Abstract
RNA secondary structure is critical to RNA regulation and function. We report a new N3-kethoxal reagent that allows fast and reversible labeling of single-stranded guanine bases in live cells. This N3-kethoxal-based chemistry allows efficient RNA labeling under mild conditions and transcriptome-wide RNA secondary structure mapping.
Knowledge of RNA folding is critical to understand the function of various RNA species1. Chemical probes have played key roles in transcriptome-wide RNA secondary structure studies2. Increasing number of methods have been developed in recent years for high-throughput RNA structure mapping3–11. Two notable classes of chemical probes, DMS and SHAPE, enable transcriptome-wide in vivo RNA structurome mapping12. Both methods are effective, but with significant space for improvement. DMS is toxic at high concentration and mostly methylates the Hoogsteen face of bases; SHAPE molecules are hydrolytically unstable and label the 2’-OH of sugar instead of the bases13,14. A more specific, non-toxic reagent that rapidly labels the Watson-Crick interface under mild conditions will offer additional advantage for in vivo RNA labeling and RNA secondary structure probing.
EDC (ethyl-3-(3-dimethylaminopropyl)carbodiimide), NAz (Nicotinoyl azide), glyoxal and its derivatives, were recently developed to expand the toolbox of probing RNA secondary structures in a low-throughput manner15–18. Kethoxal (1,1-dihydroxy-3-ethoxy-2-butanone) is known to react with guanines in single-stranded RNA (ssRNA) under mild conditions, which induces reverse transcription (RT) stop19. It could also react with inosine to form an unstable hemiacetal adduct20. However, lack of synthetic routes to modified kethoxal hampered its use for transcriptome-wide study. Here, with a new synthetic design (Supplementary Note 1), we report the preparation of azido-kethoxal (N3-kethoxal, 1) for the specific labeling of the N1 and N2 positions at the Watson-Crick interface of guanines in ssRNA (Figure 1a). The azido group offers a bioorthogonal handle that can be modified with a biotin or dyes for enrichment or other applications10. In addition, the reversibility of the kethoxal-guanine reaction under alkaline or heating conditions19 provides an additional advantage in the RT-stop-based RNA structure mapping via producing read-through controls after removing the kethoxal labels.
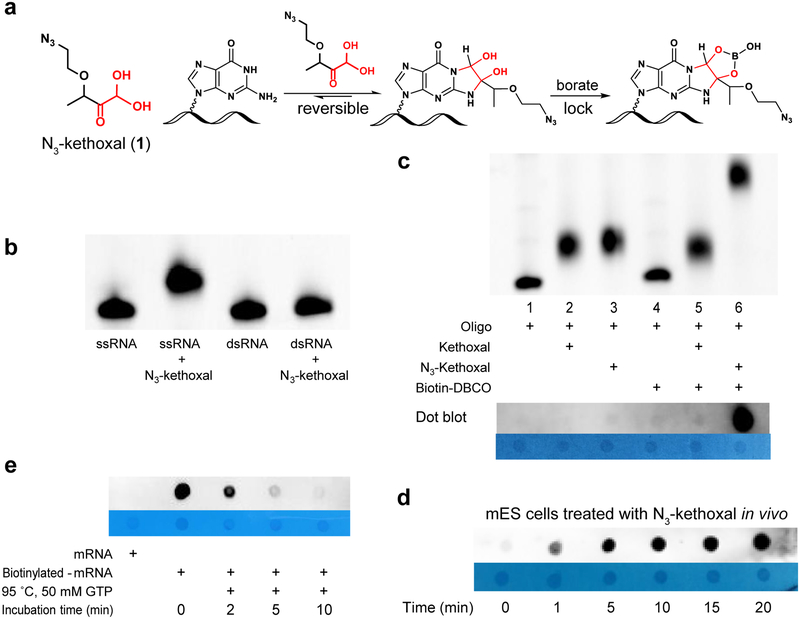
Figure 1 |. N3-kethoxal and experimental evaluation of its selectivity, cell permeability and reversibility.
