Abstract
Protein interactions guide the majority of cellular processes. Orthogonal hetero-specific protein-protein interaction domains may facilitate better control of engineered biological systems. Here, we report a tunable de novo designed set of orthogonal coiled-coil (CC) peptide heterodimers (the NICP set) and its application for the regulation of diverse cellular processes, from cellular localization to transcriptional regulation. We demonstrate the application of CC pairs for multiplex localization in single cells and exploit the interaction strength and variable stoichiometry of CC peptides for tuning of gene transcription strength. A concatenated CC peptide tag (CCC-tag) was used to construct highly potent CRISPR/dCas9-based transcriptional activators and to amplify the response of light- and small-molecule inducible transcription in cell culture as well as in vivo. The NICP set and its implementations represent a valuable toolbox of minimally disruptive modules for the recruitment of versatile functional domains and regulation of cellular processes for synthetic biology.
Introduction
Natural proteins often comprise multiple functional domains that encode different functions, including interactions with other proteins. Protein-protein interactions guide the hetero- or homo-oligomerization of proteins, which plays a key role in protein signaling, trafficking, scaffolding, and numerous other cellular processes. The modularity of protein interaction domains enables new combinations of functionalities and facilitates the design of biological systems.1 The availability of a set of orthogonal interaction domains, functional in the complex environment of mammalian cell cytosol, could enable diverse and more efficient regulation modalities. Proteins can interact either through globular domains or through shorter peptides that bind to the folded recognition domains—for example, SH3, WW, or TRAP domains.2 Coiled-coil dimers (CCs) are among the smallest peptide interaction domains, in which polypeptide helices interact along the complementary surface.3 CC homodimers (such as variants of the dimerization domain of a yeast transcription factor GCN4 and leucine zippers) have already been used to guide protein interactions.4–8 However, heterodimerization domains are more precise and efficient in guiding interactions between different protein partners. The most frequently-used CC heterodimeric peptide pair is based on peptide partners that are positively (basic or K-peptide) and negatively charged (acidic or E-peptide).9–11 The number of available orthogonal peptide pairs limits the range of different interactions that may be simultaneously introduced into the cell. In addition, highly positively charged peptides can interact non-specifically with biological polyanions, such as nucleic acids and charged polysaccharides.
CC binding specificity is based on a combination of complementary electrostatic and hydrophobic interactions between the e, g and a, d positions of the characteristic heptad repeat of CCs, respectively.12 The implementation of these interaction rules enables the de novo design of CC dimers, while the non-interacting residues (at b, c, and f positions) enable additional tuning of the interaction strength.13,14 In addition to the ability to control interaction strength and stoichiometry, multiple CC pairs must be mutually orthogonal (i.e., interact only with their designated binding partners) in order to enable multiple simultaneous molecular assemblies. Designed orthogonal CC dimers have been used in vitro for the construction of biomaterials and nanostructures.15 A set of designed heterodimeric CC peptides has been tested in yeast using the yeast-two-hybrid assay with a small set of identified orthogonal pairs,4 while four-helical bundle orthogonality in yeast cells demonstrated six orthogonal combinations with relatively low transcriptional response fold change.16 Recently, CC peptides have been used to direct the specificity of the extracellular recognition domain of therapeutic T-cells.17
Here, we report the evaluation and application of a set of orthogonal heterodimeric CC peptides in mammalian cells. Rational combinatorically designed peptides representing a set of six CC pairs were found to be functional and mutually orthogonal in mammalian cells. Interacting peptide pairs were used to simultaneously control localization of several different proteins within a single cell. CC peptides were further used to potently upregulate gene transcription, where tunability was demonstrated in a wide range of response amplitude based on the modification of the affinity of peptides. Further, concatenation of CC repeats (CCC-tag) enabled precise control of the stoichiometry of the bound transcriptional activation domains, thereby resulting in enhanced transcriptional activity. The latter strategy was used to construct some of the most potent CRISPR/dCas9- and TAL effector-based transcriptional activators known thus far. In addition, strong amplification of light- and small molecule-inducible transcriptional regulation was demonstrated in vitro as well as in vivo. The described NICP set of orthogonal heterodimeric CC peptides represents a valuable toolbox for bioengineering and cell/molecular biology.
Results
Orthogonality of designed CC peptides in mammalian cells
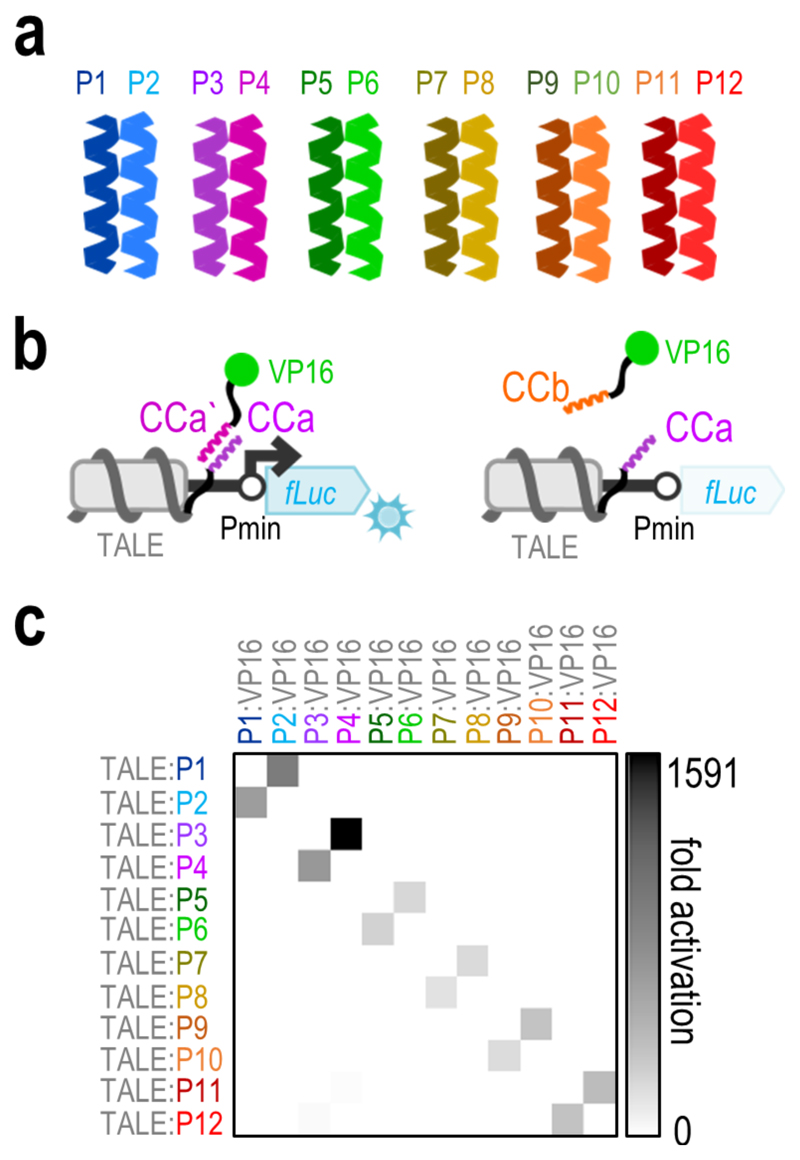
