Abstract
Objective
Exosomes derived from cancer cells can alter the microenvironment and enhance cancer malignancy through the regulation of peripheral cell functions. The present study focused on the crosstalk between chondrosarcoma cells and human umbilical vein endothelial cells (HUVECs) mediated by exosomes derived from chondrosarcoma cells and aimed to explore the potential molecular mechanism.
Materials and Methods
Chondrosarcoma cell-derived exosomes were isolated and characterized. Cell proliferation assay, tube formation assay and transwell migration assay were performed to characterize the effects of exosomes on HUVECs. The lncRNA microarray was used to select differentially expressed lncRNAs in HUVECs treated with or without exosomes. Serum samples of patients with chondrosarcoma were collected to analyze the correlation between the RAMP2-AS1 level and the clinicopathological features. Online databases were used to predict the target microRNA of RAMP2-AS1. Dual luciferase reporter assay, Western blotting and qRT-PCR assays were performed to verify the interactions among RAMP2-AS1, miR-2355-5p and VEGFR2. Rescue experiments were conducted to validate the existence of the RAMP2-AS1/miR-2355-5p/VEGFR2 axis.
Results
The exosomes secreted by chondrosarcoma cells could enhance HUVECs proliferation, migration and tube formation. LncRNA microarray analysis revealed that exosomes carried lncRNA RAMP2-AS1, and further verification showed that the level of RAMP2-AS1 was increased in the serum of chondrosarcoma patients and was closely related to local invasiveness, distant metastasis and poor prognosis. Subsequent experiments demonstrated that RAMP2-AS1 knockdown could partly abrogate the promoting effects on angiogenesis induced by exosomes derived from chondrosarcoma cells. Moreover, dual luciferase reporter assay and rescue experiments suggested that the RAMP2-AS1/miR-2355-5p/VEGFR2 axis was responsible for exosome-induced angiogenesis of HUVECs.
Conclusion
Chondrosarcoma cell-derived exosomes carry lncRNA RAMP2-AS1, which acts as a ceRNA of miR-2355-5p to regulate VEGFR2 expression, thereby positively regulating the angiogenic ability of HUVECs. Thus, exosomal RAMP2-AS1 has the potential as a novel biomarker and therapeutic target for chondrosarcoma.
Keywords: exosome, lncRNA, angiogenesis, chondrosarcoma, HUVEC
Introduction
Chondrosarcoma is a common primary malignant bone tumor, accounting for approximately 20% of the malignant bone tumors. Most of the patients are 30–60 years old, and the incidence in males is higher than that in females.1 Chondrosarcoma often involves the scapula, rib, long tubular bone and pelvis.2 Chondrosarcoma is not sensitive to radiotherapy and chemotherapy and has a high tendency of distant metastasis, leading to a poor prognosis.3 The lack of effective adjuvant therapy highlights the importance of developing new therapeutic strategies.
Angiogenesis is a critical hallmark of cancer progression. Tumor-induced angiogenesis not only provides nutrition for tumor cells but can also trigger the metastatic spread of tumor cells to distant organs.4 Therefore, the regulation of angiogenesis has become an attractive target for antitumor therapy. Tumor angiogenesis is regulated by vascular endothelial growth factor (VEGF), angiopoietin (Ang), transforming growth factor (TGF), tumor necrosis factor (TNF), interleukin and fibroblast growth factor (FGF),5 of which VEGFA and its receptor (vascular endothelial growth factor receptor, VEGFR) are the most essential modulators of angiogenesis.6 VEGFA can induce endothelial cell proliferation, vascular tube formation and increase vascular permeability by binding to VEGFR.7 VEGFR is a transmembrane protein receptor family that includes VEGFR1, VEGFR2 and VEGFR3. VEGFR2 is located in endothelial cells and is the central receptor regulating angiogenesis.8 The high expression level of VEGFR2 on the surface of vascular endothelial cells provides more binding sites for VEGFA, thus enhancing tumor angiogenesis.
Exosomes are vesicles with lipid bilayer molecular structures that are secreted by almost all cells and function by carrying biomolecules including lipids, proteins, DNAs, mRNAs, and non-coding RNAs to participate in local or systemic intercellular communication.9 Recent studies have shown that cancer cell-derived exosomes play a significant role in tumor growth, invasion, angiogenesis, local inflammation, and immune remodeling.10,11 As the main information molecules in exosomes, non-coding RNAs are primary mediators of the functions of exosomes.12 However, the role of chondrosarcoma cell-derived exosomes in the progression of chondrosarcoma remains unelucidated.
Long non-coding RNAs (lncRNAs), which are a subset of non-coding RNAs with more than 200 bases, have been indicated to regulate protein expression through various pathways to modify the physiological functions of cells. Recently, it has been proven that lncRNAs can regulate tumor angiogenesis by modulating the expression of related factors. For example, silencing lncRNA SRRM2-AS exerts suppressive effects on angiogenesis in nasopharyngeal carcinoma by activating the MYLK-mediated cGMP-PKG pathway,13 which prompted us to wonder whether lncRNAs also assume a role in angiogenesis in chondrosarcoma. Moreover, since lncRNA can be carried by exosomes, then whether exosomes secreted by chondrosarcoma cells package a specific lncRNA that plays a crucial part in the regulation of angiogenesis.