(a) The structure of N3-kethoxal and the reaction with guanine. (b) Denaturing gel electrophoresis demonstrating N3-kethoxal only react with single-strand RNA (ssRNA). (c) Upper: Denaturing gel electrophoresis analysis of the labelling reaction of kethoxal and N3-kethoxal with FAM-RNA oligo (5’-FAM-GAGCAGCUUUAGUUUAGAUCGAGUGUA, lane 1–3) and biotinylation with biotin-DBCO (lane 5, 6). Only N3-kethoxal labelled RNA can be biotinylated (lane 6). Bottom: Dot blot of RNA after labelling and Biotinylation reactions. Methylene blue dot results are listed as control. (d) Dot blot of isolated total RNA from mES cells which were treated by N3-kethoxal with different periods, 1, 5, 10, 15, 20 min. (e) Dot blot analysis of reversibility of N3-kethoxal labelled mRNA in present of 50 mM GTP at 95 °C. The N3-kethoxal modification in mRNA was removed thoroughly after 10 min incubation. Experiments were independently repeated twice with similar results obtained. Uncropped scans for b, c, d, and e are provided in Supplementary Figure 15.
N3-kethoxal only reacts with guanine in ssRNA and is inert with other nucleic bases (Supplementary Figure 1, Figure 1b). Among chemically modified guanines, N3-kethoxal does not react with m1G and m2G but can label m7G, verifying that N3-kethoxal specifically modifies the N1 and N2 positions of guanine (Supplementary Figure 1). In synthetic RNA oligos, all guanines in the guanine-containing oligos were labeled by N3-kethoxal, while the oligo without guanine showed no reaction (Supplementary Figure 2). N3-kethoxal exhibits higher RNA labeling activity compared with other reported RNA secondary structure probes, including DMS, NAI, glyoxal and EDC (Supplementary Figure 3). As shown by gel electrophoresis, dot blot, and mass spectrum analysis (Figure 1c, Supplementary Figure 4), N3-kethoxal-modified RNAs can be successfully biotinylated, which can then be enriched by streptavidin-conjugated beads, in order to increase the signal-to-noise ratio in biological applications.
We evaluated cell-based labeling efficiency by adding N3-kethoxal into the culture medium of mouse embryonic stem cells (mESC) directly. Dot blotting of biotinylated RNA indicated that N3-kethoxal could permeate into living cells efficiently in one minute, with the signal saturated in five minutes, suggesting a quick cell penetration and high labeling efficiency of N3-kethoxal (Figure 1d). The fast labeling is also confirmed by high-throughput sequencing results with G-stop ratio increased from 1 min and reached the maximum after 2.5 min (Supplementary Figure 5). The rapid labeling property enables N3-kethoxal to be used in transient events such as stress response, signaling, etc.
The kethoxal-guanine adduct is unstable under alkaline conditions19. By adding excessive guanine monomers to trap dissociated N3-kethoxal, the labeling can be removed to yield unmodified RNAs in a neutral buffer within a shorter period of time (Supplementary Figure 6a). The excessive GTP almost completely remove the N3-kethoxal modification on the labeled RNA within 8 hours at 37 °C (Supplementary Figure 6b) or within 10 min at 95 °C (Figure 1e, Supplementary Figure 6c). The labeling adduct of kethoxal-guanine could be stabilized in borate buffer as previously reported19, providing flexibility to manipulate the N3-kethoxal adduct on RNA.
We next combined N3-kethoxal probing with deep sequencing (Keth-seq) to probe RNA secondary structures in mESC. In each experiment, we constructed three different RNA libraries, including an N3-kethoxal-modified RNA, a no-treatment control sample, and an N3-kethoxal-removal sample made by erasing the N3-kethoxal labeling before the reverse transcription (Supplementary Figure 7). We observed a high correlation at both RPKM (Supplementary Figure 8a) and RT-stop level between Keth-seq replicates (Figure 2a), indicating that Keth-seq is highly reproducible. Additionally, in the N3-kethoxal sample, guanine (>80%) dominates the RT-stopped sites among all reads, with no RT stop bias across all four bases in the no-treatment control sample (Supplementary Figure 8b), confirming that N3-kethoxal is highly selective to guanine. RT-stopping sites in N3-kethoxal-removal samples decreased dramatically to a similar level to the no-treatment control, indicating that N3-kethoxal modification was almost completely removed during the reversal process (Supplementary Figure 8b). In mRNA mt-Atp8, for instance, we observed more full-length RNA fragments in the N3-kethoxal-removal sample than that in the no-treatment control sample, suggesting that the RT stopped sites could be more confidently identified using the N3-kethoxal-removal sample as the ‘background’ (Supplementary Figure 9).