A set of 12 peptides forming 6 parallel heterodimeric CC pairs (the NICP set, Figure 1a) was designed based on combinations of heptads with hydrophobic and electrostatic matches between the desired pairs and mismatches between the non-desired pairs (Supplementary Table 1). All peptides comprise 4 heptads and are 33 amino acids in length. A few peptides from the set (P1-P8) with KDs in the nano- to micromolar range have previously been characterized in vitro18 and used for the self-assembly of CC protein origami cages,15,19 while the NICP set has been extended to 12 peptides here. The additional pairs of the peptide set (P9-P10 and P11-P12) were designed following the same combinatorial principle of four heptad CC parallel homodimers as that for the P1-P8 peptides,18 based on combining the electrostatic patterns and Asn/Ile residues patterns at position a. In order to experimentally assess the activity and orthogonality of the designed set in mammalian cells, each of the 12 designed peptides was genetically fused either to the C-terminus of a transcription activator-like effector (TALE) DNA-binding domain (DBD) or to the N-terminus of the VP16 transcriptional activation domain (TAD). The TALE:CC fusion protein was designed to bind its DNA target site upstream of a minimal promoter (Pmin), which drives the expression of a reporter gene. Reporter expression is expected only in the event of interaction of the TALE-CC chimera with the corresponding matching CC:VP16 fusion (Figure 1b). The formation of functional transcription factors upon interaction of the corresponding CC peptide pairs was determined both with luciferase assays and flow cytometry (Supplementary Figure 1). The results demonstrated strong transcriptional activity only with the correct peptide pair combination, thereby confirming the functionality of the designed peptide pairs in the environment of mammalian cells. Off-target activation by non-desired pairs was several orders of magnitude weaker (Figure 1c, Supplementary Figure 2). Further, the P11-P12 pair exhibited some cross-talk with the P3-P4 pair (Supplementary Figure 2), probably due the similarity in their hydrophobic and electrostatic patterns (Supplementary Table 1); however, even in this case, the ratio between the correct and mismatched pairs remained above ~100. The strength of transcriptional activation with the P3-P4 pair, which was slightly more efficient than other pairs, was compared to the E- and K-peptides—which have frequently been used to guide protein interactions20,21—and to two pairs from the SYNZIP set4, where the P3-P4 pair was shown to be the most potent (Supplementary Figure 3). The NICP set was designed de novo and we were interested in their propensity to interact with other proteins in the human cell proteome. Therefore, we performed a computational analysis of interactions of the NICP set with the human proteome and compared it to SYNZIPs and several other frequently used CC peptides (E/K pair and GCN4 homodimerizing peptide), where off-target interactions with the human genome were analyzed. The results demonstrate a large number of possible off-target interactions for most of the SYNZIP, E/K, and GCN4 peptides, while the NICP set exhibits minimal off-target binding propensity (Supplementary Figure 4), thereby supporting their orthogonality. Low number of interaction partners means that the CC peptide is less likely to interfere with cellular processes and at the same time that cellular proteins do not hinder interactions between the CC peptide pair.
Figure 1. Orthogonality of the designed NICP peptide set in HEK293T cells.
(a) Schematic presentation of a set of six pairs of designed coiled-coil (CC) peptides. (b) Schematic presentation of the transcriptional activation system designed to assess the activity and orthogonality of CC peptides. Upon interaction of the CC peptides, the activation domain (VP16) is brought to the promoter region on a reporter plasmid guided by TALE fused to the corresponding CC, triggering expression of the reporter gene (left). When a non-matching peptide pair is co-transfected, no expression of the reporter gene is observed (right). Pmin represents the minimal promoter and fLuc the firefly luciferase reporter gene. (c) Orthogonality of the NICP set was tested in HEK293T cells by co-transfection of the [a]10_Pmin-fLuc reporter plasmid (50ng), a TALE:CC fusion encoding plasmid (25ng) and a CC:VP16 fusion encoding plasmid (25ng). Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean of replicates within two independent experiments (n = 3 and n = 4). Fold activation was calculated by normalizing the RLU values of each sample to the RLU value of the reporter only control within the same experiment. Individual data points are presented in Supplementary figure 2.
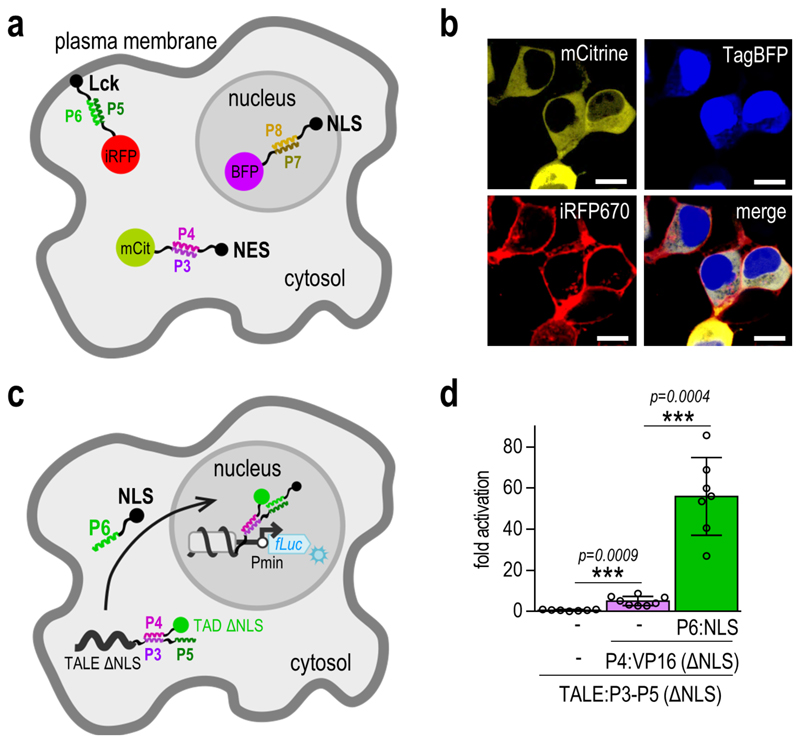
The mutual orthogonality of three of the peptide pairs was further demonstrated by CC-mediated localization of fluorescent proteins to selected cellular compartments within single cells. Three fluorescent proteins (mCitrine, TagBFP, and iRFP670) were each fused to a different CC peptide, while the complementary peptides were fused to signal peptides targeting the plasma membrane (Lck), cell nucleus (NLS), or the cytosol (NES). First, we demonstrated that the CC-signal peptide fusions are able to direct the localization of iRFP670 to selected compartments (Supplementary Figure 5). Next, the simultaneous localization of three fluorescent reporters within single cells was demonstrated upon introduction of the three corresponding CC peptides, which were fused to different localization signals (Figure 2a-b, Supplementary Figure 6), thereby demonstrating that the CC peptide toolbox is capable of multiplexing different interactions and processes within the same cell.