In this study, we selected human umbilical vein endothelial cells (HUVECs) as the vascular model of in vitro experiments because HUVECs are relatively easy to obtain and have the most important characteristics and properties of vascular endothelial cells. We demonstrated that exosomes derived from chondrosarcoma cells could promote the proliferation, migration, and tube formation of HUVECs. Through lncRNA microarray analysis, we identified lncRNA RAMP2-AS1 in exosomes as a vital regulator of angiogenesis. Chondrosarcoma-derived exosomes transport RAMP2-AS1 into HUVECs, competitively bind miR-2355-5p, and further promote the expression of VEGFR2, eventually leading to angiogenesis. Additionally, elevated serum RAMP2-AS1 levels were correlated with advanced and metastatic characteristics and poor prognosis of chondrosarcoma patients, suggesting that RAMP2-AS1 in exosomes as a novel therapeutic target of chondrosarcoma.
Materials and Methods
Clinical Samples
Forty-five chondrosarcoma patients who underwent surgical resection at the Department of Tumor Orthopedics of Wuhan Union Hospital and Zhengzhou Orthopaedics Hospital from 2014 to 2015 were selected. Chondrosarcoma tissues and blood samples were collected. The clinical and pathological information was recorded, and all patients were followed up after diagnosis until October 2019 after diagnosis. The stage of chondrosarcoma was according to the Enneking staging criteria. None of the patients received radiotherapy, chemotherapy or other adjuvant treatment after the diagnosis was confirmed. In addition, thirty healthy volunteers participated in this study as the control group. All patients and healthy volunteers signed the informed consent forms, and the Clinical Ethics Committee of Tongji Medical College and the Clinical Ethics Committee of Zhengzhou Orthopaedics Hospital approved the study.
Cell Culture
The human chondrosarcoma cell line SW1353 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). SW1353 cells were cultured in DMEM/F12 medium (Gibco, CA, USA) containing 10% exosome-free serum (System Biosciences, CA, USA) and 1% penicillin-streptomycin (Biosharp, Beijing, China). Primary HUVECs were purchased from Cyagen Biosciences (Suzhou, China) and cultured in the complete culture medium of HUVECs (Cyagen Biosciences, Suzhou, China). The culture conditions were 37°C, 5% CO2 and 99% relative humidity.
Isolation and Identification of Exosomes
Cell culture medium was collected and centrifuged at 2000×g for 10 minutes. The supernatant was collected and filtered with a 0.22 μm filter to remove cell fragments. The obtained filtrate was centrifuged at 12,000×g for 45 minutes to remove large vesicles. Then, the supernatant was collected and filtered by a 0.45 μm filter. The filtrate was ultracentrifuged at 110,000×g for 70 minutes. Removed the supernatant, resuspended in 10 mL precooled PBS and ultracentrifuged at 110,000×g for another 70 minutes. The supernatant was removed and resuspended in 1 mL precooled PBS to obtain the exosome sample. The shape of the exosomes was observed under a transmission electron microscope (HT7800, Hitachi, Tokyo, Japan). The Nanoparticle tracking analysis was performed by Flow NanoAnalyzer (Xiamen, China). The exosome markers CD63, TSG101 and HSP90 were detected by Western blot.
Exosome Labeling
To track the internalization of exosomes by recipient cells, we labeled exosomes with the PKH26 Red Fluorescent Cell Linker Kit (Sigma-Aldrich, MO, USA) according to the manufacturer’s protocol. Briefly, 2μL PKH26 solution was added to 100μg exosomes and incubated for 15 minutes at room temperature. Then the mixture was added to 18mL PBS and centrifuged at 120,000g for 2 hours at 4°C. The supernatant was removed, and the pellet was resuspended in 20mL PBS and centrifuged at 120,000g for another 2 hours at 4°C. The PKH26-labeled exosomes were resuspended in 200μL PBS and then added to the cells. After incubation for 24 h, the cells were washed twice with PBS and fixed in polyformaldehyde for 10 minutes, and internalization of the exosomes was observed by a fluorescence microscope (Olympus, Tokyo, Japan).
Cell Transfection
Small interfering RNA targeting RAMP2-AS1 (si-RAMP2-AS1) and negative control (si-NC), miR-2355-5p mimics, inhibitors and negative controls (miR-NC and inhibitors control) were synthesized by GenePharma (Shanghai, China). VEGFR2 overexpression was achieved by transfecting with the pcDNA3.1-VEGFR2 plasmid (GenePharma). In short, on the day before the cell transfection, cells were seeded in 6-well plates at 1×105 cells per well. On the following day, transfection was performed when the confluence of cells reached approximately 70%. The final concentrations of plasmid and miRNA/siRNA were 2.0μg/mL and 50nM, respectively. Transfections were conducted with Lipofectamine 3000 reagent (Invitrogen, CA, USA) accordance with the manufacturer’s guidelines. Transfection efficiency was determined by qRT-PCR and Western blot assays.
Western Blot
Cells or exosomes were lysed with RIPA lysis buffer containing cocktail protease inhibitor (Beyotime, Shanghai, China) to extract total protein. The protein concentration was determined by the BCA method, and then equal amounts of protein lysates were fractionated using 8% SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked with 5% non-fat milk for 2 hours, and incubated with the following primary antibodies overnight at 4°C: anti-CD63 (1:500, ab134045, Abcam), anti-TSG101 (1:500, ab125011, Abcam), anti-HSP90 (1:500, ab13492, Abcam), anti-VEGFR2 (1:500, ab39256, Abcam), and anti-GAPDH (1:500, ab181602, Abcam). After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 hour to develop blots. Blots were detected and recorded by and ECL device (Bio-Rad Laboratories, CA, USA).
qRT-PCR
Total RNA was extracted by TRIzol reagent and used to synthesize cDNA with the One-Step RT-PCR Kit (ThermoFisher Scientific, Shanghai, China). The cDNA samples were used for qRT-PCR analysis by using the SYBR Primer-Script RT-PCR kit (Takara, Shiga, Japan) with a CFX96 Touch quantitative PCR system (Bio-Rad Laboratories, CA, USA). The amplify conditions were: denaturation at 95°C for 10 minutes, and at 95°C for another 10 seconds, for a total of 40 cycles, and in the end, at 60°C for 30 seconds. Three replicates were performed for each specimen, and GAPDH served as internal references. The ΔΔCt method was utilized for quantification. The sequence information of primers is listed in Table 1.
