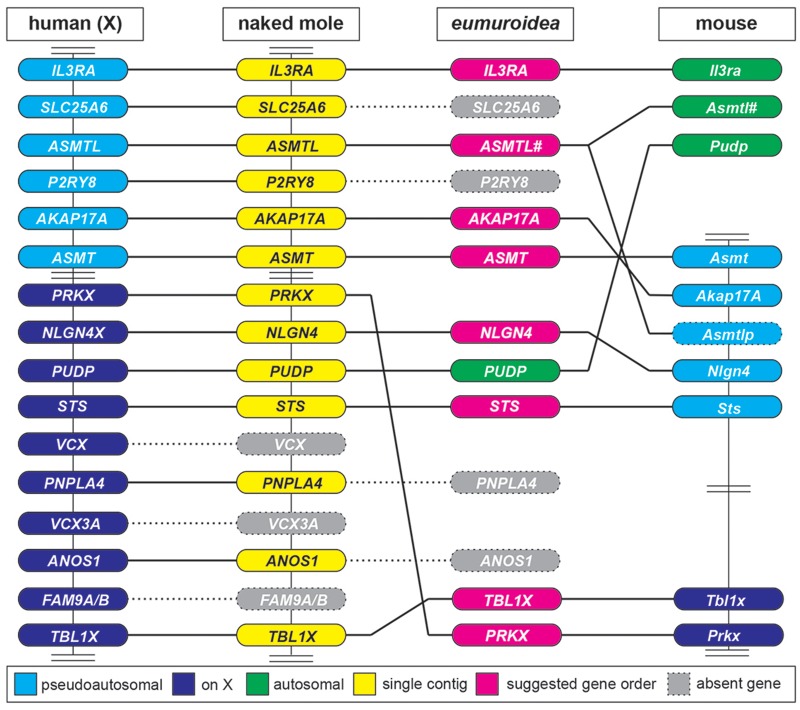
In subsequent analyses, we examined whether the striking changes in the NLGN4 gene within the infraorder myomorpha are exclusive to neuroligin-4 or a reflection of more general changes in the respective genomes. Based on our FISH data indicating a PAR localization of Nlgn4 in mice (fig. 1A) and in view of the fact that the PAR boundary in laboratory mice was mapped to an intron of the Mid1 gene (Palmer et al. 1997), we constructed an overview of the mouse PAR by integrating all information available from genomic builds of 16 rodent species and 5 other mammalian reference species available from the NCBI database (www.ncbi.nlm.nih.gov/gene, last accessed December 2019, Gene ID and information, see supplementary data sheet 3, Supplementary Material online, fig. 6, and fully detailed in supplementary fig. S3, Supplementary Material online). Many of the corresponding genes have not yet been sufficiently annotated and lack formal chromosomal assignments, but given the phylogenetic relationship of the rodent order (Wilson and Reeder 2005), we were able to identify several key changes in the PAR that distinguish rodents from other mammalian orders, such as primates (e.g., human) or carnivores (e.g., cat, dog). The X-specific and PAR regions of cats, dogs, elephants, dolphins, and humans showed an overall similar organization with almost identical gene content and order. However, cats, dogs, elephants, and dolphins as well as all rodents consistently lacked the genes FAM9A/B, VCX, VCX3A, and CXorf28. Additionally, ARSF was absent among all presumptive rodent PAR genes. Except for the absence of these genes, the PAR organization and gene content in most rodents were comparable with those of dogs, cats, elephants, dolphins, and humans (fig. 6 and supplementary fig. S3, Supplementary Material online).
Fig. 6.
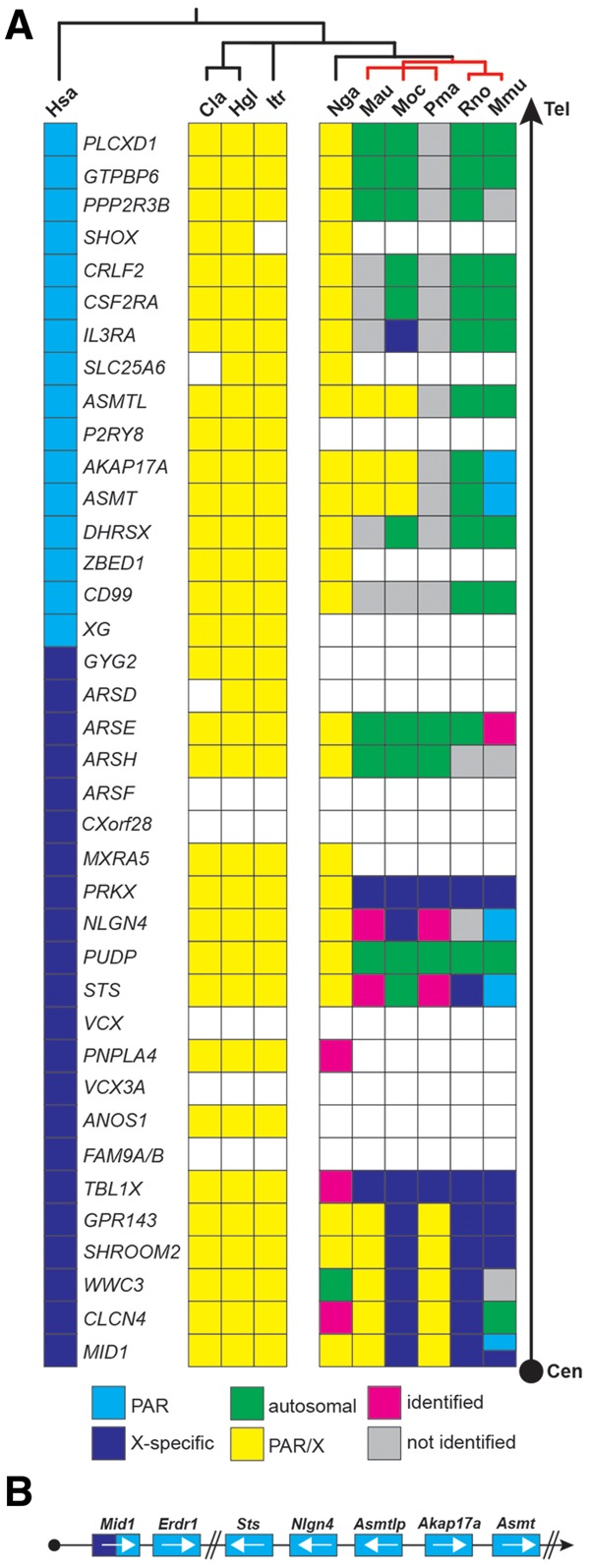
The PAR genes in rodents. (A) Schematic of the localization of the genes of the X-specific and pseudoautosomal region (PAR) of the human X-chromosome compared with selected rodents (for full annotation, see supplementary fig. S3, Supplementary Material online). All genes are listed starting with MID1, a gene that is known to harbor the PAR boundary in mice, to the most telomeric gene in humans, PLCXD1 (Tel, telomere; Cen, direction to centromere). The human PAR boundary, in contrast, is located between GYG2 and XG. As the PAR boundary in most rodents has not been determined yet, the genes are depicted in yellow without formal assignment to either X-specific (indigo) or PAR-specific regions (blue). Genes that are missing within an available contig are shown as open boxes indicating their possible absence from the respective PAR. Genes that have not been identified yet but are likely present in accordance with the overall evolutionary genomic comparison are shown as gray boxes. In case of translocation from pseudoautosomal genes to autosomes the genes are shown in green. Magenta boxes represent genes that were identified in the respective genomes but could not be clearly assigned due to the lack of flanking sequence information. Red bars represent genomes that belong to the eumuroidea clade. Cla, Chinchilla lanigera; Hgl, Heterocephalus glaber; Hsa, Homo sapiens; Itr, Ictidomys tridecemlineatus; Mau, Mesocricetus auratus; Mmu, Mus musculus; Moc, Microtus ochrogaster; Nga, Nannospalax galili; Pma, Peromyscus maniculatus; Rno, Rattus norvegicus. (B) Relative position of the PAR genes (not drawn to scale) in mouse including the mouse-specific Erdr1 gene downstream of Mid1 and the Asmtl pseudogene, Asmtlp. White arrows indicate the orientation of the respective genes on the contig and in the PAR.
