Abstract
Purpose
To evaluate the association of single nucleotide polymorphisms (SNPs) and the intronic expansion of a trinucleotide repeat (TNR) in the TCF4 gene with Fuchs endothelial corneal dystrophy (FECD) in a Thai population.
Methods
In total, 54 Thai FECD patients and 54 controls were recruited for the study. Five SNPs (rs613872, rs2123392, rs17089887, rs1452787, and rs1348047), previously reported to be associated with FECD, were genotyped by direct sequencing. The repeat length was determined by direct sequencing of PCR-amplified DNA (a short tandem repeat; STR assay) and by triplet repeat primed PCR (TP-PCR).
Results
Only one of the 54 patients with FECD harboured rs613872 (1.9%). Four SNPs (rs2123392, rs17089887, rs1452787, and rs1348047), which are not rare polymorphisms in the Thai population, were found in approximately half of the patients. Of the 54 patients, 21 (1 homozygous and 20 heterozygous patients; 39%) harboured a TNR ≥ 40, while 33 patients (61%) harboured a TNR < 40.
Conclusions
The association of TNR expansion in TCF4 with FECD is shown for the first time in the Thai population. The intronic TNR expansion identified in various ethnic groups underlines the importance of expansion as a potent pathophysiological cause of FECD.
Subject terms: Corneal diseases, Diseases
Introduction
Fuchs endothelial corneal dystrophy (FECD) is a bilateral corneal disease characterised by a progressive loss of corneal endothelial cells that typically begins after the age of 40 [1, 2]. The corneal endothelium is responsible for maintaining corneal transparency by regulating the amount of water in the corneal stroma; therefore, a severe loss of corneal endothelial cells results in vision loss due to corneal haziness.
FECD initially showed an autosomal dominant inheritance pattern with variable penetrance and expressivity in investigations of large families conducted in the 1970s and 1980s [3–5]. Early screening studies for functional candidate genes and investigations of causal genes of other corneal endothelial dystrophies identified four genes (SLCA411, TCF8, LOXHD1, and AGBL1) that were associated with late-onset FECD (the typical form of FECD) [6–11]. However, these genes were not frequently encountered in the general population of FECD patients; therefore, researchers have continued searching for a more common genetic cause.
In 2010, a genome-wide association study (GWAS) reported a significant association between the intronic single nucleotide polymorphism (SNP) in the transcription factor 4 (TCF4) and late-onset FECD [12]. Subsequently, Wieben et al. reported a strong association between FECD and a cytosine–thymine–guanine (CTG) repeat expansion in the non-coding region of TCF4 and found a sensitivity and specificity of ≥50 repeats of 79% and 96%, respectively, for identifying FECD patients in their cohort [13]. Subsequent studies have replicated these novel findings in various populations, although the frequency of the repeat expansion in FECD tends to be lower in non-Caucasian (i.e., Chinese, Indian, and Japanese) cohorts [14–19]. These studies have provided evidence to support the concept, proposed by Wieben et al. [13], that FECD is a trinucleotide repeat (TNR) expansion disorder.
This common genetic basis of FECD has revealed a novel therapeutic target for the development of new therapies, such as pharmacotherapy and gene therapy. However, the TNR expansion is not always identified in patients with FECD, probably due to the heterogeneity of this disease. Therefore, we were motivated to investigate the TCF4 variants of FECD in a patient cohort from Thailand. In the current study, we examined the previously reported SNPs found in various ethnic groups, as well as the CTG repeat expansion, to provide baseline genetic data for FECD in Thailand.
Materials and methods
Ethics statement
The human tissues used in this study were handled under the guidelines based on the ethical principles of the Declaration of Helsinki. This study was performed according to a protocol approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (IRB number 543/55), Doshisha University, and Kyoto Prefectural University of Medicine. Informed consent was obtained from the patients with FECD who participated in the study.