Table 1.
The Primers Used in This Study
| Primers | Sequence (5ʹ-3ʹ) |
|---|---|
| RAMP2-AS1 Forward | GAACTCAGGCCAGATTTACAAG |
| RAMP2-AS1 Reverse | TTGGGTCCTACAGCAACCAT |
| VEGFR2 Forward | GTGATTGCCATGTTCTTCTGGC |
| VEGFR2 Reverse | TTCATCTGGATCCATGACGA |
| GAPDH Forward | CAAGGCTGAGAACGGGAAG |
| GAPDH Reverse | TGAAGACGCCAGTGGACTC |
| miR-2355-5p Forward | ATTGTCCTTGCTGTTTGGAGAT |
| miR-2355-5p Reverse | GCGAGCACAGAATTAATACGAC |
| U6 Forward | CGCTTCGGCAGCACATATAC |
| U6 Reverse | TTCACGAATTTGCGTGTCAT |
Cell Proliferation Assay
Cell proliferation assay was performed with Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan). Briefly, cells were plated on 96-well plates with 5×103 cells per well. After the indicated times, removed the medium and added 100 μL serum-free medium and 10 μL CCK-8 solution into each well. The absorbance at 450 nm was measured by a microplate reader (ThermoFisher Scientific) after 2 hours incubation at 37°C.
Migration Assay
The migration assay was conducted with the 8-μm pore transwell chamber (BD Falcon, NJ, USA). Approximately 1×104 cells were resuspended in 200 μL serum-free medium and added to the upper chamber, and 500 μL complete medium was placed into the lower chamber. Twenty-four hours later, cells that did not pass through the polycarbonate membrane were wiped off, and chambers were fixed with polyformaldehyde for 15 minutes followed by staining in 0.1% crystal violet solution for several minutes. The migrated cells were observed and counted by an inverted microscope (Olympus, Tokyo, Japan).
Tube Formation Assay
Approximately 2×104 HUVECs were seeded on 96-well plates, which were precoated with 60 μL Matrigel (BD Falcon, NJ, USA). After 24 hours in normal cell culture conditions, tube formation was imaged by an inverted microscope, and tube branch length was quantified by ImageJ v6.0 software.
LncRNA Microarray Analysis
We conducted lncRNAs microarray analysis of HUVECs treated with or without exosomes derived from SW1353 cells by utilizing Arraystar Human LncRNA Microarray V5.0 (Agilent Technology, CA, USA). Initially, we exacted total RNA from HUVECs by using TRIzol reagent, and the purity and quantity of RNA were determined by NanoDrop 2000. Then, the cDNA was synthesized and labeled with biotin. The labeled probe was hybridized with high-density microarray under standard conditions. After hybridization, the fluorescence intensity of microarray was scanned by GenePix-4000B scanner, and the results were converted into digital ones for preservation. Agilent Feature Extraction Software was used to analyze the original data. Significantly differentially expressed lncRNAs with fold change > 2.0 and p < 0.05 were selected.
Luciferase Reporter Assay
The wild type (WT) RAMP2-AS1 or VEGFR2 3′-UTR vectors which contained miR-2355-5p specific binding sites, and vectors which contained mutant (Mut) miR-1184 binding sites were constructed by GenePharma (Shanghai, China). Cells were seeded in 24-well plates, and Lipofectamine 3000 reagent was utilized for cell co-transfection. After 24 hours, the luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, WI, USA).
Statistical Analysis
GraphPad Prism 7.0 software was used for all data analyses in this study. Student’s t-test and ANOVA were used to analyze differences between two groups or more than two groups, respectively. The chi-square test was used to analyze the correlations between the lncRNA RAMP2-AS1 expression level and clinicopathological features. Overall survival curves were analyzed by the Log rank test. Data are represented as the mean ± SD. P-value <0.05 was considered statistically significant.