Figure 2. CC-directed localization of multiple proteins.
(a) Schematic presentation of the directed localization of fluorescent reporter proteins upon addition of the corresponding CC peptide fusions. ‘mCit’, ‘BFP’ and ‘iRFP’ represent the mCitrine, TagBFP and iRFP670 fluorescent proteins, respectively. ‘NLS’ represents the nuclear localization signal, ‘NES’ the nuclear export signal and ‘Lck’ the Lck membrane localization signal. (b) Experimental evaluation of fluorescent protein localization. HEK293T cells were co-transfected with 25 ng of each fluorescent reporter-CC encoding plasmid, and 125 ng of each of the plasmids encoding the corresponding CC peptides, fused to different localization signals. Images are representative of two independent experiments. Scale bar = 10 µm. (c) Schematic presentation of the expected localization of the NLS deficient TALE:CC-CC:TAD complex. Upon addition of the P6:NLS fusion peptide, the complex is brought to the nucleus, where it triggers activation of reporter gene transcription. ‘Pmin’ represents the minimal promoter and ‘fLuc’ the firefly luciferase reporter gene. (d) The NLS deficient TALE:CC-CC:TAD complex activates transcription of the reporter gene only upon co-transfection of the P6:NLS fusion peptide. HEK293T cells were co-transfected with 50 ng of the [a]10_Pmin-fLuc reporter plasmid, 25 ng of the TALE:CC(ΔNLS) encoding plasmid, 10 ng of the CC:VP16(ΔNLS) encoding plasmid and 25ng of the CC:NLS encoding plasmid. Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean and standard deviation of replicates within two independent experiments (n = 4). Fold activation was calculated by normalizing the RLU values of each sample to the RLU value of the reporter only control within the same experiment. Statistical analysis was performed on pooled data from both experiments by the two-sample F-test and the two-tailed two-sample t-test assuming equal or unequal variances as determined by the F-test. The p-values are indicated above the bars.
CC peptides for the control of multiple functionalities
Due to their mutual orthogonality, combinations of different peptides could be employed to deliver different functional modules to the same target protein. The fusion of two different CC peptides to the mCitrine fluorescent protein enabled simultaneous control of its localization and its association with the BFP fluorescent protein (Supplementary Figure 7). This strategy was used to demonstrate localization-dependent transcriptional regulation. A TALE-DBD with a deleted NLS was fused to two different CC peptides (P3 and P5). Low activation of reporter expression was observed upon addition of the NLS-deficient VP16 activation domain, fused to the P4 peptide. Nevertheless, transcription occurred upon co-transfection of the NLS-fused P6 peptide due to the translocation of the TALE:CC-CC:VP16 complex to the nucleus (Figure 2c-d). An alternative mode of regulation of CC-mediated interactions was demonstrated using a CC linker peptide consisting of peptides P4 and P5. This linker peptide was able to mediate the interaction between the TALE-DBD and the VP16-TAD, each fused to a CC peptide from a non-corresponding pair (P3 and P6, respectively) (Supplementary Figure 8). These results further demonstrate the possibilities of the application of the CC toolbox for the control of cellular processes, such as transcription of multiple genes and protein localization to different compartments.
Tunability of CC module-based interactions and process regulation
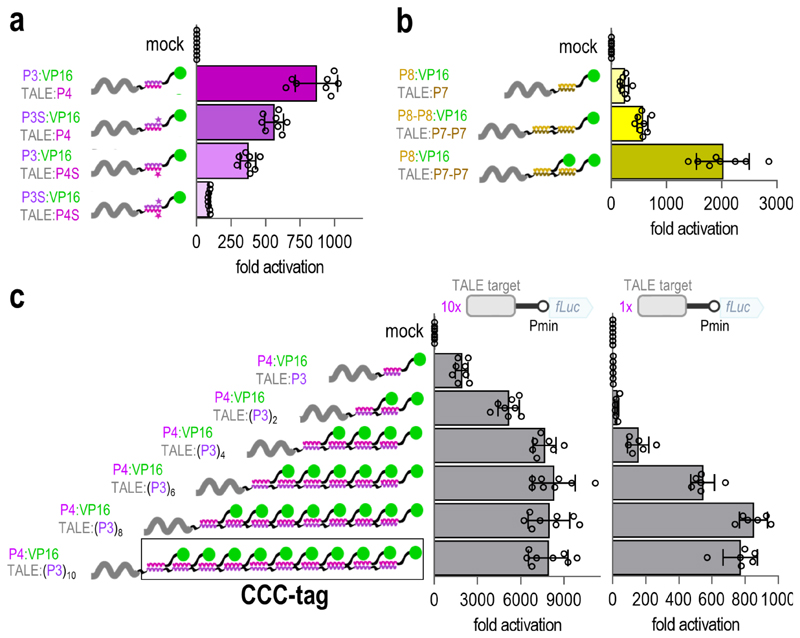
Based on the observed differences in the strength of transcriptional activation governed by the affinity of different CC pairs (Figure 1, Supplementary Figures 1 and 2), we further investigated whether modifications in the affinity of the same CC pair could be used to modulate the strength of transcriptional activation. For this purpose, we used variants of the P3 and P4 peptides (P3S and P4S), which retained the specificity determined by interacting residues at positions a, d, e, and g, while the helix-forming propensity of non-interacting residues at positions b, c, and f was varied by replacing high helical propensity Ala residues by low helical propensity Ser or Gln residues (Supplementary Figure 9a-b). This strategy has been previously used to tune the in vitro stability of CC pairs.14 These modifications resulted in the transcriptional activation strength having a wide range, depending on the combination of CC peptide variants (Figure 3a), thereby demonstrating physiological tunability of the same CC pair in the range > 1:10. The modified peptides retained orthogonality in the context of the set (Supplementary Figure 9c) and, interestingly, eliminated the cross-talk between the P3-P4 and the P11-P12 peptide pairs (Supplementary Figure 9d). This is likely due to the decreased overall stability with a maintained energy difference between the on- and off-target pairs.