Strikingly, within the infraorder myomorpha—and, here, more specifically within the clade eumuroidea, which together with the family of spalacidae is grouped as the super-family muroidea (Steppan et al. 2004; Steppan and Schenk 2017)—significant gene loss as well as multiple X-chromosomal and autosomal translocations were detected (summarized in fig. 7). These include the selective loss of ANOS1, PNPLA4, MXRA5, P2RY8, SLC25A6, and SHOX, as well as the partial loss of ARSD, GYG2, XG, and ZBED1. The relatively remote genes TBL1X and PRKX apparently converged to neighboring positions in a new X-specific location, whereas many other genes translocated to autosomes, including PUDP (causing NLGN4 and STS to converge as neighboring genes) and a gene segment encoding ARSH and ARSE in the hamster family. Further autosomal translocations occurred within certain sublineages/families. Autosomal identity was assumed based on comparisons of the gene content of the destination chromosome with the one found on the X-chromosome, which is justified because the gene content of the X-chromosome is relatively similar in most mammals due to the necessary gene dosage regulation.
Fig. 7.
Major rearrangements within the PAR and X-chromosome of the clade eumuroidea. This is a horizontal comparison of selected genes from the human X-chromosome (on the left side, including X-specific and PAR genes) with orthologs found in the naked mole (Heterocephalus glaber) and mouse genomes. The naked mole was chosen because all depicted genes in this particular species are found to localize on a single contig (NW_004624834.1). Several critical rearrangements are suggested at the node of the eumuroidea diversification, which itself is depicted as a hypothesized intermediate. VCX and FAM9A/B genes, both found on the human X-chromosome, are absent in rodents, further loss of ANOS1, PNPLA4, P2RY8, and SLC25A6 genes occurs at the node of the eumuroidea branch. The loss of SLC25A6 leads to the truncation of ASMTL. The distantly located genes TBL1X and PRKX recombine within the X-chromosome and have become neighboring genes. PUDP translocates to an autosome allowing STS to join NLGN4. In mice, Il3ra and Asmtl have translocated to different autosomes leaving a copy of Asmtl behind that has subsequently diverged into the pseudogene Asmtlp in the PAR of laboratory mice. Vertical bars connecting genes indicate that based on sequence information all genes have been shown to be present on one strand. Double bars represent gaps that exclude additional genes that are of no immediate relevance in this comparison. #, truncated version of ASMTL upon genomic recombination.
The Mouse PAR Retains Only Four of the Original PAR Genes
Over the past two decades, the genes localized to the mouse PAR have been mapped exclusively by FISH analyses. They include Sts (Salido et al. 1996), Asmt (Kasahara et al. 2010), and as shown here, Nlgn4 (fig. 1B). Furthermore, data on recombination events indicate that Asmt must be positioned between Sts and the telomer (Kasahara et al. 2010). Based on recent data on the expression of Y-chromosomal genes, Akap17a (or Sfrs17a) and Mid1 were added to this group of mouse PAR genes, albeit without further detailed explanations or data (Bellott et al. 2014).