Results
Exosomes Derived from Chondrosarcoma Cells Promote Angiogenesis of HUVECs
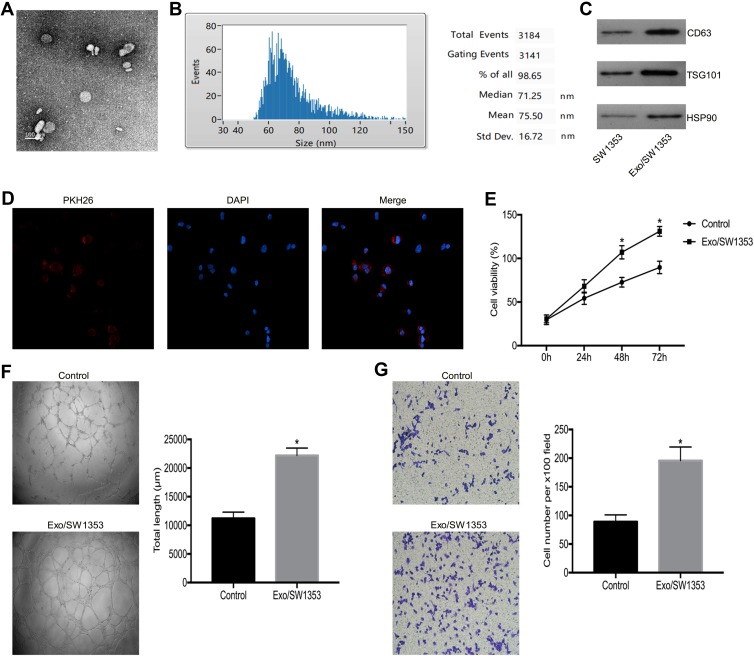
We isolated extracellular vesicles by ultracentrifugation and then characterized them by TEM analysis and NTA analysis. The TEM analysis showed that the vesicles had typical morphological characteristics of exosomes, the diameter of the vesicles was between 30 and 150 nm, and the vesicles exhibited a spherical-shaped appearance (Figure 1A). The NTA analysis demonstrated that the peak size range was 60–100 nm and the mean size was 75.50 nm. The proportion of vesicle in the size range of 30–150 nm is 98.65% (Figure 1B). Western blot analysis showed that the extracellular vesicles expressed the exosomal marker molecules CD63, TSG101 and HSP90 (Figure 1C). Therefore, we concluded that we have successfully isolated SW1353 cell-derived exosomes (Exo/SW1353).
Figure 1.
Exo/SW1353 promotes angiogenesis of HUVECs. (A) Transmission electron microscopy image of Exo/SW1353. (B) Nanoparticle tracking analysis to measure the size distribution of Exo/SW1353. (C) Western blot analysis of the expression of exosomal markers CD63, TSG101 and HSP90. (D) PKH26 labeled Exo/SW1353 was internalized by HUVECs. (E) Cell proliferation was detected in HUVECs treated with or without Exo/SW1353. (F) Tube formation ability of HUVECs treated with or without Exo/SW1353. (G) Transwell migration assay was used to determine the migration capacity of HUVECs treated with or without Exo/SW1353. *P<0.05.
In general, cancer cell-derived exosomes affect the biological functions of recipient cells by releasing functional molecules after they have been internalized by the recipient cells. Initially, we tracked exosomes and found that PKH26 labeled Exo/SW1353 could be better internalized by HUVECs (Figure 1D). Subsequent cell proliferation assay confirmed that Exo/SW1353 could enhance the proliferation activity of HUVECs (Figure 1E). Moreover, transwell migration and tube formation assay indicated that migration and tube formation capacities of HUVECs were improved after incubated with exosomes (Figure 1F and G). These results illustrated that SW1353 cell-derived exosomes could promote angiogenesis of HUVECs.
Exo/SW1353 Transport lncRNA RAMP2-AS1 to HUVECs and Improve the Angiogenic Capacity of HUVECs
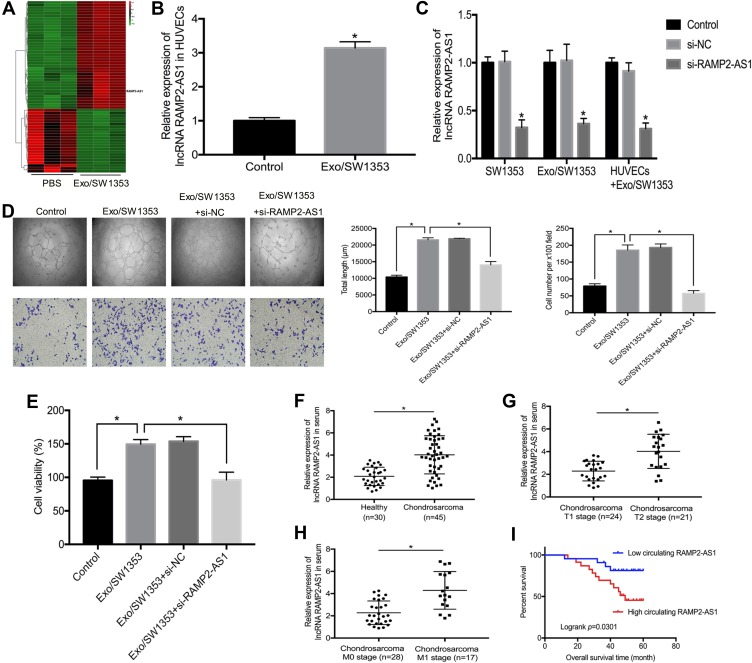
In addition to microRNAs, abundant lncRNAs are also carried by exosomes and transferred into recipient cells to exert regulatory functions. To identify the key lncRNAs that mediate the promotion of angiogenesis by Exo/SW1353, we performed lncRNA microarray analysis on HUVECs with or without Exo/SW1353 incubation and determined that lncRNA RAMP2-AS1 with the most significant expression difference (Figure 2A). Through qRT-PCR verification, we observed that the expression level of RAMP2-AS1 was significantly increased in HUVECs treated with Exo/SW1353 compared with untreated cells (Figure 2B). Additionally, we found that knockdown of RAMP2-AS1 in SW1353 cells efficiently decreased RAMP2-AS1 levels not only in Exo/SW1353 but also in the recipient HUVECs (Figure 2C). Taken together, these data demonstrated that Exo/SW1353 carried lncRNA RAMP2-AS1 and transferred it into HUVECs.
Figure 2.