Preparation of genomic DNA
The 54 patients with FECD and 54 control subjects were recruited between February 2013 and September 2018 at King Chulalongkorn Memorial Hospital. All FECD cases were examined with slit-lamp biomicroscopy and diagnosed by cornea specialists. Noncontact specular microscopy (CEM530; Nidek Co., Ltd., Aichi, Japan) was also used to confirm guttae. The inclusion criteria for the FECD cases were: (1) Presence of bilateral guttae, with or without corneal oedema; or (2) Presence of unilateral guttae, with or without corneal oedema, and the presence of bullous keratopathy in the fellow eye; as well as (3) Absence of history of endotheliitis, previous peripheral iridotomy, cataract surgery, or any intraocular surgery; and (4) Presence of a minimum grade of 1 (12 or more central nonconfluent guttae), based on noncontact specular microscopy grading, as proposed by Krachmer et al. [4], or histopathologic evidence of Descemet’s membrane thickening and guttae for patients who had undergone keratoplasty. The control subjects were selected based on the following criteria: (1) >60 years old, (2) no guttae observed by slit-lamp microscopy, and (3) absence of previous peripheral iridotomy, cataract surgery, or any intraocular surgery. After obtaining informed consent, peripheral blood leucocytes were obtained from the patients and genomic DNA was extracted using a Puregene blood kit (Qiagen, Hilden, Germany). The amount and quality of DNA were analysed by UV spectrophotometry (NanoDrop; NanoDrop Technologies, DE).
Measurement of SNPs
The primers used for PCR are listed in Supplementary Table 1. PCR reactions were performed with Ex Taq DNA polymerase (Takara Bio Inc., Otsu, Japan) under the following conditions: denaturation at 94 °C for 30 s, 32–35 cycles of annealing at 54–60 °C for 30 s, and elongation at 72 °C for 30 s. The PCR products were separated by electrophoresis on 2% agarose gels in Tris-acetate buffer, stained with ethidium bromide, and analysed with an LAS4000S luminescence imager (Fuji Film, Tokyo, Japan). The PCR products were extracted from the agarose gels using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI). Direct sequencing was carried out with a TaqDyeDeoxy Terminator Cycle Sequencing Kit and a SeqStudio Genetic Analyzer (Life Technologies, Carlsbad, CA), according to the manufacturer’s instructions.
Evaluation of the CTG repeat length
PCR reactions were performed with Ex Taq DNA polymerase (Takara Bio Inc., Otsu, Japan) under the following conditions: denaturation at 94 °C for 30 s, 32–35 cycles of annealing at 54–60 °C for 30 s, and elongation at 72 °C for 30 s. Primers used for PCR were a sense primer (5′-CAGATGAGTTTGGTGTAAGATGCATTTG-3′) and an anti-sense primer (5′-ACAAGCAGAAAGGGGGCTGCAAAGC-3′). The PCR products were separated by electrophoresis on 2% agarose gels in Tris-acetate buffer, stained with ethidium bromide, and analysed with an LAS4000S luminescence imager (Fuji Film, Tokyo, Japan). The 305-bp bands were then extracted from the agarose gels using the Wizard SV Gel and PCR Clean-Up System. Direct sequencing was performed with a TaqDyeDeoxy Terminator Cycle Sequencing Kit and a 373A DNA sequencer (Applied Biosystems, Foster City, CA), according to the manufacturer’s instructions.
Triplet repeat primed PCR (TP-PCR) was also performed to detect the CTG repeat lengths, as previously reported [14]. Briefly, the PCR was performed with the following conditions: 20 ng of genomic DNA, 0.2 μmol/L of locus-specific (forward) fluorescent primer P1, 0.006 μmol/L of repeat-specific (reverse) primer P4, 0.2 μmol/L of primer P3, 200 μmol/L dNTPs, 1 mmol/L MgCl2, and 1.25 U of TksGflex DNA Polymerase (Takara Bio Inc., Otsu, Japan). The primers were those previously reported by Mootha and colleagues. PCR was performed under the following conditions: initial denaturation of 9 min at 95 °C, 10 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, and elongation at 68 °C for 4 min, followed by 25 cycles of denaturation at 95 °C for 45 s, annealing at 62 °C for 45 s, elongation at 68 °C for 4 min 15 s, and final elongation at 68 °C for 10 min. The TP-PCR products were mixed with GeneScan 500 LIZ dye Size Standard (Applied Biosystems, Foster City, CA), subjected to capillary electrophoresis on an ABI 3730 Genetic Analyzer, and analysed with a SeqStudio Genetic Analyzer.
