Abstract
Background and Aims
Resistance to the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) in wild radish (Raphanus raphanistrum) appears to be due to a complex, multifaceted mechanism possibly involving enhanced constitutive plant defence and alterations in auxin signalling. Based on a previous gene expression analysis highlighting the plasma membrane as being important for 2,4-D resistance, this study aimed to identify the components of the leaf plasma membrane proteome that contribute to resistance.
Methods
Isobaric tagging of peptides was used to compare the plasma membrane proteomes of a 2,4-D-susceptible and a 2,4-D-resistant wild radish population under control and 2,4-D-treated conditions. Eight differentially abundant proteins were then targeted for quantification in the plasma membranes of 13 wild radish populations (two susceptible, 11 resistant) using multiple reaction monitoring.
Key Results
Two receptor-like kinases of unknown function (L-type lectin domain-containing receptor kinase IV.1-like and At1g51820-like) and the ATP-binding cassette transporter ABCB19, an auxin efflux transporter, were identified as being associated with auxinic herbicide resistance. The variability between wild radish populations suggests that the relative contributions of these candidates are different in the different populations.
Conclusions
To date, no receptor-like kinases have been reported to play a role in 2,4-D resistance. The lectin-domain-containing kinase may be involved in perception of 2,4-D at the plasma membrane, but its ability to bind 2,4-D and the identity of its signalling partner(s) need to be confirmed experimentally. ABCB19 is known to export auxinic compounds, but its role in 2,4-D resistance in wild radish appears to be relatively minor.
Keywords: 2,4-Dichlorophenoxyacetic acid; dicamba; herbicide resistance; iTRAQ; multiple reaction monitoring; plasma membrane; Raphanus raphanistrum; receptor-like kinases; wild radish
INTRODUCTION
The plant hormone indole-3-acetic acid (IAA), or auxin, is a simple molecule that elicits a complicated array of responses in plants and is involved in every aspect of plant growth and development (Vanneste and Friml, 2009). Synthetic auxins, first developed in the 1940s, have long been used as plant growth regulators to promote cell division in tissue and cell culture (Lloyd et al., 1980) or to regulate post-harvest fruit ripening (Chamarro et al., 2001), and as herbicides to selectively remove dicotyledonous weeds from cereal crops (Peterson et al., 2016). Although many synthetic auxin molecules do not superficially resemble IAA, the precise distance between their negatively charged carboxyl group and the weak positive charge on their aromatic ring determines auxin activity, whilst the concentration at which the auxin is applied, and the sensitivity of the plant species in question, determine whether the auxin acts as a plant growth promoter (low concentrations) or a herbicide (high concentrations) (reviewed in Grossmann, 2003).
With the advent of modern molecular tools, the previously obscure pathways of auxin signalling and perception are becoming clearer, also providing insights into the mode of action of synthetic auxin herbicides such as 2,4-dichlorophenoxyacetic acid (2,4-D) and 3,6-dichloro-2-methoxybenzoic acid (dicamba) (Grossmann, 2010). The great complexity surrounding auxin action is partially due to the involvement of large gene families in auxin perception, signalling, transport and metabolism (Matthes et al, 2019), which provides a high level of redundancy and allows different metabolic and tropic pathways to be activated or deactivated depending on the particular family members involved. In arabidopsis, there are six auxin receptors (exemplified by the canonical TIR1) with different specificities for different types of natural and synthetic auxin (e.g. Walsh et al., 2006); 29 co-receptors of the Aux/IAA transcriptional repressor family, which prevent expression of auxin-responsive genes until auxin levels become high enough to facilitate binding of the Aux/IAAs to TIR1 and their consequent degradation via the 26S proteasome (Ljung, 2013); and 23 auxin response factors (ARFs) which mediate expression of auxin-responsive genes once the Aux/IAA repressors have been removed (Chandler, 2016). Added to this, there are four auxin influx transporters, eight PIN-type auxin efflux transporters (most involved in polar transport) and 22 ABCB-type efflux transporters (reviewed in Matthes et al., 2019), of which at least four are involved in auxin transport (Geisler et al., 2017), as well as various proteins localized to intracellular membranes which show auxin transport activity (Skalický et al., 2018).
Given this complexity, it is not surprising that the assignment of particular members of the auxin signalling pathway to specific physiological functions has been difficult. Similarly, the identification of the molecular mechanisms of resistance to auxinic herbicides has proceeded slowly because there are so many herbicide ‘target sites’ that could potentially have been altered in weeds with evolved resistance. It is only very recently that the first report of an auxin-resistant weed with a rigorously characterized alteration in auxin signalling has emerged: a mutation in the degron region of the Aux/IAA repressor IAA16 confers dicamba and 2,4-D resistance in Kochia scoparia because the protein can no longer be degraded by the 26S proteasome and continues to repress auxin-responsive genes in the presence of the herbicide (LeClere et al., 2018).
The 2,4-D resistance mechanism(s) in wild radish (Raphanus raphanistrum), a major weed of regions with a Mediterranean-type climate such as southern Australia, has remained elusive, although most of the classical non-target site herbicide resistance mechanisms (reduced herbicide uptake, metabolic detoxification and vacuolar or apoplastic sequestration) have been ruled out (Goggin et al., 2016). Similarly, the restricted long-distance 2,4-D transport observed in some resistant populations, apparently mediated by loss of function of an ABCB transporter, does not obviously correspond to the resistance level (Goggin et al., 2018). A transcriptomic study indicated that the rapid upregulation of expression of the Aux/IAA repressors IAA29 and IAA30 and the key signalling protein mitogen-activated protein kinase (MAPK) kinase kinase 1 (MEKK1) may contribute to resistance in one particular wild radish population, but transcript levels of IAA30 and MEKK1 did not correlate with resistance when measured in other populations (Goggin et al., 2018). However, the constitutive level of phosphorylation of MAPK was correlated with resistance, suggesting that post-transcriptional processes may be equally as important as gene expression. The same study highlighted the plasma membrane as a site of potential importance for 2,4-D resistance, particularly with respect to expression of receptor-like kinases involved in plant defence and immunity. Therefore, the proteome of the wild radish plasma membrane was investigated in the current study using isobaric tags for relative and absolute quantitation (iTRAQ) to discover potential resistance proteins in one population, followed by multiple reaction monitoring (MRM) to assess whether differential constitutive expression of these proteins in a suite of wild radish populations could explain 2,4-D resistance. An additional study on the soluble proteomes of a susceptible vs. a resistant population was performed using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE).
MATERIALS AND METHODS
Plant material and treatments
The 13 Raphanus raphanistrum populations (two susceptible to 2,4-D, named S1 and S2, and 11 resistant populations named R1–R11) characterized in Goggin et al. (2018) were used in the current study. Global comparisons on the effect of 2,4-D on a susceptible vs. a resistant population were made between populations S1 and R1, and selected proteins were quantified in untreated samples of all populations. Wild radish seedlings were grown in potting mix under controlled conditions (20/15 °C with a 12 h photoperiod of white fluorescent and LED light at 90 µmol min–1 m–2) as previously described (Goggin et al., 2016). When seedlings had reached the three-leaf stage, they were sprayed with 5 mm technical-grade 2,4-D in 0.2 % (v/v) Tween-20 using a custom-built cabinet sprayer (Owen et al., 2015) at a rate equivalent to 500 g 2,4-D ha–1, the recommended field rate in Australia. Control plants were sprayed with the same volume of 0.2 % (v/v) Tween-20, and treatments were imposed at the beginning of the light cycle. Leaves were harvested, and immediately processed for protein extraction, after 24 h for the soluble protein experiment (selected because the S1 and R1 plants first showed visible differences in 2,4-D symptoms at 24 h post-treatment) and after 12 h for the plasma membrane protein experiment (selected because the transcriptomic study in Goggin et al., 2018 indicated that differences in expression of genes encoding plasma membrane proteins between S1 and R1 are manifested at least as early as 2 h post-treatment). Each replicate for the soluble protein 2D-PAGE analysis (four replicates per sample type) consisted of three leaves, one taken from each of three different individuals. Plasma membranes were prepared from leaves taken from approx. 48 individuals for the iTRAQ analysis of populations S1 and R1, and from 12 individuals for the MRM study on all 13 populations. There were three replicates per population for both iTRAQ and MRM.