Exosomal lncRNA RAMP2-AS1 improves angiogenesis of HUVECs and serum RAMP2-AS1 level is associated with clinical characters of chondrosarcoma. (A) The heatmap represented the differential expression of lncRNAs as determined by lncRNA microarray analysis in HUVECs treated with or without Exo/SW1353. Red indicates relatively higher expression, and green indicates relatively lower expression. (B) qRT-PCR analysis of RAMP2-AS1 level in HUVECs treated with or without Exo/SW1353. (C) Suppression efficiency of RAMP2-AS1 knockdown in SW1353 cells, Exo/SW1353 and HUVECs treated with Exo/SW1353. (D) RAMP2-AS1 knockdown abolished the promotion effects of Exo/SW1353 on migration and tube formation ability of HUVECs. (E) RAMP2-AS1 knockdown reversed the proliferative effect of Exo/SW1353 on HUVECs. (F) Relative RAMP2-AS1 levels in serum of chondrosarcoma patients and healthy volunteers. (G) Relative RAMP2-AS1 levels in serum of T1 stage and T2 stage chondrosarcoma patients. (H) Relative RAMP2-AS1 levels in serum of M0 stage and M1 stage chondrosarcoma patients. (I) Patients with high RAMP2-AS1 level had poorer prognosis. *P<0.05.
To further verify that the pro-angiogenic effect of Exo/SW1353 is RAMP2-AS1-dependent, we silenced RAMP2-AS1 expression in Exo/SW1353 treated HUVECs. The results of the cell proliferation assay, migration assay and tube formation assay proved that inhibition of RAMP2-AS1 in HUVECs could impair the pro-angiogenic effect of Exo/SW1353 (Figure 2D and E). Collectively, exosomal RAMP2-AS1 was responsible for the pro-angiogenic effect of Exo/SW1353.
Elevated lncRNA RAMP2-AS1 Is Associated with the Progression and Poor Prognosis of Chondrosarcoma
Angiogenesis is a significant manifestation of tumor growth and metastasis, so the molecules that affect angiogenesis are often closely related to the prognosis of patients. We collected blood samples from forty-five patients with chondrosarcoma and thirty healthy volunteers. The qRT-PCR results showed that the expression level of RAMP2-AS1 was higher in most chondrosarcoma patients than that in healthy volunteers (Figure 2F). In addition, the level of RAMP2-AS1 in patients with T1 stage chondrosarcoma was lower than that in patients with T2 stage chondrosarcoma (Figure 2G), and the level of RAMP2-AS1 in patients with distant metastasis (M1 stage) was significantly higher than that in patients without distant metastasis (M0 stage) (Figure 2H). To further analyze the correlation between the level of RAMP2-AS1 and prognosis, patients were divided into two groups based on the median expression level of RAMP2-AS1. Kaplan-Meier analysis revealed that chondrosarcoma patients with high RAMP2-AS1 levels had poorer overall survival than patients with low RAMP2-AS1 levels (Figure 2I). Additionally, the level of RAMP2-AS1 was independent of patient age, gender, and anatomical location of the tumor (Table 2). In summary, these data indicated that lncRNA RAMP2-AS1 is a promising biomarker for chondrosarcoma.
Table 2.
The Relationship Between RAMP2-AS1 Level and Clinical Characters of Chondrosarcoma
| Variables | Low Circulating RAMP2-AS1 | High Circulating RAMP2-AS1 | p value |
|---|---|---|---|
| (n=22) | n=(23) | ||
| Age (years) | 0.458 | ||
| < 50 | 10 (45.45%) | 13 (56.52%) | |
| ≥50 | 12 (54.55%) | 10 (43.48%) | |
| Gender | 0.609 | ||
| Male | 15 (68.18%) | 14 (60.87%) | |
| Female | 7 (38.82%) | 9 (39.13%) | |
| Anatomical location | 0.463 | ||
| Limb bone | 13 (59.09%) | 16 (69.57%) | |
| Axial bone | 9 (40.91%) | 7 (30.43%) | |
| Ennecking stage | 0.002 | ||
| T1 stage | 17 (77.27%) | 7 (30.43%) | |
| T2 stage | 5 (22.73%) | 16 (69.57%) | |
| Distal metastasis | <0.001 | ||
| Absent | 20 (90.91%) | 8 (34.78%) | |
| Present | 2 (9.09%) | 15 (65.22%) |
LncRNA RAMP2-AS1 Regulates VEGFR2 Expression by Sponging miR-2355-5p in HUVECs
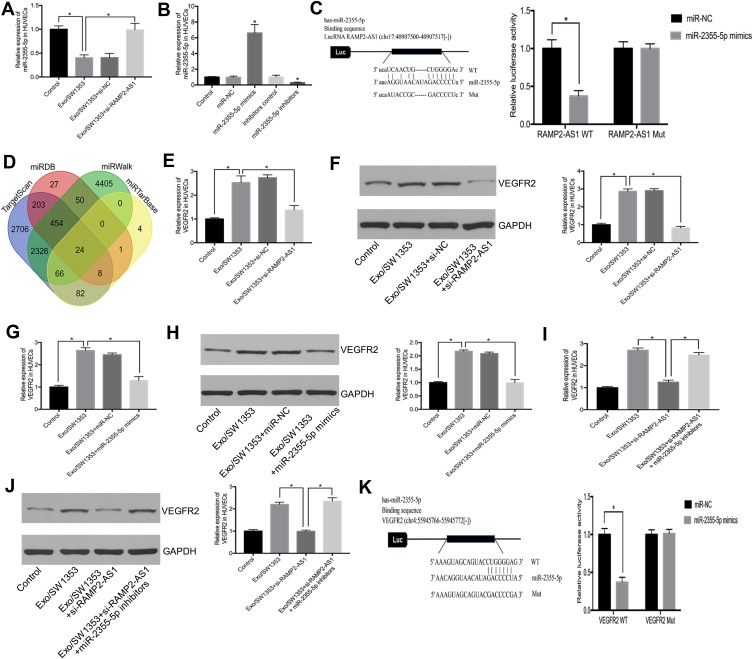
To investigate the potential mechanism of RAMP2-AS1 in angiogenesis, we speculated that RAMP2-AS1 acts as a microRNA sponge to regulate target gene expression. StarBase v3.0 (http://starbase.sysu.edu.cn/) was used to predict microRNAs that may bind to RAMP2-AS1, and we found that RAMP2-AS1 contains a potential binding site for miR-2355-5p. Then, qRT-PCR results showed that miR-2355-5p expression was reduced after HUVECs were treated with Exo/SW1353, while miR-2355-5p expression was restored after silencing RAMP2-AS1 (Figure 3A). To clarify the role of miR-2355-5p, we transfected miR-2355-5p mimics or inhibitors into HUVECs to regulate miR-2355-5p expression (Figure 3B). The luciferase reporter assay showed that HUVECs co-transfected with miR-2355-5p mimics and vector containing the RAMP2-AS1 wild-type sequence had decreased luciferase reporter activity compared with the cells transfected with vector containing the RAMP2-AS1 mutant sequence (Figure 3C).