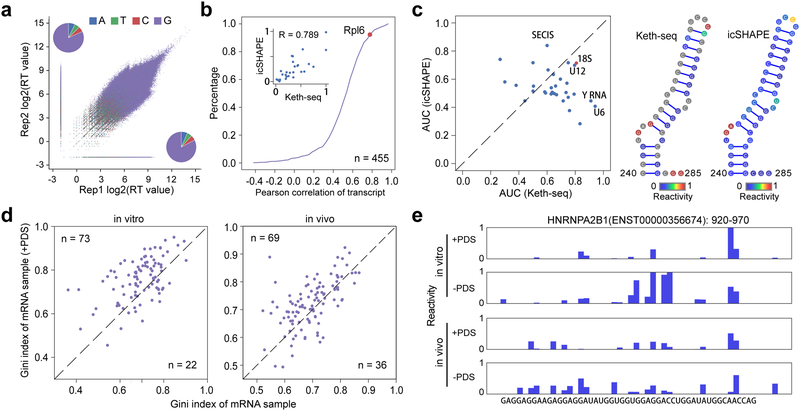
Figure 2 |. Keth-seq method and the profile around rG4 regions.
(a) Scatter plot of reverse transcription (RT) stop reads distribution between replicates for N3-kethoxal sample. The inset pie plots show RT stopped base distribution for replicate 2 (upper left, A: 604,222; T: 497,602; C: 481,596; G: 7,204,998) and replicate 1 (bottom right, A: 703,486; T: 586,297; C: 551,962; G: 8,683,824). (b) Accumulation plot of correlation coefficient between Keth-seq and icSHAPE for all transcript. For each common transcript, we calculate the Pearson correlation coefficient for structural signal of guanine bases. The inset plot shows all guanine reactivity between Keth-seq and icSHAPE for Rpl6 (a gene encoding ribosomal protein) transcript with a high correlation (Pearson correlation coefficient R: 0.789). (c) Left: scatter plot of AUC between Keth-seq and icSHAPE for RNAs with known structure model (18S ribosomal RNA from RNA STRAND database and others from Rfam database, 32 RNAs in total). Right: A fragment (240–285) of 18S ribosomal RNA with both Keth-seq and icSHAPE reactivity filled in the structure model. (d) Gini index of known rG4 regions (based on previously identified by Kwok et.al., 2016, Nature method) between +PDS treatment and native sample for in vitro (left) and in vivo (right). Only regions with structural information in both + PDS treatment and native conditions are retained for plotting (extended to 50-nucleotide long). (e) An example of Keth-seq profile around previously identified in vitro rG4 regions.
To validate Keth-seq, we analyzed guanine signals from Keth-seq and compared with icSHAPE both globally and at the transcript level10. For every common transcript (n = 455), we calculated a correlation coefficient between Keth-seq and icSHAPE by using their reactivity profile on all guanines, and plotted the whole distribution as an accumulative curve (Figure 2b). About 80% of the transcripts show a positive correlation (Pearson correlation coefficient >= 0.4, Figure 2b), indicating that Keth-seq agrees well with the established icSHAPE technology. To directly evaluate the accuracy of Keth-seq in determining RNA secondary structure, we compared the reactivity profile of Keth-seq with icSHAPE on all guanines in the mouse 18S ribosomal RNA with the known structure model from the RNA STRAND database (id: CRW_00356)21. Keth-seq reactivity profile achieves a higher AUC than icSHAPE in fitting the 18S ribosomal RNA model (Keth-seq = 0.81, icSHAPE = 0.71) (Figure 2c). More specifically, Keth-seq shows a higher reactivity score than icSHAPE for single-stranded G nucleotides and thus more accurately revealing unpaired Gs (Supplementary Figure 10a), though both methods similarly agree well with the 18S model on its double-stranded areas (Figure 2c).
We then extended the comparison by using all available mouse RNA secondary structure models from Rfam database, and found Keth-seq achieves a higher AUC than icSHAPE for most of these RNAs (Figure 2c). In addition, we compared Keth-seq and DMS-seq6 by evaluating their performance on human 18S RNA (id: CRW_00347) and showed that Keth-seq achieve a comparable accuracy as DMS-seq with similar AUCs obtained (Supplementary Figure 10b). Furthermore, we applied Keth-seq to probe RNA structure both in vivo and in vitro for mESCs and used Gini index to measure the structural evenness of RNAs6. Consistent with previous findings, we observed that RNAs in vitro showed higher Gini index than that in vivo (Supplementary Figure 10c–d), validating the folding complexity of cellular RNAs and the feasibility of Keth-seq for in vivo detection.
