Abstract
Astrocytes (AC) are the most abundant cells in the central nervous system. In the retina, astrocytes play important roles in the development and integrity of the retinal neurovasculature. Astrocytes dysfunction contributes to pathogenesis of a variety of neurovascular diseases including diabetic retinopathy. Recent studies have demonstrated the expression of Cyp1b1 in the neurovascular cells of the central nervous system including AC. We recently showed retinal AC constitutively express Cyp1b1, and global Cyp1b1-deficiency (Cyp1b1-/-) attenuates retinal ischemia-mediated neovascularization in vivo and the pro-angiogenic activity of retinal vascular cells in vitro. We also demonstrated that Cyp1b1 expression is a key regulator of retinal AC function. However, the underlying mechanisms involved need further investigation. Here we determined changes in the transcriptome profiles of Cyp1b1+/+ and Cyp1b1-/- retinal AC by RNA sequencing. We identified 585 differentially expressed genes, whose pathway enrichment analysis revealed the most significant pathways impacted in Cyp1b1-/- AC. These genes included those of axon guidance, extracellular matrix proteins and their receptors, cancer, cell adhesion molecules, TGF-β signaling, and the focal adhesion modulation. The expression of a selected set of differentially expressed genes was confirmed by RT-qPCR analysis. To our knowledge, this is the first report of RNAseq investigation of the retinal AC transcriptome and the molecular pathways impacted by Cyp1b1 expression. These results demonstrated an important role for Cyp1b1 expression in the regulation of various retinal AC functions, which are important in neurovascular development and integrity.
Introduction
The cytochrome P450 superfamily consists of many heme-containing monooxygenases. They are best known for their roles in drug metabolism. CYP1B1 is involved in many processes in the body, such as assisting with reactions that break down drugs and produce certain fats (lipids). It is expressed in both adult and fetal human extrahepatic tissues, including most of the parenchymal and stromal tissues from brain, kidney, prostate, breast, cervix, uterus, ovary, lymph nodes [1], and ocular tissues [2, 3]. Mutations in this enzyme are a risk factor for the development of primary congenital glaucoma in humans [4]. However, the underlying cellular and molecular mechanisms are not fully revealed.
We previously showed expression of Cyp1b1 is essential for ischemia-mediated retinal neovascularization as occurs in retinopathy of prematurity, and the proangiogenic function of retinal vascular cells in culture [5–7]. However, how Cyp1b1 expression impacts these processes remained largely unknown. We showed Cyp1b1 is constitutively expressed in vascular endothelial cells and perivascular supporting cells from vascular beds of many organs including retina [5, 7]. Recently, we also demonstrated that Cyp1b1 is expressed in retinal astrocytes (AC), and Cyp1b1-/- retinal AC are more proliferative and migratory [8]. These cells produced increased amounts of fibronectin and its receptors αvβ3 and α5β1 integrins. However, production of inflammatory mediators such as BMP-7 and MCP-1 were decreased in Cyp1b1-/- AC. In addition, we observed a significant increase in CD38 expression when Cyp1b1-/- AC were challenged with H2O2 compared with Cyp1b1+/+ cells. Cyp1b1-/- AC also showed enhanced connexin 43 phosphorylation compared with Cyp1b1+/+ cells [8]. Thus, Cyp1b1-deficiency in AC was associated with increased resistance towards oxidative stress.
Astrocytes are the major cell type in the optic nerve head and are vital to the development and maintenance of the retinal astrocytic network and angiogenesis [9, 10]. Under pathological conditions, AC become reactive and contribute to various ocular pathologies including glaucoma and diabetic retinopathy [11]. However, there still much more to delineate regarding Cyp1b1 expression and function in AC. The few studies available to date have confirmed Cyp1b1 expression in AC and neurons, and its upregulation in a variety of gliomas [1, 12]. To our knowledge, we were first to report the impact of Cyp1b1 expression on retinal AC function. However, the intracellular pathways that mediate these activities of Cyp1b1 in AC remain elusive.
RNAseq is one transcriptomic approach where the total complement of RNAs from a given sample is isolated and sequenced using high-throughput technologies [13]. RNAseq technology has the potential to provide very useful, detailed information on the intracellular pathways impacted by Cyp1b1 expression, and identify the networks of genes involved. The purpose of the current study was to utilize this powerful technique to delineate the detailed molecular mechanisms of Cyp1b1 action in retinal AC by determining the changes in patterns of gene expression networks impacted by Cyp1b1 expression. The identification of genes whose expression is affected by the presence or absence of Cyp1b1 will provide additional clues to the intracellular mechanisms of Cyp1b1 action and function in retinal AC. This knowledge should lead to the discovery of new targets for modulation of Cyp1b1 activity and their potential therapeutic use.
Results
RNAseq analysis and global gene expression profiles of Cyp1b1+/+ and Cyp1b1-/- retinal AC
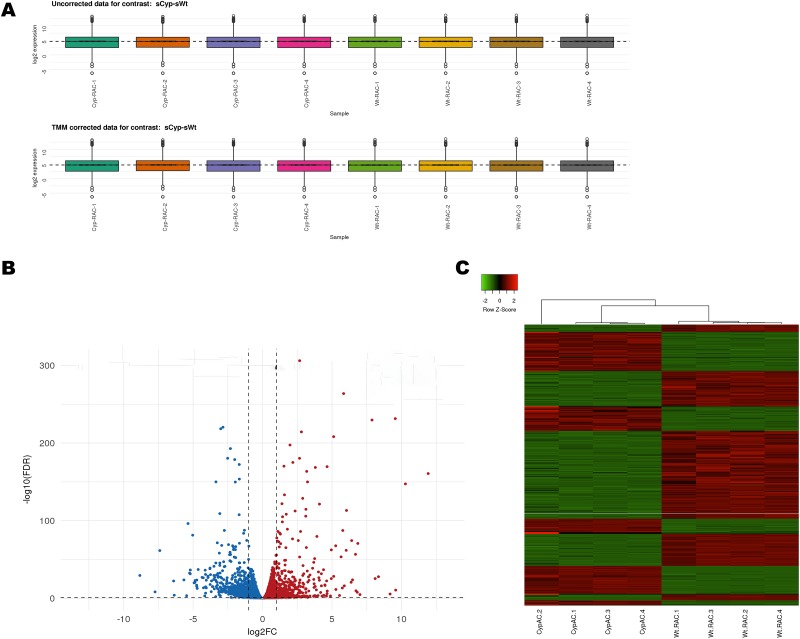
In order to investigate the impact of Cyp1b1 expression on the transcriptome profile of retinal AC, we performed RNAseq analysis of Cyp1b1+/+ and Cyp1b1-/- AC. All samples were uniquely barcoded, pooled and sequenced in one lane on an Illumina HiSeq 2500 platform. The average number of reads for Cyp1b1+/+ AC was 2.34x107 and for Cyp1b1-/- AC was 2.32x107. All sequence reads were mapped to the reference mouse genome using STAR (Spliced Transcripts Alignment to a Reference) [14]. To determine the expression level of various genes, mapped paired-end reads for genes were counted in each sample using RSEM (RNASeq by Expectation Maximization) [15]. Gene expression was normalized by the method of trimmed mean of M-values (TMM), where the product of this factor and the library sizes defines the effective library size [16]. For each sample contrast, simple boxplots per sample expression distributions were constructed before and after TMM normalization (Fig 1A). Overall TMM values were similar in each sample, indicated by the uniform distributions.
Fig 1. Gene expression profiles of the Cyp1b1+/+ and Cyp1b1-/- retinal AC.
(A) Box plot showing overall TMM expression values for the Cyp1b1-/- AC and control samples. (B) Volcano plot showing differentially express genes. For each plot, the X-axis represents log2 FC and the Y-axis represents -log10 (FDR). The differentially expressed genes (DEG) are shown as red indicating increased expression and blue indicating decreased expression. (C) Hierarchical clustering of DEG. Red indicates increased expression and green indicates decreased expression. The DEG were defined as having absolute FC> 1.5 and an FDR< 0.05.