Figure 3.
Exosomal lncRNA RAMP2-AS1 regulates VEGFR2 expression by sponging miR-2355-5p. (A) The relative expression of miR-2355-5p in HUVECs was measured by qRT-PCR. (B) The transfection efficiency of miR-2355-5p mimics or inhibitors were measured by qRT-PCR. (C) Luciferase reporter assay validated the interaction between RAMP2-AS1 and miR-2355-5p. (D) Venn diagram shows candidate targets that were predicted by four online databases. (E, F) qRT-PCR and Western blot analyzed the relative mRNA level and protein level of VEGFR2 in HUVECs treated with Exo/SW1353 and si-RAMP2-AS1. (G, H) qRT-PCR and Western blot analyzed the relative mRNA level and protein level of VEGFR2 in HUVECs treated with Exo/SW1353 and miR-2355-5p mimics. (I, J) qRT-PCR and Western blot analyzed the relative mRNA level and protein level of VEGFR2 in HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and miR-2355-5p inhibitors. (K) Luciferase reporter assay validated the interaction between VEGFR2 and miR-2355-5p. *P<0.05.
It is well known that microRNA can regulate gene expression by binding to the 3ʹ-UTR of the specific mRNAs. To confirm the targets of miR-2355-5p, we used four online databases TargetScan (http://www.targetscan.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), miRDB (http://mirdb.org/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) to predicted the candidate gene of miR-2355-5p (Figure 3D). Among the 24 overlapping prediction targets, we selected VEGFR2 as the candidate gene. Moreover, the results of qRT-PCR and Western blot showed that the expression of VEGFR2 was repressed after knockdown of RAMP2-AS1 (Figure 3E and F) or overexpression of miR-2355-5p in Exo/SW1353 treated HUVECs (Figure 3G and H). Similarly, knockdown of miR-2355-5p reversed the inhibitory effects of RAMP2-AS1 silencing on the expression of VEGFR2 (Figure 3I and J). The luciferase reporter assay validated the interaction between VEGFR2 and miR-2355-5p (Figure 3K). Taken together, these results confirmed that exosomal RAMP2-AS1 acted as a microRNA sponge by competitively binding miR-2355-5p to regulate the expression of VEGFR2 in HUVECs.
Exosomal lncRNA RAMP2-AS1 Promotes Angiogenesis via Modulation of the miR-2355-5p/VEGFR2 Axis in HUVECs
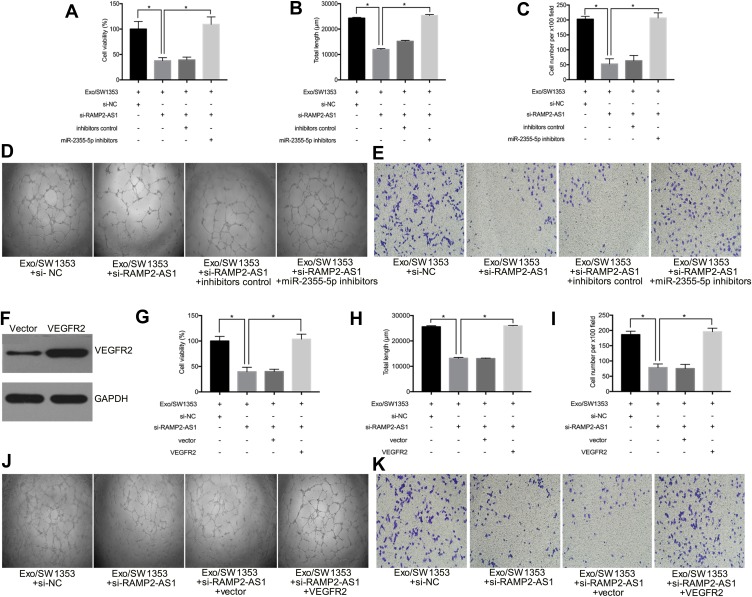
Based on the above findings, we further explored whether RAMP2-AS1 affects angiogenesis by regulating the miR-2355-5p/VEGFR2 axis. The result of cell proliferation assay indicated that silencing miR-2355-5p could reverse the inhibitory effects on cell proliferation caused by RAMP2-AS1 knockdown in Exo/SW1353 treated HUVECs (Figure 4A). The tube formation assay and transwell migration assay showed that miR-2355-5p inhibitors abrogated the inhibition effects on tube formation ability (Figure 4B–D) and migration ability (Figure 4C–E) induced by RAMP2-AS1 knockdown in Exo/SW1353 treated HUVECs. To determine the involvement of VEGFR2 in the pro-angiogenic role of RAMP2-AS1, VEGFR2 was upregulated by transfecting with VEGFR2 overexpression plasmid (Figure 4F). Subsequently, VEGFR2 overexpression reversed the inhibitory effects of RAMP2-AS1 knockdown on cell proliferation, migration and tube formation in Exo/SW1353 treated HUVECs (Figure 4G–K). Taken together, these results suggested that exosomal lncRNA RAMP2-AS1 promotes angiogenesis via the miR-2355-5p/VEGFR2 pathway in HUVECs.
