Abstract
Human leukocyte antigen (HLA) class I molecules present antigenic peptides to cytotoxic T cells, causing lysis of malignant cells. Transplantation biology studies have implicated HLA class I molecules in cell migration, but there has been little evidence presented that they influence cancer cell migration, a contributing factor in metastasis. In this study, we examined the effect of HLA-B on pancreatic cancer cell migration. HLA-B siRNA transfection increased the migration of the S2–013 pancreatic cancer cells but, in contrast, reduced migration of the PANC-1 and MIA PaCa-2 pancreatic cancer cell lines. Integrin molecules have previously been implicated in the upregulation of pancreatic cancer cell migration, and knockdown of HLA-B in S2–013 cells heightened the expression of integrin beta 1 (ITGB1), but in the PANC-1 and MIA PaCa-2 cells HLA-B knockdown diminished ITGB1 expression. A transmembrane sequence in an S2–013 HLA-B heavy chain matches a corresponding sequence in HLA-B in the BxPC-3 pancreatic cancer cell line, and knockdown of BxPC-3 HLA-B mimics the effect of S2–013 HLA-B knockdown on migration. In total, our findings indicate that HLA-B influences the expression of ITGB1 in pancreatic cancer cells, with concurrent distinctions in transmembrane sequences and effects on the migration of the cells.
Keywords: HLA, integrin, major histocompatibility complex class I, migration, pancreatic cancer
Introduction
HLA class I molecules are composed of a heavy chain, the light chain beta 2-microglobulin, and endogenous peptide.1 These heavy chains are encoded by three separate genetic loci that encode for three similar yet not completely identical isotypes: HLA-A, -B, and - C.2 HLA class I molecules are best known for their contribution to the adaptive immune system by presenting antigenic peptides to cytotoxic T cells. Peptide presentation to T cells leads to lysis of the target cell and this allows for protection against viral infection or malignant transformation.3
In the field of transplantation biology, HLA class I molecules have also been implicated in processes such as cell migration.4 Furthermore, although HLA class I molecules can be down-regulated as a form of immune escape by cancer cells, there have been studies in various types of malignancies (including breast, gastric, and lung cancer) that have shown high expression of HLA class I molecules to be positively correlated with poor prognosis.5–7 Additionally, HLA class I molecules were found to be more highly expressed by a set of medulloblastoma tumor samples that had accompanying phenotypic markers of poor prognosis (although clinical data were not available to verify the survival outcomes for all the patients).8 HLA class I molecules were demonstrated to promote increased medulloblastoma cell migration upon addition of exogenous β2m (which stabilizes cell-surface HLA class I heavy chains that have released β2m after arrival at the plasma membrane) via an ERK1/2-mediated mechanism.8–9 However, even though pancreatic cancer is known for its high rate of metastasis, an investigation of the role HLA class I molecules may play in cell migration in pancreatic cancer has not yet been reported. Moreover, the influence of specific HLA class I isotypes and allotypes on the migration of any type of cancer cells has never been fully addressed.
Integrins are cell-surface molecules that function as heterodimers and are present on nearly all nucleated cells. There are 8 α subunits and 18 β subunits that can pair together to form 24 disparate functional heterodimers. A specific range of heterodimers can exist on particular cell types, and they function mainly to bind to certain components of the extracellular matrix.10–12 The α2β1 heterodimer (composed of integrin β1 [ITGB1] and integrin α2 [ITGA2]) binds mainly to collagen but also has the ability to bind to laminin, fibronectin, and E-cadherin.13–18 This heterodimer is overexpressed in many cancer types, including pancreatic cancer, and is associated with an aggressive disease phenotype.19–25 The α2β1 heterodimer has been shown to promote the migration of pancreatic cancer cells, as well as various other cancer cell types such as gastric cancer and prostate cancer.20,26–30 A few previous transplantation biology studies have indicated that integrins interact with HLA class I molecules and that this interaction can contribute to cell migration in transplantation models.4,31,32 Whether an interaction exists between integrins and HLA class I molecules in pancreatic cancer cells, and whether this interaction correlates with pancreatic cancer cell migration have not previously been analyzed.
Thus, the purpose of this study was to investigate whether the HLA-B isotype influences the migration of pancreatic cancer cells, and, if so, to assess the potential involvement of ITGB1 and ITGA2 in this mechanism. Our findings, as described below, indicate that HLA-B is amply expressed in pancreatic cancer cells, though with variation in expression, and influences the migration of pancreatic cancer cells. Furthermore, we demonstrated that HLA-B regulates the expression of ITGB1, which has been previously established to affect the migration of pancreatic cancer cells. However, our co-immunoprecipitation data suggests that this regulation is not dependent on HLA-B association with ITGB1. Comparison of the sequences for HLA-B heavy chains that have differing effects on pancreatic cancer cell migration revealed sequences in the HLA-B transmembrane region that correspond with ability to decrease ITGB1 expression and migration. Therefore, our findings indicate that HLA-B has unexpected and important effects on pancreatic cancer cells beyond its immunological role, and that an HLA-B transmembrane sequence may govern HLA-B effects on pancreatic cancer cell migration.
Materials and Methods
Cell lines and transfections
The S2–013, PANC-1, MIA PaCa-2, and BxPC-3 pancreatic cancer cell lines33–55 were used in this study. The S2–013 cell line (a gift from Dr. Michael A. Hollingsworth, University of Nebraska Medical Center, Omaha, NE) is a SUIT-2 subclone that has been used previously in many pancreatic cancer studies.33–53 The BxPC-3 cell line was also a gift from Dr. Hollingsworth.33 PANC-1 cells were a gift from Dr. Michel Ouellette (University of Nebraska Medical Center, Omaha, NE), and MIA PaCa-2 cells were recently purchased from the American Type Culture Collection.54,55 Both PANC-1 and MIA PaCa-2 cells were cultured in supplemented DMEM medium, and S2–013 and BxPC-3 cells were cultured in supplemented RPMI 1640 medium. The media supplementation included 10% fetal bovine serum (Atlantic Biologics S11550, heat inactivated for 30 minutes at 56°C), 1 mM sodium pyruvate (11360–070), 2 mM L-glutamine (25030–081), 10 mM HEPES (15630–080), 1X non-essential amino acids (11140–050), 100 units/ml penicillin and 100 μg/mL streptomycin (from Penicillin-Streptomycin 10,000 U/ml stock 15140–122). All the aforementioned media additives were obtained from Thermo Fisher Scientific with the exception of the fetal bovine serum. The S2–013, PANC-1, and BxPC-3 cells were authenticated by short tandem repeat deoxyribonucleic acid profiling analysis (UNMC Molecular Diagnostics Facility) and the MIA PaCa-2 cells were authenticated by the American Type Culture Collection. MycoAlert Mycoplasma Detection Kit (Lonza LT07–118) was used to determine that all pancreatic cancer cell lines used in this study were free of mycoplasma.
The HLA-B heavy chain was experimentally knocked down by transient transfection with siRNA specific to HLA-B. The ON-TARGETplus SMARTpool siRNA (containing four separate HLA-B-specific siRNAs), purchased from Dharmacon/Horizon Discovery, was used to downregulate HLA-B expression in all three pancreatic cancer cell lines. The OnTARGETplus SMARTpool non-targeting siRNA pool (Dharmacon D-001810-10-50) was used as a negative control. Briefly, pancreatic cancer cells were plated in a 6-well plate at 1×105 cells/well, and on the following morning the cells were starved in base maintenance medium for 3 hours. DharmaFECT Transfection Reagent No. 1 (Dharmacon T-2001–03) was incubated with siRNA (at 10 nM) for 30 minutes and the mixture was added to respective wells dropwise. Afterward, the cells were incubated for 4 hours, and serum-containing medium was then added. Knockdown of HLA-B was confirmed by immunoblot analysis.
Antibodies
The HC-10 monoclonal antibody (which recognizes principally HLA-B and HLA-C isotypes) was used to monitor HLA-B levels.56,57 Little to no HLA-C is expressed in the pancreatic cancer cell lines utilized in this study, so immunoblotting of lysates of these cell lines with the HC10 antibody yields a band that almost entirely consists of HLA-B (as shown by the reduction in the band’s density when the cells are transfected with HLA-B siRNA). The HC-10 antibody was kindly provided by Dr. Ted Hansen (Washington University at St. Louis). For detection of ITGB1 and ITGA2 by immunoblot, antibodies specific to ITGB1 (A303–735A-T) and ITGA2 (A305–211A-T) were obtained from Bethyl Laboratories. The pFAK (Y397)-specific antibody (8556P) and total FAK-specific antibody (130095) were purchased from Cell Signaling Technologies. A rat monoclonal antibody for Hsc70 was bought from Enzo Life Sciences (ADI-SPA-815B-F).
Assessment of pancreatic cancer cell migration
Transwell assays were used to assess changes in pancreatic cancer cell migration. Pancreatic cancer cells were seeded in 6-well plates at a density of 1×105 cells/well, and 24 hours later were transfected with siRNA for 72 hours to decrease the expression of the protein of interest. Cells were then re-seeded at 1×105 cells in base maintenance media into inserts with 8-μm pores (Falcon/Thermo Fisher Scientific 353097). Cells were incubated for 24 hours at 37°C in 5% CO2, and then the inserts were stained with Hema 3 Stat Pack (Thermo Fisher Scientific 123–869) and mounted. Three random fields per well were photographed and all the cells in each field were counted. Student’s two-tailed t-test was used to determine the level of significance for migration results, and the criterion used for significance was p<0.05.
Immunoblotting
Cells were washed once with cold phosphate-buffered saline (PBS) and collected in cell lysis buffer (1 mM EGTA [Sigma E-3889], 1 mM EDTA [Sigma E-9884], 50 mM Tris-HCl pH 7.5 [made with Trizma base Sigma T-8524], 1% Triton X-100 [Sigma T-8532], 1 mM Na3VO4 [Thermo Fisher Scientific AA8110414], 2 mM DTT [Sigma D0632], 0.1 mM PMSF [Sigma 10837091001], and 1 g/ml Halt Cocktail [Thermo Fisher Scientific 78430]). Cell lysates were stored at −80°C overnight then thawed on ice, followed by centrifugation at 13,000 rpm for 30 min at 4°C. On the day of immunoblotting, aliquots of the lysate supernatants were combined with 5X sodium dodecyl sulfate loading dye (250 mM Tris-HCl pH 6.8, 10% w/v sodium dodecyl sulfate [Tokyo Chemical Industry Company D0996], 30% v/v glycerol [Sigma G-5516], 5% v/v β-mercaptoethanol [Sigma M-7522], 0.02% w/v bromophenol blue [Sigma B-7021]), and boiled for 5 minutes before loading.
Cell lysate samples were loaded on 4–20% Invitrogen Novex Tris-glycine polyacrylamide pre-cast gels (Thermo Fisher Scientific XP04205BOX) and electrophoresis was performed at 90 V at room temperature. The proteins were transferred at 40 V for 2.5 hours at room temperature to polyvinylidene difluoride Immobilon-P Millipore membranes (PIVH00010). For 1 hour, membranes were blocked in a 5% w/v solution of nonfat dry milk and were then incubated overnight at 4°C with primary antibodies. After primary antibody incubation, 3 washes with 1% Tween-20 (Thermo Fisher Scientific BP-337) in PBS were performed for 5 minutes each. Subsequently, membranes were incubated for 1 hour in secondary antibodies, and washed 3 times for 5 minutes with 1% Tween-20 in PBS. The membranes were incubated in Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific 32106) and exposed to CareStream BioMax MR film (8941114) for protein visualization.
RNA-Seq analysis
Epithelial cells (229 samples: 203 from primary tumors+26 from low-grade PanINs) and stromal cells (125 samples, with matched epithelium: 102 from primary tumors+23 from low-grade PanINs) were microdissected. Cells were isolated to produce paired samples for each patient (>1000 cells/sample). Libraries were made with the NuGEN Ovation RNA-Seq System V2 Kit and cDNAs were sequenced with an Illumina HiSeq (3500 to 30 million 100 base-pair single-end reads). The y axis shows log2 Transcripts Per Million (TPM), calculated by dividing the read counts for HLA-B by the length of the HLA-B genetic sequence in kb, and then dividing by a scaling factor (total reads per kb for all genes in the dataset/1,000,000). The TPM unit allows comparison of the proportion of reads mapping to a particular gene between samples.
Results
HLA-B is expressed in pancreatic cancer cell lines and pancreatic tumor epithelial cells
To determine the expression of the HLA-B heavy chain in a panel of human pancreatic cancer cell lines, we immunoblotted lysates of S2–013, PANC-1, and MIA PaCa-2 pancreatic cancer cell lines in comparison with hTERT-HPNE, an immortalized but not transformed human pancreas ductal cell line that serves as a model for non-malignant pancreatic ductal cells. As shown in Figure 1A, all of the pancreatic cancer cell lines tested have ample expression of HLA-B, as does the non-transformed hTERT-HPNE cells. Our findings suggest that despite the anti-cancer immunological function of this molecule, the HLA-B isotype is not necessarily diminished in human pancreatic cancer cell lines. Figure 1B displays data acquired using samples derived from pancreatic adenocarcinoma patients. These data were obtained by laser capture microdissection of epithelial and adjacent stromal cells, respectively, followed by RNA-Seq analysis. These data indicate that, for the majority of pancreatic cancer patients, primary tumor epithelial cell expression of HLA-B expression is not downregulated, although some patients have especially low (and others particularly high) HLA-B expression by their tumor cells.
Figure 1.
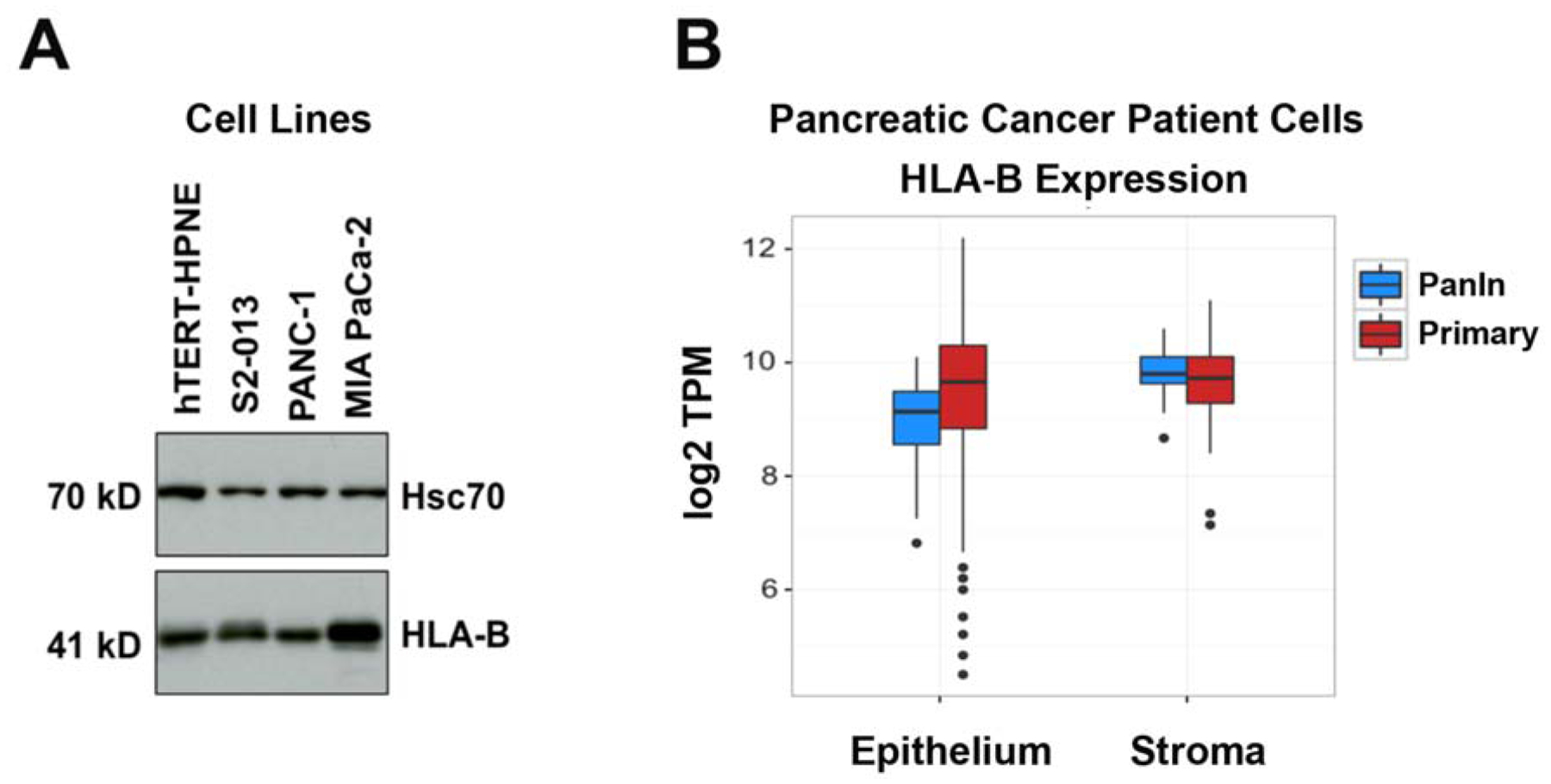
The HLA class I heavy chain isotype HLA-B is abundantly expressed in human pancreatic cancer cells. (A) Immunoblots to detect HLA-B were performed on lysates of an untransformed pancreas duct cell line (hTERT-HPNE) and S2–013, PANC-1, and MIA PaCa-2 pancreatic cancer cell lines. Hsc70 immunoblotting was included as an internal control. The results shown are representative of 3 experimental replicates to assess HLA-B expression in lysates of these cell lines. (B) HLA-B is expressed at comparable levels in primary pancreatic cancer cells from patients versus stromal cells and versus PanIn lesions from patients, as demonstrated by RNA-Seq.
HLA-B knockdown influences pancreatic cancer cell migration
To investigate whether the HLA-B isotype affects pancreatic cancer cell migration, we transfected S2–013, PANC-1, and MIA PaCa-2 pancreatic cancer cell lines with pan-HLA-B-specific siRNA (or with control scrambled siRNA). After siRNA transfection, the reduction of HLA-B protein levels was confirmed in the pancreatic cancer cell lines tested (Figure 2A). By transwell assay, the migration of the S2–013 cell line was assessed as significantly increased when HLA-B expression was knocked down (Figure 2B). In contrast, when we down-regulated the expression of HLA-B in PANC-1 and MIA PaCa-2 cells, the migration of those two cell lines was significantly impaired (Figure 2B). Thus, these findings indicate clearly that HLA-B affects the migration of pancreatic cancer cells.
Figure 2.
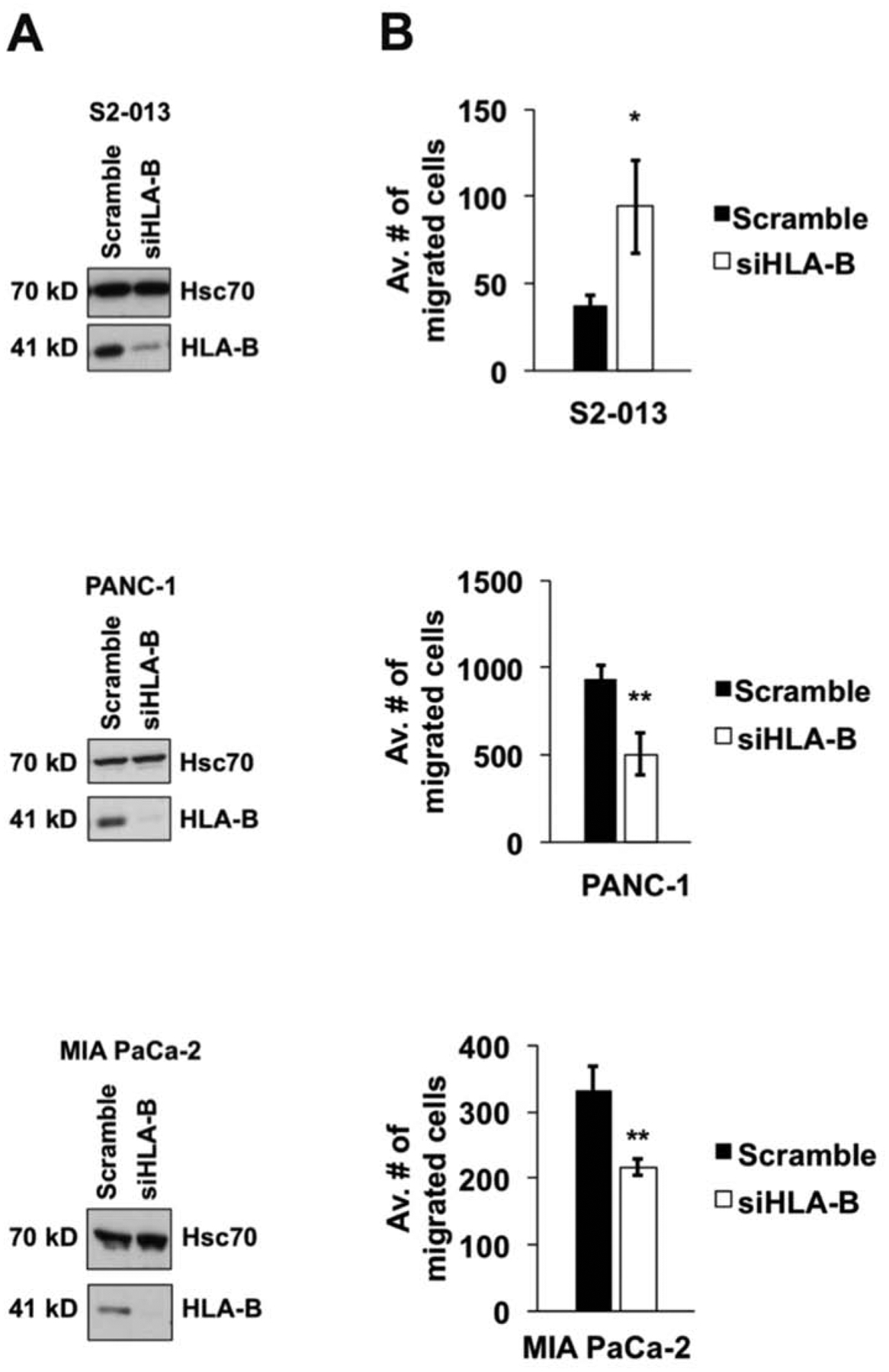
HLA-B influences pancreatic cancer cell migration rate. Upon knockdown of HLA-B expression, the migration rate of S2–013 cells was increased, and the migration of PANC-1 and MIA PaCa-2 cells was diminished. (A) S2–013, PANC-1, and MIA PaCa-2 pancreatic cancer cell lines were transfected with siRNA for pan-HLA-B (or scrambled control siRNA) and the knockdown of the HLA-B heavy chain was verified by immunoblotting (the results shown are representative of 5 repetitions for each cell line). Hsc70 was included as a loading control. (B) The migration of each pancreatic cancer cell line was assessed by transwell assay. At 72 h post-transfection, the cells were replated into 8-μm inserts and incubated for 24 h. Photographs were taken of 3 random fields, the cells in each of the fields were counted, and the results were graphed as average number of cell migrated. For each cell line, 5 repetitions were performed. The error bars represent the standard error of the mean. Student’s two-tailed t-test was used for assessment of statistical significance. The symbol * indicates p<0.05, ** indicates p<0.01, and *** indicates p<0.001.
Interference with HLA-B expression alters ITGB1 total expression but not ITGA2 expression
To examine what might be mediating the differential effects that HLA-B exerts on pancreatic cancer cell migration, integrin expression was investigated, as integrins have a major role in cancer cell migration, including the migration of pancreatic cancer cells.20,26–30 ITGB1 and ITGA2 were chosen for analysis because of their high prevalence in heterodimeric form (i.e., as integrin α2β1) in pancreatic cancer.19–25 First, we assessed the expression levels of ITGB1 and ITGA2 in pancreatic cancer cell lines. Expression of both ITGB1 and ITGA2 was confirmed in all of the pancreatic cancer cell lines tested (S2–013, PANC-1, and MIA PaCa-2) with the exception that MIA PaCa-2 cells were found to lack expression of ITGA2 (Figure 3). The expression levels for the ITGB1 and ITGA2 integrins were then assessed after HLA-B siRNA transfection of each of the cell lines. For the S2–013 cells, when HLA-B was knocked down there was little or no change in ITGA2 protein expression, but ITGB1 expression substantially increased (Figure 3A). Conversely, in PANC-1 and MIA PaCa-2 cells, there was a decrease in ITGB1 expression when HLA-B was knocked down (Figure 3B,C). ITGA2 also remained unchanged in the PANC-1 cells when HLA-B was knocked down (Figure 3B). Although the S2–013 and PANC-1 pancreatic cancer cells express ITGA2 at a higher level than the non-transformed hTERT-HPNE cells, the MIA PaCa-2 cells do not express detectable ITGA2 (Figure 3D). Notably, for each of these pancreatic cancer cell lines, the impact of HLA-B expression on ITGB1 expression exactly parallels the effect of HLA-B expression on migration (compare Figure 3 to Figure 2).
Figure 3.
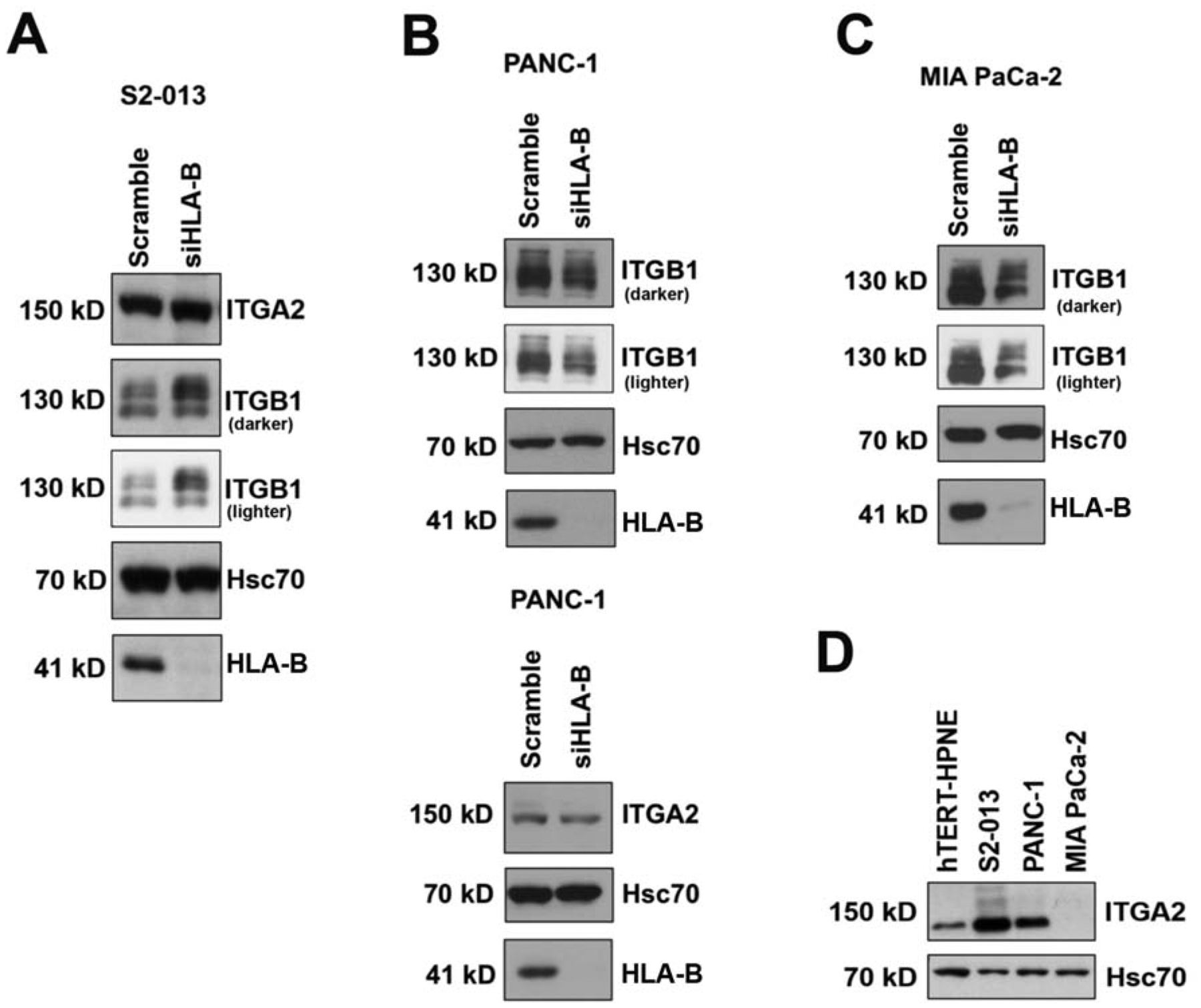
HLA-B affects the expression of ITGB1 but not ITGA2 in pancreatic cancer cell lines. HLA-B knockdown after siRNA transfection for 72 h was verified by immunoblotting with the HC-10 antibody. Control transfections with scrambled siRNA were also performed. Hsc70 immunoblotting was included as an internal control. (A) Knockdown of HLA-B in S2–013 cells increased ITGB1 expression, but did not affect ITGA2 expression. (B, top) In contrast, knockdown of HLA-B expression in PANC-1 caused a decline in ITGB1 expression. (B, bottom) ITGA2 expression was not affected by HLA-B knockdown in PANC-1 cells. (C) The knockdown of HLA-B expression in MIA PaCa-2 cells also caused a decrease in ITGB1 expression. (D) MIA PaCa-2 cells do not express ITGA2). (A,B,C) Both darker and lighter exposures of the same ITGB1 blots are shown. (A,B,D) The results shown are representative of 2 separate experiments. (C) For MIA PaCa-2, a single ITGB1 immunoblotting experiment was performed.
To evaluate whether HLA-B associates with ITGB1 in pancreatic cancer cells, we tried immunoprecipitating ITGB1 from lysates of S2–013 and PANC-1 cells and probing for co-immunoprecipitated HLA-B. Unfortunately, these experiments were indeterminate, because although there was a faint band (indicating possible weak co-immunoprecipitation), the band was not substantially darker than the weak band in the negative control obtained by performing an immunoprecipitation assay with anti-mouse IgG and immunoblotting for HLA-B (data not shown). Thus, our results do not support association of HLA-B with ITGB1 as the mechanism whereby HLA-B influences ITGB1 expression and pancreatic cancer cell migration.
FAK expression is altered in S2–013 and PANC-1 pancreatic cancer cells upon HLA-B knockdown
A major, well-studied signaling molecule downstream of integrins is FAK, a non-receptor cytoplasmic tyrosine kinase.10,58 Ligand binding leads to clustering of integrins, at which point activation of FAK by autophosphorylation at Tyr397 can occur. The Tyr397 position on FAK is a binding site for other kinases, including Src and PI3K.59–61 The integrin-FAK pathway has been associated with many aspects of pancreatic cancer metastasis, such as adhesion, migration, and invasion.62–65 To examine further downstream signaling controlled by the HLA-B class I molecule and the α2β1 heterodimer, the expression of pFAK (Y397) and total FAK was investigated in pancreatic cancer cell lines following knockdown of HLA-B. HLA-B knockdown in S2–013 pancreatic cancer cells caused an increase in pFAK (Y397) (Figure 4A), corresponding to the rise in ITGB1 expression that was observed as a result of HLA-B knockdown (Figure 3A). PANC-1 cells, which had a decrease in ITGB1 following HLA-B siRNA transfection (Figure 3B), also had a decline in pFAK (Y397) expression (Figure 4B). In S2–013 and PANC-1, the changes that occurred in the total FAK expression levels after HLA-B siRNA transfection followed the same pattern as the pFAK Y397 levels (Figure 4A,B). Total FAK protein expression has been found to be upregulated in many different cancer types (including pancreatic cancer), and diminished expression of this protein can contribute to decreased cell migration through reduction in the amount of FAK available for activation.62–66 In contrast, MIA PaCa-2 pFAK (Y397) expression was not altered by HLA-B knockdown (Figure 4C). It is possible that the absence of ITGA2 in MIA PaCa-2 cells (Figure 3D) prevents regulation of FAK phosphorylation by HLA-B in these cells. Therefore, for S2–013 and PANC-1, though not for MIA PaCa-2, the FAK protein may play a role in the migration changes resulting from HLA-B knockdown.
Figure 4.
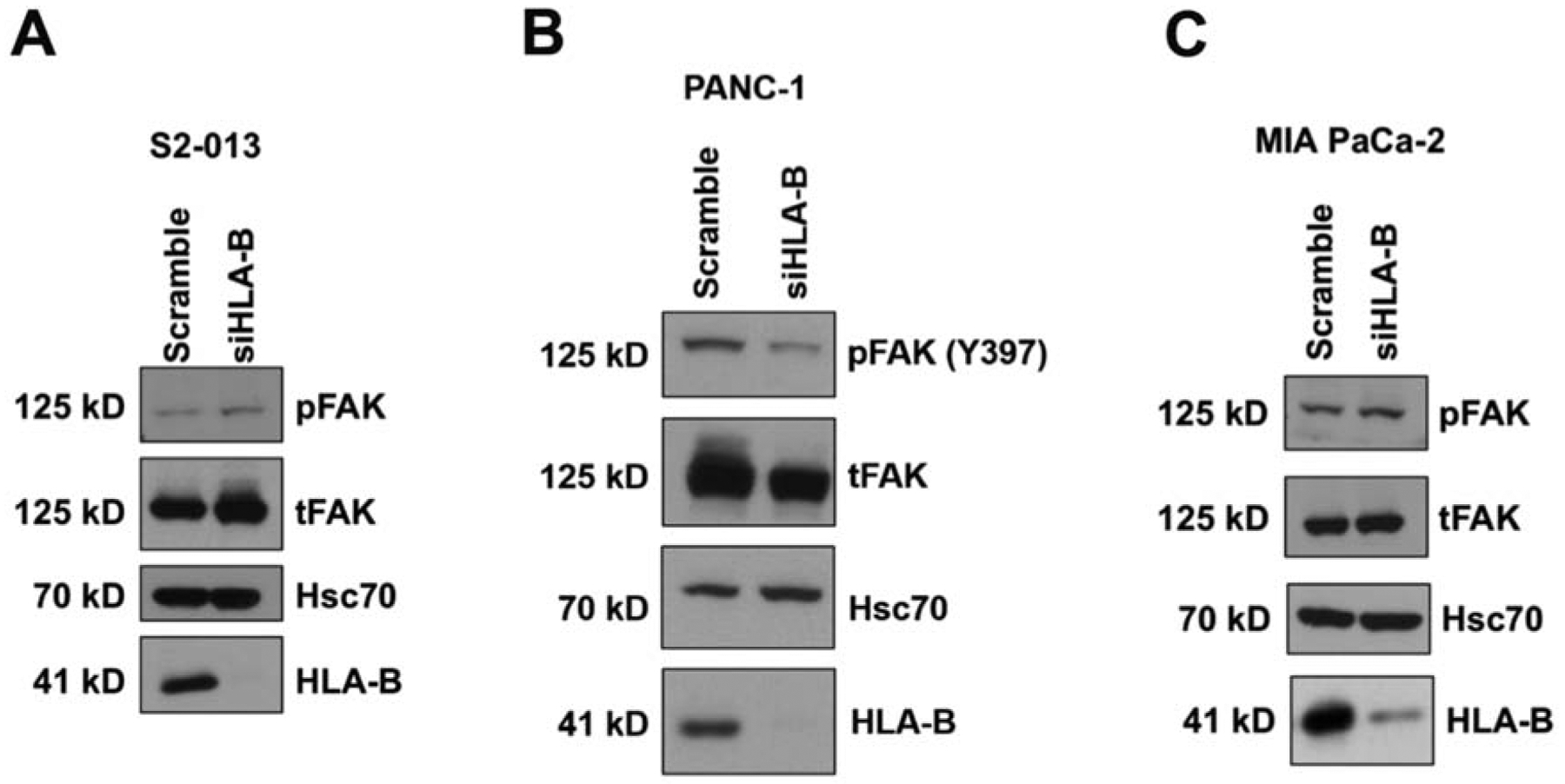
Upon HLA-B knockdown, total FAK and pFAK (Y397) expression levels were (A) increased in S2–013 cells, (B) decreased in PANC-1, and (C) unchanged in MIA PaCa-2. Cell lysates were collected from the aforementioned pancreatic cancer cell lines at 72 h post-transfection with control siRNA or HLA-B siRNA. The lysates were immunoblotted for pFAK (Y397), total FAK, Hsc70 (loading control), and HLA-B (to confirm knockdown). The results displayed are representative of multiple experiments done with 7 separate lysates for the S2–013 cell line, 2 separate lysates for the PANC-1 cell line, and 2 separate lysates for the MIA PaCa-2 cell line.
A sequence in the HLA-B transmembrane domain correlates with effects on migration
To discern what sequences in these HLA-B heavy chains might be responsible for the effects on migration, as well as the changes in ITGB1, pFAK, and total FAK expression, we compared the full sequences of the HLA-B heavy chains expressed in S2–013, MIA PaCa-2, and PANC-1. The allotypes of the HLA-B heavy chains in these cell lines were found in the TRON Cell Line Portal (www.celllines.tron-mainz.de), and the sequences for each of the allotypes were obtained from the IPD-IMGT/HLA database (www.ebi.ac.uk/ipd/imgt/hla). In this analysis, we noted that the two HLA-B heavy chains in S2–013 differed from each of the single HLA-B heavy chains expressed by MIA PaCa-2 and PANC-1 by amino acid residues in the transmembrane/cytoplasmic domain region (Figure 5). The S2–013 cell line expresses an HLA-B7 allotype (HLA-B★07:02) that has a cytoplasmic region cysteine residue at position 325 rather than a serine (the serine is present at this position in the other HLA-B allotypes shown in Figure 5). In addition to HLA-B★07:02, the S2–013 cell line also expresses HLA-B★59:01, which in its transmembrane domain has an isoleucine instead of a valine at position 282, and a threonine in place of an alanine at position 305, as compared to the HLA-B allotypes expressed by MIA PaCa-2 (HLA-B★14:02) and PANC-1 (HLA-B★38:01) (Figure 5). Since down-regulation of HLA-B in S2–013 cells has an effect on migration and ITGB1 expression that is opposite to that of HLA-B down-regulation in PANC-1 and MIA PaCa-2 cells, these disparities in the transmembrane/cytoplasmic domain sequences were considered as potential mediators of the differences.
Figure 5.
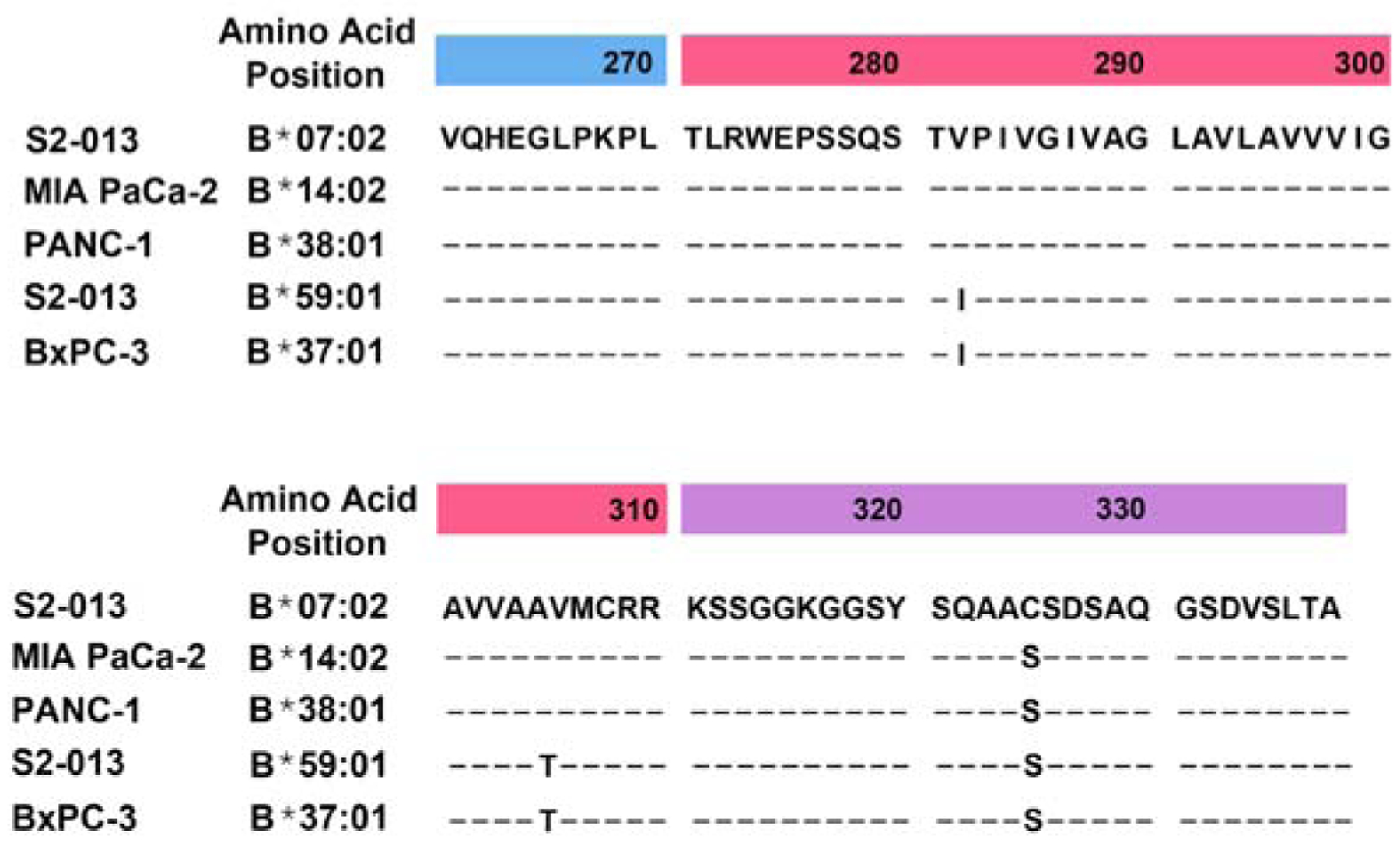
The transmembrane/cytoplasmic regions of the HLA-B heavy chains expressed by the S2–013 pancreatic cancer cell line differ from those of the MIA PaCa-2 and PANC-1 cell lines, but the transmembrane region of one of the S2–013 HLA-B heavy chains matches the transmembrane region of the HLA-B heavy chain expressed by the BxPC-3 pancreatic cancer cell line. The sequences shown from the HLA-B heavy chains expressed in the S2–013, MIA PaCa-2, PANC-1, and BxPC-3 include the final 10 amino acids in the α3 domains (positions 261–270, blue heading), the amino acids in the transmembrane domains (positions 271–310, red heading), and the amino acids in the cytoplasmic domain (positions 311–338, purple heading). Position 325 in the S2–013 B★07:02 cytoplasmic domain is a cysteine, whereas the other HLA-B heavy chains displayed have a serine at that location. Positions 282 and 305 are valine and alanine, respectively, in B★07:02 (expressed by S2–013), B★14:02 (expressed by MIA PaCa-2), and B★38:01 (expressed by PANC-1). Positions 282 and 305 are isoleucine and threonine, respectively, in B★59:01 (expressed by S2–013) and B★37:01 (expressed by BxPC-3). The sequences shown are from IPD-IMGT/HLA Release 3.38.0 2019-10-17 (https://www.ebi.ac.uk/ipd/imgt/hla/).
In searching for a pancreatic cancer cell line expressing an endogenous HLA-B heavy chain matching the sequence of an S2–013 HLA-B heavy chain in the transmembrane domain or the cytoplasmic domain, we found BxPC-3, a human pancreatic cancer cell line that expresses only HLA-B★37:01 as a single HLA-B heavy chain (Figure 5). The transmembrane sequence of HLA-B★37:01 is completely identical to the HLA-B★59:01 transmembrane sequence, sharing the isoleucine at position 282 and the threonine at position 305. In assessing the effects of downregulating HLA-B in this cell line, we found that transfection of HLA-B siRNA greatly facilitated the migration of BxPC-3 cells, and it increased the expression of ITGB1 but not ITGA2 (Figure 6A,B,C). The effect of HLA-B siRNA transfection of BxPC-3 cells on pFAK expression was also tested, and pFAK was demonstrated to rise in the transfected BxPC-3 cells (Figure 6D). Therefore, the influence of HLA-B downregulation on BxPC-3 migration, ITGB1, and pFAK was the same as the effect of HLA-B knockdown on S2–013 cells, consistent with the possible involvement of the matched transmembrane sequence in HLA-B★37:01 and HLA-B★59:01 in the observed phenotypic alterations in both S2–013 and BxPC-3.
Figure 6.
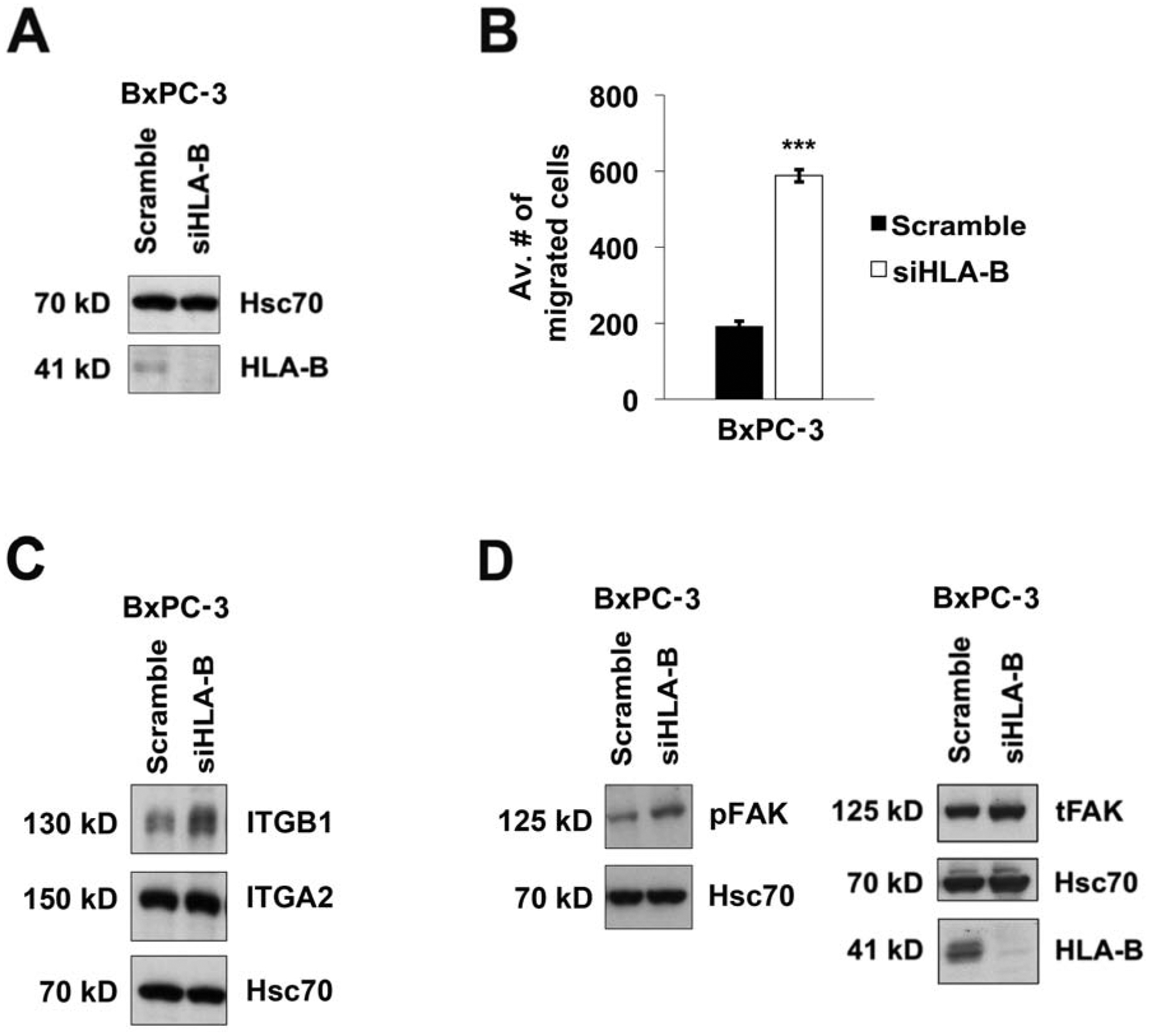
Knockdown of HLA-B★37:01 expression in BxPC-3 cells results in increased migration of BxPC-3 and elevated expression of ITGB1 (but not ITGA2). (A) BxPC-3 pancreatic cancer cells were transfected for 72 h either with siRNA to downregulate HLA-B or with scrambled control siRNA. HLA-B heavy chain knockdown was confirmed by immunoblotting, and probing the blot with antibody for Hsc70 was included as a loading control. (B) The migration of the BxPC-3 cells, transfected either with scramble control siRNA or HLA-B siRNA, was monitored by transwell assay. At the 72 h time point after transfection, the cells were plated in 8-μm inserts. After 24 h of incubation, 3 random fields were photographed. The cells in each of the photographic fields were counted, and the average numbers of cells migrated were graphed. The data shown in the graph was compiled from 4 repetitions of the experiment. The error bars indicate the standard error of the mean. Student’s two-tailed t-test was used to determine statistical significance. The symbol *** represents p<0.001. (C) Knockdown of HLA-B in BxPC-3 cells increased ITGB1 expression, but did not affect ITGA2 expression, as shown by immunoblotting for ITGB1 and ITGA2. Transfection with scramble control siRNA was performed as a negative control. Immunoblotting with an anti-Hsc70 antibody was included as a loading control. The results shown are representative of 3 experiments that included ITGB1 blots and 2 experiments that included ITGA2 blots. (D) The pFAK (Y397) and total FAK expression levels were increased in BxPC-3 cells upon transfection of HLA-B-specific siRNA, as compared to BxPC-3 cells transfected with scramble control siRNA. Hsc70 was immunoblotted as a control, and HLA-B knockdown was also confirmed by immunoblotting. The results displayed are representative of those demonstrated in 3 separate experiments.
Discussion
Our findings suggest that HLA-B, even though having the capability of presenting tumor antigens to cytotoxic T cells, remains highly (yet variably) expressed in human pancreatic cancer cell lines (Figure 1A) and human pancreatic cancer tissue samples (Figure 1B). Studies investigating HLA class I molecule expression in pancreatic cancer have mainly interrogated total HLA class I expression and not assessed differences in expression among HLA class I isotypes in normal tissue as compared to malignant tissue.67–70 Our findings indicate that the HLA-B isotype is not consistently down-regulated in pancreatic cancer (Figure 1).
Additionally, we found that HLA-B slows the migration of the S2–013 pancreatic cancer cell line while it increases the migration of the PANC-1 and MIA PaCa-2 cell lines (Figure 2). In our analysis of the mechanism by which HLA-B exerts these effects on pancreatic cancer cell migration, we assessed the possible role of integrins, since some integrin molecules have been demonstrated to interact with HLA class I molecules and contribute to cell migration in an endothelial cell model of transplantation biology.4 ITGB1 was chosen, since the integrin α2β1 heterodimer is commonly expressed in pancreatic cancer.20–21 Consistent with our theory that ITGB1 is linked to HLA-B’s effect on pancreatic cancer cell migration, we observed that ITGB1 protein expression is induced by HLA-B knockdown in S2–013 cells, yet diminished upon knockdown of HLA-B in PANC-1 and MIA PaCa-2 cells (Figure 3). The expression of the downstream effector molecule FAK (pFAK and total FAK) also followed these same trends in S2–013 and PANC-1 (but not in MIA PaCa-2, which as mentioned above may be due to its lack of ITGA2, as shown in Figure 3D).
Pancreatic cancer cells express other integrin heterodimers in addition to integrin α2β1, and therefore certain integrin heterodimers not yet explored in this study could also be contributing to the differential effects of HLA-B on pancreatic cancer cell migration. The potential influence of other integrins is particularly relevant in the case of the MIA PaCa-2 cell line, which does not express ITGA2. ITGA3, another integrin alpha chain found to heterodimerize in pancreatic cancer,71 might also be mediating effects on migration under the influence of HLA-B in MIA PaCa-2 cells. Furthermore, ITGA6, which binds to both ITGB1 and ITGB4,72–73 could also have a role in conjunction with HLA-B class I molecules in pancreatic cancer cells.
In the S2–013 cell line, HLA-B represses cell migration, as compared to the PANC-1 and MIA PaCa-2 cell lines, for which HLA-B was found to induce cell migration. S2–013 cells express two distinct allotypes of HLA-B, while PANC-1 and MIA PaCa-2 cells express only one allotype, likely having lost the other allotype during oncogenic transformation. The sequences of the particular HLA-B allotypes expressed by these pancreatic cancer cell lines must be responsible, either directly or indirectly, for differential effects on migration and ITGB1 expression.
A previous study indicated that an interaction in endothelial cells between ITGB4 and HLA class I molecules is mediated by the cytoplasmic tail of the HLA class I molecule.4 A few allotypes of HLA-B, such as HLA-B27, have previously been found to be able to form interchain disulfide bonds that cause homodimerization via cysteine residues in their extracellular sequences, and formation of “redox-induced dimers” of certain HLA class I molecules can occur that involve cysteine residues in the cytoplasmic portions of those HLA class I molecules.74–76 HLA-B★07:02, which is expressed by the S2–013 cell line (Figure 5), has not been reported to form interchain disulfide bonds, although it does share the cytoplasmic domain cysteine residue that in HLA-B27 is able to form such bonds.
In addition to HLA-B★07:02, the S2–013 cell line also expresses the HLA-B★59:01 heavy chain (Figure 5). The single HLA-B allotype expressed by the MIA PaCa-2 cell line is HLA-B★14:02, and the Panc1 cell line expresses only HLA-B★38:01 (Figure 5). HLA-B★14:02 and HLA-B★38:01 are identical throughout their transmembrane and cytoplasmic domains, and HLA-B★59:01 is also identical both to HLA-B★14:02 and to HLA-B★38:01 in the cytoplasmic domain. However, HLA-B★59:01 is different from HLA-B★14:02 and to HLA-B★38:01 at two amino acid positions (282 and 305) in the transmembrane domains (Figure 5). The HLA-B★37:01 heavy chain expressed by the BxPC-3 pancreatic cancer cell line shares with HLA-B★59:01 the isoleucine and threonine, respectively, at positions 282 and 305. Therefore, these particular sequences in the transmembrane domain of the HLA-B allotypes could be potential mediators of effects on migration and ITGB1 expression.
In seeking to determine by co-immunoprecipitation experiments whether the HLA-B heavy chains expressed by S2–013 associate with ITGB1, we found minimal to no association in comparison with control immunoprecipitation with irrelevant antibody (data not shown). It is possible that under different conditions of solubilization, or with other modifications in the immunoprecipitation parameters, association might yet be detectable; however, at least with the immunoprecipitation procedures that we have used to date we have not observed association. Thus, the identity of the protein(s) with which these transmembrane domain residues interact was not found to be ITGB1 in our study.
An interesting possibility is presented by the identification of the HLA-B transmembrane sequence 304–308, which encompasses position 305 (threonine in B★59:01 and B★37:01, but alanine in B★07:02, B★14:02, and B★38:01) as a putative PDZ ligand by Frietze et al.77 PDZ ligands are very short sequences that are bound by longer sequences called PDZ domains (which exist in hundreds of human proteins), and phosphorylation of a PDZ ligand by a serine or threonine kinase can affect the interaction of a PDZ ligand with a PDZ domain.78 Future studies can explore whether this transmembrane sequence in the HLA-B heavy chain (which, in the case of B★59:01 and B★37:01, contains a threonine) associates with a PDZ domain-containing protein in pancreatic cancer cell lines.
Together, these findings suggest that the HLA class I heavy chain isotype HLA-B influences pancreatic cancer cell migration, possibly through effects on the expression of ITGB1. By influencing cell migration, HLA-B may be playing a role in pancreatic cancer metastasis. Due to the high metastatic rate of pancreatic cancer and its persistently dismal 5-year survival rate, garnering information about factors that contribute to the migration of pancreatic cancer cells from studies such as this one will aid in the comprehension of the pathogenesis characteristic of this disease, and may potentially contribute to the generation of better treatments for pancreatic cancer and/or improved prognostic strategies based on HLA typing of patients.
Acknowledgments
This research was funded by the National Institutes of Health (R21 CA223429, Specialized Program of Research Excellence P50 CA127297, and the Fred and Pamela Buffett Cancer Center Support Grant P30 CA036727). B.H.S. received fellowships from the National Institutes of Health-funded Cancer Biology Training Program (T32 CA009476) and the UNMC Graduate Studies Office.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Krangel MS, Orr HT, Strominger JL. Assembly and maturation of HLA-A and HLA-B antigens in vivo. Cell. 1979;18:979–991. [DOI] [PubMed] [Google Scholar]
- 2.Townsend A, Ohlen C, Bastin J, Ljunggren HG, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. 1989;340:443–448. [DOI] [PubMed] [Google Scholar]
- 3.Benacerraf B Role of the MHC products in immune regulation. Science. 1981;212:1229–1238. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Rozengurt E, Reed EF. HLA class I molecules partner with Integrin β4 to stimulate endothelial cell proliferation and migration. Sci Signal. 2010;3:ra85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madjd Z, Spendlove I, Pinder SE, Ellis IO, Durrant LG. Total loss of MHC class I is an independent indicator of good prognosis in breast cancer. Int J Cancer. 2005;117:248–255. [DOI] [PubMed] [Google Scholar]
- 6.Ramnath N, Tan D, Li Q, Hylander BL, Bogner P, Ryes L, Ferrone S. Is downregulation of MHC class I antigen expression in human non-small cell lung cancer associated with prolonged survival? Cancer Immunol Immunother. 2006;55:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda Y, Ishikawa K, Shiraishi N, Yokoyama S, Kitano S. Clinical significance of HLA class I heavy chain expression in patients with gastric cancer. J Surgical Oncol. 2008;97:451–455. [DOI] [PubMed] [Google Scholar]
- 8.Smith C, Santi M, Rajan B, Rushing EJ, Choi MR, Rood BR, Cornelison R, MacDonald TJ, Vukmanovic S. A novel role of HLA class I in the pathology of medulloblastoma. J Transl Med 2009;7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith C, Santi M, Rushing EJ, Cornelison R, MacDonald TJ, Vukmanovic S. Characterization of signaling function and expression of HLA class I molecules in medulloblastoma. J Neurooncol 2011;103:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianconi D, Unseld M, Prager GW. Integrins in the spotlight of cancer. Int J Mol Sci. 2016;77:2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginsberg MH. Integrin activation. BMB Rep. 2014;47:655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2012;110:673–687. [DOI] [PubMed] [Google Scholar]
- 13.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. [DOI] [PubMed] [Google Scholar]
- 14.Gout SP, Jacquier-Sarlin MR, Rouard-Talbot L, Rousselle P, Block MR, RhoA-dependent switch between alpha2beta1 and alpha3beta1 integrins is induced by laminin-5 during early stage of HT-29 cell differentiation. Mol Biol Cell. 2001;12:3268–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275:1785–1788. [DOI] [PubMed] [Google Scholar]
- 16.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuckwell D, Calderwood DA, Green LJ, Humphries MJ. Integrin alpha 2 I-domain is a binding site for collagens. J Cell Sci. 1995;108:1629–1637. [DOI] [PubMed] [Google Scholar]
- 18.Whittard JD, Craig SE, Mould AP, Koch A, Pertz O, Engel J, Humphries MJ. E-cadherin is a ligand for integrin α2β1. Matrix Biol. 2002;21:525–532. [DOI] [PubMed] [Google Scholar]
- 19.Danen EH. Integrins: regulators of tissue function and cancer progression. Curr Pharm Des. 2005;11:881–891. [DOI] [PubMed] [Google Scholar]
- 20.Grzesiak JJ, Bouvet M. The alpha2beta1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br J Cancer. 2006;94:1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grzesiak JJ, Ho JC, Moossa AR, Bouvet M. The integrin-extracellular matrix axis in pancreatic cancer. Pancreas 2007;35:292–301. [DOI] [PubMed] [Google Scholar]
- 22.Guo W, Giancotti FG. Integrin signaling during tumor progression. Nat Rev Mol Cell Biol. 2004;5:816–826. [DOI] [PubMed] [Google Scholar]
- 23.Madamanchi A, Santoro SA, Zutter MM. alpha2beta1 Integrin. Adv Exp Med Biol. 2014;81:941–960. [DOI] [PubMed] [Google Scholar]
- 24.McCall-Culbreath KD, Zutter MM. Collagen receptor integrins:rising to the challenge. Curr Drug Targets. 2008;9:139–149. [DOI] [PubMed] [Google Scholar]
- 25.Mizejewshi GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. [DOI] [PubMed] [Google Scholar]
- 26.Bassaganas S, Carvalho S, Dias AM, Perez-Garay M, Ortiz MR, Figueras J, Reis CA, Pinho SS, Peracaula R. Pancreatic cancer cell glycosylation regulates cell adhesion and invasion through the modulation of α2β1 integrin and E-cadherin function. PLoS ONE. 2014;9:e98595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chuang YC, Wu HY, Lin YL, Tzou SC, Chuang CH, Jian TY, Chen PR, Chang YC, Lin CH, Huang TH, Wang CC, Chan YL, Liao KW. Blockade of ITGA2 induces apoptosis and inhibits cell migration in gastric cancer. Biological Procedures Online. 2018;20:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzesiak JJ, Cao HST, Burton DW, Kaushal S, Vargas F, Clopton P, Snyder CS, Deftos LJ, Hoffman RM, Bouvet M. Knockdown of the β1 integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer. 2011;129:2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall CL, Dubyk CW, Riesenberger TA, Shein D, Keller ET, van Golen KL. Type 1 collagen receptor (alpha2beta1) signaling promotes prostate cancer invasion through RhoC GTPase. Neoplasia. 2008;10:797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C Zeisberg M, Lively JC, Nyberg P, Afdhal N, Kalluri R. Integrin alpha1beta1 and alpha2beta2 are the key regulators of hepatocarcinoma cell invasion across the fibrotic matrix microenvironment. Cancer Res. 2003;63:8312–8317. [PubMed] [Google Scholar]
- 31.Zhang X and Reed EF. HLA class I: an unexpected role in integrin β4 signaling in endothelial cells. Hum Immunol. 2012;73:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Valenzuela NM, Reed EF. HLA class I antibody-mediated endothelial and smooth muscle cell activation. Curr Opin Organ Transplant. 2012;17:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwamura T, Hollingsworth MA. Chapter 5 Pancreatic Tumors In: Masters JRW, Palsson, editors. Human Cell Culture, Vol. 1 Kluwer Academic Publishers; 1999. p. 107–122. [Google Scholar]
- 34.Taniguchi S, Iwamura T, Katsuki T. Correlation between spontaneous metastatic potential and type I collagenolytic activity in a human pancreatic cancer cell line (SUIT-2) and sublines. Clin Exp Metastasis. 1992;10:259–266. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi S, Iwamura T, Kitamura N, Yamanari H, Setoguchi T. Heterogeneities of attachment, chemotaxis, and protease production among clones with different metastatic potentials from a human pancreatic cancer cell line. Clin Exp Metastasis. 1994;12:238–244. [DOI] [PubMed] [Google Scholar]
- 36.Yamanari H, Suganuma T, Iwamura T, Kitamura N, Taniguchi S, Setoguchi T. Extracellular matrix components regulating glandular differentiation and the formation of basal lamina of a human pancreatic cancer cell line in vitro. Exp Cell Res. 1994;211:175–182. [DOI] [PubMed] [Google Scholar]
- 37.Burdick MD, Harris A, Reid CJ, Iwamura T, Hollingsworth MA. Oligosaccharides expressed on MUC1 produced by pancreatic and colon tumor cell lines. J Biol Chem. 1997;272:24198–24202. [DOI] [PubMed] [Google Scholar]
- 38.Nozawa F, Hirota M, Okabe A, Shibata M, Iwamura T, Haga Y, Ogawa M. Elastase activity enhances the adhesion of neutrophil and cancer cells to vascular endothelial cells. J Surg Res. 2000;94:153–158. [DOI] [PubMed] [Google Scholar]
- 39.Satoh S, Hinoda Y, Hayashi T, Burdick MD, Imai K, Hollingsworth MA. Enhancement of metastatic properties of pancreatic cancer cells by MUC1 gene encoding an anti-adhesion molecule. Int J Cancer. 2000;88:507–518. [DOI] [PubMed] [Google Scholar]
- 40.McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, Hayashi T, Imai K, Hollingsworth MA. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int J Cancer. 2001;94:783–791. [DOI] [PubMed] [Google Scholar]
- 41.Kohlgraft KG, Gawron AJ, Higashi M, Meza JL, Burdick MD, Kitajima S, Kelly DL, Caffrey TC, Hollingsworth MA. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63:5011–5020. [PubMed] [Google Scholar]
- 42.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278:38029–38039. [DOI] [PubMed] [Google Scholar]
- 43.Hennig R, Ventura J, Segersvard R, Ward E, Ding XZ, Rao SM, Jovanovic BD, Iwamura T, Talamonti MS, Bell RH Jr, Adrian TE. LY293111 improves efficacy of gemcitabine therapy on pancreatic cancer in a fluorescent orthotopic model in athymic mice. Neoplasia. 2005;7:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, Goto M, Yonezawa S, Hollingsworth MA. RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res. 2006 2006;12:2976–2987. [DOI] [PubMed] [Google Scholar]
- 45.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, Gendler SJ, Hollingsworth MA. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. [DOI] [PubMed] [Google Scholar]
- 46.Melstrom LG, Salabat MR, Ding XZ, Milam BM, Strouch M, Pelling JC, Bentrem DJ. Apigenin inhibits the GLUT-1 glucose transporter and the phosphoinositide 3-kinase/Akt pathway in human pancreatic cancer cells. Pancreas. 2008;37:426–431. [DOI] [PubMed] [Google Scholar]
- 47.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler SJ, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuli A, Sharma M, Capek HL, Naslavsky N, Caplan S, Solheim JC. Mechanism for amyloid precursor-like protein 2 enhancement of major histocompatibility complex class I molecule degradation. J Biol Chem. 2009;284:34296–34307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mujumdar N, Mackenzie TN, Dudeja V, Chugh R, Antonoff MB, Borja-Cacho D, Sangwan V, Dawra R, Vickers SM, Saluja AK. Triptolide induces cell death in pancreatic cancer cells by apoptotoic and autophagic pathways. Gastroenterology. 2010;139:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, Subramanian S, Saluja AK. Triptolide induces the expression of miR-142–3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12:1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nomura A, McGinn O, Dudeja V, Sangwan V, Saluja AK, Banerjee S. Minnelide effectively eliminates CD133(+) side population in pancreatic cancer. Mol Cancer. 2015;14:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey P, Rachagani S, Das S, Seshacharyulu P, Sheinin Y, Naslavsky N, Pan Z, Smith BL, Peters HL, Radhakrishnan P, et al. Amyloid precursor-like protein 2 (APLP2) affects the actin cytoskeleton and increases pancreatic cancer growth and metastasis. Oncotarget. 2015;6:2064–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakraborty A, Dorsett KA, Trummell HQ, Yang ES, Oliver PG, Bonner JA, Buchsbaum DJ, Bellis SL. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J Biol Chem. 2018;293:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieber M, Mazzetta J, Nelson-Rees W, Kaplan M, Todaro G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int J Cancer 1975;15:741–747. [DOI] [PubMed] [Google Scholar]
- 55.Yunis AA, Arimura GK, Russin DJ. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. Int J Cancer 1977;19:128–135. [DOI] [PubMed] [Google Scholar]
- 56.Carreno BM, Hansen TH. Exogenous peptide ligand influences the expression and half-life of free HLA class I heavy chains ubiquitously detected at the cell surface. Eur J Immunol. 1994;24:1285–1292. [DOI] [PubMed] [Google Scholar]
- 57.Stam NJ, Vroom TM, Peters PJ, Pastoors EB, Pleogh HL. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int Immunol. 1990;2:113–125. [DOI] [PubMed] [Google Scholar]
- 58.Parsons JT, Martin KH, Slack JK, Taylor JM, Weed SA. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene 2000;19:5606–5613. [DOI] [PubMed] [Google Scholar]
- 59.Cobb BS, Schaller MD, Leu TH, Parsons JT. Stable association of pp60src and pp59fyn with the focal adhesion-associated protein tyrosince kinase, pp125FAK. Mol Cell Biol. 1994;14:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol Cell Biol. 1994;14:1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen HC, Appeddu PA, Isoda H, Guan JL. Phosphorylation of tyrosince 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. J Biol Chem. 1996;271:26329–26334. [DOI] [PubMed] [Google Scholar]
- 62.Miyazaki T, Kato H, Nakajima M, Sohda M, Fukai Y, Masuda N, Manda R, Fukuchi M, Tsukada K, Kuwano H. FAK overexpression is correlated with tumor invasiveness and lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer. 2003;89:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Itoh S, Maeda T, Shimada M, Aishima S, Shirabe K, Tanaka S, Maehara Y. Role of expression of focal adhesion kinase in progression of hepatocellular carcinoma. Clin Cancer Res. 2004;10:2812–2817. [DOI] [PubMed] [Google Scholar]
- 64.Furuyama K, Doi R, Mori T, Toyoda E, Ito D, Kami K, Koizumi M, Kida A, Kawaguchi Y, Fujimoto K. Clinical significance of focal adhesion kinase in resectable pancreatic cancer. World Journal Surgery. 2006;30:219–226. [DOI] [PubMed] [Google Scholar]
- 65.Kanteti R, Batra SK, Lennon FE, Salgia R. FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget. 2016;7:31586–31601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang H, Hegde S, Knolhoff BL, Zhu Y, Herndon JM, Meyer MM, Nywening TM, Hawkins WG, Shapiro IM, Weaver DT, Pachter JA, Wang-Gillam A, DeNardo DG. Targeting focal adhesion kinase renders pancreatic cancer responsive to checkpoint immunotherapy. Nature Med. 2016;22:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scupoli MT, Sartoris S, Tosi G, Ennas MG, Nicolis M, Cestari T, Zamboni G, Martignoni G, Lemoine NR, Scarpa A, Accolla RS. Expression of MHC class I and class II antigens in pancreatic adenocarcinomas. Tissue Antigens. 1996;48:310–311. [DOI] [PubMed] [Google Scholar]
- 68.Ryschich E, Cebotari O, Fabian OV, Autschbach F, Kleeff J, Friess H, Bierhaus A, Buchler MW, Schmidt J. Loss of heterozygosity in the HLA class I region in human pancreatic cancer. Tissue Antigens. 2004;64:696–702. [DOI] [PubMed] [Google Scholar]
- 69.Ryschich E, Notzel T, Hinz U, Autschbach F, Ferguson J, Simon I, Weitz J, Frohlich B, Klar E, Buchler MW, Schmidt J. Control of T-Cell-Mediated immune response by HLA class I in human pancreatic carcinoma. Clin Cancer Res. 2005;11:498–504. [PubMed] [Google Scholar]
- 70.Pandha H, Rigg A, John J, Lemoine N. Loss of expression of antigen-presenting molecules in human pancreatic cancer and pancreatic cancer cell lines. Clin Exper Immunol. 2006;148:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu M, Zhang Y, Yang J, Cui X, Zhou Z, Zhan H, Ding K, Tian X, Yang Z, Fung KA, Edil BH, Postier RG, Bronze MS, Fernandez-Zapico ME, Stemmler MP, brabletz T, Li YP, Houchen CW, Li M. ZIP4 increases expression of transcription factor ZEB1 to promote integrin α3β1signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology. 2019. pii:S0016–5085(19)41529–1 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cruz-Monserrate Z, O’Connor KL. Integrin alpha 6 beta 4 promotes migration, invasion through Tiam1 upregulation, and subsequent Rac activation. Neoplasia. 2008;10:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sawai H, Funahashi H, Yamamoto M, Okada Y, Hayakawa T, Tanaka M, Takeyama H, Manabe T. Interleukin-1alpha enhances integrin alpha(6)beta(1) expression and metastatic capability of human pancreatic cancer. Oncology. 2003;65:167–173. [DOI] [PubMed] [Google Scholar]
- 74.Campbell EC, Fettke F, Bhat S, Morley KD, Powis SJ. Expression of MHC class I dimers and ERAP1 in an ankylosing spondylitis patient cohort. Immunology. 2011;133:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell EC, Antoniou AN, Powis SJ. The multi-faceted nature of HLA class I dimer molecules. Immunology. 2012;106:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lenart I, Guiliano DB, Burn G, Campbell EC, Morley KD, Fussell H, Powis SJ, Antoniou AN. The MHC class I heavy chain structurally conserved cysteines 101 and 164 participate in HLA-B27 dimer formation. Antioxid Redox Signal. 2012;16:33–43. [DOI] [PubMed] [Google Scholar]
- 77.Frietze KK, Pappy II AL, Melson JW, O’Driscoll EE, Tyler CM, Perman DH, Boulanger LM. Cryptic protein-protein interaction motifs in the cytoplasmic domain of MHCI proteins. BMC Immunol. 2016;17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee H-J, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Communication Signaling. 2010;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]