Figure 3. Tunability of the interaction strength within the NICP set toolbox.
(a) Modification of the helix forming propensity of the CC peptides attenuates the strength of the transcriptional activation with retained specificity. Residues at positions a, d, e and g of the P3 and P4 peptides were conserved, while the residues at positions b, c and f were varied to design peptides P3S and P4S (Supplementary figure 9a-b) with reduced affinity. (b) Concatenation of CC peptides results in enhanced transcriptional activation due to increased interaction strength and increased stoichiometry of the bound TADs. (c) Increasing the number of bound VP16-TADs by concatenation of CC peptides to the TALE-DBD (CCC-tag) strongly enhances transcriptional activation even on a reporter plasmid with a single TALE binding site. In all panels, HEK293T cells were co-transfected with 50 ng of the reporter plasmid and 25 ng of the TALE:CC and CC:VP16 encoding plasmids. Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean and standard deviation of replicates within two independent experiments (n = 4). Fold activation was calculated by normalizing the RLU values of each sample to one of the RLU values of the reporter only (mock) control within the same experiment.
In addition to interaction strength attenuation due to mutations, enhancement of interaction was demonstrated by the concatenation of two repeats of the P7 peptide to a TALE-DBD and two repeats of the P8 peptide to the VP16-TAD. This duplication strategy resulted in substantially stronger transcriptional activation than that in a single copy of the CC peptide (Figure 3b) due to the elongation of the interaction surface.22
As an alternative implementation of tuning the response strength, concatenated CCs can be used to increase the stoichiometry of the recruited functional domains, which provides a different strategy of tuning transcriptional response. The combination of constructs coding for the TALE-DBD with two concatenated P7 repeats and the VP16-TAD with a single P8 repeat resulted in even stronger transcriptional activation than length duplication (Figure 3b) due to the recruitment of two TADs to the TALE-P7-P7 chimera instead of a single TAD. Increased activation by duplication of the DNA target-bound TAD suggested that the concatenation of several repeats of heterodimeric CC peptides could be applied for the construction of highly potent transcriptional activators. We investigated the constructs for extended TALE-DBDs, fused to up to 10 concatenated P3 peptide repeats ((P3)10). This strategy increased the transcriptional activation up to an unprecedented 8000-fold on a reporter plasmid with 10 repeats of TALE target sites, nearing the plateau at four concatenated peptide repeats (Figure 3c). We termed this enhanced activation strategy the “concatenated coiled-coil tag” (CCC-tag).
Nevertheless, transcriptional activators are required to strongly activate transcription even when a single DNA recognition domain is bound to the promoter region, particularly for endogenous gene regulation. As expected, the TALE-P3 chimeras mediated strong transcriptional activation in the case of a single binding site in combination with the P4-VP16 construct (up to 800-fold in the case of 10 concatenated P3 repeats (Figure 3c)). The replacement of VP16 by stronger TADs, such as the tetrameric derivative VP646 and the chimeric VPR domain23 resulted in an even stronger activation (3500-fold) of reporter expression with a single DNA binding site (Supplementary Figure 10a).
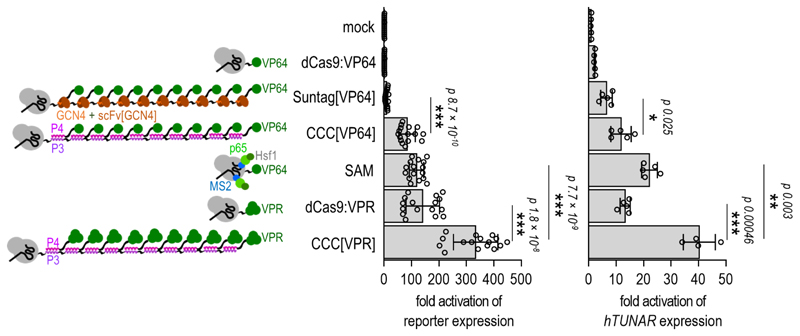
Benchmarking the CCC-tag to other designed activators
Recently, several strategies have been described for the enhancement of transcriptional activation by the CRISPR/dCas9 system.24 In order to benchmark the efficiency of the CCC-tag against other dCas9-mediated transcriptional activation strategies, the dCas9 protein was fused to the (P3)10 repeats (“CRISPR-CCC”). Potent transcriptional activation with the CRISPR-CCC strategy in combination with the VPR-TAD (CCC[VPR]) was demonstrated with flow cytometry in HEK293T cells (Supplementary Figure 10b). “Suntag”6 is a transcriptional activation strategy based on the concatenation of short peptides and targeting by scFv:TAD, with some similarity to the CRISPR-CCC system. We first benchmarked the CCC[VP64] and Suntag[VP64] systems for activation of an endogenous gene and reporters with several different gRNAs (Supplementary Figure 11), where the CCC[VP64] strategy mediated stronger activation in all tested cases. We compared our strategy to several other dCas9-based strategies, which were recently shown to be the most potent designed transcriptional activators,23,6 including the VPR-TAD,23 “Suntag,”6 and “SAM”25 (Figure 4). All the described activation approaches were tested in the HEK293T cell line on a reporter plasmid using the same gRNA target, with the CCC[VPR] strategy being the most potent one. The CRISPR/dCas9-based transcriptional activators described above were also benchmarked for activation of the TUNAR endogenous gene transcription using the same gRNA target in the HEK293T cell line. Again, the CCC[VPR] strategy was demonstrated to be the most potent compared to the other tested transcriptional activation strategies.
Figure 4. Highly potent designed CRISPR-CCC transcriptional activation platform.
Schematic presentation of the benchmarked CRISPR-based transcriptional activation strategies is shown on the left. In comparison to other transcriptional activation strategies, the CCC-tag in combination with the VPR-TAD exhibited the most potent activation of reporter gene transcription (middle) and upregulation of the genomic TUNAR gene (right) in HEK293T cells. For reporter assays, HEK293T cells were co-transfected with 50 ng of the reporter plasmid and 25ng of the corresponding gRNA encoding plasmid along with 25 ng of dCas9-fusion encoding plasmid and 25 ng of the TAD-fusion encoding plasmid. Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean and standard deviation of replicates within four independent experiments (n = 4). Fold activation was calculated by normalizing the RLU values of each sample to the RLU value of the reporter only (mock) control within the same experiment. For evaluation of hTUNAR expression with quantitative PCR, HEK293T cells were co-transfected with 250 ng of the gRNA encoding plasmid along with 250 ng of dCas9-fusion encoding plasmid and 250 ng of the TAD-fusion encoding plasmid. Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean and standard deviation of replicates within two independent experiments (n = 2 and n = 3). Fold activation was calculated by normalizing relative hTUNAR expression of each sample to the value of relative hTUNAR expression of the mock control sample (cells transfected with an empty pcDNA3 plasmid) within the same experiment. Statistical analysis was performed on pooled data from all experiments by the two-sample F-test and the two-tailed two-sample t-test assuming equal or unequal variances as determined by the F-test. The p-values are indicated above the bars.
