Abstract
How long-lived memories withstand molecular turnover is a fundamental question. Aggregates of a prion-like RNA-binding protein, cytoplasmic polyadenylation element-binding (CPEB) protein, is a putative substrate of long-lasting memories. We isolated aggregated Drosophila CPEB, Orb2, from adult heads and determined its activity and atomic structure, at 2.6 Å resolution, using cryo-EM. Orb2 formed ~75 nm-long three-fold-symmetric amyloid filaments. Filament formation transformed Orb2 from a translation repressor to an activator and “seed” further translationally active aggregation. The 31 amino acid protofilament core adopted a cross-β unit with a single hydrophilic hairpin stabilized by interdigitated glutamine packing. Unlike the hydrophobic core of pathogenic amyloids, the hydrophilic core of Orb2 filaments suggests how some neuronal amyloids could be a stable yet regulatable substrate of memory.
A memory, once formed, is maintained in a physical state that does not require continuous presence of the original experience. The substrate of memory has been extensively studied at the neuronal network level (1). However, how the underlying biochemical changes, induced by initial experience, withstand degradation to maintain altered network properties, and hence memory, remain unclear (2). One proposed solution is a self-renewing protein system that can recruit and modify newly made constituents as older ones are degraded (3). A prion-like assembly has the ability to promote the conformational conversion of other existing or newly formed monomers, creating a self-sustaining protein “conformational memory” that outlives its individual constituents (4). A prion-like protein with a defined role in memory in different species is the mRNA-binding CPEB family of proteins (5–14). In Drosophila, inhibition of Orb2 aggregation interferes with memory consolidation (6, 7, 10, 15), facilitation of Orb2 aggregation lowers the threshold for long-term memory formation (9), and inactivation of Orb2 after memory formation interferes with memory recall (9). These observations suggest that the aggregated state of Orb2 creates a protein conformational memory.
Many prion-like proteins, prevalent among RNA-binding protein (16), form amyloids, which are filamentous protein aggregates composed of cross-β cores (17, 18). However, in the nervous system, amyloids are commonly associated with disease (19). Recently, new classes of protein assemblies formed by prion-like proteins have been reported, such as the non-amyloid filaments, or the gel-like state (20, 21); physical states that are believed to be more amenable to regulation. These observations raise a fundamental question: what is the physical nature of the Orb2 aggregate in the brain that acts as a biochemical substrate of memory?
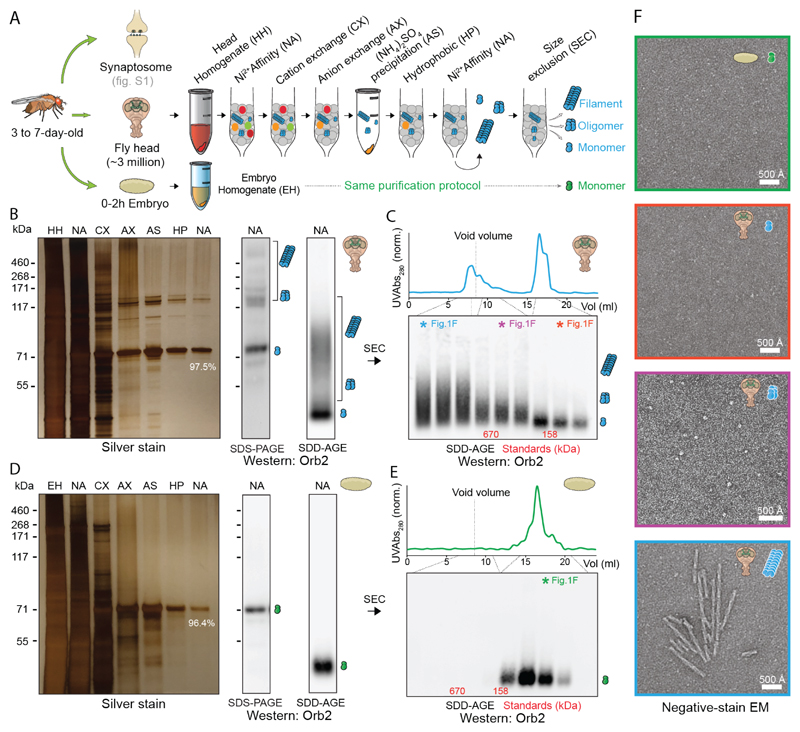
