Abstract
Mesenchymal stem or stromal cells are currently under clinical investigation for multiple diseases. While their mechanism of action is still not fully elucidated, vesicles secreted by MSCs are believed to recapitulate their therapeutic potentials to some extent. Microvesicles (MVs), also called as microparticles or ectosome, are among secreted vesicles that could transfer cytoplasmic cargo, including RNA and proteins, from emitting (source) cells to recipient cells. Given the importance of MVs, we here attempted to establish a method to isolate and characterize MVs secreted from unmodified human bone marrow derived MSCs (referred to as native MSCs, and their microvesicles as Native-MVs) and IFNγ stimulated MSCs (referred to as IFNγ-MSCs, and their microvesicles as IFNγ-MVs). We first describe an ultracentrifugation technique to isolate MVs from the conditioned cell culture media of MSCs. Next, we describe characterization and quality control steps to analyze the protein and RNA content of MVs. Finally, we examined the potential of MVs to exert immunomodulatory effects through induction of regulatory T cells (Tregs). Secretory vesicles from MSCs are promising alternatives for cell therapy with applications in drug delivery, regenerative medicine, and immunotherapy.
Keywords: Mesenchymal Stem Cells (MSCs), cell-free therapy, extracellular vesicles (EVs), drug delivery, microvesicles (MVs)
1. Introduction
The promise of cell-free-therapy has led to the development of products derived from stem cells. Mesenchymal stem or stromal cells (MSCs), in particular, have been the subject of hundreds of clinical trials to date [1]. While the mechanism of action of MSCs is currently debated, some studies suggest that MSCs secrete extracellular vesicles (EVs) that may partly mediate their therapeutic efficacy [2–7]. Inspired by this mechanism, MSC derived EVs are now under clinical investigation to treat macular holes, Type 1 Diabetes, and acute ischemic stroke [8].
EVs are packages of protein, lipids, and RNAs, and are classified into three main groups, i.e. exosomes, microvesicles (MVs), and apoptotic bodies [9]. MVs are currently considered to be distinct from other EVs such as exosomes that are released partly upon exocytosis of multivesicular bodies [10, 11]. Recent studies indicate that shedding vesicles contribute to key biological processes, including membrane trafficking and horizontal transfer of cargos (RNA and proteins [12]) to recipient target cells [13]. The mechanisms involved in biogenesis of MVs remain to be fully elucidated, and some reports suggest the involvement of cholesterol-rich micro domains of plasma membrane in biogenesis of these vesicles [14, 15].
Upon release from their cell of origin, MVs may not interact randomly with cells, but rather they may interact with specific ligands on particular cell types, thereby conferring some specificity to intercellular targeting. Once MVs fuse with or are taken up by the recipient target cells, they can transfer membrane components, such as ligands and receptors, as well as different types of RNAs, such as mRNA, rRNA, snoRNA, miRNA and lincRNA. Some reports have found the exchange of RNAs between glioblastoma and endothelial cells through EVs [16]. Through such transferring capabilities, MVs have been shown to be involved in inflammation [17], blood coagulation [18] and tumor progression [19]. MVs derived from MSCs have also been exploited for their potential therapeutic uses. For instance, it has been demonstrated that recovery from acute tubular injury (ATI) in kidneys by MSCs administration is mediated by their secretory MVs [2]. In particular, MVs derived from human bone marrow MSCs activated a proliferative and anti-apoptotic program in tubular epithelial cells that survived injury [3]. MSCs derived MVs have further been implicated in treatment of severe pneumonia [20], osteoarthritis [21], and renal ischemia-reperfusion injury after cardiac death renal transplantation [22]. Similar therapeutic efficacy has been reported from other MSC sources. For example, Chen et al. have shown than MVs secreted from human Wharton’s Jelly MSCs could ameliorate acute lung injury in rat models through hepatocyte growth factor [4]. Another major area of application of EVs is as immune therapy due to their anti-inflammatory and immunomodulatory effects. In preclinical settings, MSC-derived EVs have shown immunomodulatory effects in inflammatory or autoimmune disorders such as uveoretinitis [23], graft-versus-host disease [24], and type 1 diabetes [23, 25]. We recently reported that MSC-derived exosomes could improve clinical scores and decrease neuroinflammation in treating multiple sclerosis using an experimental autoimmune encephalomyelitis (EAE) mouse model through their immunomodulatory RNA and protein cargos (e.g. indoleamine 2,3-dioxygenase (IDO)) and induction of regulatory T cells (Tregs) [7].
There are multiple established methods to isolate MVs including ultracentrifugation [26], density gradient centrifugation [3], hydrostatic filtration dialysis [27], size exclusion chromatography [28], and affinity based capturing techniques [29, 30]. A major limitation for the clinical translation of MVs is due to the difficulties and inconsistencies in their isolation. In particular, MVs should ideally be isolated in a timely and cost-effective way, and with the required quality and in a scalable fashion. Therefore, a scalable methodology that provides rapid isolation of pure MVs from cells could greatly facilitate MVs clinical translation.
Here, we attempted to optimize the isolation of MVs using the ultracentrifugation technique. Ultracentrifugation is currently a gold standard in the area of EV isolation and purification. This technique allows robust MV isolation, facile purification, and production of a high yield of mostly intact EVs. We further used different methods to characterize MV contents and proposed a method to characterize their immunomodulatory potential. We characterized the size and shape, specific markers, protein and RNA content of MVs, and finally assayed their immune potency in the induction of Tregs through in vitro co-cultures of MVs and mouse splenocytes. This work could aid future clinical translation of therapeutic use of MSCs derived MVs in a variety of diseases including treating autoimmune disorders.
2. Materials and methods
Cells preparation, culture and IFNγ activation
Human bone marrow-derived MSCs were obtained from Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine. αMEM (Gibco, CAT#: 12571048) supplemented with 4 mM L-glutamine, 1% penicillin streptomycin and 15% fetal bovine serum (FBS, Premium Grade VWR CAT#: 60150995) was used to culture MSCs and cells were passaged < 5. To stimulate MSCs, 10 ng/mL of human IFNγ (PeproTech Inc., USA, CAT#: 30002) was supplemented in the media for 48 h. For MV collection, 80% confluent cells were cultured with complete media supplemented with 15% vesicles-depleted FBS for 3 days. To deplete extracellular vesicles from FBS, it was centrifuged at 120,000 × g for 18 h using Ti45 fixed angle rotor, which is previously shown to be effective in removal of functional FBS vesicles [7, 31].