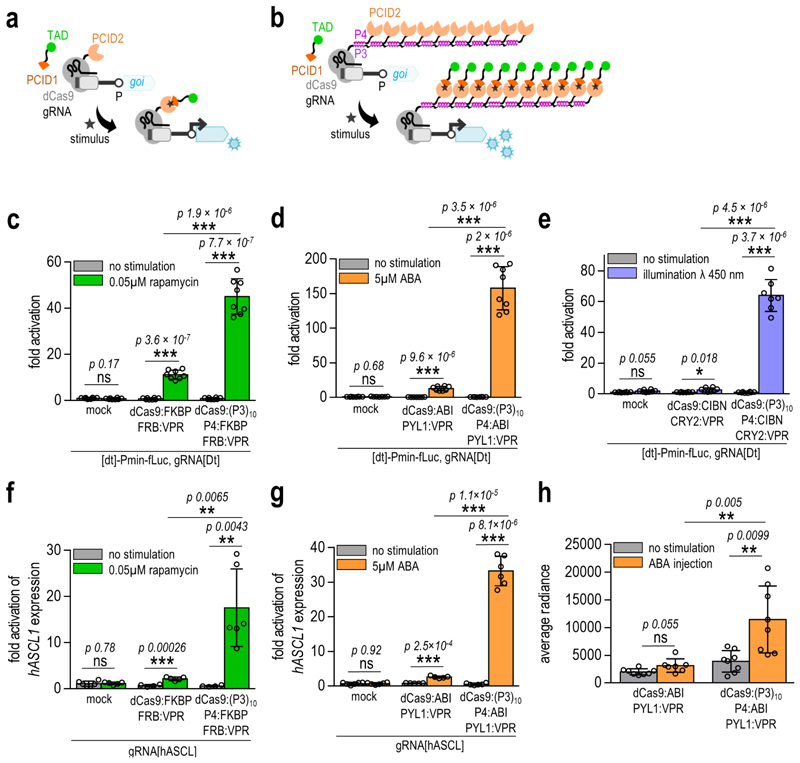
Enhancement of conditional transcriptional regulation
Conditionally regulated (i.e. inducible) transcriptional control of selected genes can be achieved by genetic fusion of a DBD and a TAD to protein domains that associate upon the presence of a chemical or physical signal (Figure 5a). The performance of physico-chemically regulated systems is extremely important as it enables external regulation of the system, particularly important for therapeutic control.26–28 Nevertheless, conditionally regulated transcriptional activation based on conventional design may typically be rather low compared to covalent DBD:TAD fusion.29 We anticipated that the response could be enhanced by the genetic fusion of one of the physico-chemically induced dimerization domains (PCID1) to a TAD (as in the conventional design) and the other (PCID2) to the P4 peptide. In turn, the CCC-tagged DBD can be designed to bind multiple P4:PCID2 chimeric proteins, which interact with the PCID1:TAD fusion upon stimulation and could, thus, recruit up to 10 activation domains (Figure 5b). Indeed, strongly enhanced transcriptional activation was demonstrated by the CRISPR-CCC system upon rapamycin, abscisic acid (ABA), and blue light stimulation using a single target site for the dCas9:gRNA complex on a reporter plasmid with up to 150 fold increase (Figure 5c-e, Supplementary Figures 12–14) as well as for the ASCL1 endogenous gene in HEK293T cells with up to 30 fold increase (Figure 5f-g). The ABA-inducible CRISPR-CCC system was also able to strongly upregulate transcription in the murine NIH-3T2 and Neuro2A cell lines, as demonstrated on the reporter plasmid and on the murine IGF1 endogenous gene (Supplementary Figure 15). In order to validate the applicability of small molecule-regulated transcriptional activation based on the CCC-tag in vivo, the ABA-inducible CRISPR-CCC system was analyzed in mice using subcutaneous implantation of engineered HEK293T cells. CCC[ABA] engineered cells exhibited a high upregulation of reporter gene expression, detected with bioluminescence imaging four to six hours after intraperitoneal ABA administration, which was significantly stronger than the conventional ABI1-PYL1 system based on the same DBD, TAD, and PCID domains (Figure 5h, Supplementary Figure 16). Bioluminescence in mice with CCC[ABA] engineered cells was also detectable 24 hours after implantation with ABA re-injection four hours prior to imaging (Supplementary Figure 16).
Figure 5. Enhancement of conditionally-regulated transcription with CRISPR-CCC.
(a) Schematic presentation of the conventional CRISPR-based conditionally-regulated transcription. (b) Schematic presentation of the CRISPR-CCC-based conditionally-regulated transcription. (c-e) Conditionally-regulated CCC-tag enhances transcriptional activation of the reporter plasmid in HEK293T cells by employing rapamycin- (c), abscisic acid (ABA)- (d) and blue light (e) inducible components. Amounts of transfected plasmids are listed in Supplementary table 6. Replicates represent HEK293T cell cultures, individually transfected with the same mixture of plasmids. The values represent the mean and standard deviation of replicates within two independent experiments (n = 4). Fold activation was calculated by normalizing the RLU values of each sample to the RLU value of the reporter only (mock) control within the same experiment. (f-g) Conditionally-regulated CCC-tag enhances transcriptional activation of the reporter plasmid in HEK293T cells by employing abscisic acid (ABA)- (f) and rapamycin- (g) inducible components. Amounts of transfected plasmids are listed in Supplementary table 6. Fold activation was calculated by normalizing relative hASCL1 expression of each sample to the value of relative hASCL1 expression of the non-stimulated mock control sample (cells transfected with an empty pcDNA3 plasmid) within the same experiment. (h) Quantification of in vivo bioluminescence images of the ABA-regulated CRISPR-based system engineered HEK293T cells implanted in mice. Images from a representative experiment are available in Supplementary figure 16. Amounts of transfected plasmids are listed in Supplementary table 6. Replicates represent individual animals, injected with engineered HEK293T cells. The values represent the mean and standard deviation of replicates within three independent experiments (n = 3). Statistical data represent the difference between the non-stimulated and stimulated samples for each transfected combination. Statistical analysis was performed on the pooled data from all experiments by the two-sample F-test and the two-tailed two-sample t-test assuming equal or unequal variances as determined by the F-test. The p-values are indicated above the bars.