Analysis of differentially expressed genes was performed with a glm using the edgeR package [17]. In order to decide which genes are differentially expressed (DEG), the adjusted p-value-not the raw p-value- was defined to be 0.05. To control the false discovery rate (FDR), a Benjamini-Hochberg correction was applied [18]. Fig 1B represents a Volcano plot showing DEG as red and blue dots denoting up- and down-regulated expression, respectively, at an adjusted p-value (FDR) significance threshold of 0.05. The gray dots reflect those genes with no evidence of statistically significant changes in expression. The two solid gray lines denote the boundary of a two-fold change. We also conducted a hierarchical clustering analysis of DEG from all samples with Ward’s method of Euclidean distances [19], and created a heatmap with the heatmap function from Heatmapper: web-enabled heat mapping for all [20]. The results indicated that gene expression was similar in each group (Fig 1C).
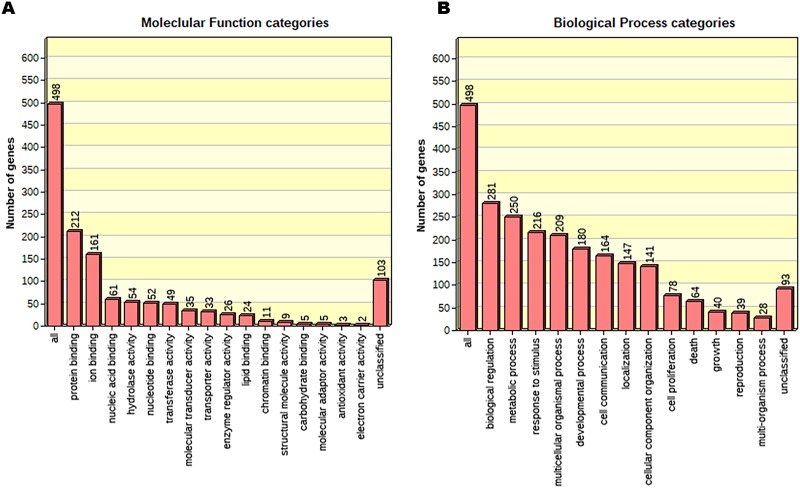
The samples from the Cyp1b1+/+ and Cyp1b1-/- AC groups were assayed for DEG. The threshold was adjusted to Log2 fold-change with an absolute value of 2.0 and a p-value <1e-7. This yielded 585 transcripts (236 up- and 349-downregulated) for the downstream pathway analysis. The 20 most up- and downregulated genes are listed in Table 1. Within the top upregulated genes, we identified the following genes: Dsp (6.81-fold), Uty (11.92-fold), Cysltr1 (8.34-fold), Cdx2 (6.47-fold) and Kdm5d (10.28-fold). These genes have important roles in different biological and molecular processes including actin-mediated cell contraction, cardiac muscle contraction, oxidoreductase activity, and demethylase activity among others [21–24]. The top downregulated genes including Ptprf (-1.01-fold), Fgf10 (-1.03-fold), Prl2c3 (-1.034), Tgfb3 (-1.04-fold), Mical1 (-1.05-fold), Ndrg2 (-1.05-fold), Ptgs1 (-1.0487-fold), Aplp1 (-1.08-fold), and Steap3 (-1.08-fold) have important roles in processes including cell proliferation, apoptosis, regulation of DNA replication, and metabolic processes among others [25–27]. DEG (585 total) were subjected to Gene Ontology to establish a connection with the biological processes and molecular functions that these genes contribute to (Fig 2).
Table 1. Top 20 up- and down-regulated genes in Cyp1b1-/-AC.
| Symbol | Description | Log2FC | p-value | FDR |
|---|---|---|---|---|
| Up-regulated | ||||
| Eif2s3y | eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | 12.01499 | 0 | 0 |
| Uty | ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | 11.91885 | 3.6E-164 | 1.9E-161 |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 11.79096 | 0 | 0 |
| Kdm5d | lysine (K)-specific demethylase 5D | 10.28087 | 8.9E-151 | 4E-148 |
| Gm43302 | predicted gene 43302 | 9.580986 | 1.21E-12 | 2E-11 |
| Rpl15-ps2 | ribosomal protein L15, pseudogene 2 | 9.550535 | 2.2E-235 | 3.8E-232 |
| Bmp8b | bone morphogenetic protein 8b | 8.847732 | 0 | 0 |
| Cysltr1 | cysteinyl leukotriene receptor 1 | 8.342675 | 1.93E-30 | 1.16E-28 |
| Gm26760 | predicted gene, 26760 | 8.102196 | 5.58E-28 | 2.97E-26 |
| Gm10020 | predicted pseudogene 10020 | 7.874941 | 1.6E-233 | 2.5E-230 |
| Gm6969 | predicted pseudogene 6969 | 6.85453 | 7.82E-74 | 1.63E-71 |
| Dsp | desmoplakin | 6.813457 | 9.78E-10 | 1.13E-08 |
| Actg-ps1 | actin, gamma, pseudogene 1 | 6.722348 | 1.08E-11 | 1.58E-10 |
| Sgsm1 | small G protein signaling modulator 1 | 6.689262 | 1.12E-59 | 1.78E-57 |
| Cdx2 | caudal type homeobox 2 | 6.474141 | 2.61E-26 | 1.27E-24 |
| Gm38312 | predicted gene, 38312 | 6.410902 | 1.62E-77 | 3.66E-75 |
| Oxct2b | 3-oxoacid CoA transferase 2B | 6.374568 | 4.13E-23 | 1.59E-21 |
| AI593442 | expressed sequence AI593442 | 6.040882 | 1.6E-116 | 6E-114 |
| Gm45315 | predicted gene 45315 | 6.018783 | 7.76E-65 | 1.3E-62 |
| Adarb2 | adenosine deaminase, RNA-specific, B2 | 5.88212 | 2.23E-18 | 6.17E-17 |
| Down-regulated | ||||
| Ptprf | protein tyrosine phosphatase, receptor type, F | -1.009044 | 3.978E-53 | 5.686E-51 |
| Gprc5a | G protein-coupled receptor, family C, group 5, member A | -1.02267 | 2.718E-12 | 4.306E-11 |
| Vax2 | ventral anterior homeobox 2 | -1.025959 | 5.515E-10 | 6.507E-09 |
| Fgf10 | fibroblast growth factor 10 | -1.028563 | 3.695E-21 | 1.264E-19 |
| Stard9 | START domain containing 9 | -1.030828 | 2.45E-13 | 4.349E-12 |
| Lmo1 | LIM domain only 1 | -1.031322 | 5.887E-17 | 1.438E-15 |
| Prl2c3 | prolactin family 2, subfamily c, member 3 | -1.034173 | 1.076E-22 | 4.025E-21 |
| Gm27177 | predicted gene 27177 | -1.034225 | 3.073E-17 | 7.67E-16 |
| Tgfb3 | transforming growth factor, beta 3 | -1.035388 | 2.308E-12 | 3.687E-11 |
| Flrt3 | fibronectin leucine rich transmembrane protein 3 | -1.038403 | 8.71E-09 | 8.981E-08 |
| Acsf2 | acyl-CoA synthetase family member 2 | -1.040813 | 9.692E-12 | 1.426E-10 |
| Ppic | peptidylprolyl isomerase C | -1.042228 | 5.977E-32 | 3.847E-30 |
| Eif3j1 | eukaryotic translation initiation factor 3, subunit J1 | -1.042647 | 4.709E-42 | 5.158E-40 |
| Mical1 | microtubule associated monooxygenase, calponin and LIM domain containing 1 | -1.046611 | 1.687E-09 | 1.899E-08 |
| Ndrg2 | N-myc downstream regulated gene 2 | -1.047743 | 2.791E-13 | 4.941E-12 |
| Ptgs1 | prostaglandin-endoperoxide synthase 1 | -1.0487 | 5.856E-29 | 3.273E-27 |
| Cbr3 | carbonyl reductase 3 | -1.050294 | 2.114E-16 | 4.933E-15 |
| Slfn8 | schlafen 8 | -1.061261 | 4.262E-23 | 1.64E-21 |
| Aplp1 | amyloid beta (A4) precursor-like protein 1 | -1.075804 | 8.932E-14 | 1.662E-12 |
| Steap3 | STEAP family member 3 | -1.08044 | 5.467E-14 | 1.046E-12 |
The cutoff criteria for this list was any DEG with a FC> 5.88 for upregulation and FC> -1.009 for downregulation.