Microvesicle and Exosome Isolation
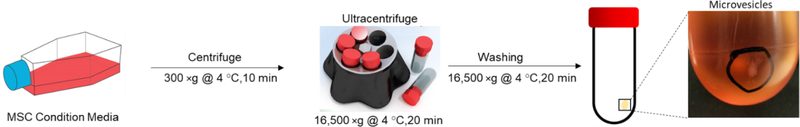
The isolation procedure is summarized schematically in Figure 1. Briefly, conditioned media from MSCs was first centrifuged at 300 ×g for 30 min at 4 °C to remove whole cells and large debris. The supernatants were then centrifuged at 16,500 ×g for 20 mins using Ti45 fixed angle rotor at 4 °C to collect the MV fraction. The obtained pellet was washed once using PBS at 16,500 ×g for 20 mins at 4 °C and subsequently resuspended in PBS and frozen at −80 °C. The remaining supernatant was then centrifuged at 120,000 ×g for 2.5 h at 4 °C to collect the exosome fraction according to our previous study [7].
Figure 1. Schematic Presentation for the Isolation Process of MSC-Derived MVs.
Condition media harvested from MSCs is first centrifuged at 300 ×g for 10 minutes. Next, the supernatant is ultracentrifuged at 16,500 ×g for 20 minutes following by a washing step. Finally, the MVs pellet is washed and resuspended in PBS and kept at −80 °C.
Microvesicle Characterization
Western blotting
MVs were homogenized with 1X RIPA buffer (Cell signaling technologies, USA, CAT#: PI89900) and underwent three 5 min consecutive sonication with vortexing in between. Protein contents were analyzed using a BCA protein assay kit (Thermo Scientific Pierce, Rockford, IL, USA). Briefly, 25 μL of sample (or 25 μL BSA standard) was added to 200 μL of working reagent (50:1 ratio of assay reagents A and B). The plate was incubated at 37 °C for 30 min and then analyzed using a SpectraMax 384 Plus spectrophotometer with the SoftMax Pro software (Molecular Devices, 1311 Orleans Drive, Sunnyvale, CA, USA). Whole protein content was then loaded onto a polyacrylamide gel (Mini-PROTEAN; Bio-Rad, USA). The separated proteins were then transferred onto a nitrocellulose membrane following by blocking via 5% Non-Fat Dry Milk (Bio-Rad Laboratories) in Tris-buffer saline (TBS) for 2 h at 4 °C. The membrane was next incubated with primary antibodies with the concentrations as suggested by the manufacturer: Calnexin (clone H-70, Santa Cruz Biotechnology, Santa Cruz, CA, USA), TSG101 (clone 4A10, Abcam, Cambridge, UK), or CD81 (clone H-121, Santa Cruz Biotechnology) in 0.25% blotting grade Blocker Non-Fat Dry Milk in TBS-Tween (TBST) overnight at 4 °C. The membrane was subsequently washed thrice with TBST for 10 min each. Secondary antibodies (for calnexin and CD81: ECL anti-rabbit IgG horseradish peroxidase-linked F(ab′)2 fragment (donkey, antirabbit); and for TSG101; ECL anti-mouse IgG horseradish peroxidase-linked F(ab′)2 fragment (sheep, anti-mouse); (GE Healthcare, Buckinghamshire, UK)) diluted in 0.25% blotting grade Blocker Non-Fat Dry Milk in TBST were incubated with the membrane for 2 h. Finally, the membranes were analyzed with ECL Prime Western Blotting Detection (GE Healthcare) and a VersaDoc 4000 MP (Bio-Rad Laboratories).
Flow Cytometry
MV solutions (15 μg, based on total protein) were incubated with 7 μL of anti-CD63-modified magnetic beads (lot OK527, Life Technologies AS, Oslo, Norway) overnight with rocking at 4 °C. The beads were washed with PBS, centrifuged at 500 ×g for 5 min, and then incubated with human IgG (Sigma-Aldrich) to block nonspecific binding for 15 min at 4 °C. Beads were then incubated with PElabeled antibodies against CD9, CD63, and CD81 or isotype control (BD Bioscience, Erembodegem, Belgium) for 40 min with gentle agitation. After another washing step and centrifugation as above, the samples were analyzed using a FACSAria (BD Bioscience), and data were processed using FlowJo Software (Tri Star, Ashland, OR, USA).
For flow cytometry experiments on splenocytes, spleens of “Foxp3-eGFP” mice (B6.CgFoxp3tm2(EGFP)Tch/J, The Jackson Laboratory) were removed and kept in ice cold RPMI supplemented with 10% heat inactivated FBS and 1% penicillin/streptomycin. Cells were then isolated by homogenization using a cell strainer, which were then centrifuged at 300 ×g for 5 min, following by 10 mins RBC lysis with ACK buffer. For Treg induction assay, 8 to 12-week-old mice from same strain were sacrificed using CO2 asphyxiation followed by cervical dislocation. Cells were isolated from spleens as described above and 106 splenocyte suspensions per ml were prepared and stimulated with anti-CD3 clone 145–2c11 (1 μg/mL) and IL-2 (25 ng/mL) without or with 10 ng/ml TGFβ1 (Biolegend, USA). The splenocytes were then co-cultured with MVs (0, 0.2, 2, or 20 μg, based on total protein) isolated from native or IFNγ-stimulated MSCs for 4 d. Cells were then harvested and stained for flow cytometry analysis of CD4+CD25+FoxP3+ T cells and CD8+CD25+FoxP3+ T cells.
