Abstract
Background and Aims
Most primary auxin response genes are classified into three families: AUX/IAA, GH3 and SAUR genes. Few studies have been conducted on Arabidopsis thaliana SAUR genes, possibly due to genetic redundancy among different subfamily members. Data mining on arabidopsis transcriptional profiles indicates that the SAUR41 subfamily members of SMALL AUXIN UP RNA genes are, strikingly, induced by an inhibitory phytohormone, abscisic acid (ABA). We aimed to reveal the physiological roles of arabidopsis SAUR41 subfamily genes containing SAUR40, SAUR41, SAUR71 and SAUR72.
Methods
Transcriptional responses of arabidopsis SAUR41 genes to phytohormones were determined by quantitative real-time PCR. Knock out of SAUR41 genes was carried out with the CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) genome editing technique. The saur41/40/71/72 quadruple mutants, SAUR41 overexpression lines and the wild type were subjected to ultrastructural observation, transcriptome analysis and physiological characterization.
Key Results
Transcription of arabidopsis SAUR41 subfamily genes is activated by ABA but not by gibberellic acids and brassinosteroids. Quadruple mutations in saur41/40/71/72 led to reduced cell expansion/elongation in cotyledons and hypocotyls, opposite to the overexpression of SAUR41; however, an irregular arrangement of cell size and shape was observed in both cases. The quadruple mutants had increased transcription of calcium homeostasis/signalling genes in seedling shoots, and the SAUR41 overexpression lines had decreased transcription of iron homeostasis genes in roots and increased ABA biosynthesis in shoots. Notably, both the quadruple mutants and the SAUR41 overexpression lines were hypersensitive to salt stress during seedling establishment, whereas specific expression of SAUR41 under the ABA-responsive RD29A (Responsive to Desiccation 29A) promoter in the quadruple mutants rescued the inhibitory effect of salt stress.
Conclusions
The SAUR41 subfamily genes of arabidopsis are ABA inducible to modulate cell expansion, ion homeostasis and salt tolerance. Our work may provide new candidate genes for improvement of plant abiotic stress tolerance.
Keywords: Arabidopsis thaliana, SMALL AUXIN UP RNA genes, abscisic acid, cell expansion, ion homeostasis, salt tolerance, CRISPR/Cas9, transcription profiling
INTRODUCTION
During plant growth and development, the phytohormone auxin modulates cell division, differentiation and elongation, largely by regulating gene expression. Most of the primary auxin response genes are classified into three families: AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA), GRETCHENHAGEN3 (GH3) and SMALL AUXIN UP RNA (SAUR) (Hagen and Guilfoyle, 2002). While great progress has been made in our understanding of the molecular mechanisms of both AUX/IAAs and GH3s, functional studies on SAURs have lagged behind, and have only begun to be performed in recent years (reviewed in Ren and Gray, 2015; Stortenbeker and Bemer, 2018).
SAUR genes are the largest family of primary auxin response genes, with plant SAUR proteins being divided into three families (Stortenbeker and Bemer, 2018), and arabidopsis SAURs phylogenetically classified into three clades (Kodaira et al., 2011). SAUR genes are regulated at both transcriptional and post-transcriptional levels in response to multiple hormonal and environmental signals (reviewed in Ren and Gray, 2015; Stortenbeker and Bemer, 2018). Some transcription factors that regulate the expression of arabidopsis SAUR genes have been identified. For example, the arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 repress many clade I and clade II SAUR genes under salt stress and abscisic acid (ABA) treatment (Kodaira et al., 2011). Similarly, the MADS-domain factor FRUITFULL represses SAUR10 in stems and inflorescence branches to control arabidopsis growth and architecture (Bemer et al., 2017). SOB3 (SUPPRESSOR OF PHYTOCHROME B4-#3) and BZR1 (BRASSINAZOLE RESISTANT 1) inactivate and activate, respectively, the transcription of SAUR19 genes to modulate hypocotyl growth in response to auxin and brassinosteroid (BR) signalling (Favero et al., 2017), while PIF3 (PHYTOCHROME INTERACTING FACTOR 3) and TCP4 (TEOSINETE BRANCHED1, CYCLOIDEA and PCF) antagonistically regulate SAUR16/50 during de-etiolation of arabidopsis cotyledons (Sun et al., 2016; Dong et al., 2019).
The SAUR proteins are diverse in their subcellular localizations and undergo complex post-translational control (reviewed in Ren and Gray, 2015). Based on results from overexpression studies, many arabidopsis SAUR proteins have been revealed to promote cell expansion, including SAUR19s (Franklin et al., 2011; Spartz et al., 2012), SAUR63s (Chae et al., 2012), SAUR41 (Kong et al., 2013), SAUR36 (Stamm and Kumar, 2013), SAUR76–78 (Li et al., 2015), SAUR14/50/65 (Sun et al., 2016) and SAUR10s (van Mourik et al., 2017). In addition, it has been demonstrated that SAUR36 functions as a positive regulator of leaf senescence (Hou et al., 2013), SAUR76 functions as a cell division regulator (Markakis et al., 2013) and an ethylene receptor modulator (Li et al., 2015), while SAUR62/75 are required for pollen tube growth (He et al., 2018). Functional studies on the SAURs of other plant species are very limited. It has been reported that rice (Oryza sativa) OsSAUR39 (Kant et al., 2009) and OsSAUR45 (Xu et al., 2017) serve as negative regulators of auxin biosynthesis/transportation, cassava (Manihot esculenta) MeSAUR1 functions as a transcriptional regulator of the gene encoding the small subunit of ADP-glucose pyrophosphorylase (Ma et al., 2017), wheat (Triticum aestivum) TaSAUR75 positively regulates drought and salt stress (Guo et al., 2017), and tomato (Solanum lycopersicum) SlSAUR69 is involved in the unripe-to-ripe phase transition of tomato fruit (Shin et al., 2018).
The mechanisms by which arabidopsis SAURs mediate cell expansion are of great interest, but remain largely unknown. It has been suggested that arabidopsis peripheral membrane SAUR19s inhibit several members of the PP2C.D-type protein phosphatases to increase Thr947 phosphorylation of plasma membrane H+-ATPases, resulting in apoplast acidification and cell expansion (Spartz et al., 2014). Arabidopsis SAUR19 constitutively expressed in S. lycopersicum also inhibited endogenous PP2C.Ds to confer auxin-independent hypocotyl elongation (Spartz et al., 2017). Further support for an antagonistic role for PP2C.D proteins against SAUR-mediated cell expansion comes from a study on loss-of-function mutants of the plasma membrane-localized sub-set of arabidopsis PP2C.Ds (Ren et al., 2018). Thus, SAURs and PP2C.Ds may be novel components for the acid growth theory that links auxin to the cell expansion process (reviewed in Haruta et al., 2015; Arsuffi and Braybrook, 2017). As an alternative mechanism, a recent study suggests that SAUR62/75 interact with ribosomal protein RPL12 family members to promote ribosome assembly and protein translation required for pollen tube growth (He et al., 2018).
Alongside auxin, the phytohormone ABA regulates many aspects of plant growth and development such as seed germination, cotyledon greening/expansion, stomatal closure, drought resistance and salt resistance. The core ABA signalling network is well understood (reviewed in Cutler et al., 2010; Hubbard et al., 2010). Briefly, ABA binds to intercellular Pyrabactin Resistance/Regulatory Component of ABA Receptor (PYR/RCAR) receptors to inhibit the PP2C.A type protein phosphatases, which leads to release of SNF1-Related Protein Kinase 2 (SnRK2) family members; these activate ion channels, transcription factors and other targets (Hauser et al., 2017). Identification of new components capable of modulating ABA sensitivity/response is of great value for engineering plant abiotic stress tolerance.
Many auxin-responsive genes, including most SAUR genes in clades I and II, were found to be commonly downregulated to establish growth inhibition and growth adaptation under abiotic stress conditions and ABA treatment (Kodaira et al., 2011). However, based on microarray and transcriptome data, the expression of the arabidopsis SAUR41 subfamily genes (SAUR40, SAUR41, SAUR71 and SAUR72), attributed to clade III, is strikingly induced by the growth-inhibiting hormone ABA, unlike most other SAUR genes (Leonhardt et al., 2004; Kodaira et al., 2011; Zeng et al., 2012; reviewed in Ren and Gray, 2015). This raises the question of whether the balanced expression of ABA-repressed and ABA-induced SAUR genes is necessary for abiotic responses. In addition, we found that SAUR41 genes have tissue-specific and developmentally regulated expression patterns during arabidopsis seedling establishment. SAUR41 was distinctively expressed in the quiescent centre and cortex/endodermis initials of root stem cell niches and in the endodermis of hypocotyls (Kong et al., 2013), whereas SAUR71 and SAUR72 were expressed in the steles of young roots and hypocotyls; in addition, SAUR71 was differentially expressed during stomatal development (Qiu et al., 2013). Taken together, further studies on SAUR41 genes may be helpful to reveal new interactions between auxin and ABA or new players in plant abiotic responses. An essential prerequisite for understanding the physiological roles of SAUR41 genes is the generation of loss-of-function mutants. In recent years, the development of CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) genome editing techniques for arabidopsis makes it possible to generate high orders of mutants for genes of interest very efficiently (Fauser et al., 2014).
Here, we verify that arabidopsis SAUR41 genes were activated by ABA. Knockout of all SAUR41 subfamily members using the CRISPR/Cas9 system led to reduced cell expansion in young arabidopsis seedlings. The quadruple mutants increased the transcription of multiple calcium homeostasis/signalling genes in seedling shoots, while the SAUR41 overexpression lines decreased the transcription of iron homeostasis genes in roots and increased ABA biosynthesis in shoots. However, both the quadruple mutants and SAUR41 overexpression were hypersensitive to salt treatment. Notably, specific expression of SAUR41 under an ABA-responsive promoter in quadruple mutants rescued the inhibitory effect of salt on seedling establishment and growth. We suggest that the SAUR41 subfamily genes of arabidopsis are new players in modulation of cell expansion, ion homeostasis and salt tolerance to fine-tune seedling growth. Our work also indicates that balanced expression of ABA-repressed and ABA-induced SAURs may be necessary for plant abiotic responses.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) were used as sources of wild-type plant material. Seeds were surface-sterilized and sown aseptically on 9 cm Petri dishes containing Gamborg’s B5 medium with 1 % (w/v) sucrose and 0.8 or 1.0 % (w/v) agar, for horizontal and vertical growth, respectively. The basal salts and vitamins of B5 medium were composed of (mg L–1): KNO3 2500, CaCl2·2H2O 150, MgSO4·7H2O 250, (NH4)2SO4 134, NaH2PO4·H2O 150, KI 0.75, H3BO3 3.0, MnSO4·H2O 10, ZnSO4·7H2O 2.0, Na2MoO4·2H2O 0.25, CuSO4·5H2O 0.025, CoCl2·6H2O 0.025, FeSO4·7H2O 27.8, Na2-EDTA 37.3, myo-inositol 100, nicotinic acid 1.0, pyridoxine HCl 1.0, and thiamine HCl 10. The plates were maintained at 4 °C for 2–3 d, then transferred to a culture room (23 °C; 30–40 % relative humidity; 80 μmol m–2 s–1 irradiance with a 16 h photoperiod).
Design of guide RNAs for CRISPR/Cas9 genomic editing
SAUR41 subfamily genes were knocked out using the CRISPR/Cas9 genome editing technique developed for arabidopsis (Fauser et al., 2014). The choice of target site motif, (N)20NGG, was based on the online tool ZIFIT Target Version 4.2 (http://zifit.partners.org/ZiFiT/Introduction.aspx). We selected three target sites for each gene. The DNA targets and the corresponding guide RNA are illustrated in Supplementary data Fig. S1.
Expression vector construction and plant transformation
Expression vectors were constructed using the GATEWAY™ cloning system (Invitrogen, Carlsbad, CA, USA). Entry vectors containing sequences of interest were created first in the pENTR™ backbone, and then they were incubated together with the corresponding destination vectors to generate the expression vectors via the LR recombination reaction (Karimi et al., 2002; She et al., 2010). For CRISPR/Cas9 genome editing, we used the Golden Gate cloning strategy (Engler et al., 2009) and two rounds of PCR to create entry clones encoding guide RNAs. Primers used for amplification of the U6-26 promoter and guide RNA coding sequences are listed in Supplementary data Table S1. Each entry vector was confirmed by DNA sequencing. To generate final expression vectors, the destination vector pDe-CAS9-ccdB (Fauser et al., 2014) was employed. For overexpression of SAUR41 genes, entry clones were created first, and suitable destination vectors for overexpression of fusion proteins were used as described previously (Kong et al., 2013; Qiu et al., 2013). For expression of SAUR41-EGFP from the RD29A promoter, the original Cauliflower mosaic virus (CaMV) 35S promoter in the protein localization construct 35S::SAUR41-EGFP (Kong et al., 2013) was replaced with a ccdB fragment including the attR1 and attR2 sites by a previously described method (Yang et al., 2012). Briefly, the ccdB fragment was PCR-amplified using pH7FWG2,0 as a template, with the primers ccdB-Up and ccdB-Dn, containing an introduced HindIII and SpeI site, respectively. The ccdB fragment was then digested to replace the 35S promoter sequence, thus forming the destination vector ccdB-SAUR41-EGFP for promoter-specific expression. PCR primers used for construction of the RD29A entry vector and the ccdB-SAUR41-EGFP destination vector are listed in Supplementary data Table S1. All expression vectors were electroporated into Agrobacterium tumefaciens strain GV3101, and plants were transformed using the vacuum infiltration method (Bechtold, 1993). Transgenic plants were selected on B5 plates with 12.5 μg mL–1 hygromycin or 10 μg mL–1 Basta depending on the selectable marker genes.
Mutation detection and plant crossing
For genome editing, DNA mutations in the leaves of the T1 primary transformants and in the young seedlings of T2 and T3 generations were detected by T7E1 endonuclease digestion or PAGE analysis. Primers for detection of CRISPR/Cas9-induced mutations in SAUR41 genes are listed in Supplementary data Table S1. Germline mutations were identified through Sanger sequencing of PCR products spanning the target sites. We first achieved single mutants for each gene of the SAUR41 subfamily, then created double, triple and quadruple mutants by crossing and screening. Briefly, gene-specific primers, listed in Supplementary data Table S1 were used in the PCR-based screen by genotyping the individual F2 progeny plants of each cross.
Tissue sections and microscopy
For transmission electron microscopy (TEM), hypocotyls of 6-day-old seedlings were cut into 0.5–1.0 cm pieces, vacuum-infiltrated and then fixed in 2.5 % glutaraldehyde overnight, followed by treatment with 1 % osmium tetroxide for 2 h. Specimens were then dehydrated with a graded series of ethanol solutions and embedded in Spurr resin. Micro-thin (1 μm) and ultra-thin (70–90 nm) sections were prepared with a LEICA EM UC7 ultratome. Micro-sections were stained with 0.1 % methylene blue in 0.1 m phosphate-buffered saline solution for 2 min at 60 °C and then washed with distilled water. Micro-sections were visualized and imaged by a Nikon Eclipse 80i microscope. Ultra-thin sections were stained with 2 % uranyl acetate in 50 % ethanol for 15 min and then with 0.3 % alkaline lead citrate (pH 12, adjusted by NaOH) for 30 min (Yang et al., 2012). Ultra-thin sections were imaged with a Hitachi Model H-7650 TEM.
RNA isolation, quantitative real-time PCR (qRT-PCR) and transcriptome analysis
For transcriptional response of SAUR41 genes to the treatments with various plant hormones, 5-day-old arabidopsis seedlings were transferred into liquid B5 medium containing 10 μm naphthaleneacetic acid (NAA), ABA or gibberellic acid (GA3), and incubated on a rotary shaker (50 rpm) for 0.5, 1.0, 2.0, 3.0 and 4.0 h. The treatments were conducted during the middle part of the light period. Control seedlings were treated with an equivalent volume of dimethylsulphoxide (DMSO), the solvent for the phytohormone stock solutions.
For RNA isolation, whole or different parts of arabidopsis seedlings were frozen and ground to powder in liquid nitrogen, followed by total RNA extraction using TRizol reagent (Takara Bio Inc., Kusatsu, Shiga, Japan). Extracted RNA was treated with RNase-free DNase I (Takara) and subjected to first-strand cDNA synthesis using AMV reverse transcriptase (Takara). The qRT-PCR was performed on the Mastercycler Realplex System (Eppendorf, Hamburg, Germany) using SYBR Green reagent (Takara). Relative mRNA levels were normalized using Ubiquitin Extension Protein 1 (UBQ1) as the standard (Yang et al., 2012). Primers for qRT-PCR measurement of SAUR41 subfamily genes, validation of RNA sequencing (RNA-seq) results and expression assay of salt tolerance genes are listed in Supplementary data Table S1.
For transcriptional profiling (RNA-seq), total RNA samples were sent to Vazyme Biotech Co., Ltd (Nanjing, China) for the following processes: library construction; Illumina HiSeq sequencing; and bioinformatics analysis, including a quality check of the raw reads, alignment of raw reads to the arabidopsis genome, assembly of gene expression from aligned reads and identification of differential gene expression, followed by Gene Ontology (GO), KEGG pathway and gene/protein network analysis. Each sample generated 4 Gb of clean data and contained three biological replicates. These data have been deposited in the Gene Expression Omnibus (GEO) at NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE134117 and GSE134119.
Determination of ABA and metal iron contents
The ABA concentration in 7-day-old seedlings was measured using a method modified from Pan et al. (2010). Tissue (0.2 g) was frozen and ground to powder in liquid nitrogen, then dissolved in a buffer containing isopropanol, hydrochloric acid and distilled water (2:0.002:1, v/v/v). Extracts were shaken at 4 °C for 30 min followed by addition of 20 mL of dichloromethane. The final extracts were dissolved in methanol containing 0.1 % methane acid. The level of ABA was determined by high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Reversed-phase HPLC was performed with a ZORBAX SB-C18 column (2.1 × 150 mm; 3.5 μm; Agilent Technologies, Santa Clara, CA, USA). The mobile phase A was 0.1 % (v/v) formic acid in distilled water, while the mobile phase B was 0.1 % (v/v) formic acid in methanol. The flow rate was 0.6 mL min–1 at 40 °C and the injection volume was 10 μL. ABA was quantified with a SCIEX-6500 Qtrap mass spectrometer (Applied Biosystems, Foster City, CA, USA) connected online to the output of the HPLC column.
Iron, calcium, sodium and potassium concentrations in 7-day-old seedlings were measured using a method described by Wild et al. (2016). Tissues were digested completely in a nitric acid–hydrogen peroxide solution (2:1, v/v) before inductively coupled plasma-atomic emission spectrometry (ICP-AES) on an Agilent 710 ICP-AES spectrometer (Agilent Technologies): power output, 1000 W; plasma gas rate, 15.0 L min–1; atomizer pressure, 200 kPa; auxiliary gas rate, 1.5 L min–1; pump speed, 15 rpm.
Growth measurement, salt tolerance, statistical analysis and image processing
Plates containing arabidopsis seedlings were digitally photographed after 5–12 d of incubation. Hypocotyl length, root length, cotyledon petiole length and cotyledon size were measured from magnified images using ImageJ software. The cell lengths of epidermal cells and cortical cells of hypocotyls were measured as described by Kong et al. (2013). Rhizosphere acidification was visualized by a method described by Spartz et al. (2014). For the salt tolerance assay, 100 and 150 mm NaCl were applied at the time of seed imbibition at 4 °C for 2 d. The seedling establishment rate was scored after 7 d of germination and growth at 23 °C. A seedling was regarded as established when the radicle was at least 5 mm long and the cotyledon was open and green (Thomas et al., 1995). The fresh weight (f. wt) of seedlings was recorded at the same time. Growth inhibition was calculated as (f. wt under salt stress – f.wt in control conditions)/f. wt in control conditions × 100 %. Each treatment contained approx. 30 seedlings or approx. 90 seeds and was replicated three times. Statistical analyses were performed using Microsoft Excel and Student’s t-test. Images were processed using Adobe Photoshop CC.
RESULTS
The SAUR41 subfamily genes are induced by ABA but not by GA and BR
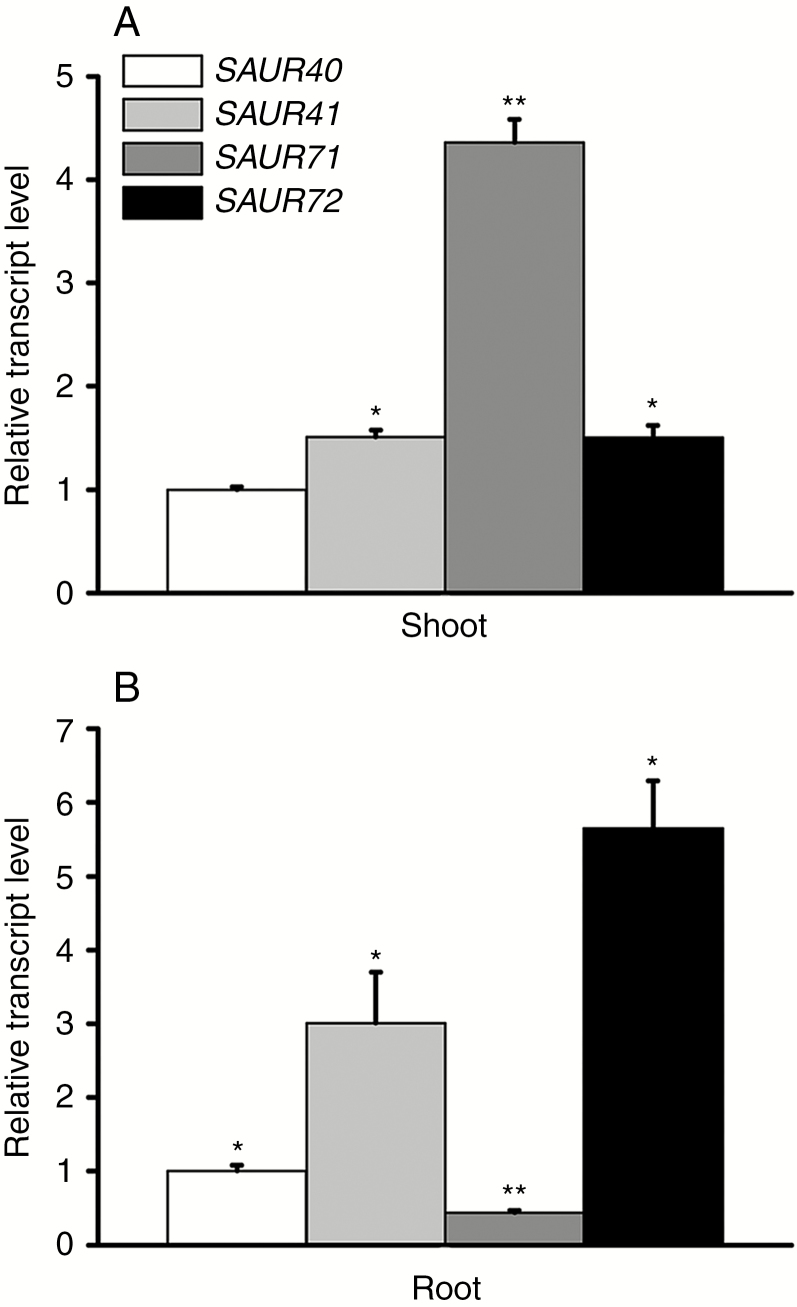
Using promoter reporter lines, we found that members of the SAUR41 subfamily had tissue-specific and developmentally regulated expression patterns in young arabidopsis seedlings (Kong et al., 2013; Qiu et al., 2013). In hypocotyls, SAUR41 was predominantly expressed in the endodermis, while SAUR71 and SAUR72 were mainly expressed in pericycles; in roots, SAUR41 was expressed in stem cell niches and the endodermis, SAUR71 were expressed in pericycles, while SAUR72 was expressed in both the endodermis and pericycles (Kong et al., 2013; Qiu et al., 2013). In an arabidopsis SAUR gene expression pattern obtained from the Bio-Analytic Resource electronic Fluorescent Pictograph browser (BAR-eFP, http://bar.utoronto.ca/efp_arabidopsis/), SAUR41 was predicted to be expressed in both shoots and roots; SAUR71 was moderately expressed in shoots; SAUR72 was highly active in roots; and data for SAUR40 were unavailable (Kodaira et al., 2011). Here, using qRT-PCR analysis, we found that the expression patterns of SAUR41, SAUR71 and SAUR72 were generally similar to the BAR-eFP prediction, with the slight difference that SAUR71 was strongly expressed instead of moderately expressed in shoots (Fig. 1A, B). SAUR40 was detectable in our qRT-PCR experiment: in the shoots/aerial parts of arabidopsis seedlings, it exhibited the lowest expression levels out of all four family members, but, in the roots, its transcription level was higher than that of SAUR71 (Fig. 1A, B).
Fig. 1.
Relative expression levels of SAUR41 genes in 5-day-old arabidopsis seedlings. (A) Relative expression levels of SAUR41 genes in seedling shoots. The transcript level of SAUR40 is set to 1.0. SAUR71 displays the highest expression level. (B) Relative expression levels of SAUR41 genes in seedling roots. The transcript level of SAUR40 is set to 1.0. SAUR41 is expressed in both shoots and roots, while SAUR72 is highly active in roots. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
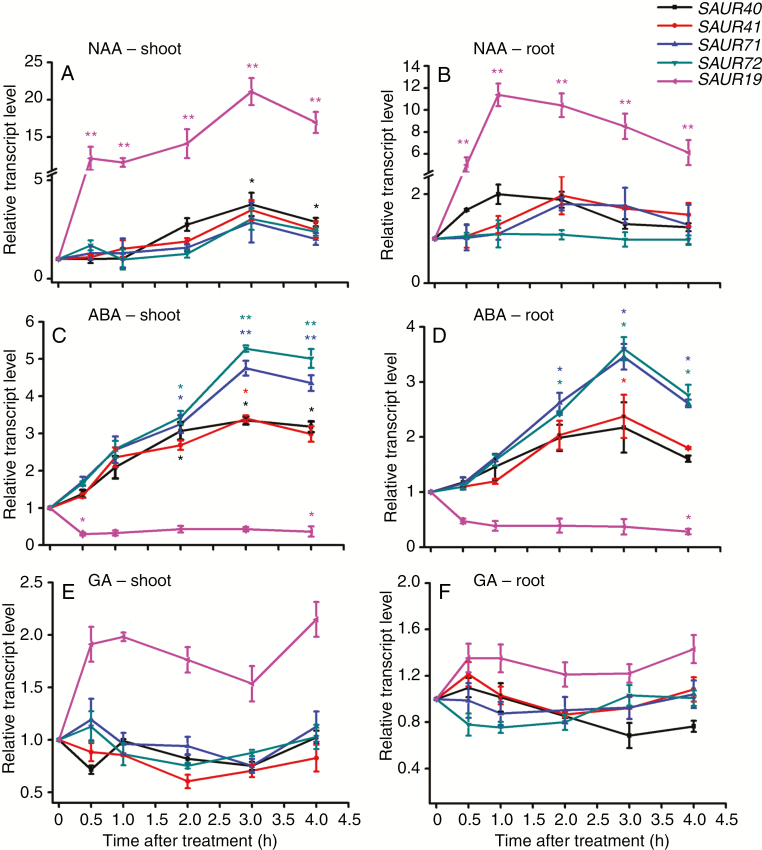
To determine plant hormone responses of SAUR41 genes, 5-day-old light-grown seedlings were treated with 10 μm NAA, ABA or GA3 for 0–4 h before qRT-PCR analysis. Results showed that, in the aerial parts, short-term NAA treatment increased the transcription levels of SAUR40 and SAUR41 by 2- to 3-fold; in contrast, the transcription levels of SAUR19, a well-documented auxin-responsive SAUR gene were increased by 23-fold (Fig. 2A). In the root parts, again, only SAUR19 displayed a very rapid and dramatic NAA response (Fig. 2B). Treatment of arabidopsis seedlings with ABA increased the transcription levels of SAUR41 genes by 3- to 5.5-fold in the aerial parts and by 2- to 3-fold in the root parts, while SAUR19 was repressed by ABA treatment in both parts (Fig. 2C, D). Notably, SAUR71 and SAUR72 were especially ABA responsive in both shoots and roots, as compared with SAUR40 and SAUR41 (Fig. 2C, D). GA3 treatment was unable to activate the expression of SAUR41 genes (Fig. 2E, F), as was EBR (2,4-epibrassinolide) treatment (Supplementary data Fig. S2).
Fig. 2.
The SAUR41 subfamily genes are induced by ABA treatment. (A) Transcriptional responses of SAUR41 genes and SAUR19 to NAA treatment in seedling shoots. Five-day-old seedlings were incubated in liquid medium containing 10 μm NAA for 0.5, 1.0, 2.0, 3.0 and 4.0 h. SAUR19 seems dramatically auxin responsive. (B) Transcriptional responses of SAUR41 genes and SAUR19 to NAA treatment in roots. (C) Transcriptional responses of SAUR41 genes and SAUR19 to ABA treatment in seedling shoots. Five-day-old seedlings were incubated in liquid medium containing 10 μm ABA for 0.5, 1.0, 2.0, 3.0 and 4.0 h. SAUR19 is repressed by ABA, unlike SAUR41 genes. (D) Transcriptional responses of SAUR41 genes and SAUR19 to ABA treatment in roots. Again, SAUR41 genes are induced by ABA but SAUR19 is repressed by ABA. (E) Transcriptional responses of SAUR41 genes and SAUR19 to GA treatment in seedling shoots. Five-day-old seedlings were incubated in liquid medium containing 10 μm GA for 0.5, 1.0, 2.0, 3.0 and 4.0 h. SAUR19 is slightly induced by GA while SAUR41 genes are not induced by GA. (F) Transcriptional responses of SAUR41 genes and SAUR19 to GA treatment in seedling roots. Each treatment contained three biological replicates. In all cases, the transcript levels of each gene without hormone treatment were set to 1.0. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
Generation of single, double, triple and quadruple mutants of the SAUR41 subfamily genes
To explore potential roles of SAUR41 genes in arabidopsis development and physiology, we first screened T-DNA insertion lines from the Arabidopsis Biological Resource Center (ABRC). Line SALK_056969 contained a T-DNA at the promoter region of SAUR41, but this insertion did not impair its expression; in addition, line SALK_121397 was a false-positive line for the SAUR41 gene (Kong et al., 2013). For SAUR40, line SALK_040036 was annotated to contain a T-DNA at the intergenic region, and lines SALK_128948, SALK_051362 and SALK_050786 all contained T-DNA at the 5'-untranslated region. Similarly, no lines contained T-DNA insertions inside the coding regions of SAUR71 and SAUR72. Thus, we used the CRISPR/Cas9 genome editing technique developed specifically for arabidopsis (Fauser et al., 2014) to generate loss-of-function mutants for SAUR 41 genes.