Fig 2. The biological process and molecular functions significantly impacted in Cyp1b1-/- AC.
(A) Bar graph of the biological process categories. (B) Bar graph of the molecular function. DEG were subjected to GO enrichment analysis having absolute FC> 2 and a FDR< 0.05.
Pathway enrichment analysis, conducted using KEGG (Kyoto Encyclopedia of Genes and Genomes) [28–30] as a mapping database, 57 pathways were identified with significance level of 0.05. To cut down on the list the P< 0.002 was applied. The pathways with multiple points of commonality and overlap among the Axon guidance, extracellular matrix-receptor interactions, pathways in cancer, cell adhesion molecules, TGF-β signaling pathways, and the focal adhesion were identified as most significantly enriched pathways (Table 2).
Table 2. Pathway enrichment analysis of significantly changed genes.
| KEGG Pathway name | Pathway Rank | GENES | FDR adjusted enrichment score p-value |
|---|---|---|---|
| Axon guidance | 1 | Sema3c, Unc5d, Sema5a, Sema3g, Ngef, Lrrc4c, Sema6b, Cxcl12, Unc5c, Sema4f, Plxnb1, Sema6a | 3.32e-07 |
| ECM-receptor interaction | 2 | Itga1, Col1a2, Lama2, Tnxb, Spp1, Lama5, Col5a3, Npnt, Thbs2 | 4.79e-06 |
| Pathways in cancer | 3 | Dapk1, Arnt2, Wnt5b, Lama2, Pgf, Hhip, Apc2, Fgf7, Pax8, Lama5, Wnt4, Tgfb3, Ar, Mmp2, Fgf10 | 1.31e-05 |
| Cell adhesion molecules (CAMs) | 4 | Vcan, Cdh4, Cd80, Cldn1, Vcam1, Cd34, Ptprf, Cdh3, Icam1 | 0.0002 |
| TGF-beta signaling pathway | 5 | Tgfb3, Gdf5, Fst, Lefty1, Bmp7, Bmp8b, Thbs2 | 0.0002 |
| Focal adhesion | 6 | Itga1, Col1a2,Lama2,Pgf, Tnxb, Spp1, Lama5, Col5a3, Myl12b,Thbs2 | 0.0002 |
| Neuroactive ligand-receptor interaction | 7 | Lepr, Adra1d, Htr1b, Gria4, Grid2, Htr2a, Cysltr1,Drd4, Calcrl, Grid1 | 0.0020 |
| Metabolic pathways | 8 | Mgat3, Itpkb, Cda, Xylb, Pla2g4b, Abat, Ckb, Ass1, Isyna1, Aldh1a7, Aldh1a1, Nnmt, Cbr2, B3gnt3, Cbr3, Gldc, Eno2, Ptgs1, Bdh2, Ldhb, Gatm, St3gal1, Sardh | 0.0041 |
| Calcium signaling pathway | 9 | Adcy7, Cacna1d, Itpkb, Cacna1h, Adra1d, Htr2a, Cysltr1 | 0.0058 |
| Cytokine-cytokine receptor interaction | 10 | Lepr, Ccl7, Bmp7, Tgfb3, Tnfsf10, Cxcl11, Cxcl12, Gdf5 | 0.0077 |
| MAPK signaling pathway | 11 | Mapk13, Tgfb3, Cacna1d, Cacna1h, Pla2g4b, Fgf10, Fgf7 | 0.0277 |
Significantly changed transcriptome. Column 1 lists the canonical KEGG pathway name, column 2 lists the pathway enrichment score rank in terms of p-value determined by hypergeometric test, column 3 lists the genes that mapped to the KEGG pathway, and column 4 shows the FDR adjusted p-value of significance of the pathway enrichment score.
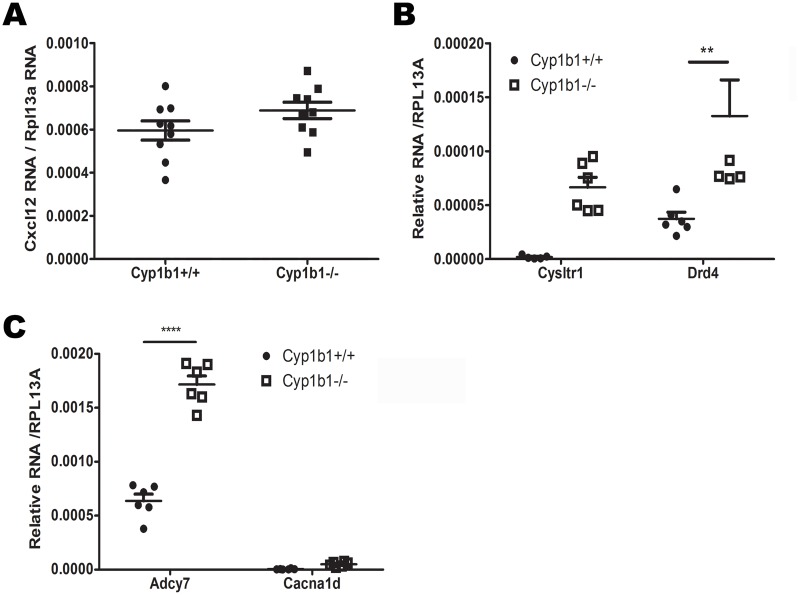
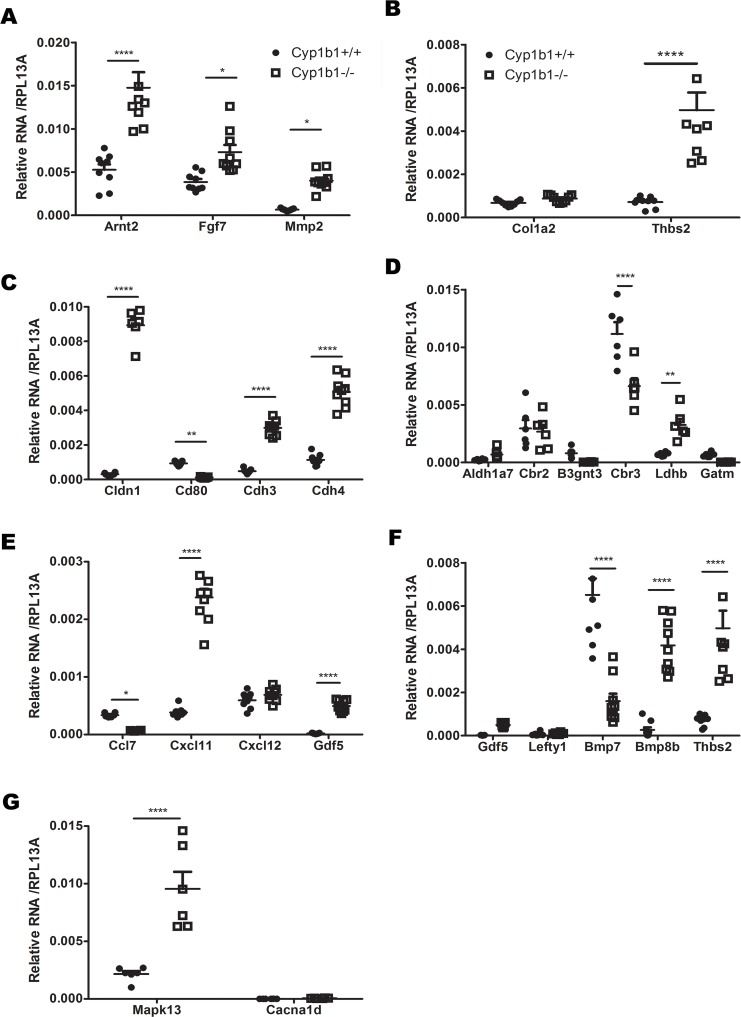
To validate the RNAseq findings, RT-qPCR were performed on newly extracted RNA from Cyp1b1+/+AC and Cyp1b1-/-AC. From each KEGG pathway, we selected key genes with important roles in angiogenesis, apoptosis, cell proliferation, cell migration, metabolism and inflammation. We selected 27 genes (Cxcl12, Cxcl11, Col1a2, Arnt2, Fgf7, Mmp2, Bmp7, Thsb2, Bmp8b, Cdh3, Cdh4, Gdf5, Cd80, Cldn1, Lefty1, Cysltr1, Drd4, Aldh1a7, Cbr2, B3gnt3, Cbr3, Ldhb, Gatm, Adcy7, Cacna1d, Ccl7, and Mapk13) involved in the pathways identified in Table 2. We examined changes in their expression by RT-qPCR (Figs 3 and 4). The results obtained with RT-qPCR showed similar changes in expression as those obtained from the RNAseq studies (Table 3). However, the changes in the expression of a number of these genes did not reach significant levels. These included Cxcl12, Cysltr1, Cacna1d, Col1a2, Aldh1a7, Cbr2, B3gnt3, Gatm, and Lefty1.