Formation of RNA G-quadruplexes (rG4) from isolated RNAs has been shown in different studies. However, the in vivo detection of rG4 remains challenging22,23. As N3-kethoxal is highly specific towards labeling N1 and N2 positions of guanine and can be enriched, Keth-seq can be sensitive on probing the potential presence of the rG4 structure in live cells. After demonstrating that N3-kethoxal can detect rG4 in vitro (Supplementary Figure 11a–c), we conducted Keth-seq using isolated HeLa RNA or in live HeLa cell in the presence or absence of PDS, which has been shown to induce rG4 formation inside cells24. We first explored the structure landscape of previously identified rG4 regions by rG4-seq under PDS treatment in vitro22, and detected 95 regions with structure information detected by Keth-seq under both native and PDS treatment conditions (Supplementary Figure 12a). In the PDS treated samples, these regions show a higher Gini index than the control sample, suggesting the formation of rG4 under PDS treatment (Figure 2d). Consistent with previous observations22, these rG4 regions preferentially occur at UTR regions (Supplementary Figure 12b) and are associated with biological pathways (Supplementary Figure 12c) including translation, transcription and metabolism, suggesting potential regulatory roles of rG4s.
To further explore whether rG4 can fold in live cells, we performed similar analysis using in vivo Keth-seq data and detected 105 previously identified rG4 regions under both native and PDS treatment conditions (Supplementary Figure 12d). 69 of these 105 regions showed higher Gini index under PDS treatment compared with the control, indicating that rG4 structure could potentially form at these regions in live cells. The genomic context distribution and top enriched biological pathways of these regions are both similar to that in vitro (Supplementary Figure 12e–f). We included examples where the signal in the defined rG4 regions in the PDS sample is lower than that in the control sample (Figure 2e, Supplementary Figure 13), indicating that PDS treatment induces rG4 formation both in vitro and in live cells.
We noted that only a small subset of rG4s possibilities from the rG4 dataset22 are detected by Keth-seq. It could be due to insufficient sequencing depth or possible that chemical labeling only detects kinetically stable structures and may miss highly dynamic rG4s25. Though rG4s detected in vitro may not fold in vivo23, our study does suggest that a portion of rG4s could exist in situ.
In summary, we showed that N3-kethoxal readily labels RNA and established Keth-seq as an effective method for transcriptome-wide RNA secondary structure mapping in live cells. Because of the high selectivity and reactivity of N3-kethoxal labeling of guanines in single-stranded RNA, Keth-seq is able to map secondary structures such as rG4 under mild conditions. The efficient live cell RNA labeling by N3-kethoxal provides an approach that could be expanded for RNA enrichment, RNA targeting and RNA proximity studies in the future.
ONLINE METHODS
Synthesis of N3-kethoxal.
The synthesis of N3-kethoxal and the characterization of compounds (1H NMR, 13C NMR and HRMS) are included in Supplementary Note 1.
General chemical and biological materials.
All chemical reagents for N3-kethoxal synthesis were purchased from commercial sources. RNA oligos were purchased from Integrated DNA Technologies, Inc. (IDT) and Takara Biomedical Technology Co., Ltd. Buffer salts and chemical reagents for N3-kethoxal synthesis were purchased from commercial sources. Superscript III, Dynabeads® MyOne™ Streptavidin C1 was purchased from Life technologies. T4 PNK, T4 RNL2tr K227Q, 5’-Deadenylase, RecJf were purchased from New England Biolabs. CircLigaseII was purchase from Epicentre® (an Illumina company). DBCO-Biotin was purchase from Click Chemistry Tools LLC (A116–10). All RNase-free solutions were prepared from DEPC-treated MilliQ-water.
The reaction of N3-kethoxal and RNA oligo.
The reaction was generally performed with following protocol: 100 pmol RNA oligo and 1 μmol N3-kethoxal was incubated in total 10 μL solution in kethoxal reaction buffer (0.1 M sodium cacodylate, 10 mM MgCl2, pH 7.0) at 37 °C for 10 min. To induce rG4 folding in vitro, RNA were denatured at 95 °C for 5 min then cooled to 4 °C for 5 min, before 1 M KCl (2 μL), 0.1 M sodium cacodylate buffer (pH 7.0) and PDS (final concentration of 5 μM) were added. The mixture was incubated at 37 °C for 10 min to achieve equilibration. N3-kethoxal was then added to the reaction mixture to react with folded RNA. The final reaction volume was 10 μL. The modified RNA was purified by Micro Bio-Spin™ P-6 Gel Columns (Biorad, 7326222). The purified labeled RNA can be used for further used for mass spectrometry, gel electrophoresis, primer extension assay and copper-free click reaction with biotin-DBCO.
Remove N3-kethoxal modification from N3-kethoxal labelled RNA.
