ABSTRACT
There is a burgeoning literature highlighting differences in health outcomes between babies born vaginally and by caesarean section (c-section) This has led to the suggestion that infants born by c-section may benefit from vaginal swabbing/seeding. Here, we discuss from an ecological perspective that it is gut-adapted, not vagina-adapted microbes that are likely to take up residence in the gut and have the most beneficial impact on the developing neonate. Further, we caution the practice of ‘vaginal seeding’ may be potentially unsafe and also give parents and health professionals a false sense of action in restoring the infant gut microbiome following c-section. Instead, we argue that restoring B. longum subsp. infantis, which has evolved to colonize the infant gut, is a safe and ecologically-sound approach to restoring the gut microbiome of infants born by c-section.
Keywords: mode of delivery, caesarean section, vaginal birth, probiotics, microbiome
Restoring the gut microbiome of infants delivered by cesarean section with B. infantis.
EVIDENCE, RISKS AND ECOLOGICAL CONSIDERATIONS OF VAGINAL SEEDING
There is a large body of literature indicating that the long-term health outcomes of babies delivered by caesarian section (c-section) are different than those delivered vaginally. The largest single study of this kind included 1.9 million infants born between 1977 and 2012. The analysis showed significantly increased risks of asthma, juvenile arthritis, inflammatory bowel disease (IBD), immune deficiencies and leukemia in the c-section group compared to babies delivered vaginally over the same time period (Sevelsted et al. 2015). It is also well known that babies born by c-section have a gut microbiome that is different from babies born vaginally (Dominguez-Bello et al. 2010; Backhed et al. 2015; Shi et al. 2018; Shao et al. 2019), which has led to the assumption that it might be beneficial to inoculate c-section delivered infants with a swab of their mother's vaginal microbiome at the time of birth in an attempt to mimic the microbial exposures encountered through a vaginal delivery. Increased awareness of the importance of the microbiome in early life has sparked considerable interest amongst parents who wish to undertake ‘vaginal seeding’. This has been paralleled recently with an interest in vaginal microbiome transplantation between women (Lev-Sagie et al. 2019). However, the scientific and medical communities remain cautious on the safety and efficacy of these yet unproven clinical practices.
One main issue with ‘vaginal seeding’ is that the proposed link between vaginally associated microbes to the infant gut microbiome has limited ecological foundation. Similar to the many different terrestrial and marine ecosystems on our planet that support their own unique variety of flora and fauna, our bodies too have different ecosystems that also support their own unique species of specialized microbes (Lloyd-Price et al. 2017). Like the plant and animal communities in Earth's biomes, microbial communities cannot be transferred to a different biological niche and be expected to flourish. The community of bacteria colonizing the vaginal canal is significantly different from the microbial communities thriving on the skin or in the gut because the environmental conditions and nutritional niches of these communities are all vastly different. This is supported by the fact that while vaginal bacterial strains can be transiently found in the infant gut soon after birth, these species tend to be in low abundance and can be readily lost or replaced by gut-adapted species. Further, assuming vaginal species do provide a health benefit, maintaining stable populations in the infant gut would require constant re-inoculation, which could not be achieved via a one-time procedure of ‘vaginal seeding’. Conversely, strains originating from the maternal gut form stable populations in the infant gut microbiome (Ferretti et al. 2018; Wampach et al. 2018; Yassour et al. 2018). This places the question on the efficacy of inoculating the infant with a purely vaginal source and suggests that providing the infant with gut-adapted strains might be more efficacious in resolving gut dysbiosis.
Providing that a one-time transfer of vaginal microbiomes is unlikely to be sufficient, the expected best outcome from ‘vaginal seeding’ would be that some of the introduced microbes would transiently colonize. However, without adequate safety screening using validated protocols, the process of vaginal swabbing carries a risk of inadvertently transferring potential pathogens, including genital herpes, Group B Streptococcus, chlamydia, and gonorrhea from an unhealthy vaginal microbiota, as well as additional nosocomial pathogens typically found in a hospital environment, directly to the baby (Huynh, Palasanthiran and McMullan 2018). Due to the insufficient evidence to show a benefit from the transient colonization of vaginal microbes and the potential risks with this process, there have been a number of medical opinion articles recommending that health professionals abstain from this practice, including the American College of Obstetricians and Gynecologists and the Danish Society of Obstetrics and Gynaecology (Committee on Obstetric Practice 2017; Haahr et al. 2018; Stinson, Payne and Keelan 2018).