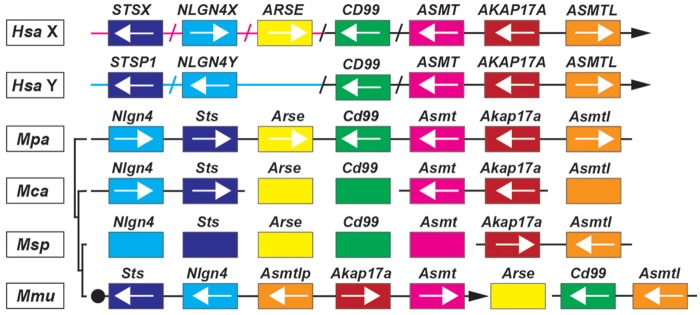
Whole-genome shotgun contigs (WGS) from laboratory mouse genomes, which were obtained from the Mouse Genomes Project (Lilue et al. 2018; Sanger Institute; http://www.sanger.ac.uk/science/data/mouse-genomes-project, last accessed December 2019, accession numbers listed in supplementary data sheet 4, Supplementary Material online) and represent sequencing reads of the Akap17a, Asmt, Nlgn4, and Sts genes, allowed us to reconstruct a substantial part of the mouse PAR, resulting in the following order: Sts–Nlgn4–Akap17a–Asmt (fig. 6B). The sequencing reads still include many unspecified stretches, mostly in parts that contain copy number variations of tandem repeats. Nevertheless, we identified a genomic region between the Nlgn4 and Akap17a genes that shows strong similarity to the Asmtl gene. However, a combination of mutated splice sites and in-frame variations, deletions, or insertions result in a loss of function of this gene, which should, thus, be named Asmtlp for “pseudo” Asmtl. Downstream of Mid1 and, therefore, formally in the PAR, the current mouse genomic build positions the Erdr1 gene. This gene seems to be mouse-specific since it cannot be found in any other species. On aggregate, our data show that the mouse PAR retains only four original genes that are found in other mammalian PARs, that is, Sts, Nlgn4, Akap17a, and Asmt. Additionally, a pseudogene version of Asmtl (Asmtlp) and the mouse-specific Erdr1 gene are found in laboratory mice, whereas Mid1 marks the PAR boundary. We cannot, however, formally exclude the presence of additional genes.
The Sanger Institute has recently added whole-genome sequences of M. pahari, M. caroli, and M. spretus to their database (Thybert et al. 2018) (references for identified PAR genes are included in supplementary data sheet 4, Supplementary Material online). A comparison of the PAR genes of laboratory strains with those other representatives of the genus Mus showed that the genome of the most distantly related mouse species, M. pahari, still contains Arse and Cd99 as well as a functional version of Asmtl in a single contig together with Nlgn4, Sts, Asmt, and Akap17a (fig. 8) indicating a more intact PAR. In addition, the M. caroli and M. spretus genomes contain a functional Asmtl gene in the immediate vicinity of the known PAR genes, indicating that the translocation of Cd99 and Asmtl to mouse chromosome 4 occurred evolutionarily most recently, leaving a copy of Asmtl in the PAR, which subsequently underwent partial destruction and inactivation to Asmtlp.
Fig. 8.
PAR gene shuffling in the genus Mus. Comparison of the gene order and localization in humans with those found in the genus Mus. Sequencing results from the Sanger Mouse Genomes Project (Thybert et al. 2018) helped us to partially resolve the relative localization of the PAR genes in different mouse species (Mus pahari, Mpa; Mus spretus, Msp; Mus caroli, Mca; Mus musculus, Mmu). Black bars (left side of schematic alignment) indicate the phylogenetic relationship of the different mouse species (not drawn to scale). PAR genes are depicted as colored boxes. Boxes on bars represent the presence of the respective genes found on a single contig and arrows their relative orientation. The closed arrowheads in Hsa and Mmu point to the telomer. Magenta- and blue-colored arrows reflect X- and Y-specific regions upstream of the human PAR boundary. Slashes are indicating the exclusion of additional human genes, which are absent in Mus, to allow a more simplified comparative overview.
Discussion
Almost two decades have passed since the first comparative analysis of the mouse genome was published (Mouse Genome Sequencing Consortium et al. 2002). The most recent update of the Genome Reference Consortium, GRCm38.p5 (2012), implies that most genes (coding or noncoding) have been annotated and their chromosomal localizations identified. Despite this progress, mouse Nlgn4 had escaped annotation prior to the present study. We previously published the sequence of the mouse neuroligin-4 protein and a partial analysis of the mouse Nlgn4 gene (“Nlgn4*” Bolliger et al. 2008; Nlgn4, Jamain et al. 2008). Triggered by the apparent divergence between the mouse Nlgn4 gene and its human NLGN4X/Y homologs, we conducted the present study to determine the chromosomal localization of Nlgn4 and to perform a comparative analysis of the Nlgn4 gene structure, sequence, and evolution.
Based on gene sequencing and comparative analyses of a substantial number of genomic sequences available for different mouse strains, rodents, and other vertebrate species, we describe an evolutionary scenario according to which the neuroligin-4 gene faced—and withstood—substantial challenges during its radiation through the rodent lineage. In essence, our study suggests three fundamental observations: 1) The mouse Nlgn4 gene is localized to the PAR, and its coding sequence differs notably from that of the neuroligin-4 gene of many other rodents, mainly due to insertions and deletions in the coding region that are observed even between closely related laboratory strains. Substantial sequence variations are further found in introns, which primarily differ in size due to copy number variations of tandem repeats. The mouse Nlgn4 gene is flanked immediately upstream by an Asmtl pseudogene and downstream by the Sts gene. 2) The considerable sequence variations between the human NLGN4X/Y variants and the mouse Nlgn4 gene are found not only in mice but also in a subset of rodents within the infraorder myomorpha, or more specifically in the clade eumuroidea. This clade comprises, among others, the hamster (cricetidae) and mouse families (muridae) (Steppan et al. 2004; Steppan and Schenk 2017). 3) The neuroligin-4 gene divergence within eumuroidea did not only affect the NLGN4 gene but has also largely impacted other genes such as STS, ASMTL, and CD99 as well as the gene content of the PAR itself.
