Dear Editor,
Among the viral-mediated pandemics that have been rapidly spread worldwide in the last twenty years, coronaviruses (CoVs) –dependent outbreaks seem to be the most severe implicated in lung pathology. CoVs represent a significant superfamily of viruses including four major sub-classifications: alpha-, beta-, gamma-, and delta-CoVs. In fact, CoVs belong in a broader viruses group – the Nidovirales order- that comprises Roniviridae, Arteriviridae, and Coronaviridae [1]. Severe Acute Respiratory Syndrome (SARS-CoV) in 2002/2003, Middle-East Respiratory Syndrome (MERS-CoV) in 2012/2013, and the current 2019/2020 Severe Acute Respiratory Syndrome-2 (SARS-CoV-2) are causes of major public health disorganization testing also the national health systems’ endurance worldwide. Extensive molecular analyses have decoded all CoVs’ genomic sequences concluding that although there are small specific differences at the RNA level referring to five only nucleotide sites, the group of 380 amino acid substitutions in 2019 novel beta-CoV (2019-nCoV) -phylogenetic and RNA sequence similarity at 96% with BatCoV RaTG13-provides not only structural divergence, but also more aggressive functional and clinic-pathogenic characteristics in infected communities [2]. Concerning its genetic substrate and protein morphology, its RNA molecule (a large positive-sense RNA genome of approximately 30 kb) is enclosed in a spherical-like glycoprotein envelope characterized by many spikes (S projections) on its surface that create a crown-like formation (corona). These modifications act as binding tools for the corresponding membrane receptors of the host epithelial cells. The main receptor-binding loci that CoVs selectively use for cell invasion are human angiotensin-converting enzyme 2 (hACE2), CD209L, and dipeptyl peptidase 4 (DPP4/CD26) [3, 4].
hACE gene is located on chromosome 17 (band 17q23.3) encoding for a specific protein acting as zinc-based enzyme (metalloenzyme) in peptide hydrolysis. It is localized on the membrane surface of lung epithelia, vascular endothelial cells, and also on Leydig and kidney epithelial cells. In contrast, hACE2 gene is located on chromosome X (band Xp22.2) and its integral membrane protein product -that acts as carboxydipeptidase-shares an approximately 50% sequence homology with hACE. It is expressed mainly in heart, vascular endothelial cells, kidney and intestine cells and also in lung epithelia, critically in type II pneumonocytes of alveoli [5]. Recently, a molecular and structural analysis detected a mechanism of SARS-CoV-2 host cell penetration based on a novel furin-like (an enzyme, member of subtilisin-like proprotein convertase family) cleavage site on S1/S2 spike functional subunits that interacts directly with hACE2 [6, 7]. Interestingly, furin (band 15q26.1) expression is elevated in lung epithelia. In fact, the S1 subunit represents the main receptor-binding domain (RBD), whereas the S2 subunit is involved in membrane fusion. hACE2 receptor recognition and binding exposes rapidly virus transmissibility in human targets. Inhibition strategies are based on SARS-CoV-2 polyclonal antibodies preventing the S1/S2 furin cleavage/ hACE2 conjunction that promotes cell entry. Similarly, furin has been found to be a significant molecule as protease for providing proteolytic cleavage of Human Immunodeficiency Virus (HIV) protein precursor subunits (GP160 to GP120-GP41) before viral particles reconstruction in the Golgi apparatus [8].
Focused on patients that suffer from hematologic malignancies and especially in hematopoietic stem cell transplant recipients (HSCT), CoVs-dependent upper and lower respiratory tract infection is a severe complication, although that there are controversial data regarding the sequence and localization of the infection. A study group based on bronchoalveolar lavage (BAL) sample strain-specific polymerase chain reaction (ss-PCR) analysis reported significant proportions of CoVs genome presence in patients with lower respiratory tract non-malignant lesions (pneumonia) [9]. Additionally, they showed that CoVs - associated mortality rates were similar to other copathogens (ie influenza virus) but an increasing need for oxygen supply was also observed reflecting the severity of the infection. In contrast to the previous published data, another study group analyzed by PCR nasopharyngeal and BAL fluid samples and detected high rates of CoVs genomes correlated with upper respiratory tract lesions compared to limited pneumonia cases [10].
Both of the previous studies are referred to the crucial oxygen supply in severe lung lesions mediated by CoV infection. And this is a key-point not only at the clinical but also at the molecular level. S furin-mediated cleavage combined with hACE interaction leads to cell membrane penetration by SARS-CoV-2 but hypoxia signaling pathways deregulation seem to be also critical for the aggressive phenotype of the current infection. Interestingly, a molecular study co-analyzing hACE2 and Mas receptor (MasR) in CD34+ hematopoietic stem/progenitor cells (HSPCs) concluded that both of them are involved and upregulated in the hypoxic stimulation of the examined cell series [11]. Additionally, another important clinicopathological and molecular (RNA/ oligonucleotide-based microarrays) analysis investigated the role of monocytes/macrophages motivation and pulmonary fibrosis development in SARS-CoV - mediated pneumonia. The study group observed that genes responsible for intracellular oxygen homeostasis (normoxia/hypoxia/hyperoxia) were over activated and highly expressed even in the first day of infection in the corresponding cell populations [12]. Interestingly, furin interacts with genes involved in hypoxia regulation regarding normal and cancerous cells [13]. Additionally, the interferon-induced Guanylate-Binding Proteins (GBPs) -especially 2/5 – inhibit furin-based envelope protein cleavage in a variety of viruses including HIV, Measles, Zika, and also Influenza A type, whereas Phoshpofurin Acidic Cluster Sorting Protein 1 (PACS1) strongly interacts with furin in trans-Golgi network [14].