Discussion
The introduction of protein interaction domains by genetic fusion is a widely used strategy for the regulation of protein complex formation, localization, construction of biosynthetic and signaling scaffolds, and the regulation of transcription. A limiting factor has been the availability of a validated orthogonal set of heterodimeric protein-protein interaction pairs. Therefore, the orthogonal set of heterodimeric interaction pairs described here (the NICP set) provides a powerful toolbox for cell engineering and enables several diverse implementations. The NICP set augmented the activation of transcription that was several orders of magnitude stronger than that of the non-cognate combinations (Figure 1, Supplementary Figures 1 and 2). The de novo designed CC pairs exhibit low similarity to cellular proteome, which is in contrast to the E:K peptide pair21 or peptides based on natural leucine zipper peptides13 (Supplementary Figure 4) and are, therefore, less likely to adversely affect cellular processes. CC dimers based on the SYNZIP set13,30 have been used in yeast cells, where the range of response between the on- and off-targets was substantially lower than reported here.4 Recently, an interaction set based on the dimeric four helical bundles has been used in yeast.16 In contrast to the large in vitro size of the orthogonal set, only six pairs were found to be orthogonal in yeast cells, thereby demonstrating additional constraints due to the complex environment of the cell cytosol. Our study validates the orthogonality of the heterodimeric NICP set in mammalian cells. Based on the modular design and adjustable stoichiometry and tunable affinity, the NICP set was used for simultaneous control of localization of different proteins and regulation of cellular processes in addition to transcriptional regulation (Figure 2, Supplementary Figures 5–8)
An additional advantage of the described peptide set is the ability to tune the affinity by varying the non-interacting residues while maintaining sequence length and pairing specificity (Figure 3, Supplementary Figure 9a-b). In fact, we observed that the decrease in the interaction strength further improved the CC orthogonality (Supplementary Figure 9c). Further expansion of the range of response intensity was provided by concatenation of several CC peptides (CCC-tag), which resulted in strongly increasing transcriptional activation in the context of TALE- and CRISPR-based designed transcription factors (Figure 3b-c, Supplementary Figure 10) with the combined dynamic range of ~100 for CC pairs. CCC-tag-based activation of transcription with the CRISPR/dCas9 system (CRISPR-CCC) was in fact superior in comparison to some of the most potent CRISPR-based activation strategies reported thus far, such as “Suntag” and “SAM” (Figure 4, Supplementary Figure 11).6,23,25,31
The CCC-tag also performed well for CRISPR-based enhancement of light-, rapamycin-, and ABA-regulated transcription in mammalian cells (Figure 5, Supplementary Figures 12–15). While concatenation of one of the induced dimerization partners on the DBD could enhance the transcriptional activity in a similar manner as the CCC-tag, the chemically-induced dimerization domains are usually larger in size. CC-forming peptides comprise only ~33 amino acid residues, thereby decreasing the genetic footprint of components in comparison to several other interaction domains. While this strategy has been demonstrated on three different conditionally regulated systems (Figure 5c-f), the CCC-tag would be likely applicable for enhancement of other types of regulation based on inducible dimerization of two protein domains. The in vivo transplantation of CCC[ABA]-engineered cells (Figure 5h, Supplementary Figure 16) demonstrates the potential use of this system for therapeutic applications.32,33 Mammalian cell-based therapeutic devices have been used to treat gout34 and Crohn’s disease28 in mice. The CCC-tag based platform could be used to trigger a strong expression of therapeutics using the selected inducible system. In addition, with appropriate delivery techniques, this system could also be used for endogenous gene regulation in vivo. While the background of the CCC[ABA]-engineered cells implanted in mice (Supplementary Figure 16) appears to be slightly higher in comparison to cell culture experiments (Supplementary Figures 12–15) it provided a substantial increase upon addition of ABA. The possible explanation for the background activation may be either the presence of ABA in the diet of the mice or, alternatively, the endogenous ABA in mammals. 35
The NICP set presented in this report represents a potent toolbox for the regulation of several concurrent interactions or processes within mammalian cells. The concatenation of CC segments potentiates interaction affinity and enables the introduction of multiple interactions and functional domains simultaneously, which could direct the formation of multicomponent complexes of precisely defined stoichiometry and introduction of different functionalities. Therefore, the above-described NICP peptide set and the CCC-tag strategy have considerable potential for use in biotechnology, bioengineering, and cell-based therapeutic applications.
Online Methods
Plasmid construction
All plasmids were constructed using the Gibson assembly method36 and are listed in Supplementary Table 2. Amino acid sequences of protein coding genes, individual protein domains, and peptides (along with their origins) are provided in Supplementary Tables 1 and 3. Nucleotide sequences of gRNAs and promoter sequences with DNA target sites are listed in Supplementary Table 4.
Cell culture, transfection and stimulation
The human embryonic kidney (HEK) 293T cell line (ATCC) and the NIH-3T3 mouse fibroblast cell line (ATCC) were cultured in DMEM medium (Invitrogen), supplemented with 10% fetal bovine serum (BioWhittaker) at 37°C in a 5% CO2 environment.The mouse neuroblastoma cell line (Neuro2A) (ATCC) was cultured in Optimem (Invitrogen), supplemented with 10% fetal bovine serum (BioWhittaker) at 37°C in a 5% CO2 environment. For luciferase experiments, 1.5-2 × 104 cells per well were seeded in CoStar White 96-well plates (Corning). For confocal microscopy experiments, 5 × 104 cells per well were seeded in eight-well tissue culture chambers (m-Slide 8 well, Ibidi). For RNA extraction and flow cytometry experiments, 1 × 105 cells per well were seeded in 24-well plates (TPP) or 2 × 105 cells per well were seeded in 12-well plates (TPP). For the subcutaneous implantation of cells into mice, 2 × 106 cells per well were seeded in petri dishes (9mm φ). At the 30–90% confluence, HEK293T cells were transfected with a mixture of DNA and PEI (6 µl/500 ng DNA, stock concentration 0.324 mg/ml, pH 7.5). At the ~70% confluence, NIH-3T3 cells were transfected with a mixture of DNA and PEI (12µl/500 ng DNA, stock concentration 0.324 mg/ml, pH 7.5). At the ~70% confluence, Neuro2A cells were transfected with a mixture of DNA and Lipofectamine LTX (Thermo Scientific), according to the manufacturer’s instructions. The amounts of transfected plasmids are indicated in the figures and figure captions or listed in Supplementary table 5. In order to normalize reporter values to transfection efficiency, different constitutively expressed control plasmids were used: i) 10ng of phRL-TK (Renilla luciferase encoding plasmid) for luciferase experiments and ii) 100ng of pTagBFP-N (BFP encoding plasmid) for flow cytometry experiments. When applicable, cells were stimulated with blue light (wavelength 450 nm, intensity 400 Lux), rapamycin or abscisic acid. The stimuli were added at 24 hours after transfection. Cells were stimulated for 20 – 24 hours.