To characterize function and structure of endogenous Orb2 aggregates, we purified Orb2 protein from approximately three million 3 to 7-day-old adult Drosophila melanogaster heads (Fig. 1A and fig. S1A). There was no qualitative difference between Orb2 purified from synaptosomes or total head extracts and we used whole-head Orb2 in subsequent experiments. Purified Orb2 contained monomers as well as heat- and SDS-resistant aggregates (Fig. 1B, fig. S1B). Quantitative mass spectrometry (LC-MS/MS) revealed that 97.5% of the purified protein is Orb2 (fig. S1C). Unlike in the adult nervous system, Orb2 protein is monomeric in the early embryo (22). Orb2 purified from 0-2h embryo (fig. S1C) remained monomeric, suggesting that the purification does not induce Orb2 aggregation (Fig. 1D, 1E). Size exclusion chromatography revealed monomeric (75-160 kDa), oligomeric (160-670 kDa), and aggregated (>1MDa) Orb2 in adult head (Fig. 1C). Monomers were not visible and oligomers appeared as heterogeneous globules (Fig. 1F, figs. S1D, S2A), reminiscent of oligomers of other amyloid-forming proteins (23). The aggregates appeared as unbranched helical filaments with an average length of ~750 Å, width of ~100 Å, and helical crossover distance of ~550 Å (Fig. 1F, figs. S1D, S2B). JJJ2, a yeast DnaJ-domain protein, enhances Orb2 aggregation and facilitate long-term memory formation in Drosophila (9). JJJ2, but not close relative JJJ3, converted Orb2 oligomers into filaments, suggesting oligomers are on-pathway precursors for filaments (fig. S3).
Fig. 1. Purification of different Orb2 species from adult fly head.
(A) Schematic representation of Orb2 purification method from fly head and embryo. (B) Silver stain analysis after each purification step (left) and western blotting after the final purification step (right) by SDS-PAGE and Semi-Denaturing Detergent-Agarose Gel Electrophoresis (SDD-AGE). The % indicates relative Orb2 purity determined by mass spectrometry. The position of monomer, oligomer and filaments are indicated schematically. (C) Western blotting of purified head Orb2 following size-exclusion chromatography. The colored asterisk fractions were visualized under negative-stain EM in F. (D) Silver staining and western blotting of Orb2 protein purified from 0-2h embryo. (E) Western blotting of purified embryonic Orb2 following size-exclusion chromatography. The colored asterisk fraction was visualized under negative-stain EM in F. (F) Negative-stain electron micrographs of purified Orb2 embryonic monomer (green box), head monomer (red box), head oligomer (magenta box) and head filament (blue box).
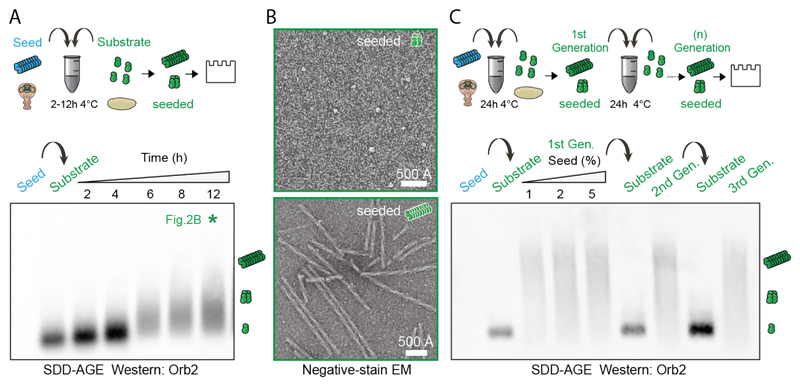
Incubation of Orb2 monomers with traces of Orb2 filaments induced monomer aggregation in a time-dependent manner (Fig. 2A, 2B) and seeded-aggregates themselves acted as a seed (Fig. 2C). Orb2 monomer alone did not aggregate in the same time period (fig. S4A) and Orb2 filaments did not cause aggregation of monomeric heterologous prion-like RNA-binding protein RBM3 (24) (fig. S4B). Both endogenous and seeded Orb2 filaments bound Thioflavin T, were recognized by the anti-amyloid OC antibody, and were resistant to protease treatment (fig. S4C-E).
Fig. 2. Endogenous Orb2 filament can seed further filament formation.