The detailed protocol of N3-kethoxal modification erasing was listed in the step 5 “N3-kethoxal-remove sample preparation” of Keth-seq protocol in supporting information. Generally, the purified N3-kethoxal modified RNA was incubated with high concentration of GTP (1/2 volume of the reaction solution, final concentration is 50 mM) at 37 °C for 6 hours or at 95 °C for 10 min. Higher temperature is benefit to remove the N3-kethoxal modification.
Fixation of N3-kethoxal modification in RNA.
The N3-kethoxal modification in RNA can be fixed in the presence of borate buffer. The solution of N3-kethoxal labelled RNA was mixed with 1/10 volume of stock borate buffer (final concentration: 50 mM; stock borate buffer: 500 mM potassium borate, pH 7.0, pH was monitored while adding potassium hydroxide pellets to 500 mM boric acid). The borate buffer fixation was used in step 2, 4, 6 of Keth-seq protocol in supporting information.
MALDI-TOF-MS analysis of N3-kethoxal labelled RNA oligo.
The N3-kethoxal labelled RNA was purified by Micro Bio-Spin™ P-6 Gel Columns. Meanwhile the buffer exchange was occurred from kethoxal reaction buffer to Tris buffer that can be directly used in MALDI-TOF-MS experiment without extra desalt step. One microliter of product solution was mixed with one microliter matrix which include 8:1 volume ratio of 2’4’6’-trihydroxyacetophenone (THAP, 10 mg/mL in 50% CH3CN/H2O) : ammonium citrate (50 mg/mL in H2O). Then the mixture was spotted on the MALDI sample plate, dried and analyzed by Bruker Ultraflextreme MALDI-TOF-TOF Mass Spectrometers.
The selectivity of N3-kethoxal to ssRNA by gel electrophoresis.
The complementary RNA oligos FS1 (Fluorescent RNA oligo) and S2 were hybridized to double-strand RNA (dsRNA) with the ratio of FS1 : S2 = 1.2 : 1 to ensure all FS1 was involved in the formation of dsRNA. After the reaction with N3-kethoxal, the purified product by Micro Bio-Spin™ P-6 Gel Columns was analyzed by denaturing gel electrophoresis (Novex™ TBE-Urea Gels, 15%, Invitrogen, EC6885BOX). Gel Imaging was collected in Pharos FX Molecular imager (Bio-Rad, USA).
RNA sequence:
FS1: 5’-FAM-GAGCAGCUUUAGUUUAGAUCGAGUGUA,
S2: UACACUCGAUCUAAACUAAAGCUGCUC
HPLC condition.
The product of N3-kethoxal with for RNA nucleic bases was analyzed using LC-6AD (Shimadzu, Japan) HPLC instrument, which equipped with an Inertsil ODS-SP column (5 μm, 250×4.6 mm) (GL Science lnc. Japan). The phase A (100 mM TEAA buffer, pH = 7.0) and phase B (CH3CN) were used as eluents with a flow rate of 1 mL/min at 35 °C (B conc.: 5-5-30% / 0-5-30 min).
The biotinylation of N3-kethoxal labelled RNA and dot blot assay.
In vitro study: The purified N3-kethoxal RNA was incubated with DBCO-Biotin at 37 °C for 2 hours in present of RNase inhibitor, borate buffer (step 2 biotinylation of Keth-seq protocol in supporting information). For RNA oligo analysis, the biotinylated product was purified by Micro Bio-Spin™ P-6 Gel Columns and subject to dot blot assay and MALDI-TOF-MS detection; for total RNA or mRNA, the product was purified by RNA clean & concentrator 5 (zymo research, R1015) and subject to further experiments.
In vivo study: 10 μL N3-kethoxal was added into the cell culture medium in 100 mm cell culture dish with nearly 80 % confluent mES cells. After incubation at 37 °C in CO2 incubator for a specific time, the medium was aspirated and the cells were washed three times by PBS. The total RNA was isolated by Trizol™ reagent (Invitrogen, 15596026) or Qiagen RNeasy plus mini kit (Qiagen, 74134). mRNA was isolated by Dynabeads™ mRNA DIRECT™ Purification Kit (Invitrogen, 61011). The biotinylation step was same as in vitro study. The biotinylated RNA was purified by RNA clean & concentrator 5.
Dot blot assay: one microliter RNA (100 ng/μL) sample was spotted onto the Amersham Hybond-N+ membrane (RPN119B, GE Healthcare) and UV crosslinked to the membrane by UVP HL-2000 hybriLinker. The membrane was washed using 1 × PBST (0.1% tween-20) and blocked with 5% nonfat dry milk in 1 × PBST overnight at 4 °C. After four times wash using 1 × PBST with ten-minute interval, the streptavidin-horseradish peroxidase (1:15000 dilutions, streptavidin-HRP, Life Technologies, S-911) in 1 × PBST with 3% BSA was added and incubated at room temperature for 40 min. Then then membrane was washed using 1 × PBST with ten-minute interval again and developed by SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, 34577). The membrane was washed by 1 × PBST again and stained by methylene blue solution (0.02% methylene blue in 0.3M sodium Acetate pH 5.2).