Rapid Evolution of the Nlgn4 Gene in the Genus Mus and Divergence of Neuroligin-4 in the Clade Eumuroidea
Our analysis of the Nlgn4 gene in C57BL/6 and CAST/Ei mice revealed numerous variations in noncoding introns, but also in the coding regions of Nlgn4. Major contributors to these changes and to the changes in total gene size are copy number variations of tandem repeats. Among these is a 31mer repeat that was previously shown to contribute to polymorphisms in the PAR of laboratory mouse strains and that is also found in the subtelomeric regions of mouse chromosomes 4, 9, and 13 (Harbers et al. 1986, 1990), hence explaining our in situ-hybridization signal on chromosome 9. Virtually identical repeats were present in a 129/Sv genomic phage clone that we had used as a probe to localize the mouse Nlgn4 gene by FISH analysis (Acc. No. EU350930, Bolliger et al. 2008) and on a BAC clone that was used by others for the chromosomal assignment of Asmt (Kasahara et al. 2010). In both cases, hybridization signals in the PAR region of the X- and Y-chromosome were observed along with signals in the subtelomeric region of chromosome 9. As numerous synonymous and also nonsynonymous changes in mouse Nlgn4 cDNAs appear to be the rule, it is not possible to judge whether the changes observed in the A/J or BTBRT+t/J strains might actually affect neuroligin-4 function and cause aspects of the autistic-like behavior that characterizes these strains (Moy et al. 2007). Indeed, none of the observed changes in A/J or BTBRT+t/J Nlgn4 is immediately reminiscent of those that were identified in human patients with ASD, such as, for instance, the NLGN4 R87W (Zhang et al. 2009), NLGN4 R704C (Yan et al. 2005), or NLGN3 R451C variants (Jamain et al. 2003).
Our comparative approach revealed that among all species analyzed, considerable changes to the neuroligin-4 gene first appeared at the node where spalacidae separated from eumuroidea. Accordingly, Nannospalax galili retains the neuroligin-4 gene organization as found in humans and the rodent infraorders castorimorpha, hystricomorpha, and sciuromorpha. Instead, a substantial reduction of the gene size and an increase in the overall GC content and GC3 content in the coding region are observed in representatives of the hamster family (deer mouse, prairie vole, Chinese hamster, golden hamster), and in mice, where a generalization to the mouse family in toto is not yet possible because neuroligin-4 has not been identified in rats. Of note in this context, the early separation of the spalacidae ∼47 Ma and a very rapid radiation of the remaining families within the eumuroidea were suggested previously based on other genes (Steppan et al. 2004; Fang et al. 2014). Current data on the number of distinct mammalian species indicate that eumuroidea comprise roughly two-thirds of all rodents, which is more than a quarter of all mammals taken together (Burgin et al. 2018).
The changes observed in the neuroligin-4 coding region in hamsters and mice resulted in a broad diversity of orthologs of the human NLGN4X/Y variants. The corresponding differences fall into two major categories, that is, single amino acid changes and insertions or deletions. Insertions were already identified as a striking feature in the first description of mouse neuroligin-4 (Bolliger et al. 2008; Jamain et al. 2008). Closer sequence inspection indicates that many insertions may have arisen in a two-step process, where short genomic DNA sequences initially duplicated and subsequently separated. Based on our comparison with the human neuroligin-4 protein structure, the insertions seem to occur in parts of the protein that likely tolerate additional “amino acid loops,” so that they might only marginally affect the overall structure of the protein and leave its physiological function intact. Indeed, the inserts in mouse neuroligin-4 do not appear to affect its synaptic localization or physiological function (Bolliger et al. 2008; Hoon et al. 2011; Hammer et al. 2015). Likewise, the substantial gain of amino acid content in the prairie vole and in the deer mouse neuroligin-4 might not change its function either, but this assumption needs to be tested for all neuroligin-4 orthologs that substantially diverge from its common structure.
Available sequencing data for the muskrat as well as our own sequencing data for the Chinese and golden hamster species revealed genomic loss of exon 3 (encoding splice site A2) of the neuroligin-4 gene. Additionally, the gene variant in the golden hamster contains a premature stop codon, which likely causes a loss of function (cf. NLGN4 loss of function in human patients with ASD, Lawson-Yuen et al. 2008). Given the link between loss of NLGN4X function and ASD, the latter finding is intriguing, because among the highly social rodents, particularly the golden (Syrian) hamsters have a solitary life style (Gattermann et al. 2001).
The PAR in Mouse
Our physical mapping results using FISH showed that the mouse Nlgn4 (fig. 1B) gene is localized to the mouse PAR, similar to what was previously shown for the Sts (Salido et al. 1996) and Asmt genes (Kasahara et al. 2010). In C57BL/6 mice, Mid1 was shown to harbor the PAR boundary, proximal to which the mouse-specific Erdr1 gene is localized (NCBI gene annotation). Analysis of whole-genome sequencing reads available from the Sanger Institute Mouse Genome Project (Lilue et al. 2018) revealed that Akap17a is localized upstream of Asmt, and that Nlgn4 and Akap17a are separated by an Asmtl pseudogene. Our integration of physical data and sequence information is in accordance with a previous study (Bellott et al. 2014), in which Mid1, Nlgn4, Asmt, Akap17a (Sfrs17a), and Sts are mentioned as likely mouse PAR candidate genes. In our study on the fate of potential rodent PAR genes, we tracked down all ancestral PAR genes found in humans downstream of MID1 to PLCXD1, irrespective of their potential gametologuous or pseudoautosomal character. Our current data indicate that of the ancestral PAR genes, only Sts, Nlgn4, Akap17a, and Asmt are left in the mouse PAR. All other genes have either been lost or recombined to X-specific or autosomal regions. The most recent recombination events must have included Arse, Asmtl, and Cd99, which are still clustered on a single contig together with Sts, Nlgn4, Akap17a, and Asmt in M. pahari. Whether these genes translocated independently, one at a time, to autosomes in ancestors of M. musculus (including the laboratory strains) or translocated as an entire segment in M. pahari needs to be determined. In rat, the mouse PAR homologs Akap17a, Asmt, and Asmtl have translocated to chromosome 12 and Sts recombined to an X-specific region, whereas Nlgn4 has not yet been identified and Erdr1 is absent.
Evolutionary Conservation of Neuroligin-4 and the Mouse PAR
In a previous study that included a partial analysis of the mouse Nlgn4 gene, we reported preliminary evidence for an apparently rapid evolution of the Nlgn4 gene (Bolliger et al. 2008). We concluded that Nlgn4 function is under less stringent evolutionary constraints than other neuroligin genes, Nlgn1-3, and that Nlgn4 might thus be relatively dispensable. The present study shows that mouse Nlgn4 is indeed evolving extremely rapidly, but that the evolutionary diversification of Nlgn4 is restricted to specific parts of the gene, such as hot spots for insertions in the coding region or enormous variations in intronic repetitive elements, whereas other parts of the gene, including distinct parts of its coding region, remain conserved. Thus, it appears that the mouse Nlgn4 gene along with the entire PAR in mice has been subject to substantial “erosion” by the invasion of repetitive sequences without changing the expression of a functional neuroligin-4 protein. This evolutionary process caused the destruction of several PAR genes and the rescue of multiple others to new chromosomal locations by translocation. Nlgn4 is one of the very few PAR genes that has withstood this erosion and remained functional in the PAR, which indicates a substantial evolutionary pressure to preserve Nlgn4 protein function.