Electron Microscopy
MVs (15 μg total protein) were first dissolved in PBS and loaded onto UV-treated formvar carbon-coated grids (Ted Pella Inc., Redding, CA, USA). Samples were then fixed in 2% electron microscopy grade paraformaldehyde before being immunostained with anti-CD63 antibody (BD Bioscience, Erembodegem, Belgium) or isotype control (Sigma-Aldrich, St Louis, MO, USA), followed by staining with a 10 nm gold-labelled secondary antibody (Sigma-Aldrich). The samples were then subsequently fixed with 2.5% glutaraldehyde and stained with 2% uranyl acetate. Grids were inserted in a LEO 912AB Omega electron microscope (Carl Zeiss NTS, Jena, Germany) and imaging performed at 70 kV.
Nanoparticle Tracking Analysis
MVs were further visualized using Nanosight system (Malvern instruments, USA) similar to our previously published method [7]. MVs were visualized by light scattering using a conventional optical microscope with perpendicular alignment to the beam, collecting light scattered from every particle within the field of view. A 60 s video recorded all events for further analysis by nanoparticle tracking analysis (NTA) software, where the Brownian motion of each particle was tracked between frames, leading to calculation of the size via Stokes-Einstein equation.
RNA Sequencing
RNAs of MVs were isolated according to a previously published method [32]. Briefly, MVs (100 μg total protein content) were re-suspended in 500 μl of lysis buffer (with 1% 2-mercaptoethanol added; miRCURY RNA isolation kit, Exiqon) prior to addition of 200 μl ethanol (95%). RNA was then isolated according to the protocol by the manufacturer. Following by RNA isolation, we created libraries for small and long RNA. We employed Illumina HiSeq 2500 for the sequencing of both the whole transcriptome (WT) and small-RNA to a read depth of approximately 50 million for the long RNA and 10 million for the small RNA according to our previous protocol [33]. Briefly, the.bcl files from the Illumina HiSeq 2500, which are raw sequence images, were converted to fastq format. We double-checked that the quality scores do not deteriorate appreciably at the read ends. For the long RNA, the fastqs were kept untrimmed, however, small-RNA were trimmed to remove the adapters. The fastqs were aligned to the reference human genome (hg19, Ensembl version 75) using Bowtie for the small-RNA and STAR v2.3.0 for the long-RNA. The STAR/Bowtie aligned.sam files were converted to .bam files. These .bam files were then sorted by coordinates using SAMtools v0.1.19.
For WT sequencing, the read counts and Fragments Per Kilobase of transcript per Million (FPKMs) mapped for genes were obtained using the hg19 gene reference from Ensembl 75. The read counts were generated using htseq-count (intersection non-empty mode) and FPKMs were produced using Cufflinks v2.2.1. A total library normalization was performed by scaling the sample libraries to the total number of reads mapping to the reference human genome. TPMs for each gene i was calculated by the following formula: TPMi= FPKMi/∑FPKMi.
For small-RNA sequencing, the samples were processed using sRNAbench, which allows mapping the reads to several RNA libraries using Bowtie. The reads were first mapped to the miRBase v21 database. Next, the reads simultaneously and competitively mapped to the other RNA libraries, i.e., rRNA (ribosomal RNA from NCBI), Mt_tRNA (Mitochondrial tRNA from Ensembl 75), tRNA (transfer RNA from UCSC http://gtrnadb.ucsc.edu/Hsapi19/hg19-tRNAs.fa), piRNA (piRBase v1.0), snoRNA (small nucleolar RNA from Ensembl 75), snRNA (small nuclear RNA from Ensembl 75), Vault RNA (Ensembl 75) and Y RNA (Ensembl 75). The results were then compiled and presented as pie-charts.
Proteomics
Samples were dissolved by addition of 20% Sodium dodecyl sulfate (SDS), 1 M Triethylammoinium bicarbonate (TEAB) (Fluka, Sigma Aldrich) and water to give a final volume between 120 and 450 μL and a concentration of 2 % SDS in 25 mM TEAB. Total protein concentration was determined with Pierce™ BCA Protein Assay (Thermo Scientific). The proteins (50 μg) were reduced by addition of 2 M DL-Dithiothreitol (DTT) to a final concentration of 100 mM and incubated at 60oC for 30 min. Trypsin digestion were performed using the filter-aided sample preparation (FASP) method modified from [34]. In short, reduced samples diluted with 400 μl 8M urea were applied on Nanosep 10k Omega filters (Pall Life Sciences) and 8M urea were used to repeatedly wash away the SDS. Alkylation was performed with methyl methane thiosulfonate (MMTS) diluted in digestion buffer (1% sodium deoxycholate (SDC) in 50 mM TEAB) and the filters were repeatedly washed with digestion buffer. Trypsin (Pierce Trypsin Protease, MS Grade, Thermo Fisher Scientific) in a ratio of 1: 100 relative to protein amount was added with digestion buffer to a pH of about 8 and the samples were incubated in 37°C over night. Another portion of trypsin were added and incubated for three hours in 37°C. The peptides were collected by centrifugation. The samples were acidified with Trifluoroacetic acid (TFA) to pH about 2 to precipitate SDC prior to desalting using PepClean C18 spin columns (Thermo Fisher Scientific) according to the manufacturer’s guidelines.
LC-MS/MS Analysis and database search
The dried desalted samples were reconstituted with 15 μl of 0.1 % formic acid (Sigma Aldrich) in 3% acetonitrile and analyzed on an Orbitrap Fusion Tribrid and a Q Exactive mass spectrometer interfaced to an Easy-nLC II (Thermo Fisher Scientific).
Peptides (2 μL injection volume) were separated using an in-house constructed analytical column (20×0.075 mm I.D.) packed with 3 μm Reprosil-Pur C18-AQ particles (Dr. Maisch, Germany). Solvent A was 0.2% formic acid in water and solvent B was 0.2% formic acid in acetonitrile. The following gradient was run at 200 nL/min; 7–27 % B over 75 min, 27–80 % B over 5 min, with a final hold at 80 % B for 10 min. Ions were injected into the Orbitrap Fusion Tribrid mass spectrometer under a spray voltage of 1.6 kV in positive ion mode. MS scans were performed at 60 000 resolution (at m/z 200), an AGC-target value of 4e5 with a mass range of m/z 400–1500. MS/MS analysis was performed in a data-dependent mode with top speed cycle of 3s for the most intense doubly or multiply charged precursor ions. Ions in each MS scan over threshold 10000 were selected for fragmentation (MS2) by collision induced dissociation (CID) for identification at 35% and detection in the ion trap. Precursors were isolated in the quadrupole with a 1.6 m/z window, an AGC-target value of 1e4 and dynamic exclusion within 10 ppm during 5 seconds was used for m/z-values already selected for fragmentation.