Fig 3. RT-qPCR validation of DEG from different pathways.
(A) Genes related to the axon guidance pathway. (B) Genes related to the Neuroactive ligand-receptor interaction pathway. (C) Genes related to the Calcium signaling pathway. (**P<0.01, ****P<0.0001, n≥ 6).
Fig 4. RT-qPCR validation of DEG from different pathways.
(A) Genes related to the Cancer pathway. (B) Genes related to the ECM-Receptor and Focal adhesion pathways. (C) Genes related to the Cell adhesion molecules pathways. (D) Genes related to the metabolic pathways. (E) Genes related to the Cytokine-Cytokine interaction pathways. (F) Genes related to the TGF-β signaling pathway. (G) Genes related to the MAPK signaling pathways. (*P<0.05, **P<0.01, ****P<0.0001, n≥ 6).
Table 3. Differentially expressed gene selected for RT-qPCR validation.
| KEGG pathway name | Symbol | Gene Description | Log2FC | p-value |
|---|---|---|---|---|
| Axon Guidance | Cxcl12 | chemokine (C-X-C motif) ligand 12 | 1.10 | 2.39E-89 |
| ECM-receptor interaction | Col1a2 | collagen, type I, alpha 2 | 5.74 | 1.69E-15 |
| Thbs2 | thrombospondin 2 | 2.67 | 4.62E-12 | |
| Pathways in cancer | Arnt2 | aryl hydrocarbon receptor nuclear translocator 2 | 1.70 | 1.28E-111 |
| Fgf7 | fibroblast growth factor 7 | 1.28 | 6.02E-86 | |
| Mmp2 | matrix metallopeptidase 2 | 5.28 | 1.77E-13 | |
| Cell adhesion molecules | Cdh4 | cadherin 4 (retinal) | 3.23 | 2.45E-153 |
| Cd80 | CD80 antigen | -3.52 | 7.99E-27 | |
| Cldn1 | claudin 1 | 4.64 | 4.05E-173 | |
| Cdh3 | cadherin 3 (placental) | 3.75 | 3.80E-54 | |
| TGF-β signaling | Gdf5 | growth differentiation factor 5 | 4.65 | 2.08E-11 |
| Lefty1 | left right determination factor 1 | 2.62 | 1.32E-12 | |
| Bmp7 | bone morphogenetic protein 7 | -2.31 | 2.04E-59 | |
| Bmp8b | bone morphogenetic protein 8b | 8.85 | 0 | |
| Thbs2 | thrombospondin 2 | 2.67 | 4.62E-12 | |
| Focal adhesion | Col1a2 | collagen, type I, alpha 2 | 5.74 | 1.69E-15 |
| Thbs2 | thrombospondin 2 | 2.67 | 4.62E-12 | |
| Neuroactive ligand- receptor interaction | Cysltr1 | cysteinyl leukotriene receptor 1 | 8.34 | 1.93E-30 |
| Drd4 | dopamine receptor D4 | 3.32 | 1.86E-11 | |
| Metabolic pathways | Aldh1a7 | aldehyde dehydrogenase family 1, subfamily A7 | 3.34 | 1.25E-25 |
| Cbr2 | carbonyl reductase 2 | 2.20 | 3.12E-10 | |
| B3gnt3 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 | -5.21 | ||
| Cbr3 | carbonyl reductase 3 | -1.05 | 2.114E-16 | |
| Ldhb | lactate dehydrogenase B | 2.15 | 7.57E-21 | |
| Gatm | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | -4.63 | 6.11E-40 | |
| Calcium signaling | Adcy7 | adenylate cyclase 7 | 1.34 | 2.44E-15 |
| Cacna1d | calcium channel, voltage-dependent, L type, alpha 1D subunit | 3.87 | 5.65E-13 | |
| Cysltr1 | cysteinyl leukotriene receptor 1 | 8.34 | 1.93E-30 | |
| Cytokine-cytokine receptor interaction | Ccl7 | chemokine (C-C motif) ligand 7 | -2.40 | 9.08E-10 |
| Bmp7 | bone morphogenetic protein 7 | -2.31 | 2.04E-59 | |
| Cxcl11 | chemokine (C-X-C motif) ligand 11 | 2.96 | 2.05E-20 | |
| Cxcl12 | chemokine (C-X-C motif) ligand 12 | 1.10 | 2.39E-89 | |
| Gdf5 | growth differentiation factor 5 | 4.65 | 2.08E-11 | |
| MAPK signaling | Mapk13 (P38-delta) | mitogen-activated protein kinase 13 | 1.66 | 5.24E-52 |
| Cacna1d | calcium channel, voltage-dependent, L type, alpha 1D subunit | 3.87 | 5.65E-13 | |
| Fgf7 | fibroblast growth factor 7 | 1.28 | 6.02E-86 |
The expression of selected genes related to the KEGG pathways were validated by RT-qPCR. LogFC and p-values were obtained from RNAseq analysis.
Discussion
In this study we used RNAseq analysis to determine the global transcriptome profile of Cyp1b1+/+ and Cyp1b1-/- AC from mouse retina. We identified 585 DEG, whose pathway analysis revealed the most significant biological functions. These included the Axon guidance, extracellular matrix (ECM)-receptor interactions, pathways in cancer, cell adhesion molecules, TGF-β signaling, and the focal adhesion regulation. We also found that some of the top downregulated genes were involved in biological and molecular processes including actin-mediated cell contraction, cardiac muscle contraction, cell proliferation, apoptosis and metabolic processes.
Our findings here were consistent with the results of our previous studies showing increased proliferation and migration of Cyp1b1-/- AC. Activation of AC proliferation and migration is important in repair of injuries in the central nervous system (CNS) and scar formation [31, 32]. AC migration is regulated by various factors, among which transforming growth factor-β (TGF-β) plays an important role [33]. In AC, TGF-β suppresses cell proliferation by inducing p15INK4B expression in a Smad3-dependent manner [34]. This is consistent with our findings that showed the downregulation of TGF-β in Cyp1b1-/- AC. The upregulation of Smad genes in Cyp1b1 -/- AC was associated with the enhanced pathways in axon guidance and cell proliferation.