(A) Time course of monomeric Orb2 aggregation seeded by filaments obtained from adult fly head. The seed (~1 ng) is undetectable in western. Within 6 hours most monomers are aggregated. (B) Negative-stain electron micrographs of seeded oligomers and filaments. (C) Propagation of seeded aggregation through successive rounds. In first round, different seed (1 ng, 2 ng, 5 ng) to monomer (100 ng) ratio was used. In subsequent rounds only 1 ng of seed was used.
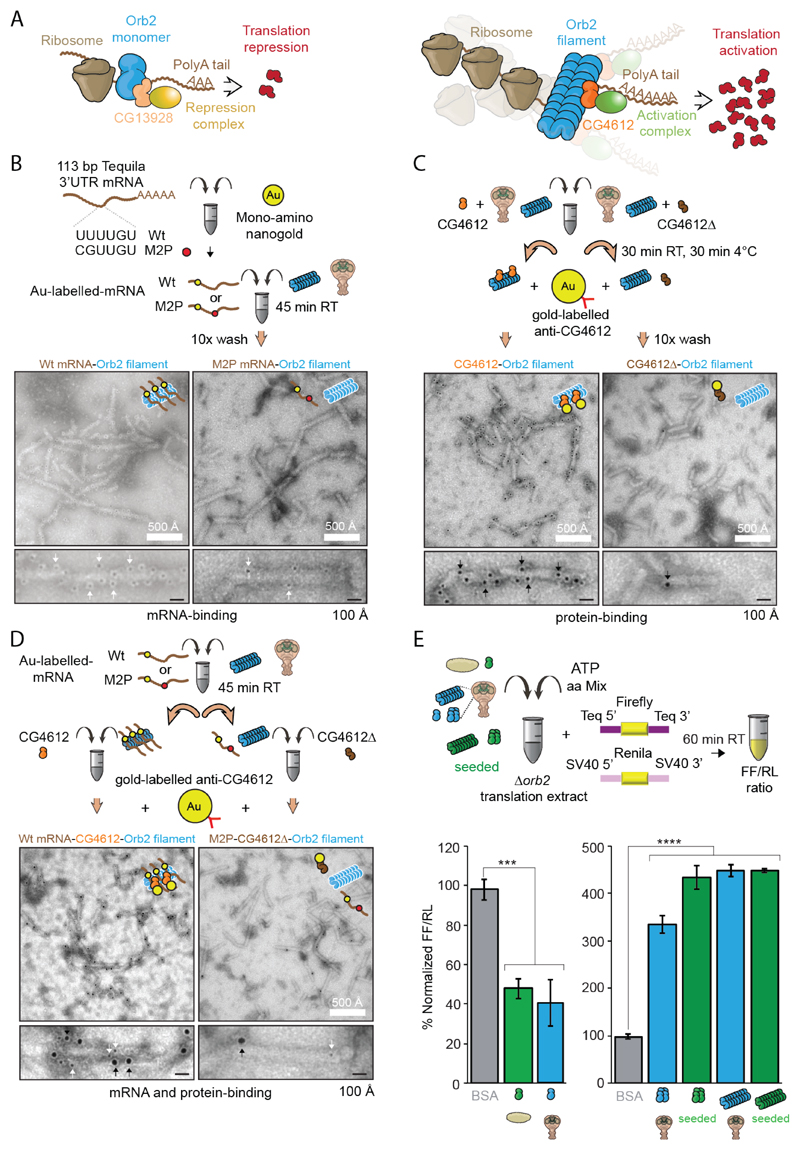
Orb2 binds to the 3' untranslated regions (UTR) of mRNAs (25, 26); monomeric Orb2, which interacts with CG13928, decreases translation, while aggregated Orb2, which interacts with CG4612, enhances translation (22) (Fig. 3A). Tequila, a protease, is one of the mRNA targets of Orb2 and required for long-term memory (26, 27). All purified Orb2 species bound to the 3'UTR of Tequila mRNA and mutations in the Orb2-binding site UUUUGU to GCUUGU reduced binding of all species (fig. S5A). Gold-labeled wild type, but not mutant (M2P) mRNA, co-localized with the oligomers and filaments (Fig. 3B, fig. S5B-D). In addition to mRNA, when incubated with recombinant binding partners (fig. S6A), Orb2 filaments bound only to GC4612. N-terminal deleted CG4612, CG4612∆, and monomer binding-partner CG13928 did not bind to Orb2 filaments (Fig. 3C, fig. S6B-D). When incubated with wildtype tequila 3'UTR mRNA and CG4612 protein, the Orb2 filaments were decorated with both mRNA and CG4612 protein (Fig. 3D, fig. S6E). In an Orb2-dependent translation assay, the addition of the Orb2 monomers reduced translation, whereas oligomers and filaments, both endogenous and seeded, enhanced translation (Fig. 3E, fig. S6F). Thus, Orb2 filament purified from fly retains aspects of its physical and functional properties.