Primer extension assay.
RNA templates (N3-kethoxal treated or not) were dissolved in 13.5 μL nuclease-free water. 1 μL of 10 μM FAM-labeled DNA primer, 2 μL reverse transcription buffer, 1 μL 0.1 M DTT, 1 μL 5 mM dNTPs and 1.5 μL RevertAid Reverse Transcriptase (200U/μL) were then added (total volume 20 μL). The G ladder was made by dideoxy sequencing method, with 2 μL of 10 mM corresponding ddNTP added to replace 2 μL of nuclease-free water. The reverse transcription was performed at 37 °C for 30 min, and then 20 μL of deionized formamide was added. The reaction mixture was immediately heated up to 95 °C for 10 min, then cooled down to 4 °C. The cDNAs were size fractionated by 20% denaturing polyacrylamide gel containing 8 M urea. The gel was scanned with Pharos FX Molecular imager (BioRad) operated in the fluorescence mode (λex = 488 nm).
Keth-seq library preparation.
The library was prepared following a previously reported procedure with slight changes10. The detailed protocol was included in Supplementary Note 2. For in vitro library preparation, RNA was isolated and refolded in RNA folding mix buffer (100 mM HEPES, pH 8.0, 100 mM NaCl, 10 mM MgCl2). The refolded RNA was treated with N3-kethoxal and then used for library construction. For in vivo study, N3-kethoxal was added into the culture medium of mES cell or HeLa cell and the RNA was isolated to be used for library construction.
Isolated N3-kethoxal labeled RNA was biotinylated with water-soluble DBCO-biotin (Click Chemistry tool, A116) by copper-free click reaction, then fragmented by sonication. The RNA Fragmentation Reagent is not suitable for the fragmentation step of Keth-seq because high temperature will affect N3-kethoxal labeling in RNA. In addition, the borate buffer is also necessary in each step before cDNA production except the kethoxal-remove experiment. Fragmented RNA was subjected to end repair by T4 PNK, 3’-adaptor ligation (3’-adaptor: /5rApp/TGGAATTCTCGGGTGCCAAGG/3ddC/) followed by 3’-adaptor removing by 5’-Deadenylase and RecJf digestion. The ligation products were separated to two fractions, with 90% used to produce the N3-kethoxal library and the rest 10% used for N3-kethoxal-remove sample library.
For the N3-kethoxal sample, RT primer, dNTPs, SuperScript III, borate solution, and RNase inhibitor were mixed with RNA to perform reverse transcription (RT primer: /5Phos/DDDNNAACCNNNNGATCGTCGGACTGTAGAACTCTGAACAT/iSp18/GGATCC/iSp18/TACCTTGGCACCC). After cDNA synthesis, the cDNA-RNA was kept cool to avoid denature and was immediately subjected to immunoprecipitation by Dynabeads® MyOne™ Streptavidin C1. Beads were washed and the truncated cDNA was eluted by RNase A/T1 and RNase H digestion. For the N3-kethoxal-remove sample, RNA was incubated with GTP in nuclease-free water at 95 °C for 10 min to remove N3-kethoxal modification. The RNA was then purified and reverse transcription was performed similarly as the N3-kethoxal sample.
cDNA from N3-kethoxal sample and N3-kethoxal-remove sample were subjected to size selection by gel electrophoresis separation, which can remove the excess RT primers and the self-ligation product of primers. The purified cDNA was used for cDNA cyclization by CircLigaseII to obtain the circDNA. The circDNA were then amplified by PCR with short primers (F: 5’-TGGCACCCGAGAATTCCA; R: 5’-TTCAGAGTTCTACAGTCCGA). In this step, qPCR was performed to evaluate the cycle numbers of each samples to avoid over-amplification. After purification and size selection, a final library PCR amplification is performed using the full sequencing primers from TruSeq® Small RNA Sample Prep Kits of (Illumina). The products were purified by low melting point agarose gel and used for deep sequencing.
For the no-treatment control sample, RNA was isolated from cells without any N3-kethoxal treatment, followed by fragmentation, adaptor ligation, reverse transcription, cDNA cyclization and PCR amplification as described above to construct the library for deep sequencing.
Sequencing data processing.