In more general terms, the erosion of the PAR in eumuroidea reveals key aspects regarding the importance and functionality of the affected genes. It seems that evolution has preserved beneficial genes by rescuing them via translocation to autosomes or X-specific regions. Important genes that still reside in the PAR are coping by tolerating variations as long as they are not disadvantageous or destroy the gene—perhaps with the exception of neuroligin-4 in the golden hamster, which may have been evolutionarily inactivated. These changes in the protein products of PAR-resident genes represent some sort of a “spill-over” of invading repetitive sequences into the coding regions of the respective genes. This phenomenon creates in the PAR an extraordinary environment to allow for gene modifications to occur, resulting in the appearance of a plethora of protein orthologs in different species. Interestingly, several PAR genes that were rescued by translocation or “protected” from the PAR erosion in mice have been studied in humans and were found to be involved in severe diseases. For instance, variants of ARSE result in X-linked recessive chondrodysplasia punctata (Braverman et al. 2008), variants of STS cause X-linked ichthyosis (for review: Reed et al. 2005), and variants of NLGN4X cause ASD (Yan et al. 2005; Lawson-Yuen et al. 2008; Zhang et al. 2009).
Materials and Methods
Nucleic Acid Resources
Live adult (>8 weeks) male mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) for the purpose of DNA and RNA isolation. The strains included C57BL/6J (No. 000664), CAST/EiJ (No. 000928), BTBRT+t/J (now BTBRT+ Itpr3tf/J, No. 002282), and A/J (No. 000646). Genomic DNA from the golden (Syrian) hamster, Mesocricetus auratus, was purchased from Envigo (Huntingdon, United Kingdom), and genomic DNA from the deer mouse, Peromyscus maniculatus bairdii, was purchased from the Peromyscus Stock Center (University of South Carolina, Columbia, SC, USA; http://www.pgsc.cas.sc.edu, last accessed December 2019). The genomic DNA from the prairie vole, Microtus ochrogaster, was a generous gift of Dr. Larry Young (Emory University, Atlanta, GA, USA). All relevant mouse procedures were performed in accordance with corresponding guidelines and after approval (National Institutes of Health Guidelines for the Care and Use of Laboratory Animals, approval by the Stanford University Administrative Panel on Laboratory Animal Care; Guidelines for the Welfare of Experimental Animals issued by the Federal Government of Germany and the Max-Planck-Society, approval by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit).
Nucleic Acid Extraction, RT-PCR, and Cloning
Genomic DNA from all mouse strains amplified and sequenced was isolated from female and male mouse tails using the Genomic DNA Isolation Kit for Tissue and Cells according to the manufacturer’s protocol (Nexttec, Hilgertshausen, Germany). For quality control, all samples were analyzed on a 1% agarose gel.
Total RNA was isolated from whole brain tissue using the TRIzol method according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed using the Superscript III reverse transcription kit (Invitrogen, Carlsbad, CA, USA). The reactions were the source to amplify full-length cDNAs (PrimeStar, TaKaRa Bio, Otsu, Shiga, Japan) of Nlgn3 and Nlgn4 using respective primers sets (IDT, Coralville, IO, USA), and the cDNAs were subcloned into pBluescript(SK+) and submitted to commercial sequencing.
Polymerase Chain Reaction Amplification
In order to identify unspecific amplifications (false positive), all PCR amplifications were performed in duplicate for each primer set used by comparing the obtained amplicons on a 1% agarose gel. In addition, at least two different master mixes were used for each primer set. The reaction mixture (21 µl), for each sample, was prepared in 96 well plates, each containing 1 µl (i.e., 40–60 ng) of mouse genomic DNA, 4 µl of primer set (mixture of forward- and reverse primer, 1 pmol each) and 16 µl of the following master mixes: 1) MG064: 0.4 µl PhireHotStart DNA Polymerase and 4.2 µl 5x-Buffer with 7.5 mM Mg2+ (Finnzymes, now Life Technologies GmbH, Darmstadt, Germany), 4.2 µl Hi-Spec Additive (Bioline GmbH, Luckenwalde, Germany), 1 µl dNTP-Set (2.5 mM each, Bioline), 6.2 µl H2O; 2) MG088: 10 µl QIAGEN Multiplex PCR Plus Kit (Qiagen, Hilden, Germany), 6 µl 5x-Q-Solution (Qiagen); 3) MG097: 0.2 µl KAPA2G Robust Hot Start Polymerase (5 units/µl), 4.2 µl 5x-KAPA2G Buffer A with 7.5 mM Mg2+, 4.2 µl 5x-KAPA Enhancer, 0.3 µl KAPA dNTP Mix (10 mM each) (KAPA BIOSYSTEMS, now Sigma–Aldrich, Munich, Germany), 7.1 µl H2O; and 4) MG093: 1 µl 7-deaza-dGTP (10 mM, Roche GmbH, Mannheim, Germany), 10 µl QIAGEN Multiplex PCR Plus Kit (Qiagen), and 6 µl 5x-Q-Solution (Qiagen).
The cycling program was carried out after an initial preheating step at 98 °C for 5 min and included 35 cycles of 1) denaturation at 98 °C for 45 s, 2) annealing at 64 °C for 2 min (or 60 °C when master mixes with 7-deaza-dGTP were used), and 3) extension at 72 °C for 2 min, followed by a final extension step at 72 °C for 20 min in a DNA Thermal Cycler (PTC-200 MJ Research, BioRad, Munich, Germany, or Veriti 96 Well Thermal Cycler, Applied Biosystems, Darmstadt, Germany).