Fig. 3. Biochemical activity of Orb2 monomer, oligomer and filament isolated from adult head.
(A) Schematic representation of the biochemical activity of Orb2. CG13928 binds to Orb2 monomer and recruit translation repression complex (indicated in yellow). CG4612 binds to aggregated Orb2 and recruit translation promoting complex (indicated in green). (B) Negative-stain EM of nanogold-labelled 3' UTR of Tequila mRNA bound to Orb2 filaments. Black dots indicated by white arrows are nanogold particle (~ 2nm) attached to target mRNA. (C) Immuno-EM of CG4612 bound to Orb2 filaments. Black dots indicated by black arrows are gold particles (~ 6nm) attached to CG4612 protein. (D) EM of Orb2 filaments bound to nanogold-labelled 3' UTR of Tequila mRNA and CG4612 protein. The larger gold particle (~ 6nm, black arrows) represents CG4612 protein and smaller gold particle (~2nm, white arrows) represents the mRNA. (E) Translation of Orb2-target mRNA in presence of different Orb2 species, obtained from embryo, adult fly head or seeding reaction. Purified Orb2 monomer represses translation while Orb2 oligomer and filament enhances translation. BSA was used as a control. ***P = 0.0003, ****P < 0.0001; Student’s t-test; two tailed. Data are expressed as mean ± s.e.m.
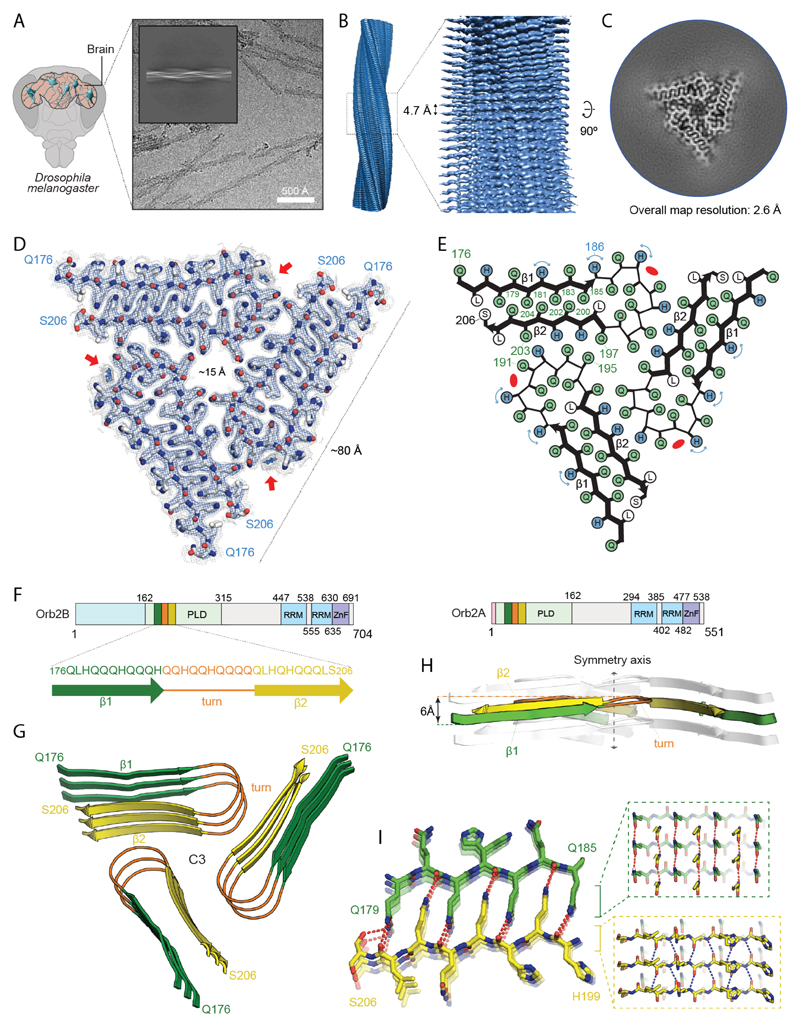
We used cryo-EM to determine the atomic structure of Orb2 filaments, which were manually picked and used for helical reconstruction in RELION (28) (Fig. 4A, fig. S7, and table S1). The final reconstruction, at an overall resolution of 2.6 Å (fig. S8, S9) showed that the Orb2 filament is composed of three identical protofilaments related by C3 symmetry with separated β-strands along the helical axis (Fig. 4B, C). The filament core has a triangular cross-section with a side of ~80 Å and a 15 Å diameter channel along helical axis (Fig. 4D). Successive rungs of three hairpin-shaped protofilaments forms extended β-sheets along the helical axis, typical of the cross-β amyloid structure (17), with a twist of -1.55° and a rise of 4.75 Å. Surrounding the stable fibril core, lower-resolution densities are visible that likely correspond to the RNA-recognition motif and protein interaction domain of Orb2 (gray density in Fig. 4C). We could identify densities that correspond to peptide-group oxygen atoms, ordered solvent molecules in each interface, and alternative conformations of histidine sidechains (fig. S10A-C). Consequently, we could build and stereochemically refine an atomic model de novo. The resulting model of the amyloid core comprised of 31 amino acid residues from the glutamine-rich prion-like domain of Orb2 (Fig. 4D, E). Orb2 protein has two isoforms in the fly brain (10), which shares the prion-like domain, but differs in their N-terminal extension (Fig. 4F). The low-abundant Orb2A seeds aggregation of prevalent Orb2B, which can subsequently seed Orb2B protein (10). 