Results
The mean age of the Thai patients with FECD was 62.7 ± 13.6 years (range, 21–85 years) (10 male and 44 female) and the mean age of controls was 68.2 ± 6.5 years (range, 60–87 years) (12 male and 42 female). No statistically significant differences were observed in the subject age and male-to-female ratio between the FECD and control groups. The rs613872, rs2123392, rs17089887, rs1452787, and rs1348047 SNPs of TCF4, which were previously reported to be correlated with FECD, were evaluated in Thai patients with FECD. The results revealed that these patients differed from the previously reported Caucasian cohorts as they rarely harboured rs613872 (2/54). By contrast, the other four SNPs (rs2123392, rs17089887, rs1452787, and rs1348047) were found in approximately half of the Thai patients. The frequencies of these SNPs were not statistically different between the FECD cases and the controls, and they were consistent with those in East Asian (EAS) populations according to the database of the 1000 Genomes Project Phase 3, based on Ensembl v80 GRCh37 (Table 1).
Table 1.
Genotyping of the reported single nucleotide polymorphisms (SNPs) in TCF4
| Chr position | rs ID | Mutation | 1000 Genomes (EUR) | 1000 Genomes (EAS) | Control | Case | P value |
|---|---|---|---|---|---|---|---|
| n = 1006 | n = 1008 | n = 54 | n = 54 | ||||
| 18:55543071 | rs613872 | T > G | G = 0.1769 | G = 0.0040 | 0/108 | 2/108 | 0.155 |
| 18:55547634 | rs2123392 | T > C | C = 0.3588 | C = 0.3611 | 45/108 | 41/108 | 0.578 |
| 18:55541025 | rs17089887 | T > C | C = 0.0885 | C = 0.4395 | 46/108 | 47/108 | 0.891 |
| 18:55539976 | rs1452787 | A > G | G = 0.2972 | G = 0.3591 | 45/108 | 41/108 | 0.578 |
| 18:55382827 | rs1348047 | G > T | T = 0.2734 | T = 0.4950 | 55/108 | 47/108 | 0.276 |
Direct sequencing and TP-PCR demonstrated that no control subjects harboured a CTG trinucleotide repeat (TNR) higher than 40 repeats. Conversely, 21 of the 54 patients (39%; 1 homozygous and 20 heterozygous) harboured CTG TNR lengths greater than 40, while 33 patients of the 54 patients (61%) did not harbour TNR lengths greater than 40 (P < 0.001). The mean age of the 5 male and 16 female patients harbouring ≥40 CTG TNR was 65.3 ± 9.6 years (range, 39–78 years), while the mean age of the 5 male and 28 female patients harbouring <40 CTG TNR was 61.1 ± 15.6 years (range, 21–85 years). No statistically significant difference was observed in either age or gender among control subjects, patients with or without CTG TNR expansion higher than 40 repeats (Table 2). In agreement with the previous study, the distribution of the TNR length was bimodal, and fourteen patients harboured ≥100 CTG TNR (Fig. 1). One patient with FECD (a 66-year-old female) was homozygous for the CTG TNR expansion, and she was considered to be a Krachmer grade 2 and exhibited a transparent cornea. Clinical manifestations of the representative Thai patients with FECD harbouring CTG TNR < 40 and ≥40 are shown in Supplementary Fig. 1.
Table 2.
Demographic data of subjects with Fuchs endothelial corneal dystrophy (FECD) with or without cytosine-thymine-guanine (CTG) trinucleotide repeat (TNR) expansion
| Control | No expansion | Expansion | ||
|---|---|---|---|---|
| (n = 54) | CTG < 40 | CTG ≥ 40 | P value | |
| (n = 33) | (n = 21) | |||
| Age, years | 0.012a | |||
| 68.2 ± 6.5 (60,87) | 61.1 ± 15.6 (21,85) | 65.3 ± 9.6 (39,78) | ||
| Sex | 0.663b | |||
| Male | 12 | 5 | 5 | |
| Female | 42 | 28 | 16 |
Data represent the mean ± SD (minimum, maximum)
aP value of analysis of variance (ANOVA)
bP value of Chi-square test
Fig. 1.