Sequencing of PCR Amplicons
The PCR amplicons were purified from unincorporated primers and dNTPs by digesting with 1 unit Shrimp Alkaline Phosphatase (SAP) und 5 units Exonuclease I (Exo) according to the manufacturer’s instructions (USB Europe GmbH, Staufen, Germany). Sequencing of the purified PCR products were carried out using the dideoxy chain terminator method with the BigDye Terminator v3.1 Cycle Sequencing Kit or a combination of the BigDye Terminator v3.1 Cycle Sequencing- and a dGTP BigDye Cycle Sequencing Terminator v3.0 Kit (Applied Biosystems, Darmstadt, Germany). Sequencing reactions (12 µl) were prepared in 96 well plates, each containing 4 µl PCR product, 4 µl sequencing primer (1 pmol/µl), and 4 µl of the before mentioned BigDye Terminator Cycle Sequencing Kit reagent. The following PCR thermal cycling profile was used after a preheating step at 94 °C for 30 s: 30–40 cycles of 1) 96 °C for 10 s, 2) 50–56 °C for 10 s, and 3) 60 °C for 2 min in Veriti 96 Well Thermal Cycler. Upon completion of the final cycling step, the sequencing reactions were precipitated using ethanol to eliminate unincorporated fluorescent-labeled nucleotides and analyzed on a 3730XL-DNA-Analyzer (Applied Biosystems). Raw data were processed with Sequencing Analysis 5.2 (Applied Biosystems) and with different modules of the DNA Star Lasergene 7.0 Software package (DNASTAR, Madison, WI, USA). For obtaining extralong sequence reads, raw data were processed online using PeakTrace Basecaller software (www.nucleics.com, last accessed December 2019).
FISH Analysis
Fluorescence in situ-hybridization was carried out by SeeDNA Biotech Inc. (Windsor, Ontario, Canada) using probes based on a 14.4-kb lambda phage clone carrying the ATG of the mouse Nlgn4 gene (for details see fig. S1 in Bolliger et al. [2008]).
Phylogenetic Analysis
The phylogenetic analysis of NLGN4 protein sequences was performed using the neighbor-joining method (Saitou and Nei 1987) with 1,000 bootstrap replications (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Evolutionary analyses were conducted in MEGA X using default settings for protein sequences (Kumar et al. 2018). Prior sequence alignment was performed using FASTA formats of respective amino acid sequences and default settings in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/, last accessed December 2019).
In Silico Analysis of Nucleotide and Amino Acid Sequence Composition
For multiple sequence alignment, we used MultAlin (Corpet 1988, http://multalin.toulouse.inra.fr/multalin, last accessed December 2019). Alignment for amino acid sequences was performed using either the preset parameters “Blosum62-12-2” or “Identity-1-0,” for nucleotide sequences: “DNA-5-0.” GC content of nucleotide sequences was calculated and plotted using the GC content calculator (https://www.biologicscorp.com/tools/GCContent/#.XRDRn3tS8qJ, last accessed December 2019) set to a window of 50 bases. The GC content in the third codon position, GC3, was calculated from codon distributions assessed with the graphical codon usage analyzer (http://gcua.schoedl.de, last accessed December 2019). Nucleotide sequences were translated into amino acid sequences using the Translate tool (http://web.expasy.org/translate/, last accessed December 2019) or reverse complement sequences were computed at http://reverse-complement.com, last accessed December 2019. Relative frequencies of nucleotide usage were displayed using the Weblogo online tool (http://weblogo.berkeley.edu/logo.cgi, last accessed December 2019). To assess theoretical biochemical characteristics, we used the compute MW/pI tool (http://web.expasy.org/compute_pi/, last accessed December 2019). Quality of presumptive signal peptides has been assessed using SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/, last accessed December 2019) with default settings for eukarya (Almagro Armenteros et al. 2019). To assess the impact of single amino acid changes in conserved stretches of the human NLGN4X protein sequence, we analyzed these changes using Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml, last accessed December 2019) (Adzhubei et al. 2010).
Identification and Characterization of NLGN4 Genes
Neuroligin-4 genes were identified in two different ways: 1) Nlgn4/NLGN4 genes were identified using NLGN4 or alias of the gene name as a search term for the NCBI databases (https://www.ncbi.nlm.nih.gov/gene/, last accessed December 2019), or 2) full-length or partial sequence elements of different available NLGN4 gene sequences were used to run BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed December 2019) against different species. The program was set to search for highly similar sequences (megablast) or somewhat similar sequences (BlastN) within the NCBI BlastN suite (search in nucleotide databases). Database parameters were set to search within the nucleotide collection (nr/nt), WGS, or high-throughput genomic sequences. The choice of organism was either unrestricted or limited to rodents. Most neuroligin-4 genes that were readily available including their Gene ID had annotations by the NCBI Eukaryotic Annotation Pipeline algorithms, and exons were classified by the status “predicted.” Although this was in most cases identical with our manual comparison of known sequence information (e.g., human and mouse) for the coding exons (i.e., exons 1 and 3–7), in numerous cases the algorithm additionally predicted potential noncoding upstream exons. The status of these exons, however, requires further validation. Some species had a predicted status of an incomplete NLGN4 that was based on sequence information with gaps. We used this information for the prairie vole, deer mouse, and golden hamster to generate PCR products from the respective genomic DNA and subsequently sequenced them to close the sequence gaps. All sequence information has been deposited in GenBank (overview of sequences, accession numbers, etc.; cf. supplementary data sheet 2, Supplementary Material online).