Extraction of soluble proteins
Polyethylene glycol (PEG) fractionation was used to remove most of the Rubisco protein present in the leaf tissue, allowing less abundant proteins to be visualized more readily. Protein extraction and fractionation was performed according to Aryal et al. (2012) except that the 15 % PEG supernatant was precipitated with methanol/ammonium acetate directly. The precipitated protein was resuspended in lysis buffer [7 m urea, 2 m thiourea, 4 % (w/v) CHAPS] (Aryal et al., 2012).
Preparation of plasma membrane proteins
Plasma membranes were prepared from wild radish leaves using aqueous two-phase partitioning with some modifications to eliminate the requirement for an ultracentrifuge. Briefly, leaves (30 g for iTRAQ samples, 7.5 g for MRM samples) were homogenized in isotonic buffer (Shimogawara and Usuda, 1993) lacking EDTA and EGTA, and the homogenate was filtered through Miracloth and centrifuged at 8200 g for 15 min. To obtain microsomes, 1 m MgCl2 was added to the supernatant to a final concentration of 50 mm and this was incubated on ice with gentle stirring for 30 min, before centrifugation at 8200 g for 30 min to pellet the microsomal membranes (Diesperger et al., 1974). The microsomal pellet was resuspended in BTP–MES buffer (10 mm bis-tris propane–MES pH 7.8, 250 mm sucrose, 50 mm NaCl, 1 mm EDTA, 1 mm EGTA) and added to a conventional 5.5 % (w/w) PEG 3500–5.5 % (w/w) dextran T500 phase system (32 mL total volume) (Shimogawara and Usuda, 1993). After the first round of partitioning, the upper phase was washed by repartitioning against fresh lower phase and the original lower phase was repartitioned against fresh upper phase. The second upper phase was also washed with fresh lower phase, and both upper phases were incubated on ice for 10 min with 0.02 % (w/v) Brij-58 to release cytosolic proteins from the plasma membrane vesicles (Elmore et al., 2012). Membranes were pelleted by diluting the Brij-58-treated suspension 1:1 with BTP–MES buffer and centrifuging 1.5 mL aliquots at 18 500 g for 60 min in a refrigerated microcentrifuge. At this point, plasma membrane and lower phase samples for marker enzyme assays were resuspended in a minimal volume of BTP–MES buffer (the lower phase was first diluted 10-fold and centrifuged to remove the dextran from the membrane pellet). Plasma membrane samples for protein extraction were washed by resuspension in water followed by centrifugation at 18 500 g for 60 min. This step was necessary to prevent precipitation of buffer components in the final acetone precipitation step. Washed membranes were resuspended in a final (combined) volume of around 30 µ L in water containing 1 mm phenylmethylsulfonyl fluoride (PMSF), warmed to room temperature, and SDS was added to a final concentration of 2 % (w/v) (Lin et al., 2012). Proteins were solubilized by vortex mixing for 20 min, and membrane residue was removed by centrifugation at 18 500 g for 15 min. After removal of an aliquot for protein assay, the solubilized proteins were precipitated with the addition of 9 vols of cold (–20 °C) acetone (Lin et al., 2012) and incubated overnight at –20 °C. Proteins were pelleted by centrifugation at 17 000 g for 20 min, and washed three times with fresh acetone. The pellets were briefly air-dried and stored at –80 °C until used for iTRAQ or MRM analysis.
Protein determination
Total soluble protein was measured using the method of Bradford (1976) with BioRad Dye Reagent Concentrate, whilst membrane protein was assayed using the BioRad RCDC kit, in a method modified from Lowry et al. (1951). Bovine serum albumin was used as a standard in both assays, and the appropriate buffer was included in the standards to account for possible interference.
Enzyme assays
In the plasma membrane preparations, vanadate-sensitive ATPase activity, a marker for the plasma membrane (Larsson et al., 1994), was measured at pH 7 over 30 min at 30 °C using the reaction conditions and malachite green-based phosphorus detection method of Schallar and deWitt (1995). NADPH-dependent cytochrome c reductase (endoplasmic reticulum marker) and cytochrome c oxidase (mitochondrial inner membrane marker) were assayed in a volume of 1 mL at 30 °C by continuously monitoring the change in absorbance of cytochrome c (Hodges and Leonard, 1974). Chlorophyll (thylakoid membrane marker) was measured according to Arnon (1949).
Soluble superoxide dismutase (SOD) activity was measured spectrophotometrically or in non-denaturing gels by homogenizing leaves in 50 mm KH2PO4 (pH 7.5) containing 1 mm EDTA, 5% (v/v) glycerol, 0.002 % (v/v) Triton X-100, 2 mm ascorbic acid, 5 mm dithiothreitol (DTT) and 1 mm PMSF (3 vols for spectrophotometry, 1 vol. for zymograms) and clarifying the extract by centrifugation at 12 000 g for 10 min (4°C). Extracts for spectrophotometric assay were then desalted into 50 mm KH2PO4 (pH 7.5) containing 1 mm EDTA using Sephadex G25 PD10 columns (GE Life Sciences), and SOD activity was measured according to Paoletti et al. (1986). SOD activity was detected on one-dimensional native PAGE (Goggin and Colmer, 2007) by loading 110 µ g of total protein per lane and running alongside 0.1–0.5 U of bovine erythrocyte SOD (Sigma-Aldrich), followed by staining using nitro blue tetrazolium/riboflavin (Beauchamp and Fridovich, 1971). Some gels were pre-incubated in 5 mm H2O2 for 30 min before staining, in order to inhibit FeSOD and Cu/ZnSOD isoforms (Asada et al., 1975). Activity band intensity was quantified from scanned gel images using Quantity One v4.2.1 (BioRad).
Soluble glutathione S-transferase (GST) activity was extracted from leaves using the same buffer as for SOD, with the additional inclusion of 5 % (w/v) polyvinylpyrrolidone (PVP) and 10 mm reduced glutathione (GSH). Extracts were desalted into 50 mm KH2PO4 (pH 8) containing 1 mm EDTA, 5 % (w/v) PVP and 10 mm GSH, and GST activity was measured spectrophotometrically using 1-chloro-2,4-dinitrobenzene (CDNB) as a substrate (Habig et al., 1974).