Figure 4.
Exosomal lncRNA RAMP2-AS1 promotes angiogenesis via modulation of miR-2355-5p/VEGFR2 axis. (A) Cell proliferation was detected in HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and miR-2355-5p inhibitors. (B, D) Tube formation assay of HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and miR-2355-5p inhibitors. (C, E) Transwell migration assay of HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and miR-2355-5p inhibitors. (F) The transfection efficiency of VEGFR2 overexpression plasmid was measured by Western blot. (G) Cell proliferation was detected in HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and VEGFR2 overexpression plasmid. (H, J) Tube formation assay of HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and VEGFR2 overexpression plasmid. (I, K) Transwell migration assay of HUVECs treated with Exo/SW1353, si-RAMP2-AS1 and VEGFR2 overexpression plasmid. *P<0.05.
Discussion
The exosomes released by cancer cells serve as a communication system by transporting a large number of functional molecules to neighboring recipient cells and remodeling the microenvironment to aggravate tumor progression. Hu et al confirmed that exosomes derived from colorectal cancer cells promote angiogenesis of HUVECs.14 Mao et al revealed that serum exosomes suppress tumor growth by inhibiting angiogenesis.15 As an important molecule carried by exosomes, lncRNAs also play a considerable role in the process of exosomes-mediated intercellular communication.16 In glioma, exosomes that contain lncRNA CCAT2 enhance angiogenesis of endothelial cells,17 and exosomal MALAT1 promotes angiogenesis in ovarian cancer.18 In this study, we chose HUVEC as a substitute for tumor vascular endothelial cell in vitro because HUVEC is convenient to obtain and has the characteristics of tube formation and migration, so. We isolated and identified exosomes derived from chondrosarcoma cells for the first time and demonstrated that Exo/SW1353 could be internalized by HUVECs and enhanced angiogenic capacity. Furthermore, through lncRNA microarray analysis, we confirmed that Exo/SW1353 carried lncRNA RAMP2-AS1 and transported it into HUVECs. Additionally, through the analysis of clinical data, we found that the level of RAMP2-AS1 was elevated in the serum of patients with chondrosarcoma, and the upregulated level of RAMP2-AS1 was positively correlated with the local invasion, distant metastasis and poor prognosis of chondrosarcoma.
Recent studies have demonstrated that lncRNAs drive angiogenesis via multiple mechanisms. Zhao et al showed that lncRNA PVT1 promotes angiogenesis by activating the STAT3/VEGFA axis in gastric cancer.19 In cervical cancer, lncRNA CCDST enhances the interaction of MDM2 with oncogenic protein DHX9 and facilitates DHX9 degradation, and eventually leading to the inhibition of angiogenesis.20 Additionally, lncRNAs modulate the expression of specific mRNAs through the ceRNA network, which is also a critical manner to regulate tumor angiogenesis. For example, Sun et al found that LINC00968 reduces angiogenesis by impeding miR-423-5p-mediated PROX1 inhibition in breast cancer.21 Through bioinformatics analysis and functional experiments, we revealed that exosomal RAMP2-AS1 functions as a ceRNA for miR-2355-5p and relieves the suppression of VEGFR2 to regulate angiogenesis.
The involvement of angiogenesis in cancer growth and metastasis is an extremely complicated process that is regulated by a series of factors.22 The expression of angiogenic factors, such as VEGFA and VEGFR, is usually increased to promote angiogenesis, thereby changing the balance between angiogenesis and anti-angiogenesis. This process is called the angiogenic switch and is generally triggered by oncogene mutations, stress state and hypoxia.23 VEGFA and VEGFR are the most essential molecules involved in the angiogenic switch. The HIF-1α expression is upregulated in tumor tissues due to hypoxia, and further promotes the expression of VEGFA as a transcription factor. VEGFA can bind to VEGFR1 and VEGFR2 on the endothelial cell surface. Although VEGFR1 has a stronger affinity for VEGFA than VEGFR2, the VEGFR1 activation of downstream signals is too weak, so VEGFR2 is the central receptor that mediates the angiogenesis-promoting effect of VEGFA.24 Recent studies have indicated that VEGFR2 expression is elevated in a variety of cancer tissues and is related to metastasis and poor prognosis. In chondrosarcoma, the expression level of VEGFR2 is associated with malignant phenotypes such as local invasion and distal metastasis.25 Small-scale clinical trials have confirmed that Apatinib, a VEGFR2 targeted inhibitor, has a positive effect on patients with advanced chondrosarcoma, proving that targeted VEGFR2 has excellent potential in the clinical treatment of chondrosarcoma.26 In this study, our data demonstrated that RAMP2-AS1 carried by Exo/SW1353 upregulated the expression of VEGFR2 in HUVECs and eventually promoted angiogenesis. More interestingly, the high level of RAMP2-AS1 was related to the aggressiveness, metastasis and poor prognosis of chondrosarcoma, which also proves that turning on the angiogenic switch is a critical link to promote the malignant progression of chondrosarcoma.