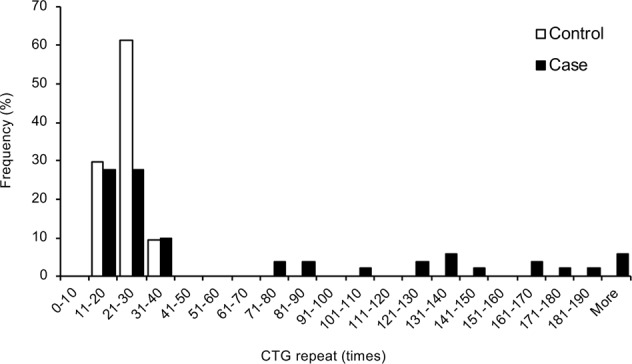
Frequency histogram of the cytosine–thymine–guanine (CTG) repeat length. The CTG trinucleotide repeat (TNR) length of the longest allele determined by direct sequencing and triplet repeat primed PCR (TP-PCR) in 54 subjects with Fuchs endothelial corneal dystrophy (FECD) and 54 non-FECD Thai controls is shown as a histogram. The distribution of TNR length in FECD cases was bimodal, and 14 patients harboured ≥100 CTG repeats
Discussion
Baratz et al. used GWAS to investigate 280 FECD cases and 410 controls and identified the minor allele (G) in rs613872 more frequently in the FECD cases (G = 0.40) than in non-FECD controls (G = 0.15); the odds ratios for this minor allele were 5.5 and 30 for being heterozygous and homozygous, respectively [12]. Subsequent reports in independent cohorts replicated the association between rs613872 and FECD. In addition to rs613872, other SNPs were identified in some populations, especially from non-Caucasian cohorts, indicating ethnic variation of SNPs in TCF4 [16, 20–23]. No previous study has examined SNPs in TCF4 in FECD in Thailand; therefore, we first looked for the reported SNPs in our Thai cohort. We found that only two among the 54 FECD subjects were polymorphic with regard to rs613872. Four other reported SNPs (rs2123392, rs17089887, rs1452787, and rs1348047) were found in our cases, but these SNPs are not rare polymorphisms, according to our findings in the controls and the 1000 Genomes Project database. We found that none of the reported SNPs in TCF4 showed an association with FECD in the Thai population.
The first report showing TNR expansion in TCF4, conducted in a Caucasian population, revealed that 52 of 66 (79%) FECD cases harboured CTG TNR ≥ 50, whereas only 2 of 63 (3%) controls had CTG TNR ≥ 50 [13]. Replication studies in an independent Caucasian cohort showed that 73% of the patients with FECD and 7% of the control subjects had CTG TNR expansion [14]. In a large hospital-based study in US, 62% of 574 FECD cases and 3.7% of 354 controls harboured TNR length >40 [18]. This association between TNR and FECD was also replicated in non-Caucasian cohorts, but the proportion of the patients harbouring TNR was lower than that observed in Caucasians. For example, in a Chinese cohort, 25 of 57 (44%) cases and 2 of 121 (1.7%) control subjects harboured CTG TNR lengths >50 [15], whereas these numbers were 15 of 44 (34%) cases and 5 of 97 (5.2%) control subjects in an Indian cohort [16], and 12 of 47 (26%) cases and 0 of 96 (0%) control subjects in a Japanese cohort [17]. In the current study, 21 of 54 patients (39%) and 0 of 54 controls (0%) in our Thai cohort harboured CTG TNR lengths ≥40. Our study in the Thai population provided further evidence that the harbouring of a CTG TNR expansion in TCF4 patients with FECD is a global finding, although the proportion was again lower in our Thai population when compared with the previously reported proportion in Caucasians.
In the current study, we utilised TP-PCR to determine very large TNR expansions, as these large expansions were difficult to detect by direct sequencing of PCR products. However, measuring very large repeats is challenging, and mosaicism within the leucocyte population further complicates these measurements. Therefore, the possibility of underestimation of patients harbouring TNR length ≥40 should be kept in mind due to the difficulty of obtaining accurate measurements of very large repeats.