2D-PAGE and peptide sequencing
Proteins (800 µ g per gel, one gel per biological replicate) were separated in the first dimension by isoelectric focusing on 13 cm IPG strips, pH 3–10 NL (GE Life Sciences) for a total of 17.5 kVh, and in the second dimension by SDS–PAGE on 12.5 % polyacrylamide gels of 16 × 18 cm. Gels were stained with colloidal Coomassie Blue G-250 (Candiano et al., 2004) and imaged on a Microtek Bio-5000 gel scanner at a resolution of 1200 dpi. Spot patterns were compared using Progenesis SameSpots v4.5 (Nonlinear Dynamics), with manual editing of the automatic spot alignment. Detected spots that appeared to be artefacts or were poorly reproducible were excluded from further analysis. Genuine spots with a ≥2-fold difference in intensity [P < 0.05, as indicated by analysis of variance (ANOVA) performed by the SameSpots software] between populations or treatments were excised from the gel for identification. Peptide sequencing of trypsin-digested spots by MALDI-TOF-MS/MS (matrix-assisted laser desorption ionization-time of flight-tanden mass spectrometry) was performed by Proteomics International, using the Mascot search engine (www.matrixscience.com) to search the Ludwig NR mass spectrometry database (taxonomy: green plants) to identify proteins. Hits with a MOWSE protein score of >50 and with two or more matched peptides were considered to be positive identifications; hits with only one matched peptide were considered indicative.
iTRAQ and MRM analysis of plasma membrane proteins
Acetone-precipitated protein pellets (100 µ g of protein per sample for iTRAQ, 40 µ g for MRM) were solubilized with sodium deoxycholate and then reduced, alkylated and digested with trypsin according to the AB Sciex iTRAQ protocol. iTRAQ samples were labelled with reagents 114, 115, 116 and 117 using the manufacturer’s protocol (AB Sciex). All labelled samples were then pooled, and peptides were desalted on a Strata-X 33 µ m polymeric reversed phase column (Phenomenex), dried, then dissolved in 2 % (v/v) acetonitrile containing 0.1 % (v/v) formic acid and separated with a high pH gradient of acetontrile on a Zorbax C18 column (2.1 × 150 mm) connected to an Agilent 1100 HPLC system. Peptides were eluted at a flow rate of 0.2 mL min–1 with a linear gradient of 2–20 % (v/v) acetonitrile in 20 mm ammonium formate, and the 95 resulting fractions were concatenated into 12 fractions and lyophilized. Each fraction was loaded onto a Zorbax 300SB-C18, 3.5 µ m column (Agilent Technologies) and peptides were separated with a linear gradient of 10–40 % acetonitrile in 0.1 % (v/v) formic acid using a Shimadzu Prominence nano HPLC system coupled to a 5600 Triple TOF mass spectrometer (AB Sciex) for electrospray ionization (ESI)-MS analysis. Spectral data were analysed using ProteinPilot v5.0 (AB Sciex) against the RadishDB database of R. raphanistrum protein sequences (version: March 2018; 38 174 sequences) (Moghe et al., 2014). The local and global false discovery rates for identified proteins were automatically calculated by the ProteinPilot software using the reversed sequences of the RadishDB database, and found to be 1.0 % and <0.1 %, respectively. Proteins were required to have at least two unique peptides identified with high confidence to be retained for analysis.
Samples for MRM were solubilized and trypsin-digested using the same method as for the iTRAQ samples. Formic acid was then added to quench trypsin activity and precipitate the deoxycholate, and peptides were desalted as above and then loaded onto an Acclaim PepMap 100 C18 LC column, 2 µ m particle size × 150 mm (Thermo Scientific) and separated with a linear gradient of 10–40 % acetonitrile in 0.1 % (v/v) formic acid on a Thermo UltiMate 3000 nanoflow UHPLC system (Thermo Scientific) coupled to a 4000 Q-TRAP mass spectrometer (AB Sciex). MRM transitions for peptides were created and searched for in the mass spectrometer, and quantitation analysis of the peptide peak area was performed using Skyline v4.1 (MacCoss Lab Software). Based on the iTRAQ analysis, three ‘housekeeping proteins’ with high numbers of detected peptides and expression ratios close to 1 between the S1 and R1 populations, and between control and 2,4-D-treated samples, were selected and analysed alongside the target proteins. The selected housekeeping proteins were an uncharacterized tetratricopeptide repeat-containing protein (RadishDB accession no. RrC1915_p3), plasma membrane ATPase 11 (RrC4992_p2) and V-type ATPase catalytic subunit A (RrC12234_p2).
Data analysis and statistics
Proteins identified in the iTRAQ experiment which had a detected protein threshold (unused ProtScore) of <1.3 (i.e. were identified with <95 % confidence) as calculated by ProteinPilot were discarded from the analysis. Expression ratios were analysed separately in each replicate, with fold changes and P-values, indicating the significance of the differential expression, being calculated by ProteinPilot. Fold changes of >1.3 (P < 0.05) in each replicate were considered to be significant, and only those proteins with significantly different levels of expression in at least two of the three replicates were retained. Nine proteins with significantly different levels of expression in the S1 vs. R1 populations were selected for MRM-based quantification in all 13 wild radish populations (untreated), with the number of quantified peptides per protein ranging from one to five. Peptide quantification data for the housekeeping proteins (two peptides per protein) were plotted to check for inconsistencies between populations and/or peptides, which resulted in only one housekeeping protein, RrC1915_p3, being used for data normalization. The peak area for each peptide of interest was normalized against the sum of the two RrC1915_p3 peptide areas in the corresponding sample. For proteins with more than one peptide quantified, the normalized peak areas were averaged to give a ‘whole protein’ value. Differences between populations (>1.3-fold) were assessed using one-factor ANOVA and Fisher’s least significant difference (LSD) test at the 5 % level of significance. ANOVA and LSD were also used to evaluate differences between measured SOD and GST activities.
Potential relationships between levels of each quantified plasma membrane protein and the herbicide resistance parameters measured in the 13 wild radish populations in Goggin et al. (2018) were assessed by weighted least-squares regression using the Real Statistics Resource Pack software (release 6.2) for Excel (Zaiontz, 2019), at the 5 % level of significance. The presence of potential outliers distorting the correlation coefficient was detected by plotting and visually inspecting the data.
RESULTS
2,4-D has little effect on expression of soluble proteins that are detectable by 2D-PAGE
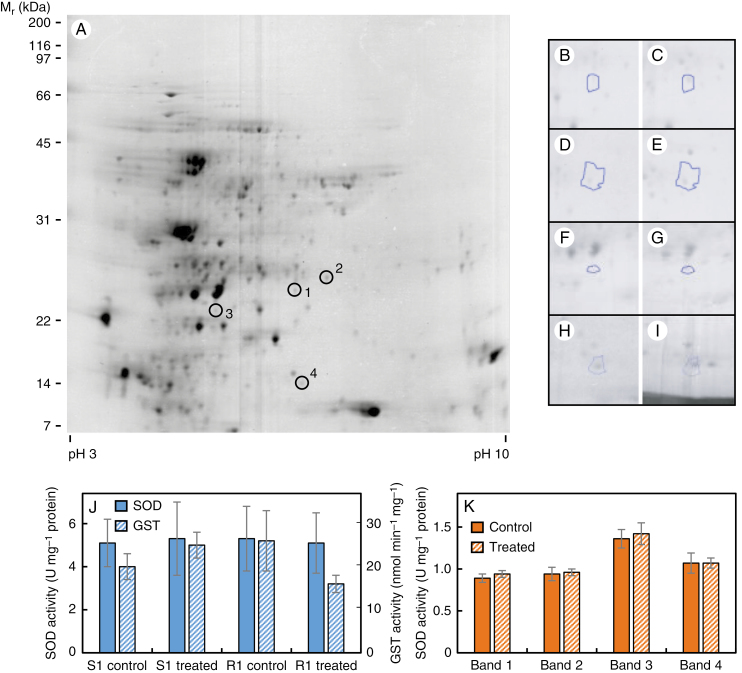
Soluble leaf proteins from 2,4-D-susceptible (S1) and -resistant (R1) populations, either treated or untreated with 500 g ha–1 2,4-D, were separated by 2D-PAGE and spot patterns were compared between populations and treatments. Twenty-four hours after 2,4-D treatment, expression of three soluble proteins [chloroplastic Fe- SOD, cytosolic GSTF9 and chloroplastic oxygen-evolving enhancer protein 2] had increased by 2- to 3-fold in S1 leaves (Fig. 1A–G; Table 1), whilst there were no significant changes in R1 leaves. Expression of one protein (chloroplastic ferredoxin-thioredoxin reductase catalytic chain) was 2-fold higher in 2,4-D-treated R1 leaves compared with treated S1 leaves (Fig. 1A, H, I; Table 1). To determine if the observed higher protein expression of SOD and GST isoforms in 2,4-D-treated vs. untreated S1 plants corresponded to higher enzyme activity, total soluble SOD and GST activity was measured in leaves from S1 and R1 plants sampled 24 h after 2,4-D (or mock) treatment. There were no significant differences between populations or treatments (Fig. 1J), and no differences in the activity of specific SOD isoforms, as illustrated by the similar band intensities on SOD zymograms (Fig. 1K).