As the library structure is similar to that of icSHAPE, we used the same strategy to process the sequencing reads by using the icSHAPE scripts at (https://github.com/qczhang/icSHAPE)10. Firstly, readCollapse.pl was used to collapse the reads with default parameter. Note that we include a barcode of random hexamer (NNNNNN) ligated to the fragments during library construction (Supplementary Figure 14). These random barcodes serve to identify PCR duplicates from real different fragments with the identical sequences. Reads with fully identical sequence (including the barcode and the insert fragment) were marked as PCR duplicates and filtered before subsequent analysis. But reads with different barcodes were retained, even they contain the identical insert fragments and subsequently mapped to the same start and end positions, as they actually represent different fragments in the library.
Then we used trimming.pl to cut potential adapter sequences (-l 13 -t 0 -c phred33 -a adapter.fa -m 0, adapter sequence: ATGGAATTCTCGGGTGCCAAGGAACTCCAGTCACATCACGATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAA). Next, we mapped the clean reads to ribosomal RNAs, and the unmapped reads were mapped to the transcriptome (Gencode, mm10 for mouse and hg38 for human) using Bowtie with default parameters. We calculated reverse transcription (RT) stop signals using the script calcRT.pl. After evaluating correlation between different replicates (correlationRT.pl), we combined RT signals of replicates (combineRTreplicates.pl) for subsequent analysis, and then normalized them for both the kethoxal and the control samples respectively (normalizeRTfile.pl -m mean:vigintile2 -d 32 -l 32). A structure reactivity score for each nucleotide in each transcript was calculated by comparing the kethoxal sample (foreground) versus the control sample (background) using the script calcEnrich.pl (-w factor5:scaling1 -x 0.25). The calculation is based on following formula: A*(RT[kethoxal] – B*RT[control])/BD[control], where RT means the RT stop count of the nucleotide in the sample, BD means the base density of the nucleotide, A and B are scaling factors to control the effects of subtraction. Our previous work on icSHAPE technology development trained the two scaling factors on mouse 5S ribosomal RNA, the structure of which has been determined by both high-throughput sequencing and low-throughput RT-stop gel analysis10,26, by maximizing the correlation between calculated reactivity scores and gel-based results. We found that although the scaling factors performs the best around A = 10 and B = 0.25, they are relatively insensitive. We thus used the same parameters in Keth-Seq and we did observe high accuracy on known structures. Finally, to obtain high-quality scores, we only kept nucleotides with adequate sequencing coverage: filterEnrich.pl -T 2 -t 200 -s 5 -e 30. Here “-T 2” requires the minimal average number of RT stops over the whole transcript to be no less than 2; “-t 200” requires the base density of a nucleotide with reactivity to be no less than 200; and “-s 5 -e 30” is to trim away the first 5 and the last 30 nucleotides as they tend to have low sequencing quality scores.
For rG4 probing, we first converted the genomic coordinated of previously reported rG4 regions in HeLa cells22 to transcriptome coordinates27. The converted regions with >=60% NULL value of structure scores from our Keth-seq experiments were filtered from subsequent analysis. The retained regions were used for comparison between +PDS and -PDS Keth-seq samples.
Comparison between Keth-seq and icSHAPE/DMS-seq.
To compare the performance of Keth-seq and icSHAPE, we collected known RNA secondary structure models from different sources, including the mouse 18S ribosomal RNA structure from the RNA STRAND database21 (CRW_00356) and the other 614 RNA structure models from the Rfam database28. Both Keth-seq and icSHAPE10 sequencing reads were remapped to these specific RNAs and the reactivity score profiles were calculated (18S rRNA and 31 other RNAs with structure information are common in the two experiments and retained). We calculated receiver operator characteristic (ROC) curves to measure to what degree the structural probing reactivity scores fits the reference structure model. Using different reactivity score cutoffs, each nucleotide can be predicted (classified) as single-stranded or double-stranded. A true positive is defined as a single-stranded base with a reactivity score higher than the cutoff. A true negative is defined as a paired base with a reactivity score lower than the cutoff. AUC is calculated using the signals of guanine nucleotides for Keth-seq while considering the signals of all four bases for icSHAPE. To compare Keth-seq with DMS-seq, we collected DMS-seq data6 for Fibroblast and K562 sample and evaluate the performance on human 18S ribosomal RNA (RNA STRAND id: CRW_00347). AUC is calculated using the signals of the adenine and cytosine nucleotides for DMS-seq. For 18S rRNA, we firstly parse the known 3D ribosome structure (PDB ID: 4V6X) and derive an accessibility score for each nucleotide. Only nucleotides with accessibility score great than 3 are retained for evaluation.