TCF4 is a member of the basic helix-loop-helix (bHLH) family of transcription factors that are expressed in various tissues, such as brain, muscle, liver, lung, testis, and cornea [24]. Associations with TCF4 have been identified for primary sclerosing cholangitis and Pitt-Hopkins syndrome, as well as FECD [25]. However, probably because of the varied roles played by TCF4 in fundamental cellular events, the mechanistic cascade that determines the contribution of TCF4 to the phenotypes of these diseases remains unclear. Du and colleagues reported that poly(CUG)n RNA accumulated as RNA foci in the corneal endothelial cells of patients with FECD, and induced RNA toxicity and missplicing [26]. The involvement of repeat-associated non-ATG translation, a protein translation mechanism that has no requirement for an initiating ATG, was also suggested to explain TNR expansion in TCF4 as part of the pathophysiology of FECD [27]. A recent study has shown that antisense oligonucleotide targeting (CAG)7 could reduce the gain-of-function RNA toxicity caused by TNR in TCF4, supporting TNR as a potent therapeutic target [28]. Our investigations of the pathophysiology of FECD have resulted in the establishment of a cellular model for the corneal endothelium of patients with FECD. We have also demonstrated that the genes that induce the epithelial–mesenchymal transition (EMT) play an important role in the production of extracellular matrix materials, such as type 1 collagen and fibronectin, which are components of guttae [29]. Sobrado et al. have also reported that overexpression of TCF4 in Madin–Darby canine kidney cells generated motile and highly invasive phenotypes that acquired EMT-related markers, and they proposed that TCF4 was an important regulator of the EMT [30]. Pathway analysis also showed that knockdown of TCF4 in neuroblastoma cells altered the expression of EMT-related genes [31]. However, the possibility that TCF4 plays a role in regulating the EMT has not been clearly elucidated in the corneal endothelium. Therefore, further studies of TCF4 function, focusing on the possible connection with the regulation of the EMT, are anticipated to elucidate the pathophysiology of FECD.
In conclusion, the findings of an association between SNPs in TCF4 and FECD differed depending on the ethnic background of the patients. The CTG repeat in intron 3 of TCF4 was always identified in all ethnic groups studied thus far, but with some variations in frequency. Here, we provide additional evidence that the CTG TNR in TCF4 is associated with FECD in the Thai population. To the best of our knowledge, this is the first genetic analysis of FECD in Thailand.
Summary
What was known before
A significant association between the SNP in the TCF4 and late-onset FECD was shown in the Caucasian population.
FECD cases frequently harbour intronic TNR expansion in TCF4 with CTG repeat ≥50 in the Caucasian population, but the proportion of the patients harbouring TNR was lower in non-Caucasians.
Ethnic variation of SNPs and TNR expansion in TCF4 is indicated, and the genetic analysis of FECD in Thai cases has not yet reported.
What this study adds
Only 2 among the 54 FECD subjects in the Thai cohort were polymorphic with regard to rs613872.
Twenty-one of 54 patients with FECD (39%) and 0 of 54 non-FECD controls (0%) in the Thai cohort harboured CTG TNR length ≥40.
This study is the first genetic analysis of FECD in Thailand which provided additional evidence that CTG repeat in TCF4 was present in all ethnic groups studied thus far with some variations in frequency.
Supplementary information
SF1: Clinical manifestation of Thai patients with FECD
Acknowledgements
The authors wish to thank Ms. Emi Ueda and Ms. Kyoko Watanabe for technical support.
Funding
This work was supported by the Programme for the Strategic Research Foundation at Private Universities from MEXT (NK and NO) and by Chulalongkorn Academic Advancement into Its 2nd Century Project and the Thailand Research Fund (BRG5980001 and DPG6180001).
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to disclose.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/3/2019
The original version of this Article contained an incorrect affiliation for Rungnapa Ittiwut. This has now been corrected in both the PDF and HTML versions of the Article.
Supplementary information
The online version of this article (10.1038/s41433-019-0595-8) contains supplementary material, which is available to authorised users.