Fig. 1.
2D-PAGE analysis of soluble leaf proteins from 2,4-D-susceptible (S1) and -resistant (R1) populations at 24 h after a foliar spray with 2,4-D or a mock solution. The positions of the four differentially expressed protein spots are indicated on the master gel image created from 16 individual gels (A). Also presented are magnified images of spot 1 (B, C); spot 2 (D, E); spot 3 (F, G); and spot 4 (H, I) in the following sample types: untreated population S1 (B, D, F); treated S1 (C, E, G, H); and treated R1 (I). Total soluble superoxide dismutase (SOD) and glutathione S-transferase (GST) activities were measured spectrophotometrically in leaf extracts from treated and untreated plants (J), and the activity of the four different SOD isozymes in S1 plants was quantified by scanning densitometry of activity bands on native gels (K). Values in (J) and (K) are means ± s.e. (n = 3); there were no significant differences between populations or treatments.
Table 1.
Identification of differentially expressed soluble proteins in leaves of 2,4-D-susceptible and -resistant wild radish plants at 24 h after 2,4-D treatment
| Spot no. | Response | Fold change | Protein match (UniProt accession no.) | MOWSE score | No. of peptides | Obs/Calc Mr (kDa) | Obs/Calc pI |
|---|---|---|---|---|---|---|---|
| 1 | Sc < St | 2.3 | Fe-superoxide dismutase (F4JRV2) | 58 | 1 | 23.0/23.8 | 5.8/6.0 |
| 2 | Sc < St | 3.0 | Glutathione S-transferase F9 (O80852) | 72 | 1 | 24.3/24.1 | 6.0/6.0 |
| 3 | Sc < St | 2.4 | Oxygen-evolving enhancer protein 2 (P11594) | 204 | 2 | 20.9/27.9 | 5.5/6.8 |
| 4 | St < Rt | 5.8 | Ferredoxin thioredoxin reductase catalytic chain (D7L839) | 127 | 4 | 13.6/16.9 | 5.8/8.1 |
Sc, susceptible control; St, susceptible treated; Rt, resistant treated; Obs, observed; Calc, calculated.
Isolation of plasma membranes from wild radish leaves
A previous transcriptomic study indicated that the plasma membrane may be an important site for 2,4-D resistance in wild radish plants, especially within the first 12 h of treatment (Goggin et al., 2018), so an investigation was made into the global expression of leaf plasma membrane proteins in 2,4-D-treated and -untreated S1 and R1 plants. Using traditional aqueous two-phase partitioning of an Mg2+-precipitated microsomal fraction, the activity of vanadate-sensitive ATPase (a plasma membrane marker) was enriched almost 9-fold in the plasma membrane preparation (upper phase) compared with the microsomal membranes collected in the lower phase, whilst mitochondrial cytochrome c oxidase and chlorophyll were enriched 25- and 73-fold, respectively, in the lower phase (Supplementary data Table S1). The endoplasmic reticulum marker, NADPH-dependent cytochrome c reductase, is known to also be present in the plasma membranes (Larsson et al., 1994) and was enriched 1.4-fold in the lower phase (Supplementary data Table S1). Based on the overall marker results, the wild radish plasma membrane preparations were deemed suitable for use in the proteomics study (Larsson et al., 1994).
Differential abundance of plasma membrane proteins in 2,4-D-susceptible and -resistant populations
Using the iTRAQ technique, a total of 407 proteins were identified (Supplementary data Table S2), of which 30 were differentially expressed (>1.3-fold, P < 0.05) between the S1 and R1 populations and/or between treated and untreated samples. Nine of these were discarded as they were annotated by the UniProt database (UniProt Consortium, 2019) as not being localized to, or associated with, the plasma membrane. Of the 21 remaining proteins, ten were differentially abundant in S1 vs. R1 under control conditions (eight were higher in R1), three under treated conditions (one was higher in R1), and eight, all in R1 only, were significantly affected by 2,4-D (seven were less abundant following treatment) (Table 2). Most of the proteins that were constitutively higher in the R1 population compared with S1 were related to plant defence and signalling. In contrast, the constitutive levels of the auxin efflux transporter ABCB19 were 3-fold lower in the R1 population compared with S1 (Table 2). Under 2,4-D treatment, the downregulated proteins in R1 were involved in diverse processes such as catalysis, nucleotide binding, signalling and transport, whilst the upregulated protein was a defence-related ABCG transporter (Table 2). The one protein that was more abundant in 2,4-D-treated R1 plants compared with S1 plants was also related to defence, being a heavy metal transporter (Table 2).
Table 2.
Identification of significantly (P < 0.05) differentially expressed plasma membrane proteins in leaves of populations S1 and R1 at 12 h after 2,4-D treatment, as identified by iTRAQ analysis
| Gene ID (RadishDB) | Response | Fold change | Protein match (UniProt accession no.) |
|---|---|---|---|
| RrC129_p7 | Sc < Rc | 8.0 ± 3.3 | Suppressor of BIR1, SOBIR1 (Q9SKB2)* |
| RrC17573_p1 | Sc < Rc | 6.7 ± 1.4 | L-type lectin domain-containing receptor kinase IV.1-like, LecRK-IV.1/LRK1 (O80939) * |
| RrC22_p12 | Sc < Rc | 16 ± 7.7 | Cationic amino acid transporter 1, CAT1 (Q84MA5) |
| RrC3859_p1 | Sc < Rc | 4.1 ± 0.9 | Putative receptor-like kinase At5g39000 (Q9FID8) |
| RrC4_p6 | Sc < Rc | 12 ± 8.2 | ABC transporter G family member 35, ABCG35 (Q7PC86)* |
| RrC4867_p2 | Sc < Rc | 6.5 ± 0.3 | Probable receptor-like kinase At1g51820 (C0LGG3)* |
| RrC538_p2 | Sc < Rc | 4.0 ± 1.7 | ABC transporter G family member 36, ABCG36 (Q9XIE2)* |
| RrC9574_p1 | Sc < Rc | 7.0 ± 2.8 | Plasma membrane-associated cation-binding protein 1, PCAP1 (Q96262)* |
| RrC22_p11 | Sc > Rc | 1.7 ± 0.6 | ADP-ribosylation factor GTPase activating protein AGD12 (Q9FVJ3) |
| RrC27p_9 | Sc > Rc | 3.0 ± 1.0 | ABC transporter B family member 19, ABCB19 (Q9LJX0)* |
| RrC1115_p3 | Rc > Rt | 7.5 ± 4.6 | Glutamate decarboxylase 2-like, GAD2 (Q42472) |
| RrC1913_p1 | Rc > Rt | 12 ± 4.8 | α-Carbonic anhydrase 4, BCA4 (Q94CE4) |
| RrC2092_p3 | Rc > Rt | 12 ± 3.4 | Remorin (O80837) |
| RrC2357_p1 | Rc > Rt | 4.8 ± 1.3 | Ca-transporting ATPase 8-like, ACA8 (Q9LF79) |
| RrC5541_p4 | Rc > Rt | 4.6 ± 0.3 | 60S ribosomal protein L23-a1, RPL23AA (Q8LD46) |
| RrC640_p7 | Rc > Rt | 22 ± 7.1 | Fasciclin-like arabinogalactan protein 8, FLA8 (O22126) |
| RrC9574_p1 | Rc > Rt | 8.1 ± 0.9 | Plasma membrane-associated cation-binding protein 1, PCAP1 (Q96262) |
| RrC1141_p3 | Rc < Rt | 26 ± 5.1 | ABC transporter G family member 40, ABCG40 (Q9M9E1)* |
| RrC1913_p1 | St > Rt | 23 ± 3.4 | α -Carbonic anhydrase 4, BCA4 (Q94CE4) |
| RrC247_p8 | St > Rt | 6.7 ± 0.9 | Uncharacterized protein At3g61260-like (Q9M2D8) |
| RrC12875_p1 | St < Rt | 15 ± 5.8 | Protein PLANT CADMIUM RESISTANCE 2-like, PCR2 (Q9LQU4)* |
Sc, susceptible control; Rc, resistant control; St, susceptible treated; Rt, resistant treated.