Luciferase assays
The cells were harvested at indicated timepoints after transfection and/or stimulation and lysed with 25 ul of 1 × Passive Lysis buffer (Promega). Firefly luciferase and Renilla luciferase expression were measured using the dual luciferase assay (Promega) on an Orion II microplate reader (Berthold Technologies). Relative luciferase units (RLU) were calculated by normalizing each sample’s firefly luciferase activity to the constitutive Renilla luciferase activity determined in the same sample.
Flow cytometry
The cells were washed with PBS and harvested with 500 ml of FACS buffer (3% fetal bovine serum in PBS). Flow cytometry analysis was performed using a CyFlow space flow cytometer (Partec). A 488-nm diode laser was used for detection of mCitrine, a 405-nm diode laser for TagBFP. In each sample, 20000–30000 cells were analyzed and gated to TagBFP transfection control. The data was processed using FlowJo software (TreeStar) and are representative of two or three independent experiments. Curves are presented as staggered histograms. Mean fluorescence intensity (MFI) in each sample was determined with the FlowJo software.
Computational analysis of interaction partners
Different CC peptides were scanned against the human proteome. Canonical and isoform protein sequences that constitute the human proteome were downloaded from Universal Protein Resource database (UniProt).37,38 The strength of binding was evaluated using the parallel CC dimer scoring function introduced by Potapov et al.30 The interaction score for a given protein-peptide pair was obtained by calculating the interaction for all possible sequence alignments of the selected pair and taking the lowest observed value as the final interaction score. If the interaction score was equal or below -8.0, the protein-peptide pairing was classified as strong. In case the interaction score was lower in comparison to the score for the designed on-target peptide pair, the interaction was labelled as off-target. The obtained distribution of interaction scores for each PNIC peptide against the human proteome was fit with a Gaussian curve:
where μ is the mean value and σ is the standard deviation.
Confocal Microscopy
Microscopic images were acquired two days after transfection, using the Leica TCS SP5 inverted laser-scanning microscope on a Leica DMI 6000 CS module equipped with a HCX PL Fluotar L 20×, numerical aperture 0.4 (Leica Microsystems). A 514-nm laser line of a 100-mW argon laser with 25% laser power was used for mCitrine excitation, and the emitted light was detected between 530 and 550 nm. A 50-mW 405-nm diode laser was used for TagBFP excitation and the emitted light was detected between 420 nm and 460 nm. A 10-mW 633-nm HeNe laser was used for iRFP670 excitation, and the emitted light was detected between 650 nm and 690 nm. Further, Leica LAS AF software was used for acquisition, and ImageJ software was used for image processing.
RNA isolation, reverse transcription, and quantitative PCR
HEK293T and Neuro2A cells were harvested at indicated timepoints after transfection and/or stimulation with 250 µl trypsin, centrifuged at 3000 rpm for 5min, resuspended in 200 µl PBS; thereafter, RNA was extracted using the High-pure RNA isolation kit (Roche). Reverse transcription was performed with the High-capacity cDNA reverse transcription kit (Applied Biosystems) with a mixture of random oligonucleotides. Quantitative PCR was performed with the LightCycler 480 SYBR Green I master mix (Roche) on the LightCycler 480 microplate reader. Oligonucleotides used for quantitative PCR are listed in Supplementary Table 6. Relative mRNA amounts were calculated using the formula mRNA = 2[(Cp(goi, mock)-Cp(GAPDH, mock))-(Cp(goi, sample)-Cp(GAPDH, sample)].39
Animal experiments
Hsd:ICR (CD-1) mice were purchased from Envigo (Italy). Eight-to-twelve-week-old male and female mice were used for experiments. All animal experiments were performed according to the directives of the EU 2010/63 and were approved by the Administration of the Republic of Slovenia for Food Safety, Veterinary, and Plant Protection of the Ministry of Agriculture, Forestry and Foods, Republic of Slovenia (Permit Number U34401-3/2017/16). Mice were injected in the right flank with 2 × 106 HEK293 cells transfected with the appropriate plasmids 24 hours prior to the experiment. After 30 minutes and after 24 hours, mice were treated intraperitoneally with 100 μl of 100 μM abscisic acid. Four to six hours after ABA injection, mice received 150 mg/kg of body weight of D-luciferin (Xenogen) subcutaneously and were in vivo imaged with IVIS® Lumina Series III (Perkin Elmer). Data were analyzed with Living Image® 4.5.2 (Perkin Elmer).
Statistics
Replicates represent HEK293T, NIH-3T3, or Neuro2A cell cultures, individually transfected with the same mixture of plasmids. Microscopic images are representative of two independent experiments and five separate observations within the same experiment. The in vivo luciferase data are representative of three independent experiments, and the replicates represent individual animals in each test group.
For each individual data point (replicate), the cells were first seeded into one well of a multiwell plate. For luciferase experiments, a mixture of plasmids was prepared in a single tube for three or four wells (replicates) of cell populations, analyzed within the same experiment (i.e. for 100ng of a plasmid per well, 300ng or 400ng of the plasmid was mixed in the tube). For flow cytometry and RNA extraction experiments, transfection mixtures were prepared individually for each well. Further, individual wells were transfected with the appropriate volume of the transfection mixture (amount of the plasmid prepared in the tube divided by the number of replicates). At selected time points, each well (replicate) was measured individually. Fold activation was calculated by normalizing the sample values from each individual experiment to the value of one of the replicates of the mock (non-stimulated) sample within the same experiment. The fold activation values from 2–4 individual experiments were pooled, and outliers were determined using the following formulas:
lower bound = Q1 – (1.5 × (Q3–Q1)), and
upper bound = Q3 + (1.5 × (Q3–Q1)),
where Q1 and Q3 represent the first and third quartiles, respectively. 40 Averages and standard deviations were calculated and statistical analysis were conducted to determine the p-values (two-sample F-test for variances and the two-tailed t-test).