97.5% of the protein in our sample is Orb2B (fig. S1C), and the protofilament core structure extends from residues 176-206 of Orb2B (Fig. 4D, E).
Fig. 4. Atomic structure of Orb2 filaments isolated from head.
(A) Cryo-electron micrograph obtained at a defocus of -1.8 µm showing individual Orb2 filaments. Inset: representative reference-free 2D class average showing an entire helical crossover. (B) Side view of the three-dimensional cryo-EM reconstruction illustrating the clear separation of the β-strands. (C) Cryo-EM reconstruction of neuronal Orb2 filament at 2.6 Å. (D) Sharpened, high-resolution cryo-EM maps with the corresponding atomic model overlaid. Unsharpened, 5 Å low-pass-filtered maps are shown as grey outlines; The weaker densities that border sidechains of H182, H186 and H189, correspond to the alternative conformation that those residues adopt. The non-proteinaceous density that runs contiguously across the rungs is highlighted by red arrows. Density maps are shown at a contour level of 2.2 σ (blue) and 1.4 σ (grey). (E) Schematic view of the filament core showing the complementary glutamine packing within the protofilament and the hydrophilic interfaces between protofilament. (F) Primary structure of Orb2A and Orb2B isoforms in fly brain. The prion-like domains are labeled PLD (residues 162-315 for Orb2B and residues 9-162 for Orb2A). The RNA-recognition motifs are labeled RRM and the zinc-finger motifs are labeled ZnF. The N-terminal amino acids preceding the prion-like domain of Orb2A (9 residues) and Orb2B (162 residues) are represented in light red and blue, respectively. Schematic depicting the sequence of the Orb2 filament core, with the observed two β-strands colored in green and yellow as well as the connecting turn in orange. (G) Rendered view of the secondary structure elements in the Orb2 fold, depicted as three successive rungs. (H) Same representation as in G, but in a view perpendicular to the helical axis, revealing the changes in height within a single molecule. (I) Close-up view of the interdigitated glutamine residues. The hydrogen bonds between main chains are shown in blue. The hydrogen bonds involving sidechains are shown in red.