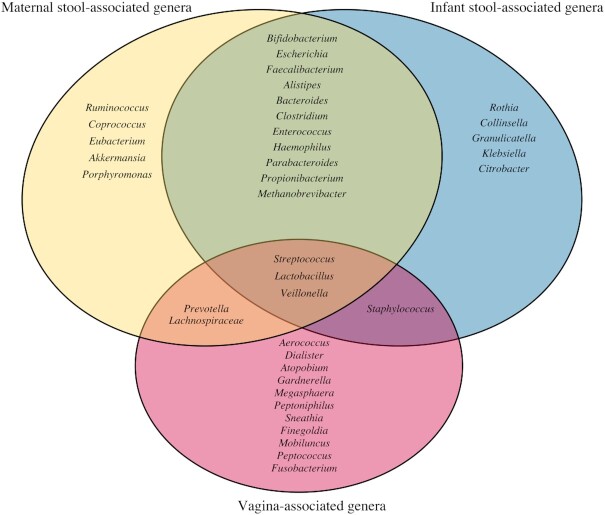
When instead we consider the ecological conditions of the gut (anaerobic, highly competitive), the nutrients available (dietary and host derived glycans), and the birthing process itself, the most obvious seeding source of the infant gut microbiome is the gut microbiome of the mother. During a vaginal delivery, the baby's head presses on and flattens the mother's colon so that is very common for the mother's stool to be expelled to some extent. The proximity of the vagina and the anus and the optimal fetal position for birthing (occiput anterior position head down, facing towards the back of the mother's pelvis) facilitates the likelihood of maternal-infant microbial transfer (Trevathan 2015). Upon birth, the newborn infant gut has unoccupied ecological niches that are environmentally similar to the mother's gut, thus allowing vertically transferred bacteria from the mother's gut microbiome to colonize and thrive in this new space, in a quintessential fecal-oral transfer, as demonstrated in multiple studies tracking the transfer of microbes from mother to infant (Asnicar et al. 2017; Ferretti et al. 2018; Wampach et al. 2018; Yassour et al. 2018). In fact, the infant gut microbiome is dominated by taxa common in the maternal gut, while microbes associated with the vagina are much less prevalent (Fig. 1.) This pattern supports that the maternal gut microbiome represents the largest contributor of the infant microbiome.
Figure 1.
Venn diagram showing a substantial overlap between genera found in the stool of infants and maternal gut, but little overlap with genera found in the vagina. The list of gut-associated genera were obtained from studies characterizing the microbiome of stools collected during gestation (Koren et al. 2012; Goltsman et al. 2018) and after birth from mother and infant pairs (Asnicar et al. 2017; Ferretti et al. 2018; Goltsman et al. 2018; Wampach et al. 2018; Yassour et al. 2018). The list of vagina-associated genera were obtained from studies characterizing the vaginal microbiome during gestation (DiGiulio et al. 2015; Goltsman et al. 2018) and the postpartum period (MacIntyre et al. 2015) of women of diverse ancestries (Fettweis et al. 2019). Note that due to space constraints these lists are designed to be exemplary, not comprehensive. Only taxa for which representatives were detected in at least two independent studies were included.