Assignment of PAR Genes
In most mammalian species, the PAR boundary has not been identified yet. When referring to PAR genes, we used as a reference all genes that are found downstream of MID1 from the human genomic build of the X-chromosome. MID1 was chosen because it contains the PAR boundary in the laboratory mouse (Palmer et al. 1997). All homologs of the respective genes that we found in the NCBI database are summarized with their Gene ID in supplementary data sheet 3, Supplementary Material online. The identification process was similar to the one described above for NLGN4 homologs. A schematic summary of our analysis is depicted in supplementary figure S3, Supplementary Material online. The classification “not identified” needs to be distinguished from “absent in contig.” The latter classification was used when a stretch of genes was present on a single contig but the genomic sequence was missing an expected gene. We note that in case of Chinchilla lanigera, Heterocephalus glaber, and Ictidomys tridecemlineatus, for instance, an entire contig is covering all genes listed, from SHROOM2 to PLCXD1. Both gene classifications do not per se rule out the existence of the respective gene elsewhere in the genome. Current species-specific genomic builds or sequence information yet to be assembled from WGS reads, however, have not yet yielded in positive BLAST results. Autosomal or X-specific assignment was selected when the combination of neighboring genes clearly identified the new locus or when the database had already assigned them to a certain chromosome. Our reconstruction of the mouse PAR region is supported by WGS sequencing results from the Mouse Genomes Project by the Sanger Institute. The reference numbers for genes and their WGS reads can be found in the supplementary data sheet 4, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank C. Harenberg, D. Warnecke, M. Schlieper, and I. Thanhäuser of the AGCT Sequencing Facility at the Max-Planck-Institute of Experimental Medicine, Göttingen, Germany, for expert technical assistance. This work was supported by the German Research Foundation (FZT103 “CNMPB” to N.B.), the European Commission (EU-AIMS FP7-115300 to N.B.), and the NIMH (R01 MH092931 to T.C.S.). S.M. is supported by a seed grant of the Saarland University Medical School (HOMFOR2017-19).
Author Contributions
S.M. designed research. S.M. and F.B. performed research. S.M., F.B., N.B., and T.C.S. analyzed the data. S.M., F.B., N.B., and T.C.S. wrote the article. All authors edited the article.
A list of all sequences referred to as well as newly sequenced ones are listed with their respective reference and/or accession numbers in supplementary data sheets 2–4, Supplementary Material online.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR.. 2010. A method and server for predicting damaging missense mutations. Nat Methods. 7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H.. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. [DOI] [PubMed] [Google Scholar]
- Araç D, Boucard AA, Ozkan E, Strop P, Newell E, Südhof TC, Brunger AT.. 2007. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron 56(6):992–1003. [DOI] [PubMed] [Google Scholar]
- Avdjieva-Tzavella DM, Todorov TP, Todorova AP, Kirov AV, Hadjidekova SP, Rukova BB, Litvinenko IO, Hristova-Naydenova DN, Tincheva RS, Toncheva DI.. 2012. Analysis of the genes encoding neuroligins NLGN3 and NLGN4 in Bulgarian patients with autism. Genet Couns. 23(4):505–511. [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508(7497):494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger MF, Pei J, Maxeiner S, Boucard AA, Grishin NV, Südhof TC.. 2008. Unusually rapid evolution of Neuroligin-4 in mice. Proc Natl Acad Sci U S A. 105(17):6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. 2015. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 16(9):551–563. [DOI] [PubMed] [Google Scholar]
- Braverman NE, Bober M, Brunetti-Pierri N, Oswald GL.. 2008. Chondrodysplasia punctata 1, X-linked In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mefford HC, Stephens K, Amemiya A, Ledbetter N, editors. Gene reviews. Seattle (WA: ): University of Washington. [PubMed] [Google Scholar]
- Budreck EC, Scheiffele P.. 2007. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci. 26(7):1738–1748. [DOI] [PubMed] [Google Scholar]
- Burgin CJ, Colella JP, Kahn PL, Upham NS.. 2018. How many species of mammals are there? J Mammal. 99(1):1–14. [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Südhof TC.. 2007. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron 54(6):919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16(22):10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Nevo E, Han L, Levanon EY, Zhao J, Avivi A, Larkin D, Jiang X, Feranchuk S, Zhu Y, et al. 2014. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat Commun. 5(1):3966. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. [DOI] [PubMed] [Google Scholar]
- Gattermann R, Fritzsche P, Neumann K, Al-Hussein I, Kayser A, Abiad M, Yakti R.. 2001. Notes on the current distribution and the ecology of wild golden hamsters (Mesocricetus auratus). J Zool. 254(3):359–365. [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM.. 2004. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119(7):1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA. 2016. Did sex chromosome turnover promote divergence of the major mammal groups?: De novo sex chromosomes and drastic rearrangements may have posed reproductive barriers between monotremes, marsupials and placental mammals. Bioessays 38(8):734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Krueger-Burg D, Tuffy LP, Cooper BH, Taschenberger H, Goswami SP, Ehrenreich H, Jonas P, Varoqueaux F, Rhee JS, et al. 2015. Perturbed hippocampal synaptic inhibition and γ-oscillations in a neuroligin-4 knockout mouse model of autism. Cell Rep. 13(3):516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbers K, Francke U, Soriano P, Jaenisch R, Müller U.. 1990. Structure and chromosomal mapping of a highly polymorphic repetitive DNA sequence from the pseudoautosomal region of the mouse sex chromosomes. Cytogenet Cell Genet. 53(2–3):129–133. [DOI] [PubMed] [Google Scholar]
- Harbers K, Soriano P, Müller U, Jaenisch R.. 1986. High frequency of unequal recombination in pseudoautosomal region shown by proviral insertion in transgenic mouse. Nature 324(6098):682–685. [DOI] [PubMed] [Google Scholar]