*These poteins were selected for MRM-based quantification in all 13 wild radish populations.
Given that the greatest difference between the S1 and R1 populations was observed under control conditions, seven proteins with constitutively different abundance in S1 vs. R1 were selected for MRM-based quantification in all 13 wild radish populations to assess their level of association with 2,4-D resistance. These were selected based on their annotated functions in UniProt and/or their potential interaction with the auxin signalling pathway, as indicated by the STRING database of functional protein interactions (Szklarczyk et al., 2019), namely: (1) SOBIR1, a receptor-like kinase promoting activation of plant defence and cell death (Liebrand et al., 2014); (2) LecRK-IV.1 (LRK1), an L-type lectin domain-containing receptor kinase which potentially interacts with at least four auxin signalling/transport-related proteins (Fig. 2A); (3) ABCG35, a defence-related transporter (UniProt Consortium, 2019); (4) At1g51820, a probable receptor-like kinase which is in the same extracellular kinase sub-network as SOBIR1 (Smakowska-Luzan et al., 2018) and may also interact with disease resistance and abscisic acid (ABA) signalling proteins (Fig. 2B); (5) ABCG36, which is required for plant immunity against pathogens (Campe et al., 2016) and mediates cadmium efflux (Kim et al., 2007); (6) PCAP1, which may be involved in calcium and phospholipid signalling (Nagata et al., 2016); and (7) ABCB19, an auxin efflux transporter (Geisler and Murphy, 2006). Two defence-related proteins which were differentially expressed under 2,4-D-treated conditions were also subjected to MRM to determine if they were constitutively different in S vs. R populations: ABCG40, which is involved in tolerance to a range of abiotic stresses (Migocka et al., 2017); and PCR2, a zinc transporter which may also be involved in cadmium resistance (Song et al., 2010). Ultimately, no suitable MRM peptides were detected for ABCG40, so this was excluded from further analysis.
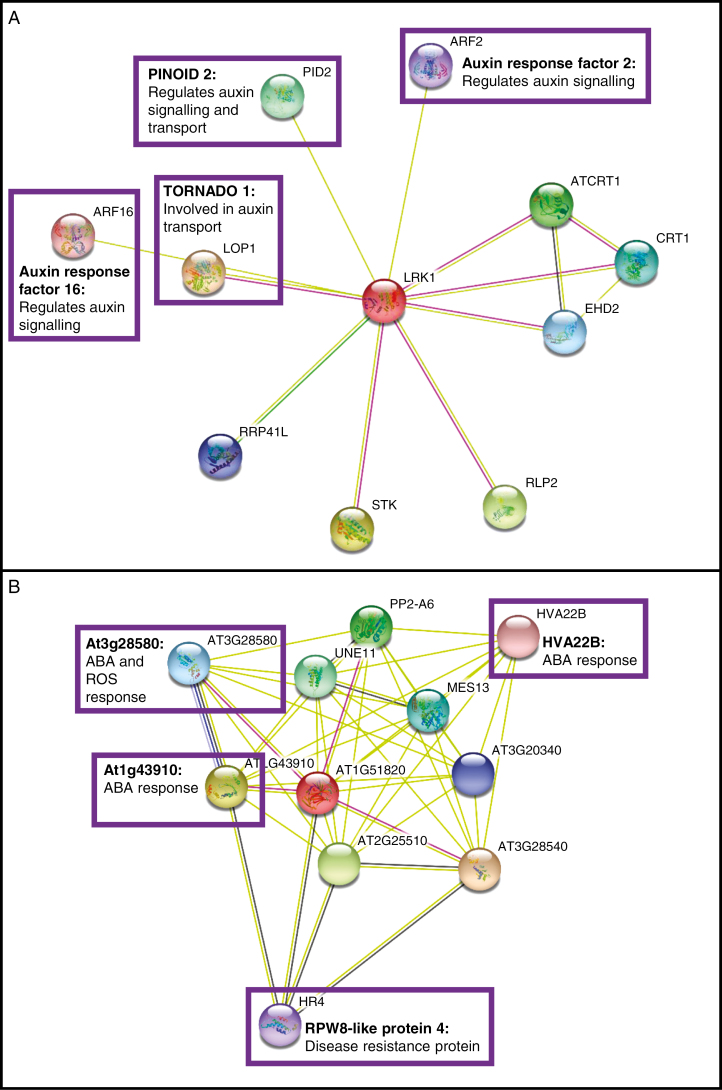
Fig. 2.
STRING diagrams of proteins potentially interacting with (A) LRK1 and (B) LRR-RLK At1g51820. Proteins that may play a role in response to auxin, abscisic acid (ABA) or reactive oxygen species (ROS), or in disease resistance, are highlighted with purple boxes. ABA and ROS are both generated in response to high auxin concentrations (Grossmann, 2010), and proteins potentially mediating disease resistance were identified in the iTRAQ study as being more highly expressed in the 2,4-D-resistant wild radish population R1.