RESTORING THE MICROBIOME OF CAESAREAN SECTION BORN INFANTS WITH B. INFANTIS
Infancy represents a unique period when humans are evolutionarily adapted to thrive on a single source of nutrition (i.e. human milk). Thus, during this time, the breastfed infant gut microbiome is mainly shaped by the only glycans reaching the infant's colon, namely human milk oligosaccharides (HMO). The importance and role of these HMO is underscored by their abundance in human milk (15% of the energy content) and the inability of these compounds to be metabolized by the infant (Turroni et al. 2018). A small, select number of gut microbes can consume or metabolize these complex glycans in human milk. The most successful bacterium is Bifidobacterium longum subsp. infantis (B. infantis) as it has several unique gene clusters to capture, transport and internally metabolize all of the approximately 200 different HMO structures (Sela et al. 2008). This competitive advantage should make this bacterium the dominant microbe in the breastfed infant gut microbiome (Tannock et al. 2016), as is the case in many populations around the world where breastfeeding rates are high and microbiome-modifying interventions are less prevalent (Huda et al. 2019; Lawley et al. 2019). Unfortunately, this is not the case in most high income nations including the United States and New Zealand (Lewis and Mills 2017; Lawley et al. 2019). Historical references indicate that this change is a recent phenomenon (Henrick et al. 2018) that has coincided with the extensive use of antibiotics and the rise in planned, non-medically required c-section deliveries, especially in countries such as Brazil, where more than half of infants are born via elective c-section (Victora et al. 2011). This generational loss of B. infantis from the general population means that many babies born today are unlikely to naturally acquire B. infantis from their mothers even if born vaginally and breastfed (Tannock et al. 2016; Lawley et al. 2019) In a clinical study, the absence of Bifidobacteriaceae from a population of breastfed infants, was associated with an increase abundance of Enterobacteriaceae, Clostridiaceae, and Bacteroidetes (Frese et al. 2017). Few species in these families can metabolize HMO, but some are vigorous mucin degraders and there is now evidence of a significant erosion of the intestinal mucous layer in infants with low abundance of Bifidobacteriaceae, which is not observed in infants colonized with B. infantis EVC001 (Karav, Casaburi and Frese 2018). Further, infants with low abundance of bifidobacteria and missing B. infantis have signatures of chronic enteric inflammation including elevated fecal TNFα, INFγ, IL1β, and calprotectin (Henrick et al. 2019), which have been linked to an increased prevalence of certain autoimmune disorders such as atopy and asthma later in life (Orivuori et al. 2015; Henrick et al. 2019). Overall, there is a repetitive trend between lower bifidobacteria in early life and a variety of chronic immune and inflammatory diseases, suggesting their presence in high abundance may have important roles in ensuring proper development during critical period of physical and immune maturation (O'Neill et al. 2017).
Feeding B. infantis EVC001 regardless of mode of delivery, robustly and persistently remodels the gut microbiome of infants and results in significantly increased levels of Bifidobacterium to up to 80% mean relative abundance of the total microbiome, as long as the infants are consuming human milk (Frese et al. 2017). These changes reflect the ecological adaptation of B. infantis to the infant gut and result in significant beneficial changes to the gut ecosystem (Duar et al. 2020), including increased production of organic acids that lower intestinal pH creating an environmental deterrent to the growth of potentially virulent enteropathogens (Casaburi and Frese 2018), antibiotic resistance taxa (Casaburi et al. 2019), and mucus eroding bacteria (Karav, Casaburi and Frese 2018). Altogether, these data suggest an evolutionary link between the mother (HMO from breast milk), the infant and B. infantis to synergistically create the ecological conditions required to protect the infant from potential infection and restore inflammatory markers to homeostatic levels (Henrick et al. 2019). These changes are durable and persist even after infants stop consuming B. infantis, which is in stark contrast to vaginal swabbing, which only partly and transiently alters the infant gut microbiome (Dominguez-Bello et al. 2016).
In conclusion, and viewed from an ecological and evolutionary perspective, it is gut-adapted, not vagina-adapted, microbes that are likely to take up residence in the infant gut (Fig. 1) and, thereby, have the most beneficial impact on the developing neonate. Thus, this and other ecological aspects of the infant gut presented in this opinion piece, should be considered when selecting probiotic strains and ingredients in infant formulas aimed to modulate the microbiome. Further, we and others (Cunnington et al. 2016; Haahr et al. 2018) concur that a continuation of the unsubstantiated practice of ‘vaginal seeding’ comes at a risk of not only being ineffective and potentially unsafe but gives parents and health professionals a false sense of action in restoring the infant gut microbiome following c-section delivery. Given the potential public health implications and economic burden associated with perturbed microbiome compositions (i.e. dysbiosis) in early life, we argue that scientific and medical focus should be placed instead on the rational design and implementation of strategies to reestablish the dominance of bifidobacteria in to the gut microbiome of infants, which shows the most promise in having a positive impact on life-long health outcomes (Duar et al. 2020). Restoring B. infantis to the infant gut microbiome represents one such approach that simply reestablishes the natural system of protection.
ACKNOWLEDGEMENTS
The authors thank Heather K. Brown (Evolve BioSystems, Inc.) for help with the preparation of Fig. 1.
Conflicts of interest
RMD and DK are employed by Evolve BioSystems, Inc. RT was the clinical lead on an infant microbiome trial at KCL sponsored by Evolve BioSystems, Inc.