Samples analyzed for a second time on the QExactive mass spectrometer under a spray voltage of 1.8 kV in positive ion mode. MS scans were performed at 70 000 resolution (at m/z 200), an AGC-target value of 1e6 with a mass range of m/z 400–1600. MS/MS analysis was performed with the top ten most abundant doubly or multiply charged precursor ions in each MS scan selected for MS2 by stepped high energy collision dissociation (HCD) of NCE-value of 30. Ions over threshold 1.1e4 were selected for MS2 with a 1.2 m/z window, an AGC-target value of 1e5 and dynamic exclusion within 10 ppm during 5 seconds was used for m/z-values already selected for fragmentation. Analyses were performed with exclusion lists generated in previous MS analysis with a five minutes retention time window for each sample.
MS raw data files for each sample were merged in the search against the Homo sapiens Swissprot Database version 2014_02 (Swiss Institute of Bioinformatics, Switzerland) using Proteome Discoverer version 1.4 (Thermo Fisher Scientific) with the Mascot search engine (Matrix Science). The MS peptide tolerance was set to 5 ppm and MS/MS tolerance for to identification of 500 millimass units (mmu). Tryptic peptides were accepted with one missed cleavage and variable modifications of methionine oxidation and cysteine alkylation. The detected peptide threshold in the software was set to 1% False Discovery Rate by searching against a reversed database and identified proteins were grouped by sharing the same sequences to minimize redundancy.
3. Results and Discussion
Isolation and characterization of Microvesicles
MSCs derived from bone marrow were cultured for 3 days with or without IFNγ stimulation, and conditioned media were collected. To isolate MVs, collected conditioned media were initially centrifuged at 300 ×g for 10 minutes to remove dead cells and debris. The conditioned media were then centrifuged at 16,500 ×g for 20 min to harvest MVs as schematically shown in Figure 1. Pellet was then washed with sterile PBS (at 16,500 ×g for 20 min) and resuspended in PBS and stored at −80 °C. Next, MVs were characterized using flow cytometry, western blotting, and nanoparticle tracking analysis (NTA).
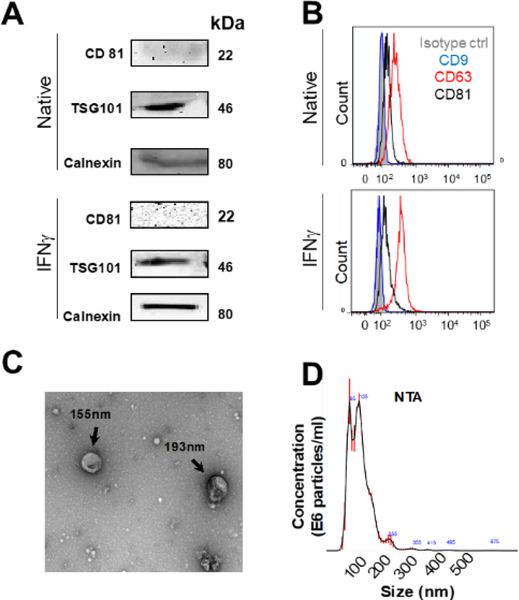
We used IFNγ to stimulate the MSCs as evidences suggest that it enhances their immunosuppressive properties [7]. Both MVs from native cells (Native-MVs) and from 10 ng/mL IFNγ stimulated MSCs (IFNγ-MVs) were characterized. An exosomal marker, CD81 [7, 35], was minimally expressed in MVs while tumor susceptibility gene 101 protein (TSG101) was present in both IFNγ-MVs and Native-MVs based on Western blotting (Figure 2A). These observations are consistent with previous reports in the literature [36]. In addition, the presence of the calnexin bands (an endoplasmic reticulum marker) suggests that at least some portion of MSCs MVs are endoplasmic reticulum-derived vesicles (Figure 2A). Minimal expression of CD81 and presence of calnexin further suggest that our method could separate populations of MVs and exosomes [7, 35]. On the surface of CD63-captured MVs, the tetraspanins CD63 was expressed (positive control) while CD81 and CD9 were minimally expressed and undetectable, respectively, based on flow cytometry analysis (Figure 2B). Similar results were observed for IFNγ-MVs (Figure 2A and Figure 2B). Because MVs are too small to be directly analyzed using standard flow cytometry technique, this CD63 bead capture method is widely used to analyze EV surface markers using flow cytometry. However, a limitation of this method is that CD63 negative EVs would be excluded in this analysis. Based on the ISEV 2018 published guidance for characterization of extracellular vesicles, presence of 3 markers and absence of one putative contaminant are minimally required to characterize EVs [9]. Here, we have provided five markers (i.e. CD9, CD63, CD81, TSG101, and Calnexin). We recently performed similar analyses for MSC derived exosomes [7]. Western blotting results of MSC derived exosomes demonstrated the presence of CD63, CD81 and TSG101, while the calnexin marker was absent. Comparing the Western blotting results from MSC derived MVs (the present study) and MSC derived exosomes [7] suggest that calnexin and CD81 may potentially be used to distinguish between exosomes and MVs.
Figure 2. Characterization of MSC-Derived MVs.
A) Western blotting of MSC-derived MVs for common EV markers i.e. CD81, TSG101 and Calnexin. B) Flow cytometry analysis of CD9, CD63 and CD81 expression on MSC derived MVs captured onto anti-CD63-coated beads. C) Transmission electron microscopy image of MSC MVs. D) Representation of particle size (nm) and concentration (particles/ml) analyzed using nanoparticle tracking analysis (NTA).