Supplementary Material
Acknowledgments
This research was supported by grants from the Slovenian Research Agency (P4-0176, J1-9173, J3-7034, N4-0080), ERC grant MaCChines to RJ, Horizon2020 CSA Bioroboost, and ERANET project MediSurf. Tina Lebar is partially supported by the UNESCO-L’OREAL national fellowship ‘For Women in Science’. We thank Helena Gradišar for providing the sequences of CC peptides and for valuable advice.
Footnotes
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author Contributions
TL and EM prepared the plasmid constructs and performed the experiments on cell culture. DL performed the experiments on mice. JA performed the bioinformatics analysis. TL and RJ designed and analyzed the experiments and wrote the manuscript. RJ conceived the study.
Competing Financial Interests Statement
The authors declare no competing financial interest.
References
- 1.Kim KH, Chandran D, Sauro HM. Design and Analysis of Biomolecular Circuits. Springer New York: 2011. Toward Modularity in Synthetic Biology: Design Patterns and Fan-out; pp. 117–138. [DOI] [Google Scholar]
- 2.Kuriyan J, Cowburn D. Modular Peptide Recognition Domains in Eukaryotic Signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 3.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–8. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 4.Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP Protein Interaction Toolbox: in Vitro and in Vivo Specifications of Heterospecific Coiled-Coil Interaction Domains. ACS Synth Biol. 2012;1:118–129. doi: 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fekonja O, Benčina M, Jerala R. Toll/Interleukin-1 Receptor Domain Dimers as the Platform for Activation and Enhanced Inhibition of Toll-like Receptor Signaling. J Biol Chem. 2012;287:30993–31002. doi: 10.1074/jbc.M112.376186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan H, Peabody NC, Vinson CR, White BH. Refined Spatial Manipulation of Neuronal Function by Combinatorial Restriction of Transgene Expression. Neuron. 2006;52:425. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selgrade DF, Lohmueller JJ, Lienert F, Silver PA. Protein scaffold-activated protein trans-splicing in mammalian cells. J Am Chem Soc. 2013;135:7713–9. doi: 10.1021/ja401689b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripet B, et al. Engineering a de novo -designed coiled-coil heterodimerization domain for the rapid detection, purification and characterization of recombinantly expressed peptides and proteins MRC Group in Protein Structure and Function, ^Department of Medical Microbiology and Immunology and the. Protein Eng. 1996;9:1029–1042. doi: 10.1093/protein/9.11.1029. [DOI] [PubMed] [Google Scholar]
- 10.Yano Y, et al. Coiled-Coil Tag−Probe System for Quick Labeling of Membrane Receptors in Living Cells. ACS Chem Biol. 2008;3:341–345. doi: 10.1021/cb8000556. [DOI] [PubMed] [Google Scholar]
- 11.Yano Y, Matsuzaki K. Live-cell imaging of membrane proteins by a coiled-coil labeling method—Principles and applications. Biochim Biophys Acta - Biomembr. 2019;1861:1011–1017. doi: 10.1016/j.bbamem.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Woolfson DN. The Design of Coiled-Coil Structures and Assemblies. Advances in protein chemistry. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan JB, Reinke AW, Keating AE. Increasing the affinity of selective bZIP-binding peptides through surface residue redesign. Protein Sci. 2014;23:940–953. doi: 10.1002/pro.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drobnak I, Gradišar H, Ljubetič A, Merljak E, Jerala R. Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity. J Am Chem Soc. 2017;139:8229–8236. doi: 10.1021/jacs.7b01690. [DOI] [PubMed] [Google Scholar]
- 15.Ljubetič A, et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat Biotechnol. 2017;35:1094–1101. doi: 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, et al. Programmable design of orthogonal protein heterodimers. Nature. 2019;565:106–111. doi: 10.1038/s41586-018-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JH, Collins JJ, Wong WW. Universal Chimeric Antigen Receptors for Multiplexed and Logical Control of T Cell Responses. Cell. 2018;173:1426–1438.e11. doi: 10.1016/j.cell.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gradišar H, Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J Pept Sci. 2011;17:100–106. doi: 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- 19.Gradišar H, et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat Chem Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: design of a heterodimeric coiled coil. Curr Biol. 1993;3:658–67. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 21.Shekhawat SS, Porter JR, Sriprasad A, Ghosh I. An Autoinhibited Coiled-Coil Design Strategy for Split-Protein Protease Sensors. J Am Chem Soc. 2009;131:15284–15290. doi: 10.1021/ja9050857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas F, Boyle AL, Burton AJ, Woolfson DN. A Set of de Novo Designed Parallel Heterodimeric Coiled Coils with Quantified Dissociation Constants in the Micromolar to Sub-nanomolar Regime. J Am Chem Soc. 2013;135:5161–5166. doi: 10.1021/ja312310g. [DOI] [PubMed] [Google Scholar]
- 23.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods. 2016;13:1043–1049. doi: 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansouri M, Strittmatter T, Fussenegger M. Light-Controlled Mammalian Cells and Their Therapeutic Applications in Synthetic Biology. Adv Sci. 2019;6:1800952. doi: 10.1002/advs.201800952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C-Y, Roybal KT, Puchner EM, Onuffer J, Lim WA. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science (80-. ) 2015;350:aab4077–aab4077. doi: 10.1126/science.aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smole A, Lainšček D, Bezeljak U, Horvat S, Jerala R. A Synthetic Mammalian Therapeutic Gene Circuit for Sensing and Suppressing Inflammation. Mol Ther. 2017;25:102–119. doi: 10.1016/j.ymthe.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lonzarič J, Lebar T, Majerle A, Manček-Keber M, Jerala R. Locked and proteolysis-based transcription activator-like effector (TALE) regulation. Nucleic Acids Res. 2016;44:1471–81. doi: 10.1093/nar/gkv1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potapov V, Kaplan JB, Keating AE, Howlett G, Schubert D. Data-Driven Prediction and Design of bZIP Coiled-Coil Interactions. PLOS Comput Biol. 2015;11:e1004046. doi: 10.1371/journal.pcbi.1004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kis Z, Pereira HS, Homma T, Pedrigi RM, Krams R. Mammalian synthetic biology: emerging medical applications. J R Soc Interface. 2015;12 doi: 10.1098/rsif.2014.1000. 20141000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2012;13:21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemmer C, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat Biotechnol. 2010;28:355–360. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 35.Wasilewska A, et al. An Update on Abscisic Acid Signaling in Plants and More …. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 36.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 37.Breuza L, et al. The UniProtKB guide to the human proteome. Database. 2016;2016 doi: 10.1093/database/bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium TU. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Kwak SK, Kim JH. Statistical data preparation: management of missing values and outliers. Korean J Anesthesiol. 2017;70:407–411. doi: 10.4097/kjae.2017.70.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.