REFERENCES
- Asnicar F, Manara S, Zolfo Met al.. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. MSystems. 2017;2:e00164–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Roswall J, Peng Yet al.. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. [DOI] [PubMed] [Google Scholar]
- Casaburi G, Duar RM, Vance DPet al.. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist In Control. 2019;8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaburi G, Frese SA. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Human Microbiome Journal. 2018;9:7–10. [Google Scholar]
- Committee on Obstetric Practice. Committee Opinion No. 725: Vaginal Seeding. Obstet Gynecol. 2017;130:e274. [DOI] [PubMed] [Google Scholar]
- Cunnington AJ, Sim K, Deierl Aet al.. “Vaginal seeding" of infants born by caesarean section. BMJ. 2016;352:i227. [DOI] [PubMed] [Google Scholar]
- DiGiulio DB, Callahan BJ, McMurdie PJet al.. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci. 2015;112:11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras Met al.. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci. 2010;107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, De Jesus-Laboy KM, Shen Net al.. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22:250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duar RM, Henrick BM, Casaburi G, et al.. Integrating the Ecosystem Services Framework to Define Dysbiosis of the Breastfed Infant Gut: The Role of B. infantis and Human Milk Oligosaccharides. Front Nutr. 2020;7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Pasolli E, Tett Aet al.. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis JM, Serrano MG, Brooks JPet al.. The vaginal microbiome and preterm birth. Nat Med. 2019;25:1012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese SA, Hutton AA, Contreras LNet al.. Persistence of Supplemented Bifidobacterium longum subsp. infantis EVC001 in Breastfed Infants. mSphere. 2017;2:e00501–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsman DSA, Sun CL, Proctor DMet al.. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28:1467–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr T, Glavind J, Axelsson Pet al.. Vaginal seeding or vaginal microbial transfer from the mother to the caesarean-born neonate: a commentary regarding clinical management. BJOG. 2018;125:533. [DOI] [PubMed] [Google Scholar]
- Henrick BM, Chew S, Casaburi Get al.. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. 2019;86:749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick BM, Hutton AA, Palumbo MCet al.. Elevated fecal pH Indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. 2018;3:e00041–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda MN, Ahmad SM, Alam MJet al.. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. 2019;143:e20181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J, Palasanthiran P, McMullan B. Potential transmission of herpes simplex virus via vaginal seeding. Pediatr Infect Dis J. 2018;37:e278. [DOI] [PubMed] [Google Scholar]
- Karav S, Casaburi G, Frese SA. Reduced colonic mucin degradation in breastfed infants colonized by Bifidobacterium longum subsp. infantis EVC001. FEBS Open Bio. 2018;8:1649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TCet al.. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley B, Otal A, Moloney-Geany Ket al.. Fecal microbiotas of indonesian and new zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol. 2019;85:e01105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Sagie A, Goldman-Wohl D, Cohen Yet al.. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat Med. 2019;25:1500–4. [DOI] [PubMed] [Google Scholar]
- Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. 2017;88:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard Get al.. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DA, Chandiramani M, Lee YSet al.. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill I, Schofield Z, Hall LJet al.. Exploring the role of the microbiota member Bifidobacterium in modulating immune-linked diseases. Emerg Top Life Sci. 2017;1:333–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orivuori L, Mustonen K, de Goffau Met al.. High level of fecal calprotectin at age 2 months as a marker of intestinal inflammation predicts atopic dermatitis and asthma by age 6. Clin Exp Allergy. 2015;45:928–39. [DOI] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya Aet al.. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci. 2008;105:18964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevelsted A, Stokholm J, Bonnelykke Ket al.. Cesarean section and chronic immune disorders. Pediatrics. 2015;135:e92–8. [DOI] [PubMed] [Google Scholar]
- Shao Y, Forster SC, Tsaliki Eet al.. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YC, Guo H, Chen Jet al.. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci Rep. 2018;8:3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson LF, Payne MS, Keelan JA. A critical review of the bacterial baptism hypothesis and the impact of cesarean delivery on the infant microbiome. Front Med. 2018;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Lee PS, Wong KHet al.. Why Don't All Infants Have Bifidobacteria in Their Stool? Front Microbiol. 2016;7:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevathan W. Primate pelvic anatomy and implications for birth. Philos Trans R Soc B Biol Sci. 2015;370:20140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Milani C, Duranti Set al.. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci. 2018;75:103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora CG, Aquino EM, do Carmo Leal Met al.. Maternal and child health in Brazil: progress and challenges. The Lancet. 2011;377:1863–76. [DOI] [PubMed] [Google Scholar]
- Wampach L, Heintz-Buschart A, Fritz JVet al.. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. 2018;9:5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Jason E, Hogstrom LJet al.. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24:146–54.. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]