- Hoon M, Soykan T, Falkenburger B, Hammer M, Patrizi A, Schmidt K-F, Sassoè-Pognetto M, Löwel S, Moser T, Taschenberger H, et al. 2011. Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc Natl Acad Sci U S A. 108(7):3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M, Satta Y, Hirai Y, Hirai H, Imai H, Takahata N.. 2003. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc Natl Acad Sci U S A. 100(9):5258–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. 2003. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 34(1):27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, et al. 2008. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci U S A. 105(5):1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MM, Lundin E, Qian X, Mirzazadeh M, Halvardson J, Darj E, Feuk L, Nilsson M, Jazin E.. 2016. Spatial sexual dimorphism of X and Y homolog gene expression in the human central nervous system during early male development. Biol Sex Differ. 7(1):5 eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara T, Abe K, Mekada K, Yoshiki A, Kato T.. 2010. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci U S A. 107(14):6412–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru V, Iyer SV, Almli LM, Stevens JS, Lori A, Jovanovic T, Ely TD, Bradley B, Binder EB, Koen N, et al. 2016. Genome-wide gene-based analysis suggests an association between neuroligin 1 (NLGN1) and post-traumatic stress disorder. Transl Psychiatry. 6(5):e820–e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Wilson HE, Thomson EJ, Lee M, Perry J, Palmer S, Ashworth A, Cooke HJ.. 1996. Structural variation of the pseudoautosomal region between and within inbred mouse strains. Proc Natl Acad Sci U S A. 93(1):171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijer KT, Schmeisser MJ, Krueger DD, Boeckers TM, Scheiffele P, Bourgeron T, Brose N, Burbach JP.. 2014. Neurobiology of autism gene products: towards pathogenesis and drug targets. Psychopharmacology 231(6):1037–1062. [DOI] [PubMed] [Google Scholar]
- Krueger-Burg D, Papadopoulos T, Brose N.. 2017. Organizers of inhibitory synapses come of age. Curr Opin Neurobiol. 45:66–77. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancioni A, Pizzo M, Fontanella B, Ferrentino R, Napolitano LM, De Leonibus E, Meroni G.. 2010. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J Neurosci. 24. 30(8):2880–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson-Yuen A, Saldivar JS, Sommer S, Picker J.. 2008. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 16(5):614–618. [DOI] [PubMed] [Google Scholar]
- Leone P, Comoletti D, Ferracci G, Conrod S, Garcia SU, Taylor P, Bourne Y, Marchot P.. 2010. Structural insights into the exquisite selectivity of neurexin/neuroligin synaptic interactions. EMBO J. 29(14):2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue J, Doran AG, Fiddes IT, Abrudan M, Armstrong J, Bennett R, Chow W, Collins J, Collins S, Czechanski A, et al. 2018. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat Genet. 50(11):1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420(6915):520–562. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Perez A, Holloway LP, Barbaro RP, Barbaro JR, Wilson LM, Threadgill DW, Lauder JM, et al. 2007. Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav Brain Res. 176(1):4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Nomura J, Ji X, Tamada K, Arai T, Takahashi E, Bućan M, Takumi T.. 2017. Functional significance of rare neuroligin 1 variants found in autism. PLoS Genet. 13(8):e1006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Perry J, Kipling D, Ashworth A.. 1997. A gene spans the pseudoautosomal boundary in mice. Proc Natl Acad Sci U S A. 94(22):12030–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente DJ, Garriga C, Baskin B, Douglas G, Cho MT, Araujo GC, Shinawi M.. 2017. Neuroligin 2 nonsense variant associated with anxiety, autism, intellectual disability, hyperphagia, and obesity. Am J Med Genet A. 173(1):213–216. [DOI] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. 2009. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63(5):628–642. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP.. 2016. The Eutherian pseudoautosomal region. Cytogenet Genome Res. 147(2–3):81–94. [DOI] [PubMed] [Google Scholar]
- Reed MJ, Purohit A, Woo LW, Newman SP, Potter BV.. 2005. Steroid sulfatase: molecular biology, regulation, and inhibition. Endocr Rev. 26(2):171–202. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434(7031):325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gökçe O, Lim BK, Fowler SC, Malenka RC, Südhof TC.. 2014. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158(1):198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Salido EC, Li XM, Yen PH, Martin N, Mohandas TK, Shapiro LJ.. 1996. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts). Nat Genet. 13(1):83–86. [DOI] [PubMed] [Google Scholar]
- Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ.. 1992. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 50(2):303–316. [PMC free article] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Südhof TC, Brose N.. 1999. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci U S A. 96(3):1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg KM, Ramachandran D, Patel VC, Shetty AC, Cutler DJ, Zwick ME.. 2012. Identification of rare X-linked neuroligin variants by massively parallel sequencing in males with autism spectrum disorder. Mol Autism. 3(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J.. 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 53(4):533–553. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Schenk JJ.. 2017. Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS One 12(8):e0183070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455(7215):903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. 2017. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171(4):745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC.. 2007. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318(5847):71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thybert D, Roller M, Navarro FCP, Fiddes I, Streeter I, Feig C, Martin-Galvez D, Kolmogorov M, Janoušek V, Akanni W, et al. 2018. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28(4):448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Südhof TC, Brose N.. 2006. Neuroligins determine synapse maturation and function. Neuron 51(6):741–754. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N.. 2004. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 83(9):449–456. [DOI] [PubMed] [Google Scholar]
- Wallis MC, Waters PD, Graves JA.. 2008. Sex determination in mammals–before and after the evolution of SRY. Cell Mol Life Sci. 65(20):3182–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Parikshak NN, Geschwind DH.. 2016. Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat Commun. 7(1):10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM.. 2005. Mammal species of the world. A taxonomic and geographic reference. 3rd ed Baltimore: Johns Hopkins University Press. [Google Scholar]
- Wilson MA, Makova KD.. 2009a. Evolution and survival on eutherian sex chromosomes. PLoS Genet. 5(7):e1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Makova KD.. 2009b. Genomic analyses of sex chromosome evolution. Annu Rev Genomics Hum Genet. 10(1):333–354. [DOI] [PubMed] [Google Scholar]
- Xu X, Xiong Z, Zhang L, Liu Y, Lu L, Peng Y, Guo H, Zhao J, Xia K, Hu Z.. 2014. Variations analysis of NLGN3 and NLGN4X gene in Chinese autism patients. Mol Biol Rep. 41(6):4133–4140. [DOI] [PubMed] [Google Scholar]
- Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones IR, Craddock N, et al. 2005. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 10(4):329–332. [DOI] [PubMed] [Google Scholar]
- Zhang C, Milunsky JM, Newton S, Ko J, Zhao G, Maher TA, Tager-Flusberg H, Bolliger MF, Carter AS, Boucard AA, et al. 2009. A neuroligin-4 missense mutation associated with autism impairs neuroligin-4 folding and endoplasmic reticulum export. J Neurosci. 29(35):10843–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L.. 1965. Molecules as documents of evolutionary history. J Theor Biol. 8(2):357–366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.