Our results showed changes in Cxcl12, (also known as SDF-1). Cxcl12 is one of the most studied chemokines that induce cell proliferation and migration by binding to its receptor. Under normal conditions, Cxcl12 expression in the CNS is relatively low. However, its expression is upregulated when the CNS is affected by trauma, ischemia, inflammation or infection [35]. Enhanced Cxcl12 expression promotes proliferation of radial glia like cells after traumatic brain injury in rats [36]. Others have shown its therapeutic value by promoting autophagy and migration via PI3K-AKT-mTOR pathway [37].
We recently showed that Cyp1b1 deficiency affects retinal AC ECM production and expression of integrin receptors [8]. We also showed upregulation of cadherins, laminin, and tenascin. These molecules are well known for their roles in cell adhesion [38], and were associated with changes in adhesion observed in Cyp1b1-/- retinal AC [8]. Cadherin-4 (also known as R-cadherin) is involved in retinal angiogenesis during development. Dorrell et al. [39] used antibodies or peptides to neutralize R-cadherin, which prevented the normal formation of the retinal vascular network in newborn mice. They also showed that R-cadherin plays a crucial role in the endothelial–astrocyte interactions [39, 40]. Thus, Cyp1b1 expression may impact AC interactions with EC.
Oxidative stress is implicated in many neurodegenerative diseases. Cytochrome P450 activities are generally involved in ROS production due to their involvement in the metabolism of steroids, fat-soluble vitamins, fatty acids, eicosanoids, drugs, carcinogens, and other xenobiotic chemicals [41–43]. CYP enzymes can generate superoxide and hydrogen peroxide through uncoupling reactions, for more details see [44]. We have demonstrated an important role for Cyp1b1 as a modulator of cellular redox state [45]. Studies utilizing vascular cells derived from Cyp1b1-/- mice showed an increase in oxidative stress in vascular endothelial cells and perivascular supporting cells [5, 7, 46]. In contrast, Cyp1b1-/- retinal AC did not show an increase in oxidative stress compared to Cyp1b1+/+ cells under basal conditions [8]. This is consistent with minimal changes in expression of genes that affect cellular redox state in the absence of Cyp1b1. However, incubation of these cells with known inducers of oxidative stress could reveal changes in genes that modulate oxidative stress. This notion is supported by our studies demonstrating that Cyp1b1-/- AC elicit a significantly more robust response in expression of CD38 when challenged with H2O2 compared with Cyp1b1+/+ cells [47]. Thus, the elucidation of the mechanisms behind AC resilience to oxidative stress, especially in the absence of Cyp1b1, needs further investigation.
Cytochrome P450 enzymes are involved in metabolism of drugs, and are major source of variability in drug pharmacokinetics and responses. However, in some cases they can also activate compounds consumed in food, converting pro-carcinogens to carcinogens [48]. CYP proteins, involved in steroid or retinoic acid metabolism, could promote or suppress tumors development through hormonal control [49, 50]. Genetic variability could play a role if a polymorphism affected a CYP protein involved in such processes [51]. Our study found multiple genes in the cancer pathway that are associates with the progression or suppression of cancer such as laminin, Tgfβ3, Hhip, Arnt2 and Mmp2 [52–54]. Mmp2 was upregulated in this pathway and is implicated in cancer cell migration [55]. A previous study using a microarray analysis, demonstrated that the increased expression of Mmp2 is involved in invasiveness of malignant glioma [56, 57], an observation that is consistent with our findings. Thus, Cyp1b1 expression has an important role in modulating AC migration by suppressing Mmp2 expression.
A limitation of our studies is the use of cells prepared from mix genders. Our initial in vivo and in vitro vascular cell culture studies did not demonstrate a gender bias in the noted phenotypes with Cyp1b1-deficiency. However, our gene expression studies here showed some of the differentially expressed genes in Cyp1b1 null cells are sex linked: Uty; Y-chromosome, Cysltr1; X-chromosome, and Kdm5d; Y-chromosome. Thus, Cyp1b1-deficiency impact on gene expression, and likely noted phenotypes, could be deferentially impacted by gender. Future studies will further address the gender contributions to various phenotypes noted with Cyp1b1-deficiecny.
In summary, RNAseq technology was used to investigate the transcriptome profiles of retinal AC and how Cyp1b1 expression modulates their cellular functions. A pathway analysis of DEG indicated the most significantly enriched pathways included Axon guidance, ECM-receptor interactions, as well as cancer and other pathways (cell proliferation, focal adhesion, and cell adhesion). Our transcriptomic approach in this investigation, which relied on RNAseq, was powerful and effective way to allow us to obtain a global view of genes whose expression are impacted by Cyp1b1, likely in a gender dependent manner.
Materials and methods
Ethics statement
All animal experiments were performed in accordance to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin School of Medicine and Public Health (the assurance number A3368-01). Animals were sacrificed according to an approved protocol by CO2 asphyxiation.
Isolation and culture of Cyp1b1-/- retinal AC
Retinal AC were isolated from mouse retina by collecting retinas from one litter of 4-week-old (6 to 7- mix gender) mice using a dissecting microscope, as previously described by us with greater than 98% purity [8, 58]. Briefly, retinas (12 to 14) were rinsed with serum-free Dulbecco’s Modified Eagle’s Medium (DMEM), pooled in a 60 mm dish, minced and digested for 45 min with collagenase Type I (1 mg/ml; Worthington, Lakewood, NJ) in serum-free DMEM at 37°C. Cells were rinsed in DMEM containing 10% fetal bovine serum (FBS) and centrifuged for 5 min at 400 xg. Digested cells were rinsed again in DMEM containing 10% FBS and filtered through a double layer of sterile 40 μm nylon mesh (Sefar America Inc., Fisher Scientific, Hanover Park, IL). Cells were centrifuged for 5 min at 400 xg and medium was aspirated. Cells were washed twice with DMEM containing 10% FBS, resuspended in 1 ml of DMEM containing 10% FBS in a 1.5 ml microfuge tube and incubated with rat-anti-mouse CD31 (Mec13.3; BD Biosciences) coated with sheep anti-rat magnetic beads, and were gently rocked for 1 h at 4°C. Using a Dynal magnetic tube holder, cells not bound to magnetic beads were collected and washed in DMEM containing 10% FBS. Cells were plated in growth medium in a single well of a 24 well plate coated with human fibronectin (2 μg/ml in serum-free DMEM; BD Biosciences, Bedford, MA), and incubated at 33°C with 5% CO2. The cells bound to magnetic beads are generally used to remove retinal EC as we described [58]. Retinal AC were grown in DMEM containing 10% FBS, 2 mM L-glutamine, 2 mM sodium pyruvate, 20 mM HEPES, 1% nonessential amino acids, 100 μg/ml streptomycin, 100 U/ml penicillin, freshly added heparin at 55 U/ml (Sigma, St. Louis, MO), endothelial growth supplement 100 μg/ml (Sigma), and the murine recombinant interferon-γ (R&D, Minneapolis, MN) at 44 U/ml. Cells were maintained at 33°C with 5% CO2. Cells were progressively passed to larger plates, maintained, and propagated in 1% gelatin-coated 60 mm dishes. For all experiments, cells were used at 70–80% confluence unless stated otherwise.
RNA purification
Total RNA from Cyp1b1+/+ and Cyp1b1-/- retinal AC was purified using RNeasy Mini kit according to manufacturer’s protocol with the DNAse treatment step to eliminate traces of genomic DNA (Qiagen, Germantown, MD). The quality and quantity of the total RNA were measured using an Agilent Model 2100 Bioanalyzer, and samples showing a RIN >8 were selected for further analysis. Samples were stored in RNase-free water and kept at -80°C until further processing.