References
- 1.Weisenthal R, Streeten B. Descemet’s membrane and endothelial dystrophies. Cornea Third Ed. 2011;1:845–64. doi: 10.1016/B978-0-323-06387-6.00080-5. [DOI] [Google Scholar]
- 2.Eghrari AO, Riazuddin SA, Gottsch JD. Fuchs corneal dystrophy. Prog Mol Biol Transl Sci. 2015;134:79–97. doi: 10.1016/bs.pmbts.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Cross HE, Maumenee AE, Cantolino SJ. Inheritance of Fuchs’ endothelial dystrophy. Arch Ophthalmol. 1971;85:268–72. doi: 10.1001/archopht.1971.00990050270002. [DOI] [PubMed] [Google Scholar]
- 4.Krachmer JH, Purcell JJ, Jr., Young CW, Bucher KD. Corneal endothelial dystrophy. A study of 64 families. Arch Ophthalmol. 1978;96:2036–9. doi: 10.1001/archopht.1978.03910060424004. [DOI] [PubMed] [Google Scholar]
- 5.Rosenblum P, Stark WJ, Maumenee IH, Hirst LW, Maumenee AE. Hereditary Fuchs’ dystrophy. Am J Ophthalmol. 1980;90:455–62. doi: 10.1016/S0002-9394(14)75011-1. [DOI] [PubMed] [Google Scholar]
- 6.Vithana EN, Morgan PE, Ramprasad V, Tan DT, Yong VH, Venkataraman D, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–66. doi: 10.1093/hmg/ddm337. [DOI] [PubMed] [Google Scholar]
- 7.Mehta JS, Vithana EN, Tan DT, Yong VH, Yam GH, Law RW, et al. Analysis of the posterior polymorphous corneal dystrophy 3 gene, TCF8, in late-onset Fuchs endothelial corneal dystrophy. Investig Ophthalmol Vis Sci. 2008;49:184–8. doi: 10.1167/iovs.07-0847. [DOI] [PubMed] [Google Scholar]
- 8.Riazuddin SA, Vithana EN, Seet LF, Liu Y, Al-Saif A, Koh LW, et al. Missense mutations in the sodium borate cotransporter SLC4A11 cause late-onset Fuchs corneal dystrophy. Hum Mutat. 2010;31:1261–8. doi: 10.1002/humu.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riazuddin SA, Zaghloul NA, Al-Saif A, Davey L, Diplas BH, Meadows DN, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riazuddin SA, Parker DS, McGlumphy EJ, Oh EC, Iliff BW, Schmedt T, et al. Mutations in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet. 2012;90:533–9. doi: 10.1016/j.ajhg.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riazuddin SA, Vasanth S, Katsanis N, Gottsch JD. Mutations in AGBL1 cause dominant late-onset Fuchs corneal dystrophy and alter protein-protein interaction with TCF4. Am J Hum Genet. 2013;93:758–64. doi: 10.1016/j.ajhg.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, et al. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;363:1016–24. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- 13.Wieben ED, Aleff RA, Tosakulwong N, Butz ML, Highsmith WE, Edwards AO, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012;7:e49083. doi: 10.1371/journal.pone.0049083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha VV, Gong X, Ku HC, Xing C. Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs’ endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2014;55:33–42. doi: 10.1167/iovs.13-12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing C, Gong X, Hussain I, Khor CC, Tan DT, Aung T, et al. Transethnic replication of association of CTG18.1 repeat expansion of TCF4 gene with Fuchs’ corneal dystrophy in Chinese implies common causal variant. Investig Ophthalmol Vis Sci. 2014;55:7073–8. doi: 10.1167/iovs.14-15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanda GG, Padhy B, Samal S, Das S, Alone DP. Genetic association of TCF4 intronic polymorphisms, CTG18.1 andrs17089887, with Fuchs’ endothelial corneal dystrophy in an Indian population. Investig Ophthalmol Vis Sci. 2014;55:7674–80. doi: 10.1167/iovs.14-15297. [DOI] [PubMed] [Google Scholar]