The protofilament core adopts a simple hairpin-like fold, comprised of two β-strands, β1 (residues 176-186) and β2 (residues 197-206), with a wide turn (residues 187-196) in between (Fig. 4G). Perpendicular to the helical axis, a difference in height of 6 Å between the highest point in the turn and the lowest point in the tip of the β2 allow an ordered hydrogen-bonding network of the β-stranded regions (Fig. 4H). The cross-β packing of the two β-strands is made of a tight interdigitation of glutamine residues, Q179, Q181, Q183 and Q185 from β1 and Q200, Q202, Q204 from β2 (Fig. 4I). A single leucine, L198, separates this block of seven glutamines from a cluster of four more interior glutamine residues in the hairpin turn: Q187, Q190, Q193 and Q196 (Fig. 4E). Next to H189, there is an additional non-proteinaceous density that runs contiguously across the rungs (red arrows in Fig. 4D, fig. S10D). This density is compatible with linear chains of aliphatic hydrocarbons and polyamines, which may have co-purified with Orb2. The three equivalent protofilament packing interfaces are made of eight polar residues – five glutamines and three histidines. These polar residues form an extensive network of hydrogen bonds on each side and across the interface (fig. S11). Orb2 mutations, known to disrupt Orb2 aggregation (10), were mapped on the polar interface between protofilaments and the cross-β packing that stabilizes the protofilament (fig. S12A). Moreover, removal of 54 amino acids from the glutamine-rich region specifically interfered with long-term, but not short-term, memory (fig. S12B) (6).
Several structures of functional amyloids, produced in vitro from truncated protein, have been solved (29, 30). In contrast, Orb2 filament represents a biochemically active full-length functional amyloid extracted from the endogenous source (fig. S13). The three-fold-symmetry of the ordered core of Orb2 resembles that of β-amyloid1-40 filaments seeded from Alzheimer’s disease brain tissue (31) or assembled in vitro (32). However, unlike the hydrophobic β-amyloid1-40 core, Orb2 forms a hydrophilic core stabilized by inter-digitated glutamines. When we assembled Orb2 filaments from recombinant protein, filaments were longer, morphologically distinct, less biochemically active, and had negligible seeding capacity compared to endogenous ones (fig. S14), suggesting that the same prion-like protein can adopt distinct structures in vitro and in vivo. Other proteins with glutamine-rich sequences are known to produce amyloids (33). The inter-digitated cross-β structure observed in Orb2 filaments could be extended on both sides of a parallel β-sheet made of only glutamine residues, which would allow for the formation of stable, multi-layered cross-β structures from long poly-glutamine sequences, as in pathological glutamine expansions.
There are number of ways “molecular memories” can be created: increase in the amount of a protein, where the kinetics of decay to the basal state would be the duration of memory; a stable protein or protein state, the half-life of which would inform the duration of memory (34, 35); or, a feed-forward molecular circuit, whose activity could be reciprocally perpetuated across time. A self-sustaining amyloid formed by a single polypeptide employs all three mechanisms. The amyloid fold itself is not “memory”, but the amyloid fold reorganizes the rest of the Orb2 protein to create a persistent alteration in the synthesis of specific synaptic proteins. Memory is the altered synaptic state that results from the change in functional state of the Orb2 (fig. S13). However, amyloid formation is generally assumed to be irreversible in physiological conditions, then how can memory be dynamic? Or is it possible that some memories are indeed irreversible, and they appear lost merely due to an inability to retrieve them? First, amyloids are not necessarily irreversible, could exist in a dynamic equilibrium with the available monomer (36). Second, the Orb2 amyloid core is based on a hydrophilic, glutamine/histidine-rich fold and the protonation state of histidine residues could influence Orb2 amyloid stability. Indeed, lowering pH destabilized Orb2 filaments (fig. S15), suggesting functional amyloid could be amenable to modification or even dissolution. Our findings question the assumption that amyloid formation in the brain is always an unintended consequence that leads to dysfunction. We postulate that the brain fosters a cellular environment that is permissive to the formation of amyloid-like state of certain proteins in order to meet the diversity of functional requirements imposed on it.
Supplementary Material
One Sentence Summary.
How amyloid can be a substrate of memory
One Sentence Summary.
A synaptic protein-synthesis regulator, important for persistence of memory, forms an amyloid in the adult Drosophila nervous system that activates protein synthesis.
Acknowledgments
We thank D. Laurents, J. Oroz, W. Redwine, K. Patton, R. Halfmann, and S. Garcia Alcantara for comments; M. Miller for illustrations; C. Zhang, P. Leal, A. Machen, and A. Rodriguez Gama for experimental assistance; A. Saraf for assistance in mass spectrometry; K. Xi, F. Guo, and T. Parmely for support with electron microscopy; and G. Murshudov and R.Warshamanage for help with REFMAC. This work is dedicated to the memory of Mark T. Fisher, Ph.D (University of Kansas Medical Center).