RNA sequencing
Eight samples of purified RNA (4 from Cyp1b1+/+ retinal AC and 4 from Cyp1b1 -/- retinal AC) were subsequently subjected to a double round of poly-A mRNA purification, fragmented, and primed for cDNA library synthesis using the TruSeq RNA sample preparation kit (RS-122-9004). All procedures were carried out according to the manufacturer’s instructions (Illumina, San Diego, CA). Following validation (Agilent 2100 Bioanalyzer, DNA 1000) and normalization, samples were clustered (TruSeq pairedend cluster kit v3-cBot-HS, PE-401-3001) followed by paired-end sequencing (100 bp; TruSeq SBS kit v3-HS 200 cycles, FC-401-3001) on a HiSeq2500. The following quality control statistics were used to evaluate the technical quality of the experiments; (1) combined per cycle base quality, (2) per cycle base frequencies, (3) per cycle average base quality, (4) relative 3k-mer diversity, (5) Phred quality distribution, (6) mean quality distribution, (7) read length distribution and (8) read occurrence distribution using the trimming software skewer [59]. Low-abundance genes with a read count below a threshold of 1.0 in two or more samples were excluded. To compare gene expression between Cyp1b1+/+AC and Cyp1b1-/-AC, samples were normalized by trimmed mean of M-values (TMM) using Edge R (version 2.5 of Bioconductor) software [16]. After the inspection of preliminary data, transcript reads were aligned to the preassembled selected reference genome sequence using STAR (Spliced Transcripts Alignment to a Reference) using the default settings [14]. Transcript abundance were performed by using RSEM (RNASeq by Expectation Maximization) [15]. Subsequently, differential analysis of significant changes in gene expression was performed with a glm using the edgeR package [17] in the different genotype pairs (e.g. Cyp1b1+/+ vs. Cyp1b1 -/-). All sequence data have been deposited in the Gene Expression Omnibus with accession number GSE145103.
Quantitative RT-PCR
Total RNA from retinal AC was extracted using mirVana PARIS kit (Invitrogen). The cDNA synthesis was performed from 1 μg of total RNA using Sprint RT Complete-Double PrePrimed kit (Clontech, Mountain View, CA). One μl of each cDNA (dilution 1∶10) was used as template in qPCR assays, performed in triplicate of three biological replicates on Mastercycler Realplex (Eppendorf) using the SYBR-Green qPCR Premix (Clontech). Amplification parameters were as follows: 95°C for 2 min; 40 cycles of amplification (95°C for 15 sec, 60°C for 40 sec); dissociation curve step (95°C for 15 sec, 60°C for 15 sec, 95°C for 15 sec). Standard curves were generated from known quantities for each of the target gene of linearized plasmid DNA. Ten times dilution series were used for each known target, which were amplified using SYBR-Green qPCR. The linear regression line for ng of DNA was determined from relative fluorescent units (RFU) at a threshold fluorescence value (Ct) to quantify gene targets from cell extracts by comparing the RFU at the Ct to the standard curve, normalized by the simultaneous amplification of RpL13a, a housekeeping gene. All primer sequence used are listed in Table 4.
Table 4. Primers to validate differentially express genes.
| Gene | Amplicon size (bp) | primer | Primer Sequence (5'->3') | Length | Gene accession |
|---|---|---|---|---|---|
| Cxcl12 | 116 | F | ctgtgcccttcagattgttg | 20 | NM_001012477.2 |
| R | ctctgcgccccttgttta | 18 | |||
| Cxcl11 | 93 | F | tgctgagatgaacaggaaggt | 21 | NM_019494.1 |
| R | cgcccctgtttgaacataag | 20 | |||
| Col1a2 | 69 | F | ctggtgcacagggtgtga | 18 | NM_007743.3 |
| R | ctcctgcttgacctggagtt | 20 | |||
| Arnt2 | 104 | F | tgcacttcggaaaactccat | 20 | NM_007488.3 |
| R | cgagagcccatacacatgc | 19 | |||
| Fgf7 | 78 | F | ttactccatagttctgcaaccagt | 24 | NM_008008.4 |
| R | tgttgcccttcccttcataa | 20 | |||
| Mmp2 | 75 | F | gggcttctgtcctgacca | 18 | NM_008610.3 |
| R | aagttcttggtgtaggtgtagatcg | 25 | |||
| Bmp8b | 70 | F | ctgtatgaactccaccaaccac | 22 | NM_028189.3 |
| R | ggggatgatatctggcttca | 20 | |||
| Cdh3 | 75 | F | aggcccagctaacacatgac | 20 | NM_001037809.5 |
| R | acaaggccacggtgtctc | 18 | |||
| Cdh4 | 60 | F | ttcctggctgctgacaatg | 19 | NM_009867.3 |
| R | gtagatctgcagggtcccagt | 21 | |||
| Gdf5 | 73 | F | tttattgacaaagggcaagatg | 22 | NM_008109.3 |
| R | aggcactgatgtcaaacacg | 20 | |||
| Cd80 | 127 | F | ttcgtctttcacaagtgtcttca | 23 | NM_009855.2 |
| R | tgccagtagattcggtcttca | 21 | |||
| Cldn1 | 92 | F | actccttgctgaatctgaacagt | 23 | NM_016674.4 |
| R | ggacacaaagattgcgatcag | 21 | |||
| Lefty1 | 92 | F | actcagtatgtggccctgcta | 21 | NM_010094.4 |
| R | aacctgcctgccacctct | 18 | |||
| Cysltr1 | 95 | F | aaggtgctgaggtaccagatagag | 24 | NM_021476.5 |
| R | aatcacagcccttgagaagc | 20 | |||
| Drd4 | 137 | F | cccaccaactacttcatcgtg | 21 | NM_007878 |
| R | gccatgagcgtgtcacag | 18 | |||
| Aldh1a7 | 85 | F | gtttgcagatgccgacttg | 19 | NM_011921.2 |
| R | cgctgcaacacaaatctgac | 20 | |||
| Cbr2 | 109 | F | gcccatgtcacctttcctaa | 20 | NM_007621.2 |
| R | ttacccggatcttgtgtgg | 19 | |||
| B3gnt3 | 115 | F | gcaaatacaaccgactgctg | 20 | NM_028189.3 |
| R | cactccaggaaaaggacctg | 20 | |||
| Cbr3 | 60 | F | aacgttagcgggagagatga | 20 | NM_173047.3 |
| R | cccttgatgtgggaaagaatc | 21 | |||
| Ldhb | 86 | F | gtagtgggcgttggacaagt | 20 | NM_001316322.1 |
| R | acatccaccagggcaagtt | 19 | |||
| Gatm | 103 | F | ggtgcactacatcggctctc | 20 | NM_025961.5 |
| R | acaggaatttcgggaggaa | 19 | |||
| Cacna1d | 60 | F | gaagctgcttgaccaagttgt | 21 | NM_001302637.1 |
| R | aacttccccacggttacctc | 20 | |||
| Ccl7 | 91 | F | ttctgtgcctgctgctcata | 20 | NM_013654.3 |
| R | ttgacatagcagcatgtggat | 21 | |||
| MAPK13 | 63 | F | caggctggccttgagtctt | 19 | NM_011950.2 |
| R | ccagggctacacagtaagatcc | 22 | |||
| Adcy7 | F | gagccttccagacgtccat | 19 | NM_007406.2 | |
| 70 | R | aggaggataacggcattgg | 19 |
Biological interaction network and KEGG pathway enrichment analysis
Pathway Enrichment Analysis was performed with WEBGESTALT web analysis software (http://bioinfo.vanderbilt.edu/wg2/) by mapping significantly changed genes to corresponding KEGG enrichment pathways, Gene Ontology enrichment and conducting a hypergeometric statistical test with significant level <0.05 after multiple testing correction [60–62]. Significance for pathway level enrichment was defined as having an enrichment score False Discovery Rate (FDR) corrected p-value < 0.05.