We next attempted to visualize MVs and characterize their size, size distribution, morphology, and count. TEM revealed round shaped vesicles within the size range (~150 nm) of MVs (Figure 2C), which is in agreement with other published literature on the size of MVs [4, 10]. MVs were further characterized by NTA (Figure 2D), showing that the MV quantity was 6.15×108 ± 9.7×106 MVs (equivalent to 41.5 ± 10.5 μg of total protein; isolated from 2.1–3.3 ×108 MSCs) with size is in the range of 147 ± 76 nm (Figure 2D). Using same isolation procedure, IFNγ-MV quantity was 1.31×109 ± 6.7×106 MVs (equivalent to 47.7 ± 21.9 μg of total protein; isolated from 2.1–3.3 ×108 IFNγ-MSCs) with the size range of 134 ± 46 nm (data not shown). Together, these data indicated the reliability of our method for isolation of MVs from MSCs conditioned media. While we attempted to isolate relatively pure MVs, it should be noted that complete separation of pure MVs from other fractions of EVs (e.g., exosomes) can be difficult via centrifugation techniques [37].
Deep RNA sequencing revealed that microvesicles contain different types of coding and non-coding RNAs
Both coding and non-coding RNAs have extensive implications in therapeutic designs, however, efficient delivery of RNA is still a major hurdle [38, 39]. Encapsulation of mRNA within EVs lipoprotein membrane offers a promising delivery platform [7, 19, 40]. Coding RNAs, could be translated into functional and therapeutic proteins upon translation within the recipient cells. These RNA classes could offer safety benefits as they do not modify the genome. In addition, non-coding RNAs can play regulatory roles and downregulate mRNA translation within the cell [41].
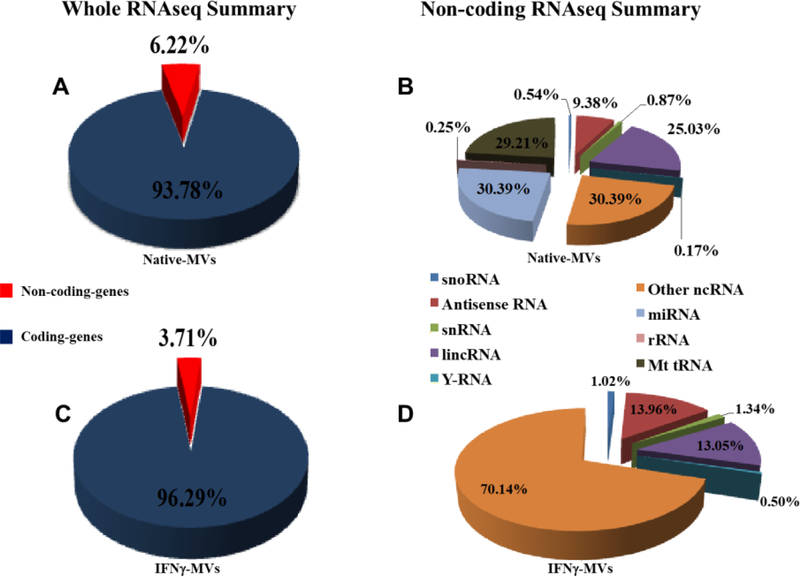
To identify the RNA content of the MVs and investigate the effect of IFNγ stimulation on RNA profile of MV, we used RNAseq technique. The quality and concentration of the RNA was examined using a nano-chip on a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Illumina HiSeq 2500 next generation sequencing platform was used for all sequencing experiments. Normalized counts of RNA show that both Native-and IFNγ-MV are enriched in coding and non-coding RNAs (Figure 3 A, C). Of note, the coding RNA portion was 93.78% and 96.29% for Native-and IFNγ-MVs, respectively. The remaining portion of RNA was non-coding, and due to important roles of noncoding RNAs in regulation of gene expression, we further analyzed the non-coding RNAs. We found that the non-coding portion contains many small regulatory RNAs including miRNAs, small (or short) interfering (siRNAs), long non-coding lincRNAs and others for Native- (Figure 3 B) and IFNγ-MVs (Figure 3 D). These observations reveal that similar to exosomal cargo [7], the MV cargo composition could be tailored through stimulation of the parental cells. In particular, IFNγ is a known cytokine that is involved in various processes of the immune response and has been widely used to promote immunosuppressive effects of MSCs through different transcriptional mechanisms [42]. IFNγ activation is also believed to change the cargo loading (both RNA and protein) into EVs during their biogenesis [43]. Our group previously observed the effect of IFNγ activation of MSC on differential non-coding RNA loading in exosomes [7]. Other groups have also reported the effect of pro-inflammatory mediators in production of potent anti-inflammatory extracellular vesicles [43, 44]. Furthermore, analysis of the miRNAs of MV showed that they have distinct profiles of miRNAs compared to exosomes [7], demonstrating that there might be a selective pathway to load particular miRNAs into MVs vs other EV fractions.
Figure 3. RNA summary of MVs.
A) Whole RNA sequence of Native-MVs showing coding and non-coding percentages. B) Non-coding RNA sequence summary (percentage of different types of non-coding RNAs) of Native-MVs. C) Whole RNA sequence of IFNγ-MVs showing coding and non-coding percentage. D) Non-coding RNA sequence summary (percentage of different types of non-coding RNAs) of IFNγ-MVs.
Proteomics
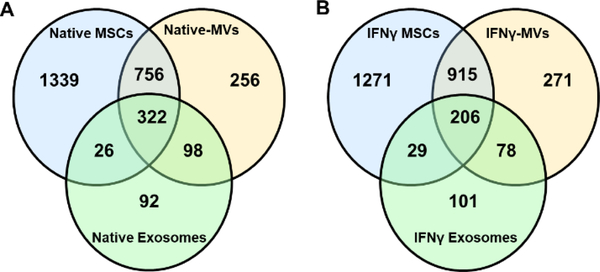
We then sought to characterize the whole protein signature of MVs and compare it with their parental MSCs as well as exosomes secreted from MSCs. We also attempted to find out the effect of IFNγ stimulation on the protein content of MVs. Comparison between Native-MVs with its parental MSCs reveals that 254 proteins are enriched in MVs while they have 756 common proteins with MSCs. Native-MVs share 98 proteins with native exosomes (Figure 3A). Moreover, 271 proteins are enriched in IFNγ-MVs while they share 915 and 78 proteins with IFNγ MSCs and IFNγ exosomes, respectively (Figure 3A). Interestingly, MVs, MSCs, and exosomes share 322 similar proteins, and this number is 206 for the IFNγ stimulated ones. This observation suggests differential enrichment of proteins in MV populations compared to their MSCs and exosome counterparts. This differential protein enrichment could be partly associated with the protein content of parent cells before and after IFNγ treatment. The effect of IFNγ stimulation on MSC protein expression has been described previously [45–47]. Proteomic analysis of IFNγ stimulated MSCs have shown to alter the protein landscape of MSCs into more immunosuppressive content [48]. For instance, surface proteins including major histocompatibility complex II (MHCII) and programmed death-ligand 1 (PD-L1) [7] and secreted proteins including IDO are upregulated for MSCs treated with IFNγ [49]. The biogenesis of EVs is either through endosomal budding or plasma membrane shedding [50]. During their biogenesis, EVs get enriched with transmembrane and intracellular proteins from the parent cells. Therefore, it is expected that protein content of stimulated vs. native MVs to be different.