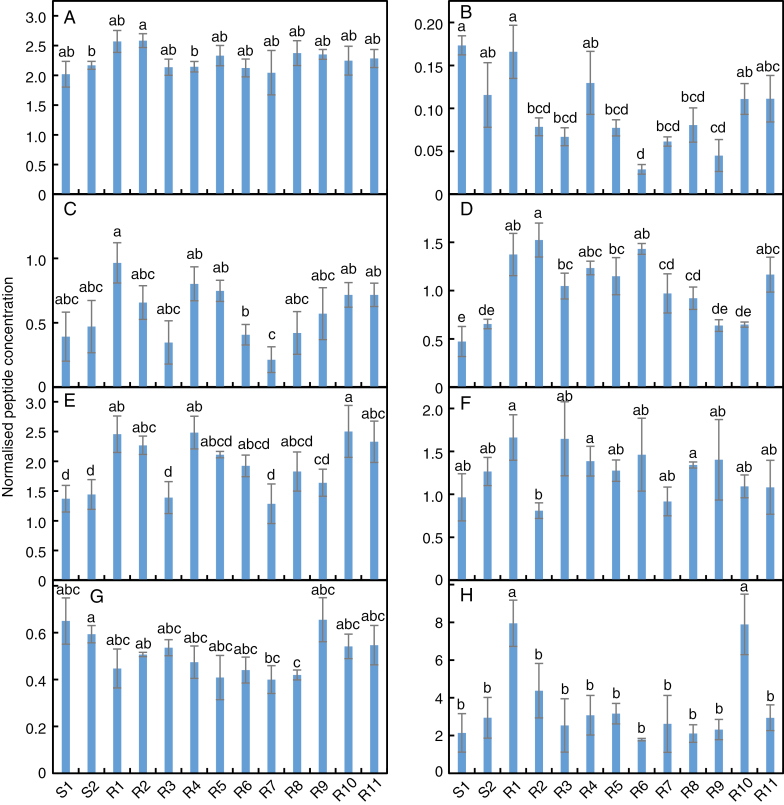
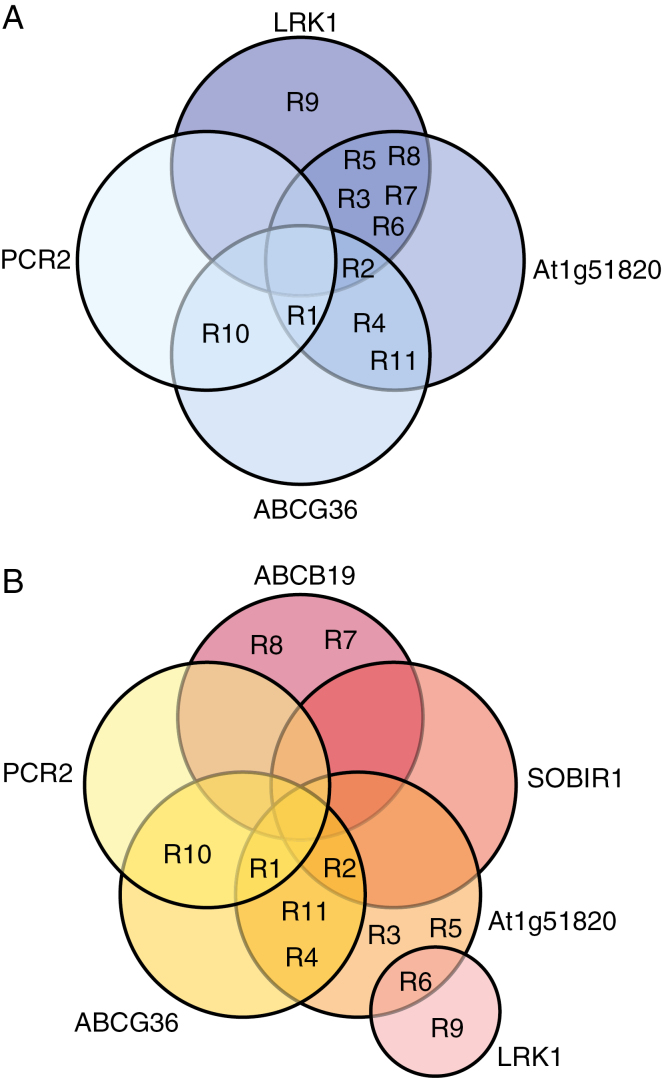
All proteins except for ABCG40 were detected in each population. In the comparison between populations S1 and R1, the fold change differences in protein levels were lower in the MRM experiment than in iTRAQ, and not all the differences were significant (Fig. 3). Among all S and R populations, there were no significant differences in protein level for ABCG35 or PCAP1, and few differences between populations for SOBIR1 (Fig. 3A, C, F). Most of the R populations had a lower abundance of LRK1 and higher levels of At1g51820 than did population S1, but there were fewer differences between the R populations and S2 (Fig. 3B, D). Five R populations had higher levels of ABCG36 than both of the S populations, and two (populations R1 and R10) had a 3-fold greater abundance of PCR2 (Fig. 3E, H). The Venn diagrams in Fig. 4 summarize the pattern of differential protein expression between the S populations and each of the R populations.
Fig. 3.
Quantification of plasma membrane proteins in two 2,4-D-susceptible (S1 and S2) and eleven 2,4-D-resistant (R1–R11) wild radish populations. Peptides from (A) SOBIR1, (B) LRK1, (C) ABCG35, (D) At1g51820, (E) ABCG36, (F) PCAP1, (G) ABCB19 and (H) PCR2 were quantified by MRM in plasma membranes isolated from the leaves of each population in the absence of 2,4-D treatment, and the data were normalized against the housekeeping protein RrC1915_p3. Values are means ± s.e. (n = 3), with different letters above bars indicating significant (P < 0.05, >1.3-fold) differences between populations.
Fig. 4.
Venn diagrams showing the pattern of differential abundance of plasma membrane proteins in each of the 2,4-D-resistant (R1–R11) populations compared with the susceptible S1 (A) and S2 (B) populations.
Although there were a greater number of R populations with differential expression of At1g51820 than of LRK1 when compared with the S populations (Fig. 4), the regression analysis showed that whilst survival of auxinic herbicide treatment (expressed as the herbicide dose required to kill 50 % of the population, LD50) was not correlated with At1g51820 levels, there were very strong, and moderately strong, negative correlations between LRK1 abundance and the LD50 values for 2,4-D and dicamba, respectively (Table 3). The ability of dicamba survivors to grow following treatment was, however, strongly positively correlated with At1g51820 abundance (Table 3). There was no relationship between 2,4-D translocation ability (Goggin et al., 2018) and ABCB19 abundance, in spite of the fact that ABCB19 is a known auxin efflux transporter (Geisler et al., 2017), but there was a moderate positive correlation between the level of this protein and the ability of 2,4-D survivors to grow following treatment (Table 3). Levels of the other proteins were not significantly correlated to any resistance parameter or to 2,4-D translocation ability (Supplementary data Table S3).
Table 3.
Weighted least-squares regression analysis of potential associations (expressed as the correlation coefficient, r, between auxinic herbicide resistance and constitutive expression of plasma membrane proteins)
| Protein | 2,4-D LD50 | Dicamba LD50 | 2,4-D biomass | Dicamba biomass | Translocation |
|---|---|---|---|---|---|
| LRK1 | –0.915*** | –0.668* | +0.354n.s. | –0.398n.s. | –0.239n.s. |
| At1g51820 | –0.219n.s. | –0.143n.s. | +0.105n.s. | +0.804*** | +0.192n.s. |
| ABCB19 | –0.174n.s. | +0.239n.s. | +0.567* | –0.244n.s. | –0.021n.s. |
Resistance to 2,4-D and dicamba was quantified in Goggin et al. (2018) in terms of the herbicide dose required to kill 50 % of the population (LD50) and the ability of survivors to grow following herbicide treatment (biomass). The ability of populations to translocate 2,4-D (expressed as % retention in the treated leaf) was quantified in the same study.
Asterisks denote significant r values (*P < 0.05; ***P < 0.001; n.s., not significant).
The full correlation matrix is presented in Supplementary data Table S3.
DISCUSSION
The high level of genetic diversity within and between wild radish populations (Bhatti et al., 2016), and the results of the current and previous investigations of the mechanism of 2,4-D resistance in 11 distinct populations, are leading ever more strongly to the conclusion that each of them may possess a slightly different resistance mechanism. The difficulty and expense involved in discovery-type studies of global gene and protein expression (e.g. RNAseq and iTRAQ) means that, in general, only a few populations can be compared. In the previous transcriptomic study (Goggin et al., 2018) and the current proteomic study, many of the genes and proteins of interest in population R1 appear to be less important in the other populations when targeted by quantitative RT-PCR and MRM. One possible exception is the plasma membrane L-type lectin domain-containing receptor kinase IV.1 (LecRK-IV.1 or LRK1), whose constitutive protein levels were strongly negatively correlated with 2,4-D and dicamba resistance (Table 3).
The 45 members of the lectin receptor kinase (LecRK) family have closely related structures broadly consisting of an extracellular lectin-like ligand-binding domain and a cytosolic serine/threonine kinase domain used to transmit the perceived extracellular signals, but their physiological roles and substrate specificities are somewhat poorly characterized (Barre et al., 2002). A few members have been identified as being important for plant immunity, bacterial and fungal resistance, pollen development, regulation of ABA response or ATP perception (Wang et al., 2014). Interestingly, the extracellular domain of LecRK proteins lacks the essential asparagine residue permitting monosaccharide binding in legume lectins, but the hydrophobic cavity is conserved (Barre et al., 2002). It has long been hypothesized that LecRK proteins may therefore have the capacity to bind to hydrophobic molecules such as tryptophan, IAA and other plant hormones (Hervé et al., 1996). A meta-analysis of LecRK expression demonstrated that LRK1 is expressed in all tissues (unlike most LecRKs) and its expression does not respond to IAA, ABA or ethylene treatment, but is greatly upregulated by salicylic acid (Bouwmeester and Govers, 2009). Its transcriptional response to a range of pathogens and elicitors was not particularly strong when compared with many other LecRK genes (Bouwmeester and Govers, 2009), giving little clue to the physiological function of LRK1. It is tempting to speculate that although LRK1 expression is not modified by IAA, it may be capable of binding this molecule, and thus also synthetic auxins such as 2,4-D, on the apoplastic side of the plasma membrane.
A search of the STRING database of protein interactions (Fig. 2A) indicates that LRK1 may interact with two signalling proteins that regulate the auxin transport machinery, namely PINOID2 (Haga et al., 2014) and TORNADO1 (Cnops et al., 2006). Recent studies have pointed to a role for certain receptor-like kinases in mediating the localization and activity of the PIN efflux transporters (reviewed in Kubeš and Napier, 2019), so LRK1 may belong to this group. LRK1 is also annotated as interacting with two auxin response factors (ARF2 and ARF16) that repress transcription of auxin-responsive genes (Fig. 2A). ARF2 is a positive regulator of leaf senescence (Lim et al., 2010) whose DNA binding activity is known to be modulated by phosphorylation (Zhao et al., 2016), whilst ARF16 positively regulates ABA signalling (Liu et al., 2013). Overall, the observed negative correlation between constitutive LRK1 levels and auxinic herbicide resistance in wild radish suggests that the resistant populations are less receptive to the presence of these herbicides at the plasma membrane, but this hypothesis needs to be tested by performing in vitro 2,4-D and dicamba binding assays with heterologously expressed LRK1, and by assessing the auxin response of mutants engineered to have high or low levels of this protein.
Even if LRK1 is a mediator of 2,4-D (and dicamba) resistance in wild radish, the variability in auxin response between populations (Goggin et al., 2018) means that it is unlikely to be the sole determinant of resistance. In spite of the fact that the levels of no other proteins were significantly correlated with 2,4-D or dicamba survival, nine of the 11 resistant populations had higher amounts of the leucine-rich repeat receptor-like kinase At1g51820 than one or both of the susceptible populations, and higher At1g51820 levels coincided with a greater ability of dicamba survivors to grow following treatment with this herbicide (Table 3). The precise function of At1g51820 is currently unknown, so it is difficult to hypothesize specifically on how it could be involved in auxinic herbicide resistance (e.g. direct perception of the herbicide molecule; or direct/indirect interaction with the primary perceiving protein) until functional characterization studies are performed.
In contrast, ABCB19, whose levels were positively correlated with the ability of survivors to grow following 2,4-D treatment (Table 3), is very well characterized as an auxin efflux transporter (Geisler et al., 2017), although it should be noted that its ability to transport 2,4-D, specifically, has not been experimentally demonstrated (Bailly et al., 2012). It was originally hypothesized that the greatly reduced 2,4-D translocation observed in two resistant wild radish populations was due to decreased levels, or loss of function, of an ABCB-type auxin efflux transporter (Goggin et al., 2016). However, subsequent work with a larger number of populations showed that reduced translocation was not correlated with higher resistance (Goggin et al., 2018), and that lower ABCB19 levels were not correlated with reduced translocation (current study). Unexpectedly, higher amounts of this protein were instead associated with the ability of surviving plants to grow after a treatment with 2,4-D. ABCB19 is thought to maintain long-distance auxin transport in the phloem by preventing the insertion of IAA molecules into the membranes of adjacent cells as the IAA travels through the phloem stream (reviewed in Reemmer and Murphy, 2014). Following 2,4-D treatment of wild radish seedlings, the mature leaves of both susceptible and resistant plants are slowly killed, but the apical meristem survives in the resistant plants and new growth is formed after a week or two (Supplementary data Fig. S1). A higher amount of ABCB19 may therefore enhance 2,4-D transport from the apical meristem towards the mature, ultimately senescing leaves, ‘diluting’ the amount of herbicide in the surviving plant parts. As noted above, however, it is currently still unknown whether 2,4-D is a substrate of the ABCB auxin efflux transporters.
Largely negative results were obtained in the study of the soluble proteomes of populations S1 and R1 (Table 1) and also in the preliminary data from a nuclear magnetic resonance (NMR)-based analysis of soluble metabolites (data not shown). The (detectable) soluble proteome of R1 was essentially unresponsive to 2,4-D and the few changes in the S1 population, most notably an increase in the levels of the antioxidant enzymes SOD and GST, were not accompanied by increases in enzyme activity. Overall, the similarity of the soluble proteomes (and probably also metabolomes) of populations S1 and R1 strengthens the idea that 2,4-D resistance in wild radish is primarily mediated by signalling and transport at the plasma membrane and by low-abundance signalling proteins in the symplast.
CONCLUSIONS
It is likely that auxinic herbicide resistance in wild radish is mediated by multiple processes that may be present in different proportions in different populations or even between individuals within a population. Along with the previously identified potential resistance mechanisms involving upregulation of the Aux/IAA repressors IAA29 and IAA30 in population R1 and increased constitutive MAPK phosphorylation across the 11 R populations studied (Goggin et al., 2018), the current investigation has used shotgun and targeted proteomics to identify candidate resistance proteins in the plasma membrane. The receptor-like kinases LRK1 and At1g51820, never previously implicated in the 2,4-D response, are of particular interest as potential 2,4-D-perceiving proteins and/or as kinases that may operate upstream of the MAPK pathway. The putative 2,4-D exporter ABCB19 may also function in enhancing tolerance to 2,4-D and influencing long-distance shoot transport of this herbicide, but its precise role needs to be clarified. The contributions of LRK1, At1g51820 and ABCB19 to auxinic herbicide resistance could be confirmed and quantified by heterologous expression studies followed by biochemical and physiological characterization of the transformants.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: appearance of 2,4-D-susceptible and -resistant plants at 14 d after 2,4-D treatment. Table S1: marker enzyme activities in plasma membrane and microsome fractions. Table S2: list of proteins identified in plasma membranes by mass spectrometry. Table S3: full correlation matrix of resistance parameters and protein levels.
ACKNOWLEDGEMENTS
The mass spectrometry analyses were performed in the Western Australian Proteomics Facility, supported by Lotterywest and Bioplatforms Australia, at the Harry Perkins Institute for Medical Research. We thank the anonymous reviewers of this and a previous version of the manuscript for their helpful comments.
FUNDING
This work was supported by the Australian Research Council (grant no. LP150100161) in partnership with Nufarm Australia.
LITERATURE CITED
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiology 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal UK, Krochko JE, Ross ARS. 2012. Identification of phosphoproteins in Arabidopsis thaliana leaves using polyethylene glycol fractionation, immobilized metal-ion affinity chromatography, two-dimensional gel electrophoresis and mass spectrometry. Journal of Proteome Research 11: 425–437. [DOI] [PubMed] [Google Scholar]
- Asada K, Yoshikawa K, Takahashi M-A, Maeda Y, Enmanji K. 1975. Superoxide dismutases from a blue-green alga, Plectonema boryanum. Journal of Biological Chemistry 250: 2801–2807. [PubMed] [Google Scholar]
- Bailly A, Yang H, Martinoia E, Geisler M, Murphy AS. 2012. Plant lessons: exploring ABCB functionality through structural modeling. Frontiers in Plant Science 2: 108. doi: 10.3389/fpls.2011.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre A, Hervé C, Lescure B, Rougé P. 2002. Lectin receptor kinases in plants. Critical Reviews in Plant Sciences 21: 379–399. [Google Scholar]
- Beauchamp C, Fridovich I. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry 44: 276–287. [DOI] [PubMed] [Google Scholar]
- Bhatti MA, Cocks PS, Bennett SJ, Malik AU. 2016. Adaptive significance of within-site variation in morphological and reproductive traits of naturalized wild radish (Raphanus raphanistrum) populations in south-western Australia. International Journal of Agriculture and Biology 18: 975–982. [Google Scholar]
- Bouwmeester K, Govers F. 2009. Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. Journal of Experimental Botany 60: 4383–4396. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Campe R, Langenbach C, Leissing F, et al. 2016. ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytologist 209: 294–306. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, et al. 2004. Blue-silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Chamarro J, Östin A, Sandberg G. 2001. Metabolism of indole-3-acetic acid by orange (Citrus sinensis) flavedo tissue during fruit development. Phytochemistry 57: 179–187. [DOI] [PubMed] [Google Scholar]
- Chandler JW. 2016. Auxin response factors. Plant, Cell and Environment 39: 1014–1028. [DOI] [PubMed] [Google Scholar]
- Cnops G, Neyt P, Raes J, et al. 2006. The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. The Plant Cell 18: 852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesperger H, Müller CR, Sandermann H Jr. 1974. Rapid isolation of a plant microsomal fraction by Mg2+-precipitation. FEBS Letters 43: 155–158. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, Coaker G. 2012. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Molecular and Cellular Proteomics 11: M111.014555. doi: 10.1074/mcp.M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. 2006. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Letters 580: 1094–1102. [DOI] [PubMed] [Google Scholar]
- Geisler M, Aryal B, di Donato M, Hao P. 2017. A critical view on ABC transporters and their interacting partners in auxin transport. Plant & Cell Physiology 58: 1601–1614. [DOI] [PubMed] [Google Scholar]
- Goggin DE, Colmer TD. 2007. Wheat genotypes show contrasting abilities to recover from anoxia in spite of similar anoxic carbohydrate metabolism. Journal of Plant Physiology 164: 1605–1611. [DOI] [PubMed] [Google Scholar]
- Goggin DE, Cawthray GR, Powles SB. 2016. 2,4-D resistance in wild radish: reduced herbicide translocation via inhibition of cellular transport. Journal of Experimental Botany 67: 3223–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin DE, Kaur P, Owen MJ, Powles SB. 2018. 2,4-D and dicamba resistance mechansims in wild radish: subtle, complex and population-specific? Annals of Botany 122: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K. 2003. Mediation of herbicide effects by hormone interactions. Journal of Plant Growth Regulation 22: 109–122. [Google Scholar]
- Grossmann K. 2010. Auxin herbicides: current status of mechanism and mode of action. Pest Management Science 66: 113–120. [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. 1974. Glutathione-S-transferases: the first enzymatic step in mercapturic acid formation. Journal of Biological Chemistry 249: 7130–7139. [PubMed] [Google Scholar]
- Haga K, Hayashi K-I, Sakai T. 2014. PINOID AGC kinases are necessary for phytochrome-mediated enhancement of hypocotyl phototropism in Arabidopsis. Plant Physiology 166: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Galaud J-P, Rougé P, Lescure B. 1996. Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. Journal of Molecular Biology 258: 778–788. [DOI] [PubMed] [Google Scholar]
- Hodges TK, Leonard RT. 1974. Purification of a plasma membrane-bound adenosine triphosphatase from plant roots. Methods in Enzymology 32: 392–406. [DOI] [PubMed] [Google Scholar]
- Kim D-Y, Bovet L, Maeshima M, Martinoia E, Lee Y. 2007. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. The Plant Journal 50: 207–218. [DOI] [PubMed] [Google Scholar]
- Kubeš M, Napier R. 2019. Non-canonical auxin signalling: fast and curious. Journal of Experimental Botany 70: 2609–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S. 1994. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods in Enzymology 228: 451–469. [Google Scholar]
- LeClere S, Wu C, Westra P, Sammons RD. 2018. Cross-resistance to dicamba, 2,4-D, and fluroxypyr in Kochia scoparia is endowed by a mutation in an AUX/IAA gene. Proceedings of the National Academy of Sciences, USA 105: E2911–E2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Burg HA, Joosten MHAJ. 2014. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends in Plant Science 19: 123–132. [DOI] [PubMed] [Google Scholar]
- Lim PO, Lee IC, Kim J, et al. 2010. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. Journal of Experimental Botany 61: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Liu H, Liu Z, Wang X, Liang S. 2012. Shotgun analysis of membrane proteomes using a novel combinative strategy of solution-based sample preparation coupled with liquid chromatography–tandem mass spectrometry. Journal of Chromatography B 901: 18–24. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, et al. 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 110: 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. 2013. Auxin metabolism and homeostasis during plant development. Development 140: 943–950. [DOI] [PubMed] [Google Scholar]
- Lloyd CW, Lowe SB, Peace GW. 1980. The mode of action of 2,4-D in counteracting the elongation of carrot cells grown in culture. Journal of Cell Science 45: 257–268. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry 193: 265–275. [PubMed] [Google Scholar]
- Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P. 2019. Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Molecular Plant 12: 298–320. [DOI] [PubMed] [Google Scholar]
- Migocka M, Papierniak A, Rajsz A. 2017. Cucumber PDR8/ABCG36 and PDR12/ABCG40 plasma membrane proteins and their up-regulation under abiotic stresses. Biologia Plantarum 61: 115–126. [Google Scholar]
- Moghe GD, Hufnagel DE, Tang H, et al. 2014. Consequences of whole-genome triplication as revealed by comparative genomic analysis of the wild radish Raphanus raphanistrum and three other Brassicaceae species. The Plant Cell 26: 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata C, Miwa C, Tanaka N, et al. 2016. A novel-type phosphatidylinositol phosphate-interactive, Ca-binding protein PCaP1 in Arabidopsis thaliana: stable association with plasma membrane and partial involvement in stomatal closure. Journal of Plant Research 129: 539–550. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Martinez NJ, Powles SB. 2015. Multiple herbicide-resistant wild radish (Raphanus raphanistrum) populations dominate Western Australian cropping fields. Crop and Pasture Science 66: 1079–1085. [Google Scholar]
- Paoletti F, Aldinucci D, Mocali A, Caparrini A. 1986. A sensitive spectrophotometric method for the determination of superoxide dismutase activity in tissue extracts. Analytical Biochemistry 154: 536–541. [DOI] [PubMed] [Google Scholar]
- Peterson MA, McMasters SA, Riechers DE, Skelton J, Stahlmann PW. 2016. 2,4-D past, present and future: a review. Weed Technology 30: 303–345. [Google Scholar]
- Reemmer J, Murphy A. 2014. Intercellular transport of auxin. In: Zažimalová E, Petrasek J, Benková E, eds. Auxin and its role in plant development. Vienna: Springer-Verlag, 75–100. [Google Scholar]
- Schallar GE, deWitt ND. 1995. Analysis of the H+-ATPase and other proteins of the Arabidopsis plasma membrane. Methods in Cell Biology 50: 129–148. [PubMed] [Google Scholar]
- Shimogawara K, Usuda H. 1993. A concentrating two-phase partitioning: its application to isolation of plasma membrane from maize roots. Analytical Biochemistry 212: 381–387. [DOI] [PubMed] [Google Scholar]
- Skalický V, Kubeš M, Napier R, Novák O. 2018. Auxins and cytokinins – the role of subcellular organization on homeostasis. International Journal of Molecular Sciences 19: 3115. pii: E3115. doi: 10.3390/ijms19103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, et al. 2018. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Y, Choi KS, Kim DY, et al. 2010. Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. The Plant Cell 22: 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research 47–D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research 47: D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, et al. 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiology 142: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, Beseh P, Shan W, Govers F. 2014. Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Molecular Plant-Microbe Interactions 27: 1390–1402. [DOI] [PubMed] [Google Scholar]
- Zaiontz C. 2019. Real statistics using Excel. http://www.real-statistics.com. Accessed: 11 June 2019.
- Zhao S, Zhang M-L, Ma T-L, Wang Y. 2016. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. The Plant Cell 28: 3005–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.