Funding
This work was supported by the UK Medical Research Council (MC_UP_A025_1013, to S.H.W.S.) and Stowers Institute for Medical Research (to K.S);
Footnotes
Author contributions: Conceptualization, K.S., and R.H.; Investigation, R.H., M.J.R., Y.P., W.Z., A.G.M., and S.H.W.S.; Resources, K.S, J.A.J.F., and S.H.W.S.; Software, S.H.W.S.; Supervision, K.S., Writing – Original Draft, K.S., R.H.; Writing – Review & Editing, all authors;
Competing interests: The authors declare no competing financial interests.;
Data and materials availability: Cryo-EM density map for Orb2 has been deposited in the Electron Microscopy Data Bank (EMDB) under accession number EMD-21316. Refined atomic model has been deposited in the Protein Data Bank (PDB) under accession number 6VPS. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MassIVE repository with the dataset identifier PXD016266. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at https://www.stowers.org/research/publications/LIBPB-1486.
References
- 1.Josselyn SA, Tonegawa S. Memory engrams: Recalling the past and imagining the future. Science. 2020;367 doi: 10.1126/science.aaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch G, Baudry M. The biochemistry of memory: a new and specific hypothesis. Science. 1984;224:1057–1063. doi: 10.1126/science.6144182. [DOI] [PubMed] [Google Scholar]
- 3.Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- 4.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 5.Fioriti L, et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron. 2015;86:1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 7.Kruttner S, et al. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron. 2012;76:383–395. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruttner S, et al. Synaptic Orb2A Bridges Memory Acquisition and Late Memory Consolidation in Drosophila. Cell Rep. 2015;11:1953–1965. doi: 10.1016/j.celrep.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, et al. A Putative Biochemical Engram of Long-Term Memory. Curr Biol. 2016;26:3143–3156. doi: 10.1016/j.cub.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Raveendra BL, et al. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol. 2013;20:495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephan JS, et al. The CPEB3 Protein Is a Functional Prion that Interacts with the Actin Cytoskeleton. Cell Rep. 2015;11:1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- 13.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 15.Hervas R, et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016;14:e1002361. doi: 10.1371/journal.pbio.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iglesias V, et al. In silico Characterization of Human Prion-Like Proteins: Beyond Neurological Diseases. Front Physiol. 2019;10:314. doi: 10.3389/fphys.2019.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 20.Wu H, Fuxreiter M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell. 2016;165:1055–1066. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes MP, et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan MR, et al. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell. 2015;163:1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 24.Peretti D, et al. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 2015;518:236–239. doi: 10.1038/nature14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepien BK, et al. RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc Natl Acad Sci U S A. 2016;113:E7030–E7038. doi: 10.1073/pnas.1603715113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci U S A. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didelot G, et al. Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science. 2006;313:851–853. doi: 10.1126/science.1127215. [DOI] [PubMed] [Google Scholar]
- 28.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 30.Mompean M, et al. The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell. 2018;173:1244–1253 e1210. doi: 10.1016/j.cell.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu JX, et al. Molecular structure of beta-amyloid fibrils in Alzheimer's disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 34.Heo S, et al. Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc Natl Acad Sci U S A. 2018;115:E3827–E3836. doi: 10.1073/pnas.1720956115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsien RY. Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc Natl Acad Sci U S A. 2013;110:12456–12461. doi: 10.1073/pnas.1310158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiang W, Kelley K, Tycko R. Polymorph-specific kinetics and thermodynamics of beta-amyloid fibril growth. J Am Chem Soc. 2013;135:6860–6871. doi: 10.1021/ja311963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohi MD, et al. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 39.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 42.Chien P, DePace AH, Collins SR, Weissman JS. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature. 2003;424:948–951. doi: 10.1038/nature01894. [DOI] [PubMed] [Google Scholar]
- 43.Kayed R, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheres SHW. Amyloid structure determination in RELION-3.1. bioRxiv. 2019 doi: 10.1107/S2059798319016577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afroz T, et al. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev. 2014;28:1498–1514. doi: 10.1101/gad.241133.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.