Functional assay
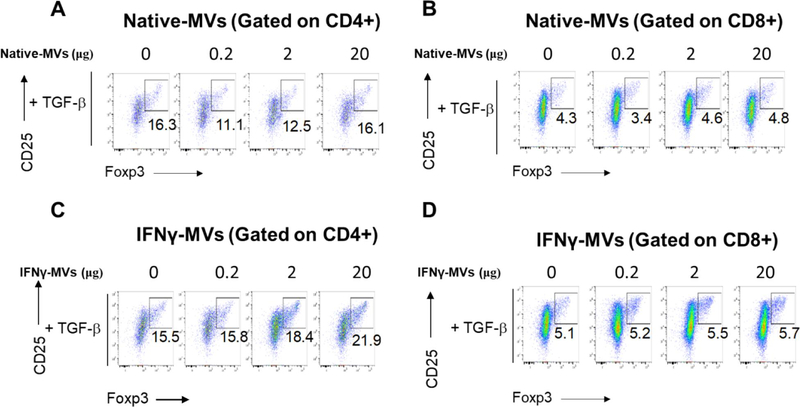
One important criterion in assessing the potency of extracellular vesicles is a robust functional biological assay. Of particular interest to us is MV’s immunomodulatory effects in treating autoimmune disorders. We have previously established an in vitro assay to study the immune-regulatory potency of exosomes [7]. We are particularly interested in regulatory T cells (Tregs) due to their critical roles in the pathogenesis and treatment of many autoimmune disorders [51] including multiple sclerosis [7, 52], Type 1 Diabetes [53, 54], psoriasis [55] and systemic lupus [56].We therefore attempted to test the potency of MVs in induction of CD4+CD25+FoxP3+ and CD8+CD25+FoxP3+ Tregs. Murine splenocytes from Foxp3-eGFP ‘Treg reporter’ mice were stimulated with anti-CD3 + IL-2 with or without TGFβ1 and were further co-cultured with the indicated concentrations of IFNγ- or Native-MVs. Tregs are marked by expression of their master transcription factor, Foxp3 and high levels of IL-2 high affinity receptor alpha chain (IL-2Rα, or CD25). Our preliminary data revealed that IFNγ-MVs may be able to enhance CD4 but not CD8 Treg production in the presence of TGFβ1, whereas both Native-MVs are less effective in inducing Tregs (representative data shown in Figure 5). Note that a minimal of 3–5 biological replicates and 3 technical replicates are generally required to robustly determine EV’s effect in Treg induction in vitro, as we established in our previous exosome study [7]. However, the purpose of the Figure 5 was to focus on the method and this representative data is meant to be illustrative rather than drawing biological conclusions. Future extensive studies using this Treg assay as an in vitro surrogate for MSC MV’s immunomodulatory functions will be performed with more replicates and statistical analyses. If MSC MV can robustly induce Tregs, we believe that they could serve as therapeutics for treatment of autoimmune disorders.
Figure 5. Treg induction of MVs.
Foxp3-eGFP mice splenocytes were stimulated with anti-CD3 + IL-2 + TGFβ and were further cultured in the presence of indicated concentrations of MSC MVs. A, B) Native MVs for CD4+ and CD8+ gate. C, D) IFNγ-MVs for CD4+ and CD8+ gate.
4. Conclusion
EVs are emerging cell-free biologics to treat variety of health disorders. Recent studies suggest that MVs secreted by MSCs are potent drug delivery agents that are effective treatment in severe pneumonia [20], osteoarthritis [21], and renal ischemia-reperfusion injury after cardiac death renal transplantation [22]. These observations motivate further research development and clinical translation of MVs. Robust production methods, reproducible and reliable characterization methods, and quality/potency control are yet to be fully established to effectively translate EVs into therapeutics. To that end, we developed methods to isolate and characterize MVs derived from human bone marrow MSCs.
We chose to isolate and purify MVs through ultracentrifugation technique. MVs could also be isolated through other methods including density gradient centrifugation [3], hydrostatic filtration dialysis [27], size exclusion chromatography [28], and affinity based capturing techniques [29, 30]. An ideal MV isolation technique should be scalable without undermining the required quality and purity, with a timely and cost-effective manner. A scalable procedure that rapidly isolates pure MVs from cells could greatly facilitate MVs clinical translation. We therefore used ultracentrifugation technique, which offers a facile and rapid technique to isolate MVs. We next used established markers and characterization techniques to study the yield and purity of our MVs, which were consistent with other published literature [2, 9, 21].
It has been shown that EVs could deliver proteins [57] and RNAs [58]. MVs contain nucleic acids and proteins [41, 59], and are composed of lipid bilayers containing transmembrane proteins. To fully characterize MVs, we analyzed their protein and RNA contents. We found that, while MVs host specific proteins compared to their exosome and cell counterparts, they also share similarities. We also profiled their encapsulated RNA profiles and found that more than ~93% of genomic cargos are proteincoding. It has been reported that RNA profiles from MVs are distinct from those of the producer cells [41, 60]. This offers that RNA encapsulation in these vesicles is likely to occur through a selective process.
We also found that stimulation of parental MSCs could be employed to differentially engineer the MV cargo to some extent. Further stimulation of parental cells and protocols for MV isolation, modification and characterization may enable MVs in the future to become new classes of therapeutics and diagnostics consisting of multiple biologics on the membranes and the interior of natural nano- to micro-scale vesicles.
Figure 4. Proteomics analysis of MVs.
A) Comparison of protein composition of Native-MVs with its parental cell and exosomes. B) Comparison of protein composition of IFNγ-MVs with its parental cells and exosomes.
Highlights.
A facile method to isolate microvesicles secreted from Mesenchymal Stem cells
We also introduced complete characterization methods for microvesicles including their proteins and RNAs
Potency of microvesicles also were analyzed by a Treg induction method
Acknowledgement
We thank the Cahalan lab at UC Irvine for providing eGFP-FOXP3 mice for this study. Some of the materials employed in this work were provided by the Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine through a grant from ORIP of the NIH, Grant # P40OD011050. The authors would like to acknowledge microscopy support from the Centre for Cellular Imaging at the University of Gothenburg and the National Microscopy Infrastructure, NMI (VR-RFI 2016–00968). We thank the Proteomics Core Facility at Sahlgrenska Academy, University of Gothenburg, for performing the proteomic analysis. The Proteomics Core Facility is grateful to the Inga-Britt and Arne Lundbergs Forskningsstiftlese for the donation of the Orbitrap Fusion Tribrid MS instrument.
Funding
This work was supported by the National Institute of Health (DP2CA195763). M.R. was supported by a National Institute of Neurological Disorders and Stroke (NINDS/NIH) Training Grant (award no. NS082174).
Footnotes
Conflict of interest
W.Z. is the founder of Velox Biosystems Inc. and a co-founder of Amberstone Biosciences Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1. https://clinicaltrials.gov/ct2/results?cond=mesenchymal+stem+cells&term=&cntry=&state=&city=&dist=.
- 2.Bruno S, et al. , Mesenchymal Stem Cell-Derived Microvesicles Protect Against Acute Tubular Injury. Journal of the American Society of Nephrology, 2009. 20(5): p. 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collino F, et al. , Exosome and Microvesicle-Enriched Fractions Isolated from Mesenchymal Stem Cells by Gradient Separation Showed Different Molecular Signatures and Functions on Renal Tubular Epithelial Cells. Stem Cell Reviews and Reports, 2017. 13(2): p. 226–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, et al. , Microvesicles derived from human Wharton’s Jelly mesenchymal stem cells ameliorate acute lung injury partly mediated by hepatocyte growth factor. The International Journal of Biochemistry & Cell Biology, 2019. 112: p. 114–122. [DOI] [PubMed] [Google Scholar]
- 5.Zhu YG, et al. , Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin-induced acute lung injury in mice. Stem Cells, 2014. 32(1): p. 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laso-García F, et al. , Therapeutic potential of extracellular vesicles derived from human mesenchymal stem cells in a model of progressive multiple sclerosis. PLOS ONE, 2018. 13(9): p. e0202590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riazifar M, et al. , Stem Cell-Derived Exosomes as Nanotherapeutics for Autoimmune and Neurodegenerative Disorders. ACS Nano, 2019. 13(6): p. 6670–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiklander OPB, et al. , Advances in therapeutic applications of extracellular vesicles. Science Translational Medicine, 2019. 11(492): p. eaav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C, et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 2018. 7(1): p. 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratajczak J, et al. , Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia, 2006. 20(9): p. 1487–95. [DOI] [PubMed] [Google Scholar]
- 11.Beaudoin AR and Grondin G, Shedding of vesicular material from the cell surface of eukaryotic cells: different cellular phenomena. Biochim Biophys Acta, 1991. 1071(3): p. 203–19. [DOI] [PubMed] [Google Scholar]
- 12.Guescini M, et al. , Microvesicle and tunneling nanotube mediated intercellular transfer of Gprotein coupled receptors in cell cultures. Exp. Cell Res, 2012. 318(5): p. 603–613. [DOI] [PubMed] [Google Scholar]
- 13.Muller G, et al. , Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell Signal, 2011. 23(7): p. 1207–23. [DOI] [PubMed] [Google Scholar]
- 14.Bissig C and Gruenberg J, Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol, 2013. 5(10): p. a016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbeil D, et al. , Prominin-1: a distinct cholesterol-binding membrane protein and the organisation of the apical plasma membrane of epithelial cells, in Cholesterol Binding and Cholesterol Transport Proteins: Structure and Function in Health and Disease. 2010. p. 399–423. [DOI] [PubMed] [Google Scholar]
- 16.Skog J, et al. , Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 2008. 10: p. 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzas EI, et al. , Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol, 2014. 10(6): p. 356–364. [DOI] [PubMed] [Google Scholar]
- 18.Engelmann B and Massberg S, Thrombosis as an intravascular effector of innate immunity. Nat. Rev. Immunol, 2013. 13(1): p. 34–45. [DOI] [PubMed] [Google Scholar]
- 19.Antonyak MA, et al. , Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. U. S. A, 2011. 108(12): p. 4852–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monsel A, et al. , Therapeutic Effects of Human Mesenchymal Stem Cell–derived Microvesicles in Severe Pneumonia in Mice. American Journal of Respiratory and Critical Care Medicine, 2015. 192(3): p. 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tofiño-Vian M, et al. , Microvesicles from Human Adipose Tissue-Derived Mesenchymal Stem Cells as a New Protective Strategy in Osteoarthritic Chondrocytes. Cellular Physiology and Biochemistry, 2018. 47(1): p. 11–25. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, et al. , Micro-vesicles derived from human Wharton’s Jelly mesenchymal stromal cells mitigate renal ischemia-reperfusion injury in rats after cardiac death renal transplantation. Journal of Cellular Biochemistry, 2018. 119(2): p. 1879–1888. [DOI] [PubMed] [Google Scholar]
- 23.Shigemoto-Kuroda T, et al. , MSC-derived extracellular vesicles attenuate immune responses in two autoimmune murine models: type 1 diabetes and uveoretinitis. Stem cell reports, 2017. 8(5): p. 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. , Extracellular vesicles released from human umbilical cord-derived mesenchymal stromal cells prevent life-threatening acute graft-versus-host disease in a mouse model of allogeneic hematopoietic stem cell transplantation. Stem cells and development, 2016. 25(24): p. 1874–1883. [DOI] [PubMed] [Google Scholar]
- 25.Nojehdehi S, et al. , Immunomodulatory effects of mesenchymal stem cell–derived exosomes on experimental type-1 autoimmune diabetes. Journal of Cellular Biochemistry, 2018. 119(11): p. 9433–9443. [DOI] [PubMed] [Google Scholar]
- 26.Cha JM, et al. , Efficient scalable production of therapeutic microvesicles derived from human mesenchymal stem cells. Scientific Reports, 2018. 8(1): p. 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musante L, et al. , A Simplified Method to Recover Urinary Vesicles for Clinical Applications, and Sample Banking. Scientific Reports, 2014. 4: p. 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corso G, et al. , Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Scientific Reports, 2017. 7(1): p. 11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai W, et al. , A novel affinity-based method for the isolation of highly purified extracellular vesicles. Scientific Reports, 2016. 6: p. 33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma P, et al. , Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. Journal of Extracellular Vesicles, 2018. 7(1): p. 1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelke GV, et al. , Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. Journal of Extracellular Vesicles, 2014. 3(1): p. 24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cvjetkovic A, Lötvall J, and Lässer C, The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. Journal of Extracellular Vesicles, 2014. 3(1): p. 23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lässer C, et al. , Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biology, 2017. 14(1): p. 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox J and Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology, 2008. 26: p. 1367. [DOI] [PubMed] [Google Scholar]
- 35.Kowal J, et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences, 2016. 113(8): p. E968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Au - Menck K, et al. , Isolation and Characterization of Microvesicles from Peripheral Blood. JoVE, 2017(119): p. e55057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieu M, et al. , Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nature Cell Biology, 2019. 21(1): p. 9–17. [DOI] [PubMed] [Google Scholar]
- 38.Yoshinaga N, et al. , Bundling mRNA Strands to Prepare Nano-Assemblies with Enhanced Stability Towards RNase for In Vivo Delivery. Angewandte Chemie International Edition, 2019. 0(0). [DOI] [PubMed] [Google Scholar]
- 39.Lieberman J, Tapping the RNA world for therapeutics. Nature Structural & Molecular Biology, 2018. 25(5): p. 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valadi H, et al. , Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 2007. 9: p. 654. [DOI] [PubMed] [Google Scholar]
- 41.Temoche-Diaz MM, et al. , Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. bioRxiv, 2019: p. 612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vigo T, et al. , IFN-γ orchestrates mesenchymal stem cell plasticity through the signal transducer and activator of transcription 1 and 3 and mammalian target of rapamycin pathways. Journal of Allergy and Clinical Immunology, 2017. 139(5): p. 1667–1676. [DOI] [PubMed] [Google Scholar]
- 43.Domenis R, et al. , Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cells-derived exosomes. Scientific Reports, 2018. 8(1): p. 13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y, et al. , Exosomal miR-146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin-1beta-Primed Mesenchymal Stem Cells Against Sepsis. 2017(1549–4918 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 45.Kim DS, et al. , Enhanced Immunosuppressive Properties of Human Mesenchymal Stem Cells Primed by Interferon-γ. EBioMedicine, 2018. 28: p. 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polchert D, et al. , IFN-γ activation of mesenchymal stem cells for treatment and prevention of graft versus host disease. European Journal of Immunology, 2008. 38(6): p. 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klinker MW, et al. , Morphological features of IFN-γ–stimulated mesenchymal stromal cells predict overall immunosuppressive capacity. Proceedings of the National Academy of Sciences, 2017. 114(13): p. E2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Q, et al. , Proteomic Analysis of Reprograming of Mesenchymal Stem/Stromal Cells (MSC) Following Interferon Gamma Identifies Pathways That Are Upregulated in Suppression. Blood, 2015. 126(23): p. 384. [Google Scholar]
- 49.Croitoru-Lamoury J, et al. , Interferon-γ Regulates the Proliferation and Differentiation of Mesenchymal Stem Cells via Activation of Indoleamine 2,3 Dioxygenase (IDO). PLOS ONE, 2011. 6(2): p. e14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pegtel DM and Gould SJ, Exosomes. Annual Review of Biochemistry, 2019. 88(1): p. 487–514. [DOI] [PubMed] [Google Scholar]
- 51.Dominguez-Villar M and Hafler DA, Regulatory T cells in autoimmune disease. Nature Immunology, 2018. 19(7): p. 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zozulya AL and Wiendl H, The role of regulatory T cells in multiple sclerosis. Nat Clin Pract Neurol, 2008. 4(7): p. 384–398. [DOI] [PubMed] [Google Scholar]
- 53.Lindley S, et al. , Defective suppressor function in CD4+ CD25+ T-cells from patients with type 1 diabetes. Diabetes, 2005. 54(1): p. 92–99. [DOI] [PubMed] [Google Scholar]
- 54.Marek-Trzonkowska N, et al. , Administration of CD4<sup>+</sup>CD25<sup>high</sup>CD127<sup>−</sup> Regulatory T Cells Preserves β-Cell Function in Type 1 Diabetes in Children. Diabetes Care, 2012. 35(9): p. 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartwig T, et al. , Regulatory T Cells Restrain Pathogenic T Helper Cells during Skin Inflammation. (2211–1247 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 56.Yang J, et al. , Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis & Rheumatism, 2009. 60(5): p. 1472–1483. [DOI] [PubMed] [Google Scholar]
- 57.Toh WS, et al. , MSC exosome works through a protein-based mechanism of action. Biochemical Society Transactions, 2018: p. BST20180079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanada M, et al. , Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proceedings of the National Academy of Sciences, 2015. 112(12): p. E1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colombo M, Raposo G, and Théry C, Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annual Review of Cell and Developmental Biology, 2014. 30(1): p. 255–289. [DOI] [PubMed] [Google Scholar]
- 60.Janas T, et al. , Mechanisms of RNA loading into exosomes. FEBS Letters, 2015. 589(13): p. 13911398. [DOI] [PubMed] [Google Scholar]