In conclusion, this study demonstrated the role of chondrosarcoma cell-derived exosomal lncRNA RAMP2-AS1 in angiogenesis by mediating the miR-2355-5p/VEGFR2 axis. Thus, exosomal lncRNA RAMP2-AS1 has the potential as a novel biomarker and therapeutic target for chondrosarcoma.
Acknowledgments
This work was supported by grants from the Major Research Plan of the National Natural Science Foundation of China (grant numbers: 91649204 and 81572203) and the Hubei Natural Science Foundation (grant number: 2018CFB118).
Disclosure
All authors declare that they have no competing interests.
References
- 1.Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol. 2018;36(2):188–193. doi: 10.1200/JCO.2017.75.1743 [DOI] [PubMed] [Google Scholar]
- 2.Leddy LR, Holmes RE. Chondrosarcoma of bone. Cancer Treat Res. 2014;162:117–130. [DOI] [PubMed] [Google Scholar]
- 3.Speetjens FM, de Jong Y, Gelderblom H, Bovee JV. Molecular oncogenesis of chondrosarcoma: impact for targeted treatment. Curr Opin Oncol. 2016;28(4):314–322. doi: 10.1097/CCO.0000000000000300 [DOI] [PubMed] [Google Scholar]
- 4.Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–426. doi: 10.1007/s10456-017-9562-9 [DOI] [PubMed] [Google Scholar]
- 5.Ribatti D, Vacca A, Nico B, Sansonno D, Dammacco F. Angiogenesis and anti-angiogenesis in hepatocellular carcinoma. Cancer Treat Rev. 2006;32(6):437–444. doi: 10.1016/j.ctrv.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 6.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–127. doi: 10.1111/joim.12019 [DOI] [PubMed] [Google Scholar]
- 7.Versleijen-Jonkers YM, Vlenterie M, van de Luijtgaarden AC, van der Graaf WT. Anti-angiogenic therapy, a new player in the field of sarcoma treatment. Crit Rev Oncol Hematol. 2014;91(2):172–185. doi: 10.1016/j.critrevonc.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 8.Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem. 2013;153(1):13–19. doi: 10.1093/jb/mvs136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480–6488. doi: 10.1158/0008-5472.CAN-17-0994 [DOI] [PubMed] [Google Scholar]
- 10.Moore C, Kosgodage U, Lange S, Inal JM. The emerging role of exosome and microvesicle- (EMV-) based cancer therapeutics and immunotherapy. Int J Cancer. 2017;141(3):428–436. doi: 10.1002/ijc.30672 [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laffleur B, Basu U, Lim J, Exosome RNA. Non-coding RNA-coupled mechanisms in AID-mediated genomic alterations. J Mol Biol. 2017;429(21):3230–3241. doi: 10.1016/j.jmb.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Lv L, Zhan Z, et al. Silencing of long noncoding RNA SRRM2-AS exerts suppressive effects on angiogenesis in nasopharyngeal carcinoma via activating MYLK-mediated cGMP-PKG signaling pathway. J Cell Physiol. 2019. doi: 10.1002/jcp.29382 [DOI] [PubMed] [Google Scholar]
- 14.Hu HY, Yu CH, Zhang HH, et al. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470–477. doi: 10.1016/j.ijbiomac.2019.03.221 [DOI] [PubMed] [Google Scholar]
- 15.Mao L, Li X, Gong S, et al. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018;25(9–10):248–259. doi: 10.1038/s41417-018-0032-3 [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Li L, Han ZY, Wang ZX, Qin LX. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am J Cancer Res. 2019;9(7):1367–1381. [PMC free article] [PubMed] [Google Scholar]
- 17.Lang HL, Hu GW, Zhang B, et al. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep. 2017;38(2):785–798. doi: 10.3892/or.2017.5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ. Exosomal metastasis-associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int J Biol Sci. 2018;14(14):1960–1973. doi: 10.7150/ijbs.28048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109. doi: 10.1038/s41388-018-0250-z [DOI] [PubMed] [Google Scholar]
- 20.Ding X, Jia X, Wang C, Xu J, Gao SJ, Lu C. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ. 2019;26(9):1750–1765. doi: 10.1038/s41418-018-0242-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X, Huang T, Zhang C, et al. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle. 2019;18(16):1908–1924. doi: 10.1080/15384101.2019.1632641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6 Suppl 16):15–18. doi: 10.1053/sonc.2002.37263 [DOI] [PubMed] [Google Scholar]
- 23.Fagiani E, Christofori G. Angiopoietins in angiogenesis. Cancer Lett. 2013;328(1):18–26. doi: 10.1016/j.canlet.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 24.Peach CJ, Mignone VW, Arruda MA, et al. Molecular pharmacology of VEGF-A isoforms: binding and signalling at VEGFR2. Int J Mol Sci. 2018;19(4):1264. doi: 10.3390/ijms19041264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yukata K, Matsui Y, Goto T, Kubo T, Yasui N. Differential expression of VEGF isoforms and VEGF receptors in cartilaginous tumors. Anticancer Res. 2005;25(2A):955–957. [PubMed] [Google Scholar]
- 26.Xie L, Guo W, Wang Y, Yan T, Ji T, Xu J. Apatinib for advanced sarcoma: results from multiple institutions’ off-label use in China. BMC Cancer. 2018;18(1):396. doi: 10.1186/s12885-018-4303-z [DOI] [PMC free article] [PubMed] [Google Scholar]