Calculating Gini index for regions.
We followed a previously reported method to calculate the Gini index to measure the level of RNA structure formation6. In principle, a Gini index represents data evenness and a completely unfolded and a completely base-paired RNA would both have a low Gini index. However, in reality, a RNA usually has a comparable number of single-stranded and double-stranded nucleotides, and within a normal range, the Gini index of a RNA well correlates with its structure formation. To explore the correlation between RNA structureness and Gini index, we collected all mouse RNAs from the Rfam database (n = 614), the secondary structures of which have been determined. We performed a simulation analysis, where a random reactivity score is assigned to each nucleotide in the RNA, with a score in 0.5–1.0 for single stranded nucleotide, and a score in 0.0–0.5 for double-stranded nucleotide. Then a Gini index is calculated from the simulated reactivity profile. The random simulation was repeated 100 times. Clearly, we observed that, the higher the Gini index the more structured (higher double stranded nucleotides ratio) the RNA is (Supplementary Figure 8c). Also note that another measurement of RNA structure level is the average of reactivity scores, and we have repeated and confirmed all conclusions in the main text concerning Gini index with reactivity score average.
Assume the reactivity profile of a region is: , where xn is the reactivity score for base n. We calculate a Gini index value as follows: 1) Sort the reactivity value of the region in ascending order and take the summation and accumulation 2) Calculate the accumulating area and fair area ; 3) Calculate the Gini index value .
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21572172, 21778040, 21822704 to X.W.; 21432008, 91753201, and 21721005 to X.Z.; 31671355, 91740204, and 31761163007 to Q.C.Z.) and National Institutes of Health HG008935 (C.H.). C. H. is an investigator of the Howard Hughes Medical Institute. X. W. is supported by China Scholarship Council (CSC) during his visit to the University of Chicago. We acknowledged. S. Frank, MA, who edited the manuscript.
Footnotes
Data availability
All genomic data sets have been deposited in the Gene Expression Omnibus under accession number GSE 122096. Other data and materials are available from the authors upon reasonable request.
Code availability
All custom codes used in this study are available at https://github.com/Tsinghua-gongjing/Keth-seq.
Competing financial interests:
C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc., and a shareholder of Epican Genetech.
References
- 1.Wan Y, Kertesz M, Spitale RC, Segal E & Chang HY Nat. Rev. Genet 12, 641–655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubota M, Tran C & Spitale RC Nat. Chem. Biol 11, 933–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kertesz M et al. Nature 467, 103–107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Underwood JG et al. Nat. Methods 7, 995–1001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucks JB et al. P. Natl. Acad. Sci. USA 108, 11063–11068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouskin S, Zubradt M, Washietl S, Kellis M & Weissman JS Nature 505, 701–705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Y et al. Nature 505, 696–700 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Talkish J, May G, Lin Y, Woolford JL & McManus CJ RNA 20, 713–720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y et al. Nature 505, 706–709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitale RC et al. Nature 519, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zubradt M et al. Nat. Methods 14, 75–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z & Chang HY Curr. Opin. Struct. Biol 36, 142–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Toxicology Program. Dimethyl sulfate. Rep Carcinog. 12, 174–175 (2011). [PubMed] [Google Scholar]
- 14.Merino EJ, Wilkinson KA, Coughlan JL & Weeks KM J. Am. Chem. Soc 127, 4223–4231 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Mitchell D et al. RNA 24, 114–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell D et al. RNA 25, 147–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang PY, Sexton AN, Culligan WJ & Simon MD RNA 25, 135–146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng C et al. Nat. Chem. Biol 14, 276–283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z & Culver GM Method. Enzymol 468, 147–165 (Academic Press, 2009). [DOI] [PubMed] [Google Scholar]
- 20.Morse DP & Bass BL Biochemistry 36, 8429–8434 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Andronescu M, Bereg V, Hoos HH & Condon A BMC Bioinformatics 9, 340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok CK, Marsico G, Sahakyan AB, Chambers VS & Balasubramanian S Nat. Methods 13, 841–844 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Guo JU & Bartel DP Science 353, aaf5371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biffi G, Di Antonio M, Tannahill D & Balasubramanian S Nat. Chem 6, 75–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwok CK, Marsico G, & Balasubramanian S Cold Spring Harb. Perspect. Biol, 10, a032284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spitale RC et al. Nat. Chem. Biol 9, 18–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Z et al. Cell, 165, 1267–1279 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalvari I et al. Nucleic Acids Res. 46, D335–D442 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.