- 17.Nakano M, Okumura N, Nakagawa H, Koizumi N, Ikeda Y, Ueno M, et al. Trinucleotide repeat expansion in the TCF4 Gene in Fuchs’ endothelial corneal dystrophy in Japanese. Investig Ophthalmol Vis Sci. 2015;56:4865–9. doi: 10.1167/iovs.15-17082. [DOI] [PubMed] [Google Scholar]
- 18.Vasanth S, Eghrari AO, Gapsis BC, Wang J, Haller NF, Stark WJ, et al. Expansion of CTG18.1 trinucleotide repeat in TCF4 Is a potent driver of Fuchs’ corneal dystrophy. Investig Ophthalmol Vis Sci. 2015;56:4531–6. doi: 10.1167/iovs.14-16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foja S, Luther M, Hoffmann K, Rupprecht A, Gruenauer-Kloevekorn C. CTG18.1 repeat expansion may reduce TCF4 gene expression in corneal endothelial cells of German patients with Fuchs’ dystrophy. Graefes Arch Clin Exp Ophthalmol. 2017;255:1621–31. doi: 10.1007/s00417-017-3697-7. [DOI] [PubMed] [Google Scholar]
- 20.Li YJ, Minear MA, Rimmler J, Zhao B, Balajonda E, Hauser MA, et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One. 2011;6:e18044. doi: 10.1371/journal.pone.0018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thalamuthu A, Khor CC, Venkataraman D, Koh LW, Tan DT, Aung T, et al. Association of TCF4 gene polymorphisms with Fuchs’ corneal dystrophy in the Chinese. Investig Ophthalmol Vis Sci. 2011;52:5573–8. doi: 10.1167/iovs.11-7568. [DOI] [PubMed] [Google Scholar]
- 22.Wang KJ, Jhanji V, Chen J, Law RW, Leung AT, Zhang M, et al. Association of transcription factor 4 (TCF4) and protein tyrosine phosphatase, receptor type G (PTPRG) with corneal dystrophies in southern Chinese. Ophthalmic Genet. 2014;35:138–41. doi: 10.3109/13816810.2013.804098. [DOI] [PubMed] [Google Scholar]
- 23.Oldak M, Ruszkowska E, Udziela M, Ozieblo D, Binczyk E, Sciezynska A, et al. Fuchs endothelial corneal dystrophy: strong association with rs613872 not paralleled by changes in corneal endothelial TCF4 mRNA level. Biomed Res Int. 2015;2015:640234. doi: 10.1155/2015/640234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5’ exon usage and splicing. PLoS One. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. The emerging roles of TCF4 in disease and development. Trends Mol Med. 2014;20:322–31. doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Du J, Aleff RA, Soragni E, Kalari K, Nie J, Tang X, et al. RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J Biol Chem. 2015;290:5979–90. doi: 10.1074/jbc.M114.621607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soragni E, Petrosyan L, Rinkoski TA, Wieben ED, Baratz KH, Fautsch MP, et al. Repeat-associated non-ATG (RAN) translation in Fuchs’ endothelial corneal dystrophy. Investig Ophthalmol Vis Sci. 2018;59:1888–96. doi: 10.1167/iovs.17-23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarouchlioti C, Sanchez-Pintado B, Hafford Tear NJ, Klein P, Liskova P, Dulla K, et al. Antisense therapy for a common corneal dystrophy ameliorates TCF4 repeat expansion-mediated toxicity. Am J Hum Genet. 2018;102:528–39. doi: 10.1016/j.ajhg.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura N, Minamiyama R, Ho LT, Kay EP, Kawasaki S, Tourtas T, et al. Involvement of ZEB1 and Snail1 in excessive production of extracellular matrix in Fuchs endothelial corneal dystrophy. Lab Investig. 2015;95:1291–304. doi: 10.1038/labinvest.2015.111. [DOI] [PubMed] [Google Scholar]
- 30.Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, Portillo F, et al. The class I bHLH factors E2-2A and E2-2B regulate EMT. J Cell Sci. 2009;122:1014–24. doi: 10.1242/jcs.028241. [DOI] [PubMed] [Google Scholar]
- 31.Forrest MP, Waite AJ, Martin-Rendon E, Blake DJ. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8:e73169. doi: 10.1371/journal.pone.0073169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SF1: Clinical manifestation of Thai patients with FECD