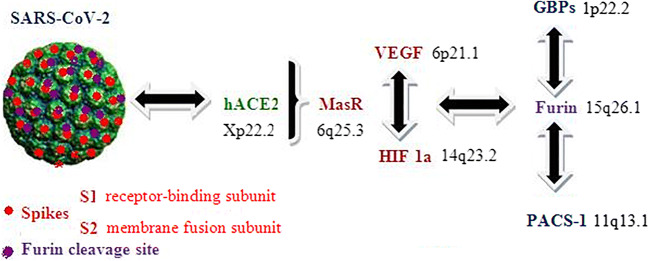
Based on these continually updated molecular data we suggest that Vascular Endothelial Growth Factor/receptor (VEGF- band 6p21.1 /VEGFR), Mas receptor (MasR-6q25.3) and Hypoxia - Inducible Factor 1-alpha (HIF-1a - band14q23.2) should be analyzed in conjunction with hACE2 and Furin/GBP/PACS 1 complex for understanding the complete mechanism of SARS-CoV-2 - dependent severe pneumonia complicated or not with pulmonary embolism in the general population and especially in immunocompromised patients with hematologic malignancies and HSCTs (Fig. 1). Specific polymorphisms (ie single nucleotide polymorphisms –SNPs) in these genes could potentially lead to an optimal discrimination of SARS-CoV-2 infected patients based on their genetic profile explaining the differences in clinical features (resistance and good prognosis) to viral mediated pneumonia and response rates to specific therapeutic anti-viral strategies.
Fig. 1.
Schematic presentation of SARS-CoV-2 cell entry mechanism. Genes that regulate oxygen metabolism (normoxia/hypoxia/hyperoxia) could be useful biomarkers for co-analysis with hACE2 and Furin/GBP/PACS 1 complex at the gene polymorphism level in order to find patients that demonstrate resistance to viral infectivity and mediated pneumonia (SARS-CoV-2: Severe Acute Respiratory Syndrome-Coronovirus-2, hACE2: human angiotensin-converting enzyme 2, Vascular Endothelial Growth Factor (VEGF), MasR: Mas receptor, HIF-1a: Hypoxia - Inducible Factor 1-alpha, GBPs: Guanylate-Binding Proteins, PACS 1: Phoshpofurin Acidic Cluster Sorting Protein 1).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
For PubMed citation:
Tsiambas E, Papanikolaou V, Chrysovergis A, Mastronikolis N, Ragos V, Kavantzas N, Lazaris AC, Kyrodimos E
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu A, Peng Y, Huang B, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microb. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. Structure, function, and evolution of coronavirus spike proteins. Ann Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:E59–E63. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;180:1–12. doi: 10.1016/j.cell.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742–247 [DOI] [PMC free article] [PubMed]
- 8.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W (1992) Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp 160. Nature 360(6402):358–361 [DOI] [PubMed]
- 9.Ogimi C, Waghmare AA, Kuypers JM et al (2017) Clinical Significance of Human Coronavirus in Bronchoalveolar Lavage Samples From Hematopoietic Cell Transplant Recipients and Patients With Hematologic Malignancies 64(11):1532–1539 [DOI] [PMC free article] [PubMed]
- 10.Hakki M, Rattray RM, Press RD (2015) The clinical impact of coronavirus infection in patients with hematologic malignancies and hematopoietic stem cell transplant recipients. J Clin Virol 68:1–5 [DOI] [PMC free article] [PubMed]
- 11.Joshi S, Wollenzien H, Leclerc E, Jarajapu YP (2019) Hypoxic regulation of angiotensin- converting enzyme 2 and Mas receptor in human CD34+ cells. J Cell Physiol 234(11):20420–20431 [DOI] [PMC free article] [PubMed]
- 12.Hu W, Yen YT, Singh S, Kao CL, Wu-Hsieh BA (2012) SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol 25(4):277–288 [DOI] [PMC free article] [PubMed]
- 13.Fu J, Zhang J, Gong Y, Testa CL, Klein-Szanto AJ (2015) Regulation of HIF-1 alpha by the proprotein convertases furin and PC7 in human squamous carcinoma cells. Mol Carcino 54(9):698–706 [DOI] [PMC free article] [PubMed]
- 14.Braun E, Hotter D, Koepke L et al (2019) Guanylate-Binding Proteins 2 and 5 Exert Broad Antiviral Activity by Inhibiting Furin-Mediated Processing of Viral Envelope Proteins. Cell Rep 27(7):2092–2104 [DOI] [PubMed]