Statistical analysis
RNA-seq data were analyzed and gene expressions were normalized by the method of trimmed mean of M-values (TMM) and glm using the edgeR package, respectively. This yielded a total of 13,575 genes. KEGG pathway enrichment analysis cut-off criteria of FDR<0.05 with a |FC|>2 was apply. RT-PCR data were analyzed with student's unpaired t-test (2-tailed) or one-way ANOVA with post-Bonferroni’s test for multiple comparisons. P≤ 0.05 was considered significant. Data are presented as Mean ± SEM from cells with n≥ 6 (as indicated in figure legends). All data analysis was done in GraphPad Prism or Microsoft Excel.
Acknowledgments
The authors thank the University of Wisconsin- Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility for providing library preparation and next generation sequencing services. We thank Sammed Mandape for assistance with the bioinformatics analysis.
Data Availability
All relevant data are within the paper.
Funding Statement
The work in NS lab is supported by an unrestricted award from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences, Retina Research Foundation, P30 EY016665, P30 CA014520, and R01 EY026078. CMS is supported by the RRF/Daniel M. Albert Chair. NS is a recipient of RPB Stein Innovation Award. JFP is supported by F31 EY027987 training grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Muskhelishvili L, Thompson PA, Kusewitt DF, Wang C, Kadlubar FF. In situ hybridization and immunohistochemical analysis of cytochrome P450 1B1 expression in human normal tissues. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2001;49:229–36. 10.1177/002215540104900210 . [DOI] [PubMed] [Google Scholar]
- 2.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6(4):641–7. 10.1093/hmg/6.4.641 . [DOI] [PubMed] [Google Scholar]
- 3.Stoilov I, Akarsu AN, Alozie I, Child A, Barsoum-Homsy M, Turacli ME, et al. Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am J Hum Genet. 1998;62(3):573–84. 10.1086/301764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Sorenson CM, Sheibani N. Cytochrome P450 1B1 and Primary Congenital Glaucoma. J Ophthalmic Vis Res. 2015;10(1):60–7. Epub 2015/05/26. 10.4103/2008-322X.156116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Y, Scheef EA, Wang S, Sorenson CM, Marcus CB, Jefcoate CR, et al. CYP1B1 expression promotes the proangiogenic phenotype of endothelium through decreased intracellular oxidative stress and thrombospondin-2 expression. Blood. 2009;113(3):744–54. Epub 2008/11/14. 10.1182/blood-2008-03-145219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Scheef EA, Gurel Z, Sorenson CM, Jefcoate CR, Sheibani N. CYP1B1 and endothelial nitric oxide synthase combine to sustain proangiogenic functions of endothelial cells under hyperoxic stress. Am J Physiol Cell Physiol. 2010;298(3):C665–78. Epub 2009/12/25. 10.1152/ajpcell.00153.2009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palenski TL, Sorenson CM, Jefcoate CR, Sheibani N. Lack of Cyp1b1 promotes the proliferative and migratory phenotype of perivascular supporting cells. Lab Invest. 2013;93(6):646–62. Epub 2013/04/10. 10.1038/labinvest.2013.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falero-Perez J, Sorenson CM, Sheibani N. Cyp1b1-deficient retinal astrocytes are more proliferative and migratory and are protected from oxidative stress and inflammation. Am J Physiol Cell Physiol. 2019;316(6):C767–c81. Epub 2019/03/21. 10.1152/ajpcell.00021.2019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruttiger M. Development of the retinal vasculature. Angiogenesis. 2007;10(2):77–88. 10.1007/s10456-007-9065-1 . [DOI] [PubMed] [Google Scholar]
- 10.Stone J, Makarov F, Hollander H. The glial ensheathment of the soma and axon hillock of retinal ganglion cells. Vis Neurosci. 1995;12(2):273–9. Epub 1995/03/01. 10.1017/s0952523800007951 [DOI] [PubMed] [Google Scholar]
- 11.Fruttiger M, Calver AR, Kruger WH, Mudhar HS, Michalovich D, Takakura N, et al. PDGF mediates a neuron-astrocyte interaction in the developing retina. Neuron. 1996;17(6):1117–31. 10.1016/s0896-6273(00)80244-5 [DOI] [PubMed] [Google Scholar]
- 12.Barnett JA, Urbauer DL, Murray GI, Fuller GN, Heimberger AB. Cytochrome P450 1B1 expression in glial cell tumors: an immunotherapeutic target. Clin Cancer Res. 2007;13(12):3559–67. Epub 2007/06/19. 10.1158/1078-0432.CCR-06-2430 [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. Epub 2008/11/19. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. Epub 2012/10/30. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 Epub 2011/08/06. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25 Epub 2010/03/04. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. Epub 2009/11/17. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–75. Epub 2003/02/14. 10.1093/bioinformatics/btf877 [DOI] [PubMed] [Google Scholar]
- 19.Ward JH. Hierarchical Grouping to Optimize an Objective Function. Journal of the American Statistical Association. 1963;58(301):236–44. 10.1080/01621459.1963.10500845 [DOI] [Google Scholar]
- 20.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–53. Epub 2016/05/18. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rungger-Brandle E, Achtstatter T, Franke WW. An epithelium-type cytoskeleton in a glial cell: astrocytes of amphibian optic nerves contain cytokeratin filaments and are connected by desmosomes. J Cell Biol. 1989;109(2):705–16. Epub 1989/08/01. 10.1083/jcb.109.2.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piromkraipak P, Sangpairoj K, Tirakotai W, Chaithirayanon K, Unchern S, Supavilai P, et al. Cysteinyl Leukotriene Receptor Antagonists Inhibit Migration, Invasion, and Expression of MMP-2/9 in Human Glioblastoma. Cell Mol Neurobiol. 2018;38(2):559–73. Epub 2017/06/11. 10.1007/s10571-017-0507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao T, Gan Q, Stokes A, Lassiter RN, Wang Y, Chan J, et al. beta-catenin regulates Pax3 and Cdx2 for caudal neural tube closure and elongation. Development. 2014;141(1):148–57. Epub 2013/11/29. 10.1242/dev.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atala A. Re: Resistance to Docetaxel in Prostate Cancer is Associated with Androgen Receptor Activation and Loss of KDM5D Expression. J Urol. 2017;197(1):154–5. Epub 2016/12/17. 10.1016/j.juro.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Alexander PB, Wang XF. TGF-beta Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb Perspect Biol. 2017;9(4). Epub 2016/12/07. 10.1101/cshperspect.a022145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian X, Yang C, Yang L, Sun Q, Liu N. PTPRF as a novel tumor suppressor through deactivation of ERK1/2 signaling in gastric adenocarcinoma. OncoTargets and therapy. 2018;11:7795–803. Epub 2018/11/23. 10.2147/OTT.S178152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng W, Wang Y, Zhao S, Zhang Y, Chen Y, Zhao X, et al. MICAL1 facilitates breast cancer cell proliferation via ROS-sensitive ERK/cyclin D pathway. J Cell Mol Med. 2018;22(6):3108–18. Epub 2018/03/11. 10.1111/jcmm.13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol. 2007;396:71–91. Epub 2007/11/21. 10.1007/978-1-59745-515-2_6 [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101; discussion -3, 19–28, 244–52. Epub 2003/01/24. [PubMed] [Google Scholar]
- 30.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. Epub 1998/12/10. 10.1093/nar/27.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res Bull. 1999;49(6):377–91. Epub 1999/09/14. 10.1016/s0361-9230(99)00072-6 . [DOI] [PubMed] [Google Scholar]
- 32.Saadoun S, Papadopoulos MC, Watanabe H, Yan D, Manley GT, Verkman AS. Involvement of aquaporin-4 in astroglial cell migration and glial scar formation. J Cell Sci. 2005;118(Pt 24):5691–8. Epub 2005/11/24. 10.1242/jcs.02680 [DOI] [PubMed] [Google Scholar]
- 33.Huang XQ, Zhang XY, Wang XR, Yu SY, Fang SH, Lu YB, et al. Transforming growth factor beta1-induced astrocyte migration is mediated in part by activating 5-lipoxygenase and cysteinyl leukotriene receptor 1. Journal of neuroinflammation. 2012;9:145 Epub 2012/06/28. 10.1186/1742-2094-9-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rich JN, Zhang M, Datto MB, Bigner DD, Wang XF. Transforming growth factor-beta-mediated p15(INK4B) induction and growth inhibition in astrocytes is SMAD3-dependent and a pathway prominently altered in human glioma cell lines. J Biol Chem. 1999;274(49):35053–8. Epub 1999/11/27. 10.1074/jbc.274.49.35053 [DOI] [PubMed] [Google Scholar]
- 35.Trettel F, Di Castro MA, Limatola C. Chemokines: Key Molecules that Orchestrate Communication among Neurons, Microglia and Astrocytes to Preserve Brain Function. Neuroscience. 2019. Epub 2019/08/04. 10.1016/j.neuroscience.2019.07.035 [DOI] [PubMed] [Google Scholar]
- 36.Mao W, Yi X, Qin J, Tian M, Jin G. CXCL12 promotes proliferation of radial glia like cells after traumatic brain injury in rats. Cytokine. 2019;125:154771 Epub 2019/08/11. 10.1016/j.cyto.2019.154771 [DOI] [PubMed] [Google Scholar]
- 37.Gao D, Tang T, Zhu J, Tang Y, Sun H, Li S. CXCL12 has therapeutic value in facial nerve injury and promotes Schwann cells autophagy and migration via PI3K-AKT-mTOR signal pathway. Int J Biol Macromol. 2019;124:460–8. Epub 2018/11/06. 10.1016/j.ijbiomac.2018.10.212 [DOI] [PubMed] [Google Scholar]
- 38.Hillen AEJ, Burbach JPH, Hol EM. Cell adhesion and matricellular support by astrocytes of the tripartite synapse. Prog Neurobiol. 2018;165–167:66–86. Epub 2018/02/15. 10.1016/j.pneurobio.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 39.Dorrell MI, Aguilar E, Friedlander M. Retinal vascular development is mediated by endothelial filopodia, a preexisting astrocytic template and specific R-cadherin adhesion. Invest Ophthalmol Vis Sci. 2002;43(11):3500–10. . [PubMed] [Google Scholar]
- 40.Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, et al. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148(3):579–90. Epub 2000/02/09. 10.1083/jcb.148.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hrycay EG, Bandiera SM. Involvement of Cytochrome P450 in Reactive Oxygen Species Formation and Cancer. Adv Pharmacol. 2015;74:35–84. Epub 2015/08/04. 10.1016/bs.apha.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 42.Hassan HM, Guo H, Yousef BA, Guerram M, Hamdi AM, Zhang L, et al. Role of Inflammatory and Oxidative Stress, Cytochrome P450 2E1, and Bile Acid Disturbance in Rat Liver Injury Induced by Isoniazid and Lipopolysaccharide Cotreatment. Antimicrob Agents Chemother. 2016;60(9):5285–93. Epub 2016/06/22. 10.1128/AAC.00854-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu D, Ma Y, Zhang W, Bao D, Dong W, Lian H, et al. Knockdown of cytochrome P450 2E1 inhibits oxidative stress and apoptosis in the cTnT(R141W) dilated cardiomyopathy transgenic mice. Hypertension. 2012;60(1):81–9. Epub 2012/06/06. 10.1161/HYPERTENSIONAHA.112.191478 [DOI] [PubMed] [Google Scholar]
- 44.Veith A, Moorthy B. ROLE OF CYTOCHROME P450S IN THE GENERATION AND METABOLISM OF REACTIVE OXYGEN SPECIES. Current opinion in toxicology. 2018;7:44–51. Epub 2018/03/13. 10.1016/j.cotox.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falero-Perez J, Song YS, Sorenson CM, Sheibani N. CYP1B1: A key regulator of redox homeostasis. Trends in cell & molecular biology. 2018;13:27–45. Epub 2018/01/01. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Wang S, Sorenson CM, Teixeira L, Dubielzig RR, Peters DM, et al. Cyp1b1 mediates periostin regulation of trabecular meshwork development by suppression of oxidative stress. Mol Cell Biol. 2013;33(21):4225–40. Epub 08/28. 10.1128/MCB.00856-13 Epub 2013 Aug 26. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Y, Wu D, Ding X, Ying W. CD38 plays key roles in both antioxidation and cell survival of H2O2-treated primary rodent astrocytes. International journal of physiology, pathophysiology and pharmacology. 2014;6(2):102–8. Epub 2014/07/25. [PMC free article] [PubMed] [Google Scholar]
- 48.Mittal B, Tulsyan S, Kumar S, Mittal RD, Agarwal G. Cytochrome P450 in Cancer Susceptibility and Treatment. Adv Clin Chem. 2015;71:77–139. Epub 2015/09/29. 10.1016/bs.acc.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 49.Gajjar K, Martin-Hirsch PL, Martin FL. CYP1B1 and hormone-induced cancer. Cancer Lett. 2012;324:13–30. 10.1016/j.canlet.2012.04.021 . [DOI] [PubMed] [Google Scholar]
- 50.Bolton JL, Thatcher GR. Potential mechanisms of estrogen quinone carcinogenesis. Chem Res Toxicol. 2008;21(1):93–101. Epub 2007/12/07. 10.1021/tx700191p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–41. 10.1016/j.pharmthera.2012.12.007 . [DOI] [PubMed] [Google Scholar]
- 52.Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells. Cancer Lett. 2005;223(1):1–10. Epub 2005/05/14. 10.1016/j.canlet.2004.08.030 [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–72. Epub 2009/01/21. 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun H, Ni SJ, Ye M, Weng W, Zhang Q, Zhang M, et al. Hedgehog Interacting Protein 1 is a Prognostic Marker and Suppresses Cell Metastasis in Gastric Cancer. Journal of Cancer. 2018;9(24):4642–9. Epub 2018/12/28. 10.7150/jca.27686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Wang Y, Chen Z, Sternlicht MD, Hidalgo M, Steffensen B. Matrix metalloproteinase-2 contributes to cancer cell migration on collagen. Cancer Res. 2005;65(1):130–6. Epub 2005/01/25. [PubMed] [Google Scholar]
- 56.Hur JH, Park MJ, Park IC, Yi DH, Rhee CH, Hong SI, et al. Matrix metalloproteinases in human gliomas: activation of matrix metalloproteinase-2 (MMP-2) may be correlated with membrane-type-1 matrix metalloproteinase (MT1-MMP) expression. J Korean Med Sci. 2000;15(3):309–14. Epub 2000/07/15. 10.3346/jkms.2000.15.3.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramachandran RK, Sorensen MD, Aaberg-Jessen C, Hermansen SK, Kristensen BW. Expression and prognostic impact of matrix metalloproteinase-2 (MMP-2) in astrocytomas. PLoS One. 2017;12(2):e0172234 Epub 2017/02/25. 10.1371/journal.pone.0172234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheef E, Wang S, Sorenson CM, Sheibani N. Isolation and characterization of murine retinal astrocytes. Mol Vis. 2005;11:613–24. . [PubMed] [Google Scholar]
- 59.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182 Epub 2014/06/14. 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirov S, Ji R, Wang J, Zhang B. Functional annotation of differentially regulated gene set using WebGestalt: a gene set predictive of response to ipilimumab in tumor biopsies. Methods Mol Biol. 2014;1101:31–42. Epub 2013/11/16. 10.1007/978-1-62703-721-1_3 [DOI] [PubMed] [Google Scholar]
- 61.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–8. Epub 2005/06/28. 10.1093/nar/gki475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. Epub 2013/05/25. 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.