Abstract
This study investigates the performance of diagnostic methods for detection of Clostridioides difficile infection in Sweden, including impact of PCR ribotype on diagnostic performance. Between 2011 and 2016, a total of 17,878 stool samples from 26 laboratories were tested by either well-type enzyme immunoassays (EIAs), membrane bound EIAs, cell cytotoxicity neutralization assay (CTA), or nucleic acid amplification tests (NAATs) and subsequently cultured for C. difficile. Roughly half of the samples (9454/17878) were subjected to diagnostic testing both on the fecal sample and on the 1323 isolated C. difficile strains. All C. difficile isolates were typed by PCR ribotyping, and the isolates were classified as toxigenic or non-toxigenic based on the empirical knowledge of the association between toxin-positivity and ribotype. The overall sensitivity, specificity, and positive and negative predictive values were highest for NAATs and membrane EIAs. Ribotype-specific sensitivity varied greatly between methods and ribotypes. All methods had 100% sensitivity against ribotype 027 and 013. For other types, the sensitivity ranged from 33 to 85% in fecal samples and from 78 to 100% on isolates. For the most prevalent ribotypes (014, 020, and 001), the sensitivity varied between 38 and 100% in the fecal samples, with the lowest sensitivity observed for well-type EIAs and CTA. The large variation in diagnostic sensitivity implies that type distribution significantly affects the outcome when evaluating diagnostic performance. Furthermore, performing comparative studies of diagnostic tests in settings with high prevalence of ribotype 027 will overestimate the general performance of diagnostic tests.
Introduction
Clostridioides difficile infection (CDI) is one of the most common healthcare-associated infections worldwide. A European point prevalence survey accredited 48% of all healthcare-associated gastrointestinal infections to CDI with an attributable mortality of 3% [1]. Toxigenic strains of C. difficile produce one or two major toxins, enterotoxin (toxin A), and cytotoxin (toxin B) encoded by the pathogenicity locus (PaLoc) [2]. The ability of a strain to produce one or both toxins is crucial for clinical disease. The toxins cause colonic tissue damage to the enteric cytoskeletal wall and disruption of the tight junctions that connect colonic cells [3]. Toxin production is dependent by growth phase, nutritional status and can vary greatly between strains of the same ribotype [4, 5]. Some strains produce also the binary actin-ADP-ribosylating toxin that increases microtubule polymerization which might increase the adherence of C. difficile to target cells [6].
In Sweden, around 6000 new cases of CDI are reported yearly, and even though incidence has decreased in recent years [7], it remains a significant burden for the patient and the health care system. An accurate diagnosis of CDI remains a challenge, and underdiagnosis is an issue in Europe [8]. False negative CDI test results may increase the risk of transmission in addition to mistreatment of the patient, while false positive results may lead to unnecessary treatment interventions for CDI. A wide variety of diagnostic tests are available to detect C. difficile toxins, i.e., enzyme immunoassays (well-type EIAs), membrane bound enzyme immunoassays (membrane EIA), cell cytotoxicity neutralization assay (CTA), and toxigenic culture (TC), or to detect the toxin genes of C. difficile using nucleic acid amplification tests (NAATs). In 2016, the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) published updated guidelines for CDI diagnostics [9], recommending two-step algorithms that aims to detect the presence of the bacteria with a highly sensitive test and free toxins using EIAs. However, the optimal method for the detection of CDI is still under debate [10]. One drawback of many studies comparing diagnostic techniques is that the local epidemiology is not taken into consideration when assessing the performance of a diagnostic test, and several studies have been carried out in high prevalence or outbreak settings. In this study, we aim to assess the ribotype-specific sensitivity of different diagnostic methods in addition to evaluating the overall performance of each method in a non-outbreak setting with high ribotype diversity [7]. The choice of reference method or gold standard is crucial in order to assess the accuracy of a test. In this study, we use PCR ribotyping to determine if an isolate belongs to a toxigenic or non-toxigenic ribotype as gold standard instead of using the standard approach of CTA, toxigenic or cytotoxigenic culture. As the ability of a strain to produce toxins is crucial to diagnose CDI, we do not aim to evaluate the performance of glutamate dehydrogenase tests for the detection of C. difficile bacteria in this study.
Methods
Sampling, diagnostic testing, and culturing
This prospective study was conducted on a national level in Sweden and included all 26 local laboratories performing diagnostics for C. difficile. Sampling was performed twice yearly between 2011 and 2016, during 1 week in spring (w11) and 1 week in autumn (w39). Fecal samples from both hospital- and community-associated cases were included in the study. Eligible patients included those with suspected CDI, for whom unformed stool samples were sent for CDI testing regardless of age of the patient. Information about diagnostic method and/or algorithm and test performance results were registered. Fecal samples were tested at the local laboratory and C. difficile isolates were sent to the Public Health Agency of Sweden for PCR ribotyping. Multiple samples from the same patient were allowed due to use of anonymous data.
All laboratories were asked to culture every stool sample tested for CDI during the collection period and to perform CDI diagnostics both on the fecal sample as well as on the cultured isolate using their standard diagnostic test. The diagnostic tests adopted during the collection period were well-type EIAs (VIDAS (bioMeriéux, France)) and Premier Toxin A&B (Meridian bioscience, USA), membrane EIAs (C. diff Quik Check Complete (Techlab, USA)), Immunocard Toxin A&B (Meridian bioscience, USA), NAATs (Illumigene (Meridian bioscience, USA)), Gene Xpert (Cepheid, USA), GenomEra® C. difficile (Abacus diagnostica, Finland), and CTA. Several laboratories switched methods during the study period, and the tests were performed by different technicians. TCCFA selective media was used for isolation of C. difficile [11].
PCR ribotyping
PCR ribotyping between 2011 and 2012 was performed by a gel-based method according to the method of Stubbs et al. [12] with modifications described in Svenungsson et al. [13] and from 2013 and onwards by capillary gel electrophoresis [14]. The ribotypes classified as non-toxigenic have been described previously in the literature [15, 16].
Results
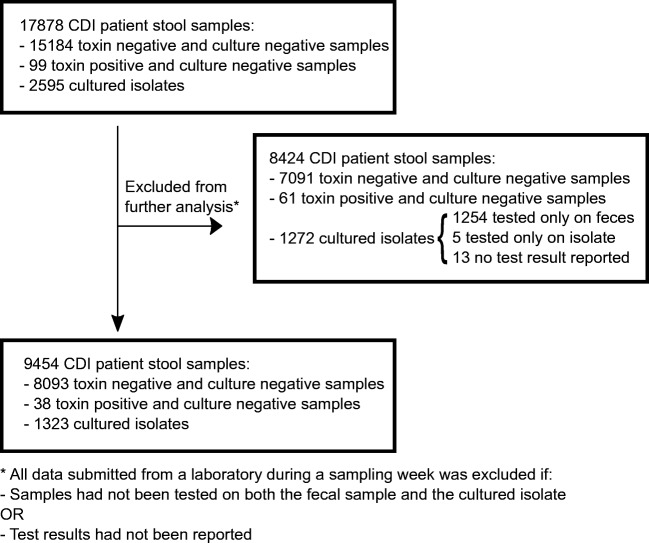
A total of 17,878 stool samples and 2595 C. difficile isolates were collected for the study, 99 fecal samples (0.6%) tested positive for the presence on toxins A/B (n = 65) or toxigenic C. difficile by NAAT (n = 34) but were C. difficile culture-negative (Fig. 1). Samples were judged to be positive or negative for CDI based on the local laboratories routine interpretation of the test result. Fifty-one percent of the isolates (1323/2595) were tested according to the given instructions i.e. tested both on the fecal sample and on the cultured isolate. For 1254/2595 isolates (48%), only the fecal samples were tested for CDI, and the isolates were never subjected to further testing due to the lack of compliance to the study instructions by the local laboratories. Furthermore, for five isolates, the results from the fecal samples test were not recorded; for 13 isolates, no test results were reported. Samples not tested according to the given instructions and samples with missing information on CDI test results were all excluded from further analysis. Samples reported with a threshold value for CDI, as determined by the manufacturer (VIDAS, bioMeriéux), were considered as negative in this study. Five different types of diagnostic methods or combination of methods were used during the collection period (Table 1).
Fig. 1.
Flowchart of selection process for the samples included in the study
Table 1.
CDI testing result from the routine interpretation of clinical laboratories for each C. difficile isolate collected according to the study protocol divided by diagnostic method
| CDI test result (feces/isolate) |
CTA | Well-type EIA | Membrane EIA | NAAT | NAAT + well-type EIA | Total |
|---|---|---|---|---|---|---|
| -/- | 11 | 93 | 28 | 1 | 133 | |
| th/- | 12 | 12 | ||||
| -/th | 12 | 12 | ||||
| -/+ | 28 | 170 | 6 | 37 | 14 | 255 |
| th/th | 6 | 6 | ||||
| th/+ | 14 | 14 | ||||
| +/- | 23 | 1 | 1 | 25 | ||
| +/+ | 67 | 358 | 33 | 393 | 8 | 859 |
| +/th | 7 | 7 | ||||
| Total | 106 | 695 | 39 | 459 | 24 | 1323 |
+, CDI test positive; -, CDI test negative; th, threshold/CDI test deemed negative; CTA, cell-cytotoxicity neutralization assay; EIA, enzyme immunoassay; NAAT, nucleic acid amplification test
Comparison of test performance
A total of 9454/17878 (53%) patient samples were tested according to the given instructions, and 1323 isolates of C. difficile were cultured. The majority of samples were analyzed either by well-type EIA (52%) or NAAT (32%), while 11% were analyzed by CTA, 3% by membrane EIA, and 2% by NAAT + well-type EIA. The prevalence of CDI in this study was defined as the number of isolates with a toxigenic ribotype divided by the number of clinical samples tested for CDI and amounted to 13% (1226/9454). The specificity values were high for all methods (> 99%), whereas the sensitivity values varied widely between methods. The NAAT + well-type EIA algorithm showed the lowest sensitivity (41%), while NAAT as a standalone test had the highest sensitivity (91%). Similarly, the lowest negative predictive value (NPV) was reported for NAAT + well-type EIA and the highest for NAAT (Table 2). NAAT + well-type EIA had the highest positive predictive value (PPV), but NAATs and membrane EIAs performed best having both high PPV and NPV.
Table 2.
Performance of diagnostic methods or algorithm on fecal samples compared with ribotype toxigenicity as the gold standard
| CTA | Well-type EIA | Membrane EIA | NAAT | NAAT + well-type EIA | |
|---|---|---|---|---|---|
| PPV % (95% CI) | 95.7 (95.5–95.9) | 93.5 (93.5–93.6) | 97.1 (96.9–97.2) | 95.1 (95.1–95.2) | 100 (100–100) |
| NPV % (95% CI) | 96.5 (96.5–96.5) | 94.2 (94.3–94.3) | 97.8 (97.7–97.8) | 98.5 (98.5–98.5) | 92.8 (92.7–92.9) |
| Specificity % (95% CI) | 99.7 (99.7–99.7) | 99.4 (99.4–99.4) | 99.6 (99.6–99.6) | 99.2 (99.2–99.3) | 100 (100–100) |
| Sensitivity % (95% CI) | 67.0 (66.7–67.3) | 59.7 (59.6–59.8) | 84.6 (84.2–85.0) | 90.6 (90.6–90.8) | 40.9 (40.2–41.6) |
PPV, positive predictive value; NPV, negative predictive value, CI, confidence interval, CTA, cell cytotoxicity neutralization assay; EIA, enzyme immunoassay; NAAT, nucleic acid amplification test
Ribotype-specific sensitivity
Results from PCR ribotyping was used to determine if an isolate belonged to a toxigenic or non-toxigenic ribotype. A total of 144 different PCR ribotypes were determined, and the 10 most common were 014 (n = 141), 020 (n = 102), 001 (n = 84), 002 (n = 84), 078/126 (n = 65), 023 (n = 59), 005 (n = 56), 220 (n = 44), 010 (n = 43), and 045 (n = 43), respectively (other ribotypes n = 645).
Seven percent (97/1323) of the isolates belonged to non-toxigenic PCR ribotypes such as 010, 009, 031, 032, 039, and 084. These were predominantly found among the isolates that tested negative in all tests, but 1% (n = 13) were positive in feces (Table 3). Toxin negativity by the absence of PaLoc locus was confirmed by whole genome sequencing of 6 arbitrarily chosen isolates of ribotype 010 that tested positive for CDI (data not shown).
Table 3.
CDI testing results per diagnostic method for 97 isolates belonging to 10 toxin-negative ribotypes, as judged from the routine interpretation at the clinical laboratories
| CDI test result (feces/isolate) |
CTA | Well-type EIA | NAAT | NAAT + well-type EIA |
|---|---|---|---|---|
| -/- | 2 | 39 | 18 | 1 |
| -/+ | 4 | 9 | 7 | 1 |
| +/- | 4 | |||
| +/+ | 3 | 4 | ||
| +/th | 2 | |||
| th/- | 3 |
+, CDI test positive; -, CDI test negative; th, threshold/CDI test negative; CTA, cell cytotoxicity neutralization assay; EIA, enzyme immunoassay; NAAT, nucleic acid amplification test
Of the 1226 isolates belonging to toxigenic ribotypes, only isolates of ribotype 027 and 013 had a sensitivity of 100% for all CDI tests on fecal samples. For other types, the overall sensitivity ranged from 33 to 85% in fecal samples and from 78 to 100% on isolates. The ribotype-specific sensitivity for each type of diagnostic test is reported in Table 4. No significant difference was found when comparing ribotype distribution between samples tested by NAAT or well-type EIAs (p = 0.98 chi-squared test).
Table 4.
Ribotype-specific sensitivity for each diagnostic method tested on fecal samples and cultured isolates. Only toxigenic ribotypes are included. Ribotypes are sorted by prevalence in descending order and ribotypes with less than 6 isolates are grouped together
| Ribotype | Overall sensitivity feces/isolate (n) | CTA sensitivity feces/isolateb (n) | Well-type EIA sensitivity feces/isolatea (n) | Membrane EIA sensitivity feces/isolatea (n) | NAAT sensitivity feces/isolate (n) | NAAT + well-type EIA sensitivity feces/isolate (n) |
|---|---|---|---|---|---|---|
| 014*ǂ | 77/92 (141) | 64/91 (11) | 64/86 (72) | 100/100 (2) | 98/100 (55) | 0/100 (1) |
| 020*ǂ | 65/89 (102) | 38/88 (8) | 48/82 (56) | 100/100 (1) | 95/100 (37) | ND |
| 001* | 76/95 (84) | 64/100 (11) | 67/90 (39) | 83/100 (6) | 93/100 (28) | ND |
| 002*ǂ | 77/88 (84) | 71/86 (7) | 68/78 (41) | 100/100 (4) | 90/100 (30) | 50/100 (2) |
| 078/126*ǂ | 62/80 (65) | 100/100 (6) | 38/67 (39) | 100/100 (2) | 94/100 (18) | ND |
| 023*ǂ | 78/88 (59) | 67/100 (3) | 68/79 (34) | 100/100 (1) | 100/100 (20) | 0/100 (1) |
| 005* | 66/93 (56) | 33/67 (3) | 57/89 (28) | 67/100 (3) | 89/100 (19) | 33/100 (3) |
| 220* | 50/86 (44) | 50/100 (2) | 40/80 (30) | 60/100 (5) | 100/100 (6) | 0/100 (1) |
| 045* | 77/86 (43) | 100/100 (4) | 62/81 (26) | 100/100 (1) | 100/100 (11) | 100/0 (1) |
| 029 | 65/95 (40) | 100/100 (1) | 57/90 (21) | 100/100 (2) | 79/100 (14) | 0/100 (2) |
| 081 | 85/94 (34) | 100/100 (2) | 50/94 (16) | ND | 80/93 (15) | 0/100 (1) |
| 012 | 76/100 (34) | 0/100 (2) | 80/100 (10) | ND | 80/100 (20) | 100/100 (2) |
| 070 | 82/100 (29) | 67/100 (3) | 77/100 (13) | ND | 92/100 (13) | ND |
| 046 | 59/93 (27) | 25/100 (4) | 61/89 (18) | ND | 80/100 (5) | ND |
| 017 | 80/88 (25) | ND | 72/83 (18) | ND | 100/100 (6) | 100/100 (1) |
| 018 | 80/100 (25) | 100/100 (4) | 64/100 (14) | ND | 100/100 (7) | ND |
| 011 | 79/96 (24) | 100/100 (2) | 67/92 (12) | ND | 89/100 (9) | 100/100 (1) |
| 003 | 61/91 (23) | 0/100 (1) | 60/87 (15) | 0/100 (2) | 100/100 (5) | ND |
| 026* | 65/85 (20) | 100/100 (2) | 17/83 (6) | 100/100 (1) | 82/82 (11) | ND |
| 570 | 69/88 (16) | 100/100 (1) | 63/100 (8) | 100/100 (1) | 67/67 (6) | ND |
| 231 | 83/83 (12) | 100/100 (1) | 67/33 (3) | ND | 88/100 (8) | ND |
| 054 | 70/100 (10) | 50/100 (2) | 0/100 (2) | ND | 100/100 (6) | ND |
| 043 | 70/90 (10) | 100/100 (1) | 50/83 (6) | ND | 100/100 (3) | ND |
| 258 | 67/100 (9) | 100/100 (1) | 67/100 (3) | ND | 75/100 (4) | 0/100 (1) |
| 027 | 100/100 (8) | 100/100 (1) | 100/100 (2) | 100/100 (1) | 100/100 (4) | ND |
| 103 | 75/100 (8) | 100/100 (1) | 33/100 (3) | ND | 100/100 (3) | 100/100 (1) |
| 015 | 75/88 (8) | ND | 75/75 (4) | 100/100 (1) | 67/100 (3) | ND |
| 013 | 100/100 (8) | 100/100 (2) | 100/100 (4) | ND | 100/100 (2) | ND |
| 087 | 71/100 (7) | ND | 75/100 (4) | ND | 67/100 (3) | ND |
| 808 | 33/83 (6) | 100/100 (1) | 0/67 (3) | ND | 50/100 (2) | ND |
| 207 | 67/83 (6) | ND | 67/67 (3) | ND | 67/100 (3) | ND |
| Others | 73/81 (159) | 53/62 (13) | 66/76 (82) | 100/100 (6) | 89/89 (54) | 25/100 (4) |
| < 6*ǂ | ||||||
| Total*ǂ | 68/90 (1226) | 67/90 (100) | 60/83 (635) | 85/100 (39) | 91/97 (430) | 41/95 (22) |
aEquivalent to toxigenic culture
bEquivalent to cytotoxigenic culture
*Difference between well-type EIA and NAAT on feces is statistically significant at p < 0.05 (Fisher’s exact test)
ǂDifference between well-type EIA and NAAT on isolates is statistically significant at p < 0.05 (Fisher’s exact test)
ND, no data; CTA, cell cytotoxicity neutralization assay; EIA, enzyme immunoassay; NAAT, nucleic acid amplification test
Six isolates of PCR ribotype 023 and two of ribotype 231 that tested negative in feces and/or on the isolate by well-type EIA were sequenced by WGS to determine the presence of toxin genes. All eight isolates carried genes for toxin A, toxin B, and the isolates of ribotype 023 carried also binary toxin genes (data not shown).
Discussion
A rapid and correct identification of a CDI case is crucial to ensure correct treatment and adequate infection control measures [17]. Reference assays for CDI have been CTA and/or TC; at times, also cytotoxigenic culture has been adopted. The major drawback of these methods is the long turnaround time, and thus they are rarely used in routine diagnostics [18]. All methods have their limit of detection and fecal samples are complex and can inhibit both antibody binding as well as nucleic acid amplification. Furthermore, a drawback of all antibody-dependent tests is the variation in affinity towards all genetic variants of toxin A and/or B, as mutations that change the antibody’s antigen binding site could cause reduced antibody affinity [19]. Similarly, if mutations should arise in the primer regions of toxin genes targeted by NAATs, this could also reduce NAAT sensitivity. By using the toxigenic status associated with a given PCR ribotype as a confirmatory test, we aimed to circumvent the problems caused by limit of detection dependent on mutations that affect affinity or low toxin production caused by nutritional status or growth phase [4, 5]. Some of the toxigenic ribotypes could have lost the ability to produce toxins by homologous recombination of the PaLoc [20], but in such a case, we would observe no difference when comparing the ribotype-specific sensitivity of the fecal samples to the ribotype-specific sensitivity of cultured isolates. Instead, the type-specific sensitivity was higher on isolates compared with fecal samples, irrespectively of diagnostic method or ribotype. By sequencing a subset of isolates, we confirmed that isolates deemed as false negatives actually carried the toxin genes and, vice versa, isolates considered as false positives lacked the toxin genes. As we observed an increased sensitivity when testing on cultured isolates, the major cause of the low sensitivity observed for well-type EIAs and CTA was most likely due to the low amount and/or degradation of toxins in stool samples.
Toxin-negative ribotypes could also have been erroneously isolated from six toxin-positive fecal samples, but this aspect was not investigated further.
By using the toxigenic properties of a ribotype as a gold standard, there is a risk of overestimating CDI prevalence, but the CDI prevalence in this study was similar to that in another Swedish study carried out during the same time period with CTA as a comparative test (prevalence 13.2% [21]). Although this was a small regional study, we believe that comparing performance against the toxigenic properties of a ribotype does not largely affect CDI prevalence.
Enrichment prior to culturing could have improved the results from this study, but the clinical relevance of samples with C. difficile concentration too low for culturing is uncertain. Preferably, a new sample should be taken for re-testing if CDI symptoms persist. In this study, all samples were included, thus, also new samples from patients that initially have tested negative.
The results showed that membrane EIAs and NAATs as standalone tests performed best with highest PPV and NPV. However, since only a small number of samples and fewer ribotypes were tested with membrane EIAs, these results should be interpreted carefully. NAAT + well-type EIA had a PPV of 100%, but due to the low sample size, just one false positive result would lower the PPV to 90%. Compared with the pooled values from the ESCMID diagnostic guidance document [9], the NAAT tests in this study had a lower sensitivity but higher specificity. The lower sensitivity may be due to the relatively high ribotype diversity in this study. Many comparative diagnostic studies have been conducted in settings with high prevalence of ribotype 027, which would have resulted in an overestimation of the true performance of a diagnostic test. The higher specificity of NAATs in this study may be due to the inclusion algorithm for patients, i.e. testing only samples based on clinical suspicion for CDI and not all diarrheal samples. Performance outcome may also be affected by the choice of comparative standard method. In this study, the classical gold standard methods, CTA, TC (testing isolates with well-type EIA), and cytotoxigenic culture (testing isolates with CTA), had a sensitivity of 67%, 83%, and 90%, respectively, and all were less sensitive than NAAT performed on feces (91% sensitivity). The poorest sensitivity for CTA and well-type EIAs was observed for some of the most common ribotypes circulating in Sweden, e.g., 014, 020, 078/126, and 005. If cytotoxigenic culture or TC (with well-type EIA) would have been used as gold standard in this study, 12% and 18% of these ribotypes, respectively, would have been reported as false positives using NAAT. The reason for the reduced sensitivity of diagnostic tests that detect toxin A and/or B against the above mentioned ribotypes may be due to the variation in toxin production. Some hypervirulent ribotypes like 027 are known to have higher toxin levels in vitro compared with other common ribotypes [22]. It is possible that toxin production in vivo may vary substantially between strains or during an ongoing infection. Furthermore, immunocompromised patients could develop CDI symptoms also when toxin levels are below the limit of detection.
Using the toxigenic status of a ribotype as a gold standard for diagnosing CDI may have some drawbacks. For example, there is still a possibility that mutations in regulatory elements or within the PaLoc can affect the ability to produce toxins. However, as we observed a higher sensitivity for tests made directly on the isolate, we believe that this is unlikely for the majority of ribotypes. This approach improves the limit of detection compared with traditional gold standard methods but is still not optimal as a gold standard for CDI diagnostics. Possibly, the gold standard for CDI diagnostic test performance would include a test result related to symptom resolution following CDI treatment, thus enabling to diagnose cases where clinical suspicion of CDI is strong despite a negative CDI test.
Although the sample size of ribotype 027 isolates was rather small (n = 8, 0.6%), our results indicate a higher performance for all diagnostic methods towards ribotype 027. Thus, caution should be made when interpreting comparative studies of diagnostic tests in settings with high prevalence of ribotype 027. To date, only one study has addressed the effect of strain type on diagnostic sensitivity, and in accordance with the results in this study, NAATs showed higher overall sensitivity compared with EIAs but similar performance against ribotype 027 [23].
Local recommendations for C. difficile diagnostic tests should be adapted to ensure the best performance according to local C. difficile epidemiology, and take into consideration sample logistics, unless refrigerated, samples should be tested within 2 h from sampling if the test targets free toxins to avoid degradation of toxins [24]. When using NAAT as a standalone test, it is preferred to select stool samples for CDI based on clinical suspicion of CDI to reduce the risk of detecting asymptomatic carriage. Although, when analyzing samples only based on clinical suspicion, there is a risk of underdiagnosis, but this has been shown not to be an issue in Sweden [8], where the current national recommendation is to test only unformed stool samples upon clinical suspicion [25].
Acknowledgments
We acknowledge all the clinical microbiology laboratories participating in the Swedish C. difficile surveillance program for their contribution of isolates. We also acknowledge I. Alefjord and J. Furuskog for the excellent technical laboratory assistance.
Funding Information
Open access funding provided by Public Health Agency of Sweden.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.European Centre for Disease Prevention and Control (2013) Point prevalence survey of healthcare associated infections and antimicrobial use in European acute care hospitals 2011–2012. ECDC. https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/healthcare-associated-infections-antimicrobial-use-PPS.pdf [DOI] [PubMed]
- 2.Janezic S, Garneau JR, Monot M. Comparative genomics of Clostridium difficile. Adv Exp Med Biol. 2018;1050:59–75. doi: 10.1007/978-3-319-72799-8_5. [DOI] [PubMed] [Google Scholar]
- 3.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 4.Zhu S, Zhang L, Zhang C, Chen X, Chen Q, Li Z. Comparison of polymerase chain reaction ribotyping, toxinotyping and nutritional aspects of toxin production of Clostridium difficile strains. Biomed Rep. 2014;2(4):477–480. doi: 10.3892/br.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann JD, Otto A, Berges M, Biedendieck R, Michel A-M, Becher D, Jahn D, Neumann-Schaal M (2018) Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front Microbiol 9(1970). 10.3389/fmicb.2018.01970 [DOI] [PMC free article] [PubMed]
- 6.Aktories K, Papatheodorou P, Schwan C. Binary Clostridium difficile toxin (CDT) - a virulence factor disturbing the cytoskeleton. Anaerobe. 2018;53:21–29. doi: 10.1016/j.anaerobe.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Rizzardi K, Noren T, Aspevall O, Makitalo B, Toepfer M, Johansson A, Akerlund T. National surveillance for Clostridioides difficile infection, Sweden, 2009-2016. Emerg Infect Dis. 2018;24(9):1617–1625. doi: 10.3201/eid2409.171658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies KA, Longshaw CM, Davis GL, Bouza E, Barbut F, Barna Z, Delmee M, Fitzpatrick F, Ivanova K, Kuijper E, Macovei IS, Mentula S, Mastrantonio P, von Muller L, Oleastro M, Petinaki E, Pituch H, Noren T, Novakova E, Nyc O, Rupnik M, Schmid D, Wilcox MH. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14(12):1208–1219. doi: 10.1016/S1473-3099(14)70991-0. [DOI] [PubMed] [Google Scholar]
- 9.Crobach MJT, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2016;22(Supplement 4):S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Fang FC, Polage CR, Wilcox MH. Point-counterpoint: what is the optimal approach for detection of Clostridium difficile infection? J Clin Microbiol. 2017;55(3):670–680. doi: 10.1128/jcm.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15(3):443–446. doi: 10.1128/JCM.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbs SLJ, Brazier JS, O’Neill GL, Duerden BI. PCR targeted to the 16S-23S rRNA gene Intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR Ribotypes. J Clin Microbiol. 1999;37(2):461–463. doi: 10.1128/JCM.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svenungsson B, Burman LG, Jalakas-Pörnull K, Lagergren Å, Struwe J, Åkerlund T. Epidemiology and molecular characterization of Clostridium difficile strains from patients with diarrhea: low disease incidence and evidence of limited cross-infection in a Swedish teaching hospital. J Clin Microbiol. 2003;41(9):4031–4037. doi: 10.1128/JCM.41.9.4031-4037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akerlund T, Alefjord I, Dohnhammar U, Struwe J, Noren T, Tegmark-Wisell K, Swedish CDSG (2011) Geographical clustering of cases of infection with moxifloxacin-resistant Clostridium difficile PCR-ribotypes 012, 017 and 046 in Sweden, 2008 and 2009. Euro Surveillance : Bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 16 (10) [DOI] [PubMed]
- 15.Carman RJ, Wickham KN, Chen L, Lawrence AM, Boone JH, Wilkins TD, Kerkering TM, Lyerly DM. Glutamate dehydrogenase is highly conserved among Clostridium difficile ribotypes. J Clin Microbiol. 2012;50(4):1425–1426. doi: 10.1128/JCM.05600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janezic S, Zidaric V, Pardon B, Indra A, Kokotovic B, Blanco JL, Seyboldt C, Diaz CR, Poxton IR, Perreten V, Drigo I, Jiraskova A, Ocepek M, Weese JS, Songer JG, Wilcox MH, Rupnik M. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol. 2014;14(1):173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108(4):478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 18.Planche T, Wilcox M. Reference assays for Clostridium difficile infection: one or two gold standards? J Clin Pathol. 2011;64(1):1–5. doi: 10.1136/jcp.2010.080135. [DOI] [PubMed] [Google Scholar]
- 19.Sela-Culang I, Kunik V, Ofran Y. The structural basis of antibody-antigen recognition. Front Immunol. 2013;4:302–302. doi: 10.3389/fimmu.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott B, Dingle KE, Didelot X, Crook DW, Riley TV. The complexity and diversity of the pathogenicity locus in Clostridium difficile clade 5. Genome Biol Evol. 2014;6(12):3159–3170. doi: 10.1093/gbe/evu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norén T, Alriksson I, Andersson J, Åkerlund T, Unemo M. Rapid and sensitive loop-mediated isothermal amplification test for Clostridium difficile detection challenges cytotoxin B cell test and culture as gold standard. J Clin Microbiol. 2011;49(2):710–711. doi: 10.1128/jcm.01824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 23.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Persing DH. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48(10):3719–3724. doi: 10.1128/jcm.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brazier JS. Role of the laboratory in investigations of Clostridium difficile diarrhea. ClinInfect Dis. 1993;16(Supplement_4):S228–S233. doi: 10.1093/clinids/16.Supplement_4.S228. [DOI] [PubMed] [Google Scholar]
- 25.Sweden PHAo (2017) Clostridium difficile-infektion Kunskapsunderlag och rekommendationer för övervakning, prevention och utbrottshantering. Public Health Agency of Sweden, Folkhälsomyndigheten. doi:https://www.folkhalsomyndigheten.se/contentassets/9f71be5f49ae427ca2e92b61b061ad73/clostridium-difficile-infektion.pdf.