Abstract
Transfusion safety relating to blood‐transmissible agents is a major public health concern, particularly when faced with the continuing emergence of new infectious agents. These include new viruses appearing alongside other known reemerging viruses (West Nile virus, Chikungunya) as well as new strains of bacteria and parasites (Plasmodium falciparum, Trypanosoma cruzi) and finally pathologic prion protein (variant Creutzfeldt‐Jakob disease). Genomic mutations of known viruses (hepatitis B virus, hepatitis C virus, human immunodeficiency virus) can also be at the origin of variants susceptible to escaping detection by diagnostic tests. New technologies that would allow the simultaneous detection of several blood‐transmissible agents are now needed for the development and improvement of screening strategies. DNA microarrays have been developed for use in immunohematology laboratories for blood group genotyping. Their application in the detection of infectious agents, however, has been hindered by additional technological hurdles. For instance, the variability among and within genomes of interest complicate target amplification and multiplex analysis. Advances in biosensor technologies based on alternative detection strategies have offered new perspectives on pathogen detection; however, whether they are adaptable to diagnostic applications testing biologic fluids is under debate. Elsewhere, current nanotechnologies now offer new tools to improve the sample preparation, target capture, and detection steps. Second‐generation devices combining micro‐ and nanotechnologies have brought us one step closer to the potential development of innovative and multiplexed approaches applicable to the screening of blood for transmissible agents.
ABBREVIATIONS:
- PSA
prostate‐specific antigen
- QD(s)
quantum dot(s)
- SPR
surface plasmon resonance
Over the past few decades, blood screening has contributed significantly to the improvement in blood transfusion safety. The emergence of nucleic acid amplification technology (NAT) for the screening of human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV) genomes marked a technological turning point not only by increasing the level of sensitivity and reducing the diagnostic window periods, but also by facilitating the parallel detection of several viruses. 1 The relationship between potential infectivity and viral load was recently evaluated to accurately estimate the residual risk from these viruses. 2 Nevertheless, blood testing must be able to adapt to the demands of microbiologic safety and thus consider the fact that the engaged offensive against the transmissible agents has not yet finished. 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12
The problems faced developing a sound approach for the simultaneous detection of multiple blood‐transmissible agents remain unresolved by currently available technologies. Advances in microtechnologies over the past few years have led to the development of miniaturized supports for the analysis of nucleic acid sequences and proteins. Developments in this area have been most marked by DNA microarrays coupled with molecular fluorophore probes; however, technical limitations for their use in clinical diagnostics became quickly apparent. The quest for improved technologies of detection has led to the development of alternative approaches avoiding target amplification and fluorescent labeling. Of these, new technologies based on biosensors promise reliable results and are the subject of increasing interest. Elsewhere, nanobiotechnologies are being applied to molecular diagnostics, offering new tools to overcome some of the technological obstacles currently faced. Exploiting the properties of nanoparticles has improved sample preparation, target capture, and multiplexing in biologic samples. Finally, the combination of established techniques with these emerging technologies is under development with the aim of designing the second generation of integrated systems for pathogen detection. Rapid, sensitive, and mutiplex diagnostic methods for detecting different transmissible agents in a single assay should have an important impact not only on blood transfusion safety but also in disease monitoring.
MICROARRAY‐BASED DETECTION OF INFECTIOUS AGENTS
Microarrays or chips offer the advantage of being able to detect in parallel multiple nucleic acid sequences (hundreds to several hundreds of thousands), antigens, or antibodies with varying specificity. 3 , 13 Numerous chip formats are available depending on the nature and the size of the chosen surface, the probe immobilization strategies, and the methods of detection used. 13 , 14 DNA microarrays using optical detection of a fluorescent signal are the most common; however, other nonplanar formats using beads also exist. 15 Highly complex and difficult to master interactions between proteins have led to a considerably reduced development of protein chips compared to that of DNA chips.
The use of chips in research for the analysis and discovery of genomes (genomic) and the study of gene expression (transcriptomic, proteomic) has seen considerable development. Consequently, in 2001, while considering their potential application in blood testing, Petrik 16 estimated that a limited number of probes (300‐400) would allow the analysis of blood groups, platelet and granulocyte antigens, and the screening of infection. The concept later exploited in immunohematology consisted of the multiplex amplification of genomic DNA and optical detection of the amplification products after their hybridization onto probes immobilized onto chips, 17 , 18 , 19 384‐well microplates, 20 or beads. 21 While protein chips are proving to be more complex to develop than DNA chips, experts agree on their likely strong impact on immunohematology allowing the detection of certain phenotypic modifications that escape genetic analysis. 22 , 23 , 24 , 25 , 26 Microtechnology‐based biochips are in rapid development for application in blood typing, 27 but improved technologies are needed to overcome the problems faced when applying these biochips in the multiplex detection of blood transmissible agents.
The variable level of infectious genomes of interest in biologic samples complicate microarray‐based analysis. Multiplex polymerase chain reaction (PCR) amplification of viral or bacterial nucleotide sequences for the simultaneous detection of transmissible agents is at present difficult to conceive. 13 , 28 , 29 , 30 , 31 Indeed, the use of multiplexing in diagnostics is currently limited to the detection of two to four viral targets. 31 , 32
Miniaturization techniques in genomics, 14 , 33 and high‐throughput sequencing, 34 , 35 , 36 were originally developed for use in research for the parallel detection of numerous viral or bacterial pathogens. In 2002, Wang and colleagues 37 published in PNAS a prototype DNA chip capable of simultaneously detecting hundreds of viruses. After the extraction of nucleic acids from the sample and their random amplification by PCR, sequences labeled by a fluorochrome were hybridized onto the microchip and optically detected. A second version carrying 10,000 oligonucleotides targeting approximately 1000 viruses permitted the identification of the SARS virus in 2003. 38 While the Virochip is a very strong research tool, it remains too complex for use in routine diagnostics. In 2007, an Austrian team proposed a DNA chip which, after genomic amplification, allowed the parallel detection of 25 different pathogens (bacteria and fungi). The observed detection limits varied from 10 bacteria (Escherichia coli) to 105 bacteria (Staphylococcus aureus) per mL of artificially spiked blood. 39 The use of DNA chips also represents an efficient approach for the rapid detection of genetic variations of a given virus, as shown by the team of Rios and colleagues 40 monitoring the genetic variations of the West Nile virus. Recently in Nature Reviews Microbiology, Ecker and coworkers 41 presented an original strategy using only six pairs of primers to amplify the genomes of 373 bacteria. The analysis of amplicons was performed by mass spectrometry on a new industrial platform (Ibis 5000). Thus, for the detection of infectious agents, the difficulties of multiplex amplification may be bypassed by developing random PCR procedures, as is the case with the Virochip approach, or generic PCR procedures targeting conserved regions of the bacterial or viral genome and allowing the simultaneous amplification of numerous sequences, which are then analyzed by hybridization on chips presenting probes specific to the target sequences. Other microarray developments have been proposed for pathogen detection and several systems are today commercialized for use in biodefense applications. 42 However, microarray technology coupled with molecular fluorophore probes has several limitations. These include the need for target amplification and fluorophore labeling, slow binding kinetics between target sequence and probe, and in the case of multiplexed analysis, overlapping spectral features of the fluorophores, nonuniform photobleaching rates, the need for multiple‐laser excitation sources, and complex, expensive instrumentation. 43 Finally, few systems have to date received official approval allowing their commercialization for use in clinical diagnostics. 13
At the protein level, all chips proposed to date for use in microbiology are mainly planar supports onto which antigens or antibodies have been attached. These systems, corresponding to immunoassays in a miniaturized format, offer the advantage of parallelism and of a reduced cost per analysis. 44 , 45 , 46 Important features for the sensitive and specific detection of proteins on microarrays include a high signal‐to‐noise ratio, compatibility with a multiplex format, and low instrumentation costs. The use of chemiluminescence has drawbacks in terms of dynamic range and multiplexing. Signal detection on microarrays using radioactivity and chemiluminescence can only be performed once and fluorescence detection requires a signal amplification technology to provide sufficient sensitivity for most applications. 44 While much progress has been made, considerable development is still required to ensure high standards of quality. 47 , 48
Another application for protein arrays is in studying the proteomic profile of a sample for the identification of biomarkers, which could potentially be useful in new and innovative diagnostic tests. 49 The SELDI‐TOF/MS ProteinChip technology enables protein capture, purification, analysis, and processing from complex biologic samples directly on ProteinChip Array surfaces. Using this innovative approach, we have compared plasma samples, respectively, from unexposed donors, donors with resolved HCV infection, and chronic HCV carriers. Apolipoprotein C‐III was identified as the first reported candidate biomarker in plasma associated with the spontaneous resolution of infection by HCV. 50 This technology therefore represents an important new tool to develop proteomic studies in human plasma.
Finally, the most commonly used methods in pathogen detection rely on culture, colony counting, immunoassays, and PCR amplification. 28 , 51 , 52 In the case of blood infections, the amount of human genomic DNA can be 1014 times higher than the target pathogen nucleic acids, an important consideration in terms of specificity and sensitivity. 28 , 53 While microarrays are useful tools for high‐throughput analysis of biomolecules, their use in clinical diagnostics is limited by the high cost and length of time associated with sample amplification and labeling. One way of overcoming these limitations is to develop improved systems for sample preparation and signal detection. New technologies based on biosensors and nanobiotechnologies potentially offer the molecular tools required for application in the detection of infectious agents.
BIOSENSORS AND PATHOGEN DETECTION
A biosensor can be defined as a compact analytical device, incorporating a biologic or biomimetic sensing element, either closely connected to or integrated within a transducer system. 54 It is used to detect, transmit, and register semiquantitative or quantitative information on a biochemical or physiologic modification. 55 The choice of biorecognition element depends on a large number of factors including specificity, sensitivity, storage conditions, and operational and environmental stability. The selection also considers the target of interest (antigen, nucleic acid, infectious agent, chemical compound, hormone, etc.). Accordingly, antibodies, enzymes, nucleic acids, receptors, microorganisms, and even whole cells and tissues have all been used as bioreceptors. Antibodies are more commonly used than DNA probes in biosensors designed for pathogen detection. 51 One of the basic requirements of a biosensor is that the bioreceptor be in close proximity to the transducer to enable the communication of physicochemical changes. Immobilization technologies have therefore played a major role. 51 , 56 Depending on the method of transduction used, the biosensors are classed as optical, electrochemical, piezoelectric, magnetic, or micromechanical, 51 the first three classes currently being those most developed for use in detecting infectious agents. 56
Optical biosensors
The optical biosensors are the most widely used in biologic analysis for their specificity and sensitivity. 51 , 57 The detection is usually based on fluorescence or on surface plasmon resonance (SPR). 56 The fluorescence offers the advantage of being able to be coupled to other technologies of reference such as PCR or enzyme‐linked immunosorbent assay (ELISA). Despite the fact that much progress has been made in terms of miniaturization and automation, future developments are still needed to enable the multiplex detection of infectious agents in blood or serum. Moreover, the use of fluorescence technologies requires reagents that are both costly and often associated with lengthy analyses. 56 SPR spectroscopy is particularly attractive for the label‐free detection of molecules. 56 , 57 A glass slide covered with a thin film of gold onto which antibodies have been immobilized is irradiated from the backside by polarized light (from a laser) via a prism and the reflectivity is measured. 51 The optical‐electronic basis of SPR is the transfer of the energy carried by photons of light to electrons at the surface of the metal. 58 Hence, this method consists in measuring changes in the angle of resonance induced by the fixing of target molecules to the surface of the biosensor. This technique, while at present not suitable for high‐throughput detection, has been applied to the detection of bacteria and viruses but is above all used to measure the affinity constants between antigen and antibody or reaction kinetics. The main drawbacks of SPR lie in its complexity, high equipment costs, and sensitivity to nonspecific binding on the surface making blood measurements difficult. 51
Electrochemical biosensors
Electrochemical biosensors are based on measurements of electrical change subsequent to interactions of the sample with the surface of the biosensor. These systems are classed according to the observed variable: current (amperometric), potential (potentiometric), impedance (impedancemetric), or conductivity (conductimetric).
The amperometric biosensors, needing a redox labeling of a secondary antibody, are the most widely used. Many studies have been published describing their application in the food and environmental industries for the rapid detection of bacteria and viruses. 51 , 59 , 60 The development of potentiometric biosensors is much more recent and is encountering many difficulties (sensitivity, reproducibility, need for secondary antibody labeling) in translating biologic binding events. 51 On the other hand, impedancemetric biosensors offer several advantages for use in diagnostics, notably by allowing the measurement of antigen‐antibody interactions in real time and without the need for labeling. The signals measured are directly proportional to the concentration of the target molecule. 51 , 56 , 59 Compared to optical biosensors, these biosensors offer the advantage of not being affected by the turbidity of the sample, by the fluorescence of certain molecules of biologic interest, or by the quenching by certain substances. In addition, the instrumentation is simple, economic, and easily miniaturized. Electrochemical impedance spectroscopy is a very efficient technique to measure biologic binding events at the surface of an electrode. 51 The chemistry at the surface of the electrode determines the sensitivity and the specificity of the biosensor. To evaluate this method for pathogen detection, we developed an immunosensor on a bacterial model using a biotinylated E. coli antibody linked to a self‐assembled monolayer at the surface of a gold electrode. A linear relationship between the electrochemical measurements and the decimal logarithmic value of E. coli concentrations was found ranging from 10 to 103 colony‐forming units (CFUs)/mL. These preliminary results obtained using a conventional electrochemical cell (pure cultures diluted in saline buffer) showed a considerable improvement against the sensitivity of 107 CFUs/mL obtained by SPR optical detection. 61 The antibodies used in this electrochemical impedance spectroscopy biosensor were then coupled to magnetic nanoparticles. 62 This coupling allowed the simultaneous purification of species of interest and their concentration on the microelectrode by the simple use of a magnet, thus leading to the functionalization of the microelectrode to detect the corresponding antigen. Methods used in this work enabling an improved detection signal 62 are currently being developed. Finally, conductimetric biosensors, characterized by their large sensitivity, represent a new pertinent class of analytical systems for diagnostic applications. 56 We have evaluated the possibility of using simple conductimetric transducers to measure antigen‐antibody interactions using E. coli detection as a model. The nanoparticles funtionalized by the E. coli antibody were immobilized onto a conductimetric transducer. Results showed an excellent sensitivity on E. coli cultures with a detection limit of 1 CFU/mL coupled with a good specificity of binding. 63 The encouraging preliminary results obtained over the past 2 years 61 , 62 , 63 , 64 have led to further developments for the application of electrochemical biosensors in viral detection. In this line, a microelectrode‐based impedance immunosensor was recently developed by Wang and coworkers 65 for the detection of avian influenza virus H5N1.
Considering the conductive properties of nucleic acids, electrochemical sensors also offer opportunities for the detection of selected DNA sequences or mutated genes associated with human disease. 53 , 66 , 67 The ultimate goal, for pathogen detection in particular, is an assay involving rapid nucleic acid isolation with the rapid detection of a few copies. Research on the application of these systems to biologic samples is in progress and is expected to produce novel alternatives for bioanalysis. 68
Piezoelectric biosensors
Piezoelectric detection is most frequently based on the measurement of variations in the resonance frequency of a quartz crystal, otherwise called quartz crystal microbalance QCM, after a change in the mass caused by the interaction with the target of interest (antigen, antibody, DNA). These biosensors appear to be less sensitive than those based on optical or electrochemical detection. 56 , 59 Piezoelectric biosensors based on nucleic probes or antibodies for the detection of viruses have been developed, but further studies are needed to evaluate the stability of the surface of these biosensors in biologic fluids. 51 , 69 Problems relating to surface regeneration, nonspecific binding of proteins, and the length of incubation time represent known limitations that need to be overcome to develop a platform adapted to diagnostics. 56
To conclude, several biosensors described in the literature for the detection of biomolecules are commercially available to date. One study carried out between 1998 and 2004 reported that more than 6000 publications and 1100 patents had been published at the time on biosensors, highlighting the importance of this research. 56 However, much research and development work is still needed before biosensors can become a true and trustworthy alternative tool for use in the detection of blood transmissible agents. The development of a fast, robust, and long‐lasting biosensor is a genuine challenge. One of the major remaining questions is whether biosensors will play an important role in the detection of pathogens in complex media such as the blood or serum. 56 Outstanding issues include 1) how to take a sample size of clinical relevance for pathogen detection and reduce it down to miniaturized size to detect the presence of pathogens, 2) how to improve the recognition and diffusion kinetics between target and probe, and 3) how to integrate and miniaturize the different diagnostic steps. Technical problems also include methods of sensor calibration, sterilization, and the reproducible production of numerous sensors. 53 , 59 Recent developments of nanobiotechnology approaches are, however, offering new tools to push the limits of biosensors for use in bioanalysis.
NANOBIOTECHNOLOGIES APPLIED TO INFECTIOUS AGENT DIAGNOSTICS
Nanotechnologies are defined as “the group of techniques allowing the production, observation and measurement of objects, structures and systems of a few nanometers in size (between 1 and 100 nm) in at least one dimension in space.” It is also the field of science concerning nanoscience applications. Thus the nanoworld is the world of molecules and atoms. In terms of scale, the difference in size between an atom and a tennis ball is equivalent to that between the tennis ball and Earth! Europe is well situated in the development of nanomedicine. Three strategic axes of research have been identified by academic experts and industry members of the European technology platforms: nanodiagnostics including imaging, targeted drug delivery systems, and regenerative medicine. Nanotechnologies offer new tools able to improve the sensitivity, specificity, and reliability of diagnostic tests. The ultimate aim is to develop fast, robust, sensitive, specific, and cost‐effective in vitro diagnostic strategies that will allow the detection of a small number of molecules in a complex sample.
A new dimension
As mentioned, the first generation of microarrays requires fluorescent labeling, a long and costly procedure. In addition, classic fluorophores are sensitive to photobleaching and present wide emission spectra that limit the possibility of multiplexing. 70 The analysis of nucleic acid target sequence requires its prior amplification. While PCR offers the advantage of reaching extremely high levels of sensitivity, it is complex, sensitive to contamination, costly, and difficult to miniaturize and multiplex for the detection of infectious genomes. Protein chips should not only be miniaturized immunoassays but also integrate the detection system. 71 While biosensors allow the use of new methods of detection, their reduced sensitivity and specificity in complex media, both essential criteria in transfusion applications for the multiplex detection of transmissible agents, limit the possibility of routine application of numerous current systems.
The nanosciences and nanotechnologies allow a better control of the organization of atoms and molecules to create nanostructures with new or improved properties. The nanobiotechnologies reach the single‐cell or molecular scale and consequently overcome certain current technological obstacles. Since DNA measures approximately 2.5 nm wide and protein molecules between 1 and 20 nm, these technologies allow the direct analysis of the target of interest. 72 They also enable different measurements to be taken in parallel and the integration of several analytical steps from sample preparation through to detection in one individual miniaturized system. They offer new perspectives in diagnostics in terms of avoiding the amplification of targets and classic fluorescent labeling. 70 , 73 A major focus in nanotechnology research is the development of multiplexed assay systems that can detect a multitude of molecules or whole viruses or bacteria simultaneously. The combination of nano‐objects and nano‐systems in current technologies offers new possibilities for applications in the detection of infectious agents. 70 , 74
Micro‐ and nanoparticles
The physicochemical characteristics of nanoparticles make them ideal for use in diagnostics. 58 , 70 , 74 , 75 , 76 , 77 , 78 For certain substances, new properties are revealed as the size of each of its individual components is reduced into the nanoworld. This is likely explained by the increase in the number of surface‐reactive atoms relative to the total number of atoms in the particle. For example, the available surface area for attaching the probes is considerably increased thus allowing more efficient binding of targets of interest. Fluorescent nanocrystals “quantum dots,” gold nanoparticles, and magnetic particles are the most commonly used 74 , 79 and offer new approaches to molecular recognition and detection (cf. Fig. 1). New methods combining several approaches are emerging for which a strong impact in diagnostics is being revealed (Table 1).
Figure 1.
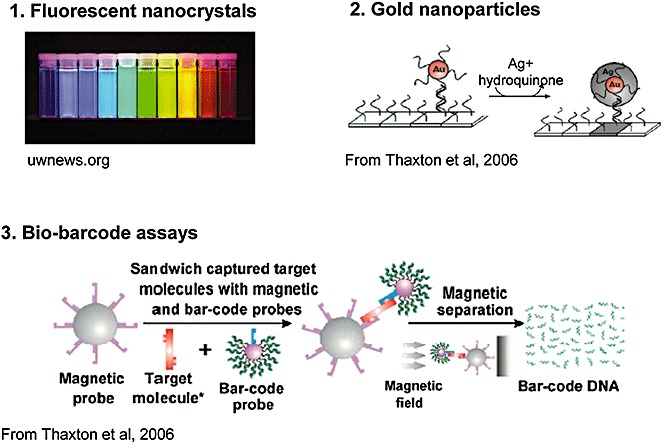
Nanoparticles: applications in diagnostics.
Table 1.
Applications of nanotechnologies to pathogen detection
| Biologic agents | Recognition | Assay format | Sensitivity | Reference |
|---|---|---|---|---|
| Viruses | ||||
| HAV | DNA/DNA | Gold nanoparticles DNA chip | 0.1 pmol/L (DNA) | Wan et al., 2005 90 |
| HBV, HIV, Ebola, variola | DNA/DNA | Bio‐barcodes scanometric detection | 0.5 pmol/L (DNA) | Stoeva et al., 2006 43 |
| HBV, HIV, Ebola, variola | DNA/DNA | Bio‐barcodes capillary analyzer | 5 pmol/L (DNA) | He et al., 2008 104 |
| P24 HIV | Ag/Ab | Bio‐barcodes scanometric detection | 4 fmol/L (p24) | Tang et al., 2007; 101 Kim et al., 2008 102 |
| HBV, HCV, HIV | Ag/Ab | QDs microfluidic | pmol/L (Ab) | Klostranec et al., 2007 114 |
| Bacteria | ||||
| B. anthracis | DNA/DNA | Bio‐barcodes scanometric detection | 550 zmol/L (DNA) | Nam et al., 2004 97 |
| S. aureus | DNA/DNA | Gold nanoparticles optical detection | 33 fmol/L (DNA) | Storhoff et al., 2004 89 |
| E. coli | Ag/Ab | Magnetic nanoparticles conductimetric detection | 1 CFUs/mL (E. coli) | Hnaien et al., 2008 63 |
| B. subtilis | DNA/DNA | Bio‐barcodes scanometric detection | 2.5 fmol/L (DNA) | Hill et al., 2007 103 |
Ag/Ab = antigen/antibody.
Fluorescent nanocrystals
Quantum dots (QDs) are inorganic fluorophores offering significant advantages over organic fluorophores conventionally used to label nucleic acids or proteins for optical detection. 58 , 80 , 81 These biocompatible semiconductor crystals are composed of a nucleus and a shell allowing the binding of ligands and thus the attachment of this fluorescent marker to the target. They are stable and highly luminescent, each offering a wide excitation spectrum and narrow emission spectrum with wavelength controlled by the size and nature of the nucleus (cf. Fig. 1). The absorption and emission spectra are well spaced thus increasing the sensitivity by reducing the phenomenon of autofluorescence. These remarkable properties are due to their nanometric size. The QDs can be used in viral diagnostics as shown by Agrawal and colleagues 82 in 2005 for the detection in real time of respiratory syncytial virus. They are also compatible with analyses of whole blood. 58 The major interest for their use in diagnostics relates to the possibility of multiplexing, thus allowing the performance of high‐throughout analyses of biomolecules. In theory, a combination of three colors and 10 different levels of intensity allow the generation of 999 excitable microbeads by one unique source of light. 80 , 83 This optical method, also called spectral barcoding, opens up new possibilities for the development of innovative diagnostic tests. Nevertheless, in practice, QDs remain difficult to synthesize, funtionalize, and integrate into miniaturized systems. 84
Gold nanoparticles
Gold nanoparticles have been used in biotechnologies for a long period of time; however, it is only over the past 10 years that they have been considered with interest for the detection of nucleic acids or proteins. Progress in the ability to control the size and funtionalize the surface of nanoparticles has allowed the production of optically and chemically defined probes for use in the detection of biomolecules, as described by Mirkin and coworkers. 77 , 85 , 86 , 87 Gold nanoparticles are useful as markers in biosensor systems since numerous optical techniques or electrical conductivity measurements can be used to detect them. One study published in Science in 2000 demonstrated the possibility of detecting hybridization on a chip of oligonucleotides labeled by gold nanoparticles with a simple scanner. 85 This “scanometric” detection is simple and selective and allows the discrimination of a one point mutation on the strand of DNA analyzed. In addition, coupling this detection to a silver amplification method led to a level of sensitivity 100 times greater compared to fluorescent systems using confocal microscopy. Indeed, in the presence of a simple solution of silver and hydroquinone, the silver ions are reduced to the metal form at the surface of the gold nanoparticles (cf. Fig. 1), thus allowing a 100,000‐fold increase in the scanned signal intensity. 77 , 85 This technique has been applied to the direct genomic detection of infectious agents. Within the context of preventing nosocomial infections, Storhoff and coworkers 88 in 2004 demonstrated the possibility of easily detecting the presence of strains of S. aureus resistant to methicillin. Results demonstrated the feasibility of detecting both specifically and directly, without PCR amplification, the DNA extracted from cultures of resistant strains. Another simple colorimetric test using gold nanoparticles was published in Nature Biotechnology by the same group, which allowed the direct detection of 20,000 copies of bacterial genomic DNA within a 1‐µL volume spotted onto a glass slide, corresponding to a femtomolar sensitivity (10−15 mol/L). 89 DNA chips using gold nanoparticle probes and silver enhancement have been developed for the rapid detection of hepatitis A 90 and E, 91 amplified genomes allowing the analysis of 100 fmol/L of amplicon. More recently, antibodies conjugated to gold nanoparticles and silver enhancement were used to develop a compact and rapid sandwich immunoassay. 92 Thus, the use of gold nanoparticles for optical detection should allow the development of new and cost‐effective tests.
Magnetic nanoparticles
While the use of fluorescent nanocrystals and gold nanoparticles aims to improve the detection signal, one reason for using magnetic particles is to optimize the capture of the analyte of interest in the analyzed sample. 64 The use of magnetic nanoparticles for the purification of nucleic acids or proteins has grown over the past several years and is exploited in a number of commercially available kits. The reduction in size of the particles toward the nanometric scale increases the surface available for attachment of the oligonucleotide or protein probes complementary to the target molecule and therefore increases the sensitivity of recognition. For example, Fuentes and colleagues 93 used magnetic nanoparticles to detect small traces of DNA. The presence of two molecules of cDNA of HCV in 1 mL of solution could then be revealed after PCR. Elsewhere, one new approach involves the use of magnetic beads not only for labeling and purification of the analyte but also for detection. Since no significant magnetic background is present in biologic samples, detection and manipulation are possible without affecting biologic interactions. 94 The paramagnetic properties of the beads lead to a modification of the magnetic field allowing their analysis by magnetic transducers. This simple and miniaturizable strategy was used to detect Yersinia pestis antigen F1 with a detection limit comparable to ELISA for detection of the F1 protein. 95 Magnetic labels offer some unique advantages, especially those relating to their physical and chemical stability, their inexpensive production, and their potential detection by a wide range of methods. 94 Magnetic particles are increasingly being used in biodetection for the conception of miniaturized systems.
Bio‐barcodes
The bio‐barcode test, originally developed by Mirkin and colleagues, 96 , 97 , 98 , 99 is an ultrasensitive system of amplification and detection of nucleic acids or proteins. It uses two types of particles to perform purification, amplification, and detection steps (cf. Fig. 1). The first particle is a magnetic microparticle consisting of a target recognition probe: in the case of DNA, this probe is an oligonucleotide complementary to the target DNA and in the case of protein the recognition probe is a monoclonal antibody. The second particle is a nanoparticle bearing a recognition element that is complementary to another region of the target molecule, the result being a sandwich‐based capture assay. This second element would be either an oligonucleotide in the case of a nucleic acid target or a polyclonal antibody if the target was a protein. In addition, this second particle is functionalized with hundreds of oligonucleotide tags or barcodes. These tags classically measure around 15‐ to 20‐mer, theoretically giving rise to 420 different possible combinations, permitting the association of one particular tag with each recognized analyte. The application of a magnetic field leads to the purification of complexes formed in the rest of the sample. The barcode tags are then released either chemically or by heating and identified by a sensitive system of detection (for example, the scanometric method). Thus, the DNA barcode acts as a reporter of the targeted molecule that corresponds to signal amplification. It is therefore possible to directly detect the DNA with zeptomolar sensitivity (10−21 mol/L or 333 DNA copies/mL) making it as sensitive as PCR but avoiding the need for enzymes. 97 With regard to proteins, this test reaches attomolar sensitivity (10−18 mol/L), corresponding to a sensitivity 1 million times higher than that of ELISA tests. 98 , 99 , 100 This technique was applied in a microplate format to the multiplex detection of four types of synthetic DNA (HBV, Ebola, HIV, and smallpox) in a concentration of 500 fmol/L (0.5 pmol/L) within 6 hours. 43 Also using this method, Tang and coworkers 101 more recently showed that it was possible to detect the antigen p24 of the virus HIV with femtomolar sensitivity. This is 150‐fold more sensitive than the conventional ELISA with the advantage of an earlier screening of seroconversion (a gain of 3 days). This modified bio‐barcode assay was developed on p24 antibody–coated microplates to capture viral antigen and streptavidin‐coated nanoparticle‐based bio‐barcode DNAs for signal amplification followed by detection using a chip‐based scanometric method. The preliminary results obtained in plasma samples demonstrated the potential applicability of nanotechnology in viral diagnostics, in particular, in resource‐limited settings where NAT is not feasible. 101 A second study based on the direct capture of HIV‐p24 antigen in 160 µL of plasma confirmed the superiority of the bio‐barcode approach compared to ELISA for the detection of the antigen p24 of HIV. A sensitivity of 100% and specificity of 99% was obtained on 112 samples of plasma from infected subjects with a detection range between 4.2 fmol/L (0.1 pg/mL approx. corresponding to 1000 virus particles/mL or 2000 RNA copies/mL) and 420 pmol/L (10 ng/mL) HIV‐p24 antigen. 102 The bio‐barcode assay was also used for the direct detection of genomic double‐stranded DNA using the bacterial model Bacillus subtilis, a model close to Bacillus anthracis (anthrax). 103 This sensitive assay (2.5 fmol/L) represents the first step in the transition of the bio‐barcode laboratory test toward a rapid and sensitive test for the detection of agents used as bioweapons. Elsewhere, the bio‐barcode test was combined with a capillary analyzer for the multiplex detection of the synthetic DNA of four viruses (HBV, Ebola, HIV, and smallpox) in a concentration of 5 pmol/L within 40 minutes. 104 Recently, antibody pairs were selected allowing the detection of the recombinant prion protein in a bio‐barcode test. 105 The concept of the bio‐barcode is particularly original and represents a potential alternative to the classic technologies of PCR and ELISA.
Microfluidic and integrated systems
One of the objectives for developing microfluidic systems is the conception of a truly integrated laboratory‐type platform on a chip. The lab‐on‐a‐chip can be imagined as a glass or silicon plate onto which microcanals and microreservoirs have been engraved within which the sample to be analyzed and the reagents circulate. Different strategies are possible for the design of integrated systems. 84 , 94 , 106 , 107 , 108 One recent review published in Nature highlights the impact of these integrated systems on public health, particularly in developing countries. Microfluidic technologies allow miniaturization and integration of complex functions, which could move sophisticated diagnostic tools out of the developed‐world laboratory. 109 When demonstrating the practicability of new techniques, an effort should be made to integrate sample preparation from “raw” clinical samples and compare detection sensitivity to conventional methods. 84 In 2005, Lin and colleagues 110 developed a miniaturized immunoassay using microchannels and gold nanoparticles, which allows the direct detection of Helicobacter pylori and E. coli antigens at levels equivalent to conventional ELISA. Another microchip‐based sensor using paramagnetic particles was proposed by Aytur and coworkers 111 for the detection of antibodies directed against the dengue virus in biologic samples. This biosensor showed a very good correlation with measurements made with ELISA; in 2008, the bio‐barcode approach was integrated into a microfluidic chip, marking an important step toward automation. 112 Here, the prostate‐specific antigen (PSA) was chosen as a protein target and was detected at concentrations as low as 0.5 fmol/L. The bio‐barcode assay for on‐chip PSA detection offers a sensitivity of detection 4 orders of magnitude higher than commercial ELISA tests. These proof‐of‐concept experiments have helped establish that this microfluidic device has the capacity to detect biomarkers at ultralow concentrations within a biologic sample. 112 Although much of the work in the past has focused on the development of sensing methods, magnetic label–based assays are now being incorporated into instruments that incorporate sample fluidics. 94 Through the convergence of nano‐ and microtechnologies, various new approaches are now in development. Liu and coworkers 113 described a disposable electrochemical immunosensor that integrates the immunochromatographic strip technique with an electrochemical immunoassay and exploits QDs as labels to amplify the signal. Immunoglobulin (Ig)G was used as a model analyte and, in this study, QDs were used as labels with which to tag anti‐IgG by electrochemical immunosensing. A sandwich immunoreaction was performed. After dissolution of the captured QDs, the released metal ions were detected by highly sensitive voltammetric measurements. This very simple miniaturized system was successfully applied to the detection of PSA in human serum and provides a new tool for protein biomarker detection. Another microfluidic immunoassay using QDs, this time as optical labels, was proposed for the multiplex analysis of viruses HBV, HCV, and HIV antibodies in the pmol/L range. 114 Lien and colleagues 115 developed a microfluidic platform to automatically collect, incubate, mix, purify, and enrich virus samples using magnetic beads and microfluidic systems. This device was used to successfully perform the purification and enrichment of dengue virus. Very recently, this research group presented an innovative fluorescent‐based microfluidic system for the rapid serologic analysis of IgM and IgG associated with the dengue virus infection by utilizing virus‐bound magnetic beads. In this microfluidic system, 100 µL of serum sample is loaded into the sample chamber of the integrated device and the entire process is performed within 30 minutes. 116 Another approach exploiting magnetic beads in a microfluidic chip was developed to discriminate the pathologic form of the prion protein from its normal form. This microfluidic enzymatic reactor based on proteinase K–grafted magnetic beads exploits the differences in enzymatic susceptibility between the two forms. 117 Magnetic nanotags have also been developed in ultrasensitive assays based on magnetic signals. Since biologic samples lack a detectable magnetic background signal and therefore do not interfere with the magnetic transduction mechanism, the magnetic nanosensor technology is well suited for multiplex protein detection in clinical samples, as described recently in PNAS 118 and Nature Medicine. 119 Osterfeld and coworkers 118 implemented magnetic nanotag‐based detection on a protein chip, which has an array of 64 sensors and a 200‐µL reaction well placed on top, allowing easy pipetting and aspiration of reagents. The multianalyte capacity, high sensitivity (reaching the low fmol/L concentration range), scalability, and ease of use of this technology in serum samples make it a strong candidate for use in multiplexed molecular diagnostics. This approach has been characterized in detail for several candidate analytes to prove its generality and its superiority to ELISA. 119 , 120 In particular, this method overcomes the drawbacks of biologic matrices concerning interference and the limited sensitivity, specificity, and multiplexing capacity of the majority of protein detection platforms. This magnetic nanosensor technology can be used for multiplexing, with 64 assays performed on the same device at sensitivities reaching 1000‐fold that of ELISA and an extensive linear dynamic range of over 6 orders of magnitude for diverse biologic fluids. 119
The microfluidic‐based devices have also been developed for nucleic acid analysis to integrate the amplification step. Recently, a sequence‐specific electrochemical DNA sensor was developed and applied to the detection of genomic DNA of Salmonella enterica with a limit of detection below 10 amol/L. 121 Electrochemical detection offers many advantages as a basis for such platforms, including portability and integration with electronics. The biggest obstacle relates to the linking of a “raw” sample to the on‐chip amplification process. 122 The complete integration of all analytical steps and in particular of sample preparation onto a miniaturized support with a view to performing quantitative analysis remains a challenge.
TOWARD THE SECOND GENERATION OF MINIATURIZED SYSTEMS
The second generation of biosensors should allow the development of highly effective platforms. The new principles of target capture and detection exploited in these systems combine micro‐ and nanotechnologies. Excellent levels of sensitivity have been reached bringing us closer to the development of cost‐effective miniaturized tests for nucleic acid– or protein‐based diagnostics. To date however, there still exist no platforms exploiting these miniaturized systems that are as effective and robust as those based on ELISA tests or PCR amplification. However, the combination of different biosensor/nanoparticle/microfluidic approaches should lead to new solutions (Fig. 2). The possible combinations are infinite considering the available multiple innovative principles. Among those remaining to be developed are combinations able to meet the requirements for use in screening for infectious agents transmissible by blood.
Figure 2.
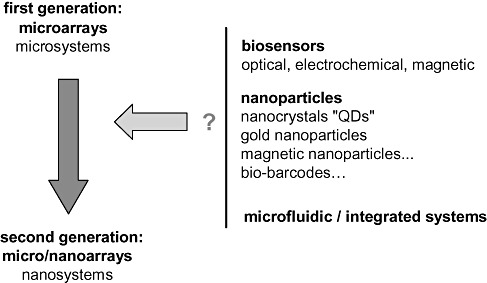
Second generation of miniaturized systems.
ETHICS AND RISK
All scientific and technological advances carry a certain risk of negative consequences and nanobiotechnologies are no exception to the rule. Public authorities are sensitive to these issues and argue in favor of a reasoned, controlled, and responsible development of nanotechnology. The potentially toxic effects of the nanoparticles generally concern their use in vivo, notably for imaging techniques or for drug delivery and less their use in in vitro diagnostics. 72 Nevertheless, these particles of a diameter less than 20 nm are able to penetrate into cells and therefore their innocuousness should be tested thoroughly. The elimination of waste potentially containing nanoparticles should be managed appropriately. Numerous reports have been published and several European and international projects are under way to evaluate the dangers linked to nanotechnologies.
CONCLUSIONS
Currently, the risk of transmission of the retroviruses HIV, human T‐lymphotropic virus, or hepatitis C or B via blood transfusion is extremely low. Monitoring should remain rigorous, however, for the early detection of any eventual new variants and emerging infectious agents (viruses, bacteria, parasites, or prions). The development of multiplex and flexible tests allowing the simultaneous analysis of pathogens presenting a transfusional risk remains a challenge. Most of the development concerning microtechnologies and in particular chips has largely been for research applications. On the other hand, few systems have been commercialized for use in diagnostics due to the consistent rapid discovery of technological and commercial constraints. Furthermore, multiplex amplification of viral or bacterial genomes is technically very challenging. DNA microarray–based technology has limitations including the need for target amplification and fluorophore labeling that increase analysis costs and the time to obtain results. Inherent autofluorescence or optical absorption of many biologic samples or reagents are also a major limiting factor in ELISAs and protein microarrays since the readout is based on a fluorescent or colorimetric signal. Technological advances over the past 10 years have enabled the conception of a second generation of biosensors combining microtechnologies and nanotechnologies, opening up new avenues to explore other potential applications. New nanotools are now available helping improve sample preparation and capture of the biologic target of interest. The properties of nanoparticles will take multiplexing far beyond what can be achieved using molecular labels. These new devices offer a potentially greater flexibility and higher multiplexing capabilities than conventional optical‐based detection methods. The signal amplification approaches could replace those of target amplification (PCR, transcription‐mediated amplification) exploited in the platforms in current use for genomic diagnostics. Other principles of detection that avoid fluorescent labeling could be applied to molecular diagnostics. Electrochemical detection is not affected by the turbidity of the sample and can be miniaturized and multiplexed relatively easily. In addition, electrochemical biosensors show good analytical performance and allow the direct detection in real time of molecular targets. As such, these biosensors are undergoing rapid development and yet much more is needed before they are adaptable for use in the detection of infectious diseases. Elsewhere, magnetic nanotags also represent a promising alternative to fluorescent labels in biomolecular detection assays. Magnetic nanosensor technology is matrix insensitive and an innovative and highly sensitive approach that can be used for multiplexing. With the evolution of nanotechnologies and our increasing understanding of the universe of the infinitesimal small, the field of possibilities is endless. Applying these new technologies to the detection of blood‐transmissible agents will require the collaboration of physicists, engineers, chemists, biochemists, and biologists. Considering the complexity of biologic fluids, all emerging innovative principles should be rigorously tested in the medium to be analyzed. While the miniaturization of the screening of transmissible agents represents a difficult challenge, the identified benefits should be enough to motivate all players concerned.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest relevant to the manuscript submitted to TRANSFUSION.
ACKNOWLEDGMENT
The authors thank C. Mirkin of the Northwestern University for permission to reprint Fig. 1.
REFERENCES
- 1. Coste J, Reesink HW, Engelfriet CP, Laperche S, Brown S, Busch MP, Cuijpers HT, Elgin R, Ekermo B, Epstein JS, Flesland O, Heier HE, Henn G, Hernandez JM, Hewlett IK, Hyland C, Keller AJ, Krusius T, Levicnik‐Stezina S, Levy G, Lin CK, Margaritis AR, Muylle L, Niederhauser C, Pastila S, Pillonel J, Pineau J, Van Der Poel CL, Politis C, Roth WK, Sauleda S, Seed CR, Sondag‐Thull D, Stramer SL, Strong M, Vamvakas EC, Velati C, Vesga MA, Zanetti A. Implementation of donor screening for infectious agents transmitted by blood by nucleic acid technology: update to 2003. Vox Sang 2005;88:289‐303. [DOI] [PubMed] [Google Scholar]
- 2. Kleinman SH, Lelie N, Busch MP. Infectivity of human immunodeficiency virus‐1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion 2009;49:2454‐89. [DOI] [PubMed] [Google Scholar]
- 3. Fournier‐Wirth C, Coste J. Fitting new technologies into the safety paradigm: use of microarrays in transfusion In: Dax EM, Farrugia A, Vyas GN, editors. Advances in transfusion safety, Vol. IV. Basel: Karger; 2007. p. 61‐70. [PubMed] [Google Scholar]
- 4. Hoummady M, Morel P. Nanotechnologies revolutionize the biological qualification of blood donations In: Hervé P, Muller JY, Tiberghien P, editors. Transfusion medicine: looking to the future. Paris: John Libbey Eurotext; 2006. p. 45‐58. [Google Scholar]
- 5. Coste J. Residual risks in transfusion: a strategic perspective In: Hervé P, Muller JY, Tiberghien P, editors. Transfusion medicine: looking to the future. Paris: John Libbey Eurotext; 2006. p. 24‐33. [Google Scholar]
- 6. Dong J, Olano JP, McBride JW, Walker DH. Emerging pathogens: challenges and successes of molecular diagnostics. J Mol Diagn 2008;10:185‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alter HJ, Stramer SL, Dodd RY. Emerging infectious diseases that threaten the blood supply. Semin Hematol 2007;44:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dodd RY. Emerging infections and global blood safety. Dev Biol (Basel) 2007;127:237‐48. [PubMed] [Google Scholar]
- 9. Dodd RY. Emerging infections, transfusion safety, and epidemiology. N Engl J Med 2003;349:1205‐6. [DOI] [PubMed] [Google Scholar]
- 10. Allain JP, Stramer SL, Carneiro‐Proietti AB, Martins ML, Lopes da Silva SN, Ribeiro M, Proietti FA, Reesink HW. Transfusion‐transmitted infectious diseases. Biologicals 2009;37:71‐7. [DOI] [PubMed] [Google Scholar]
- 11. Busch M, Walderhaug M, Custer B, Allain JP, Reddy R, McDonough B. Risk assessment and cost‐effectiveness/utility analysis. Biologicals 2009;37:78‐87. [DOI] [PubMed] [Google Scholar]
- 12. Rios M. Climate change and vector‐borne viral diseases potentially transmitted by transfusion. ISBT Sci Ser 2009;4:87‐94. [Google Scholar]
- 13. Petrik J. Diagnostic applications of microarrays. Transfus Med 2006;16:233‐47. [DOI] [PubMed] [Google Scholar]
- 14. Loy A, Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Acta 2006;363:106‐19. [DOI] [PubMed] [Google Scholar]
- 15. Dunbar SA. Applications of Luminex xMAP technology for rapid, high‐throughput multiplexed nucleic acid detection. Clin Chim Acta 2006;363:71‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrik J. Microarray technology: the future of blood testing? Vox Sang 2001;80:1‐11. [DOI] [PubMed] [Google Scholar]
- 17. Beiboer SH, Wieringa‐Jelsma T, Maaskant‐Van Wijk PA, Van Der Schoot CE, Van Zwieten R, Roos D, Den Dunnen JT, De Haas M. Rapid genotyping of blood group antigens by multiplex polymerase chain reaction and DNA microarray hybridization. Transfusion 2005;45:667‐79. [DOI] [PubMed] [Google Scholar]
- 18. Avent ND. Large scale blood group genotyping. Transfus Clin Biol 2007;14:10‐5. [DOI] [PubMed] [Google Scholar]
- 19. Bres JC, Merieux Y, Dugas V, Broutin J, Vnuk E, Jaber M, Rigal D, Martin JR, Souteyrand E, Cabrera M, Cloarec JP. New method for DNA microarrays development: applied to human platelet antigens polymorphisms. Biomed Microdevices 2005;7:137‐41. [DOI] [PubMed] [Google Scholar]
- 20. Denomme GA, Van Oene M. High‐throughput multiplex single‐nucleotide polymorphism analysis for red cell and platelet antigen genotypes. Transfusion 2005;45:660‐6. [DOI] [PubMed] [Google Scholar]
- 21. Hashmi G, Shariff T, Seul M, Vissavajjhala P, Hue‐Roye K, Charles‐Pierre D, Lomas‐Francis C, Chaudhuri A, Reid ME. A flexible array format for large‐scale, rapid blood group DNA typing. Transfusion 2005;45:680‐8. [DOI] [PubMed] [Google Scholar]
- 22. Reid ME. Applications of DNA‐based assays in blood group antigen and antibody identification. Transfusion 2003;43:1748‐57. [DOI] [PubMed] [Google Scholar]
- 23. Petrik J, De Haas M, Denomme G, Scott M, Seghatchian J. Small world—advance of microarrays: current status and future trends. Transfus Apher Sci 2007;36:201‐6. [DOI] [PubMed] [Google Scholar]
- 24. Hillyer CD, Shaz BH, Winkler AM, Reid M. Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev 2008;22:117‐32. [DOI] [PubMed] [Google Scholar]
- 25. Campbell CJ, O'Looney N, Chong Kwan M, Robb JS, Ross AJ, Beattie JS, Petrik J, Ghazal P. Cell interaction microarray for blood phenotyping. Anal Chem 2006;78:1930‐8. [DOI] [PubMed] [Google Scholar]
- 26. Robb JS, Roy DJ, Ghazal P, Allan J, Petrik J. Development of non‐agglutination microarray blood grouping. Transfus Med 2006;16:119‐29. [DOI] [PubMed] [Google Scholar]
- 27. Van Der Schoot CE, De Haas M, Engelfriet CP, Reesink HW, Panzer S, Jungbauer C, Schwartz DM, Mayr WR, Castilho L, St‐Louis M, Long A, Denomme G, Semple E, Fernandes B, Flegel WA, Wagner F, Doescher A, Poli F, Villa MA, Paccapelo C, Veldhuisen B, Nogues N, Muniz‐Diaz E, Daniels G, Martin P, Finning K, Reid ME. Genotyping for red blood cell polymorphisms. Vox Sang 2009;96:167‐79. [DOI] [PubMed] [Google Scholar]
- 28. Peters RP, Van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke‐Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 2004;4:751‐60. [DOI] [PubMed] [Google Scholar]
- 29. Tomioka K, Peredelchuk M, Zhu X, Arena R, Volokhov D, Selvapandiyan A, Stabler K, Mellquist‐Riemenschneider J, Chizhikov V, Kaplan G, Nakhasi H, Duncan R. A multiplex polymerase chain reaction microarray assay to detect bioterror pathogens in blood. J Mol Diagn 2005;7:486‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsia CC, Chizhikov VE, Yang AX, Selvapandiyan A, Hewlett I, Duncan R, Puri RK, Nakhasi HL, Kaplan GG. Microarray multiplex assay for the simultaneous detection and discrimination of hepatitis B, hepatitis C, and human immunodeficiency type‐1 viruses in human blood samples. Biochem Biophys Res Commun 2007;356:1017‐23. [DOI] [PubMed] [Google Scholar]
- 31. Candotti D, Alain JP. The utility of multiplex NAT in blood screening. Dev Biol (Basel) 2007;127:Ed Karger:71‐86. [PubMed] [Google Scholar]
- 32. Compston LI, Sarkobie F, Li C, Candotti D, Opare‐Sem O, Allain JP. Multiplex real‐time PCR for the detection and quantification of latent and persistent viral genomes in cellular or plasma blood fractions. J Virol Methods 2008;151:47‐54. [DOI] [PubMed] [Google Scholar]
- 33. Csako G. Present and future of rapid and/or high‐throughput methods for nucleic acid testing. Clin Chim Acta 2006;363:6‐31. [DOI] [PubMed] [Google Scholar]
- 34. Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. Viral discovery and sequence recovery using DNA microarrays. Plos Biol 2003;1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. Plos Pathog 2008;4:e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R. Microbiology in the post‐genomic era. Nat Rev Microbiol 2008;6:419‐30. [DOI] [PubMed] [Google Scholar]
- 37. Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray‐based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A 2002;99:15687‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen‐Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003;300:1394‐9. [DOI] [PubMed] [Google Scholar]
- 39. Wiesinger‐Mayr H, Vierlinger K, Pichler R, Kriegner A, Hirschl AM, Presterl E, Bodrossy L, Noehammer C. Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition. BMC Microbiol 2007;7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grinev A, Daniel S, Laassri M, Chumakov K, Chizhikov V, Rios M. Microarray‐based assay for the detection of genetic variations of structural genes of West Nile virus. J Virol Methods 2008;154:27‐40. [DOI] [PubMed] [Google Scholar]
- 41. Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, Hofstadler SA. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol 2008;6:553‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uttamchandani M, Neo JL, Ong BN, Moochhala S. Applications of microarrays in pathogen detection and biodefence. Trends Biotechnol 2009;27:53‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem Int Ed Engl 2006;45:3303‐6. [DOI] [PubMed] [Google Scholar]
- 44. Schweitzer B, Kingsmore SF. Measuring proteins on microarrays. Curr Opin Biotechnol 2002;13:14‐9. [DOI] [PubMed] [Google Scholar]
- 45. MacBeath G. Protein microarrays and proteomics. Nat Genet 2002;32 Suppl:526‐32. [DOI] [PubMed] [Google Scholar]
- 46. Mezzasoma L, Bacarese‐Hamilton T, Di Cristina M, Rossi R, Bistoni F, Crisanti A. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem 2002;48:121‐30. [PubMed] [Google Scholar]
- 47. Hultschig C, Kreutzberger J, Seitz H, Konthur Z, Bussow K, Lehrach H. Recent advances of protein microarrays. Curr Opin Chem Biol 2006;10:4‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kreutzberger J. Protein microarrays: a chance to study microorganisms? Appl Microbiol Biotechnol 2006;70:383‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sheridan C. Protein chip companies turn to biomarkers. Nat Biotechnol 2005;23:3‐4. [DOI] [PubMed] [Google Scholar]
- 50. Molina S, Missé D, Roche S, Badiou S, Cristol JP, Bonfils C, Dierick JF, Veas F, Levayer T, Bonnefont‐Rousselot D, Maurel P, Coste J, Fournier‐Wirth C. Identification of apolipoprotein C‐III as a potential plasmatic biomarker associated with the resolution of hepatitis C virus infection. Proteomics, Clin Appl 2008;2:751‐61. [DOI] [PubMed] [Google Scholar]
- 51. Lazcka O, Del Campo FJ, Munoz FX. Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron 2007;22:1205‐17. [DOI] [PubMed] [Google Scholar]
- 52. Schrenzel J. Clinical relevance of new diagnostic methods for bloodstream infections. Int J Antimicrob Agents 2007;30 Suppl 1:S2‐6. [DOI] [PubMed] [Google Scholar]
- 53. Teles FR, Fonseca LP. Trends in DNA biosensors. Talanta 2008;77:606‐23. [Google Scholar]
- 54. Velasco‐Garcia MN. Optical biosensors for probing at the cellular level: a review of recent progress and future prospects. Semin Cell Dev Biol 2009;20:27‐33. [DOI] [PubMed] [Google Scholar]
- 55. Scheller FW, Wollenberger U, Warsinke A, Lisdat F. Research and development in biosensors. Curr Opin Biotechnol 2001;12:35‐40. [DOI] [PubMed] [Google Scholar]
- 56. Pejcic B, De Marco R, Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst 2006;131:1079‐90. [DOI] [PubMed] [Google Scholar]
- 57. Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Sensitive optical biosensors for unlabeled targets: a review. Anal Chim Acta 2008;620:8‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jain KK. Nanotechnology in clinical laboratory diagnostics. Clin Chim Acta 2005;358:37‐54. [DOI] [PubMed] [Google Scholar]
- 59. Ivnitski D, Abdel‐Hamid I, Atanasov P, Wilkins E. Biosensors for detection of pathogenic bacteria. Biosens Bioelectron 1999;14:599‐624. [DOI] [PubMed] [Google Scholar]
- 60. Elsholz B, Nitsche A, Achenbach J, Ellerbrok H, Blohm L, Albers J, Pauli G, Hintsche R, Worl R. Electrical microarrays for highly sensitive detection of multiplex PCR products from biological agents. Biosens Bioelectron 2009;24:1737‐43. [DOI] [PubMed] [Google Scholar]
- 61. Maalouf R, Fournier‐Wirth C, Coste J, Chebib H, Saikali Y, Vittori O, Errachid A, Cloarec JP, Martelet C, Jaffrezic‐Renault N. Label‐free detection of bacteria by electrochemical impedance spectroscopy: comparison to surface plasmon resonance. Anal Chem 2007;79:4879‐86. [DOI] [PubMed] [Google Scholar]
- 62. Maalouf R, Hassen WM, Fournier‐Wirth C, Coste J, Jaffrezic‐Renault N. Comparison of two innovative approaches for bacterial detection: paramagnetic nanoparticles and self‐assembled multilayer processes. Microchim Acta 2008;163:157‐61. [Google Scholar]
- 63. Hnaien M, Hassen WM, Abdelghani A, Fournier‐Wirth C, Coste J, Bessueille F, Leonard D, Jaffrezic‐Renault N. A conductimetric immunosensor based on functionalized magnetite nanoparticles for E. coli detection. Electrochem Comms 2008;1152‐4. [Google Scholar]
- 64. Jaffrezic‐Renault N, Martelet C, Chevolot Y, Cloarec JP. Biosensors and bio‐bar code assays based on biofunctionalized magnetic microbeads. Sensor 2007;7:589‐614. [Google Scholar]
- 65. Wang R, Wang Y, Lassiter K, Li Y, Hargis B, Tung S, Berghman L, Bottje W. Interdigitated array microelectrode based impedance immunosensor for detection of avian influenza virus H5N1. Talanta 2009;79:159‐64. [DOI] [PubMed] [Google Scholar]
- 66. Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol 2003;21:1192‐9. [DOI] [PubMed] [Google Scholar]
- 67. Kuhr WG. Electrochemical DNA analysis comes of age. Nat Biotechnol 2000;18:1042‐3. [DOI] [PubMed] [Google Scholar]
- 68. Merkoci A. Electrochemical biosensing with nanoparticles. Febs J 2007;274:310‐6. [DOI] [PubMed] [Google Scholar]
- 69. Zuo B, Li S, Guo Z, Zhang J, Chen C. Piezoelectric immunosensor for SARS‐associated coronavirus in sputum. Anal Chem 2004;76:3536‐40. [DOI] [PubMed] [Google Scholar]
- 70. Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev 2005;105:1547‐62. [DOI] [PubMed] [Google Scholar]
- 71. Marquette CA, Blum LJ. State of the art and recent advances in immunoanalytical systems. Biosens Bioelectron 2006;21:1424‐33. [DOI] [PubMed] [Google Scholar]
- 72. Jain KK. Nanomedicine: application of nanobiotechnology in medical practice. Med Princ Pract 2008;17:89‐101. [DOI] [PubMed] [Google Scholar]
- 73. Liu G, Lin Y. Nanomaterial labels in electrochemical immunosensors and immunoassays. Talanta 2007;74:308‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jain KK. Applications of nanobiotechnology in clinical diagnostics. Clin Chem 2007;53:2002‐9. [DOI] [PubMed] [Google Scholar]
- 75. Fortina P, Kricka LJ, Surrey S, Grodzinski P. Nanobiotechnology: the promise and reality of new approaches to molecular recognition. Trends Biotechnol 2005;23:168‐73. [DOI] [PubMed] [Google Scholar]
- 76. Azzazy HM, Mansour MM, Kazmierczak SC. Nanodiagnostics: a new frontier for clinical laboratory medicine. Clin Chem 2006;52:1238‐46. [DOI] [PubMed] [Google Scholar]
- 77. Thaxton CS, Georganopoulou DG, Mirkin CA. Gold nanoparticle probes for the detection of nucleic acid targets. Clin Chim Acta 2006;363:120‐6. [DOI] [PubMed] [Google Scholar]
- 78. Bally M, Voros J. Nanoscale labels: nanoparticles and liposomes in the development of high‐performance biosensors. Nanomed 2009;4:447‐67. [DOI] [PubMed] [Google Scholar]
- 79. Johnson CJ, Zhukovsky N, Cass AE, Nagy JM. Proteomics, nanotechnology and molecular diagnostics. Proteomics 2008;8:715‐30. [DOI] [PubMed] [Google Scholar]
- 80. Arya H, Kaul Z, Wadhwa R, Taira K, Hirano T, Kaul SC. Quantum dots in bio‐imaging: revolution by the small. Biochem Biophys Res Commun 2005;329:1173‐7. [DOI] [PubMed] [Google Scholar]
- 81. Wang L, O'Donoghue MB, Tan W. Nanoparticles for multiplex diagnostics and imaging. Nanomed 2006;1:413‐26. [DOI] [PubMed] [Google Scholar]
- 82. Agrawal A, Tripp RA, Anderson LJ, Nie S. Real‐time detection of virus particles and viral protein expression with two‐color nanoparticle probes. J Virol 2005;79:8625‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Han M, Gao X, Su JZ, Nie S. Quantum‐dot‐tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol 2001;19:631‐5. [DOI] [PubMed] [Google Scholar]
- 84. Myers FB, Lee LP. Innovations in optical microfluidic technologies for point‐of‐care diagnostics. Lab Chip 2008;8:2015‐31. [DOI] [PubMed] [Google Scholar]
- 85. Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science 2000;289:1757‐60. [DOI] [PubMed] [Google Scholar]
- 86. Oh M, Mirkin CA. Chemically tailorable colloidal particles from infinite coordination polymers. Nature 2005;438:651‐4. [DOI] [PubMed] [Google Scholar]
- 87. Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 2002;297:1536‐40. [DOI] [PubMed] [Google Scholar]
- 88. Storhoff JJ, Marla SS, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, Cork W, Muller UR. Gold nanoparticle‐based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron 2004;19:875‐83. [DOI] [PubMed] [Google Scholar]
- 89. Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat Biotechnol 2004;22:883‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wan Z, Wang Y, Li SS, Duan L, Zhai J. Development of array‐based technology for detection of HAV using gold‐DNA probes. J Biochem Mol Biol 2005;38:399‐406. [DOI] [PubMed] [Google Scholar]
- 91. Liu HH, Cao X, Yang Y, Liu MG, Wang YF. Array‐based nano‐amplification technique was applied in detection of hepatitis E virus. J Biochem Mol Biol 2006;39:247‐52. [DOI] [PubMed] [Google Scholar]
- 92. Gupta S, Huda S, Kilpatrick PK, Velev OD. Characterization and optimization of gold nanoparticle‐based silver‐enhanced immunoassays. Anal Chem 2007;79:3810‐20. [DOI] [PubMed] [Google Scholar]
- 93. Fuentes M, Mateo C, Rodriguez A, Casqueiro M, Tercero JC, Riese HH, Fernandez‐Lafuente R, Guisan JM. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR‐ELISA. Biosens Bioelectron 2006;21:1574‐80. [DOI] [PubMed] [Google Scholar]
- 94. Tamanaha CR, Mulvaney SP, Rife JC, Whitman LJ. Magnetic labeling, detection, and system integration. Biosens Bioelectron 2008;24:1‐13. [DOI] [PubMed] [Google Scholar]
- 95. Meyer MH, Stehr M, Bhuju S, Krause HJ, Hartmann M, Miethe P, Singh M, Keusgen M. Magnetic biosensor for the detection of Yersinia pestis. J Microbiol Methods 2007;68:218‐24. [DOI] [PubMed] [Google Scholar]
- 96. Nam JM, Park SJ, Mirkin CA. Bio‐barcodes based on oligonucleotide‐modified nanoparticles. J Am Chem Soc 2002;124:3820‐1. [DOI] [PubMed] [Google Scholar]
- 97. Nam JM, Stoeva SI, Mirkin CA. Bio‐bar‐code‐based DNA detection with PCR‐like sensitivity. J Am Chem Soc 2004;126:5932‐3. [DOI] [PubMed] [Google Scholar]
- 98. Nam JM, Thaxton CS, Mirkin CA. Nanoparticle‐based bio‐bar codes for the ultrasensitive detection of proteins. Science 2003;301:1884‐6. [DOI] [PubMed] [Google Scholar]
- 99. Cheng MM, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T, Ferrari M. Nanotechnologies for biomolecular detection and medical diagnostics. Curr Opin Chem Biol 2006;10:11‐9. [DOI] [PubMed] [Google Scholar]
- 100. Nam JM, Jang KJ, Groves JT. Detection of proteins using a colorimetric bio‐barcode assay. Nat Protoc 2007;2:1438‐44. [DOI] [PubMed] [Google Scholar]
- 101. Tang S, Zhao J, Storhoff JJ, Norris PJ, Little RF, Yarchoan R, Stramer SL, Patno T, Domanus M, Dhar A, Mirkin CA, Hewlett IK. Nanoparticle‐based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr 2007;46:231‐7. [DOI] [PubMed] [Google Scholar]
- 102. Kim EY, Stanton J, Korber BT, Krebs K, Bogdan D, Kunstman K, Wu S, Phair JP, Mirkin CA, Wolinsky SM. Detection of HIV‐1 p24 Gag in plasma by a nanoparticle‐based bio‐barcode‐amplification method. Nanomed 2008;3:293‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hill HD, Vega RA, Mirkin CA. Nonenzymatic detection of bacterial genomic DNA using the bio bar code assay. Anal Chem 2007;79:9218‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. He M, Li K, Xiao J, Zhou Y. Rapid bio‐barcode assay for multiplex DNA detection based on capillary DNA analyzer. J Virol Methods 2008;151:126‐31. [DOI] [PubMed] [Google Scholar]
- 105. Brooks BD, Albertson AE, Jones JA, Speare JO, Lewis RV. Efficient screening of high‐signal and low‐background antibody pairs in the bio‐bar code assay using prion protein as the target. Anal Biochem 2008;382:60‐2. [DOI] [PubMed] [Google Scholar]
- 106. Henares TG, Mizutani F, Hisamoto H. Current development in microfluidic immunosensing chip. Anal Chim Acta 2008;611:17‐30. [DOI] [PubMed] [Google Scholar]
- 107. Xu X, Zhang S, Chen H, Kong J. Integration of electrochemistry in micro‐total analysis systems for biochemical assays: recent developments. Talanta 2009;80:8‐18. [DOI] [PubMed] [Google Scholar]
- 108. Kuswandi B, Nuriman, Huskens J, Verboom W. Optical sensing systems for microfluidic devices: a review. Anal Chim Acta 2007;601:141‐55. [DOI] [PubMed] [Google Scholar]
- 109. Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature 2006;442:412‐8. [DOI] [PubMed] [Google Scholar]
- 110. Lin FY, Sabri M, Alirezaie J, Li D, Sherman PM. Development of a nanoparticle‐labeled microfluidic immunoassay for detection of pathogenic microorganisms. Clin Diagn Lab Immunol 2005;12:418‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. A novel magnetic bead bioassay platform using a microchip‐based sensor for infectious disease diagnosis. J Immunol Methods 2006;314:21‐9. [DOI] [PubMed] [Google Scholar]
- 112. Goluch ED, Nam JM, Georganopoulou DG, Chiesl TN, Shaikh KA, Ryu KS, Barron AE, Mirkin CA, Liu C. A bio‐barcode assay for on‐chip attomolar‐sensitivity protein detection. Lab Chip 2006;6:1293‐9. [DOI] [PubMed] [Google Scholar]
- 113. Liu G, Lin YY, Wang J, Wu H, Wai CM, Lin Y. Disposable electrochemical immunosensor diagnosis device based on nanoparticle probe and immunochromatographic strip. Anal Chem 2007;79:7644‐53. [DOI] [PubMed] [Google Scholar]
- 114. Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WC. Convergence of quantum dot barcodes with microfluidics and signal processing for multiplexed high‐throughput infectious disease diagnostics. Nano Lett 2007;7:2812‐8. [DOI] [PubMed] [Google Scholar]
- 115. Lien KY, Lin JL, Liu CY, Lei HY, Lee GB. Purification and enrichment of virus samples utilizing magnetic beads on a microfluidic system. Lab Chip 2007;7:868‐75. [DOI] [PubMed] [Google Scholar]
- 116. Lee YF, Lien KY, Lei HY, Lee GB. An integrated microfluidic system for rapid diagnosis of dengue virus infection. Biosens Bioelectron 2009;25:745‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Le Nel A, Minc N, Smadja C, Slovakova M, Bilkova Z, Peyrin JM, Viovy JL, Taverna M. Controlled proteolysis of normal and pathological prion protein in a microfluidic chip. Lab Chip 2008;8:294‐301. [DOI] [PubMed] [Google Scholar]
- 118. Osterfeld SJ, Yu H, Gaster RS, Caramuta S, Xu L, Han SJ, Hall DA, Wilson RJ, Sun S, White RL, Davis RW, Pourmand N, Wang SX. Multiplex protein assays based on real‐time magnetic nanotag sensing. Proc Natl Acad Sci U S A 2008;105:20637‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS, Wang SX. Matrix‐insensitive protein assays push the limits of biosensors in medicine. Nat Med 2009;15:1327‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Fishbein I, Levy RJ. Analytical chemistry: the matrix neutralized. Nature 2009;461:890‐1. [DOI] [PubMed] [Google Scholar]
- 121. Ferguson BS, Buchsbaum SF, Swensen JS, Hsieh K, Lou X, Soh HT. Integrated microfluidic electrochemical DNA sensor. Anal Chem 2009;81:6503–8. Epub 2009 Jul 8. [DOI] [PubMed] [Google Scholar]
- 122. Chen L, Manz A, Day PJ. Total nucleic acid analysis integrated on microfluidic devices. Lab Chip 2007;7:1413‐23. [DOI] [PubMed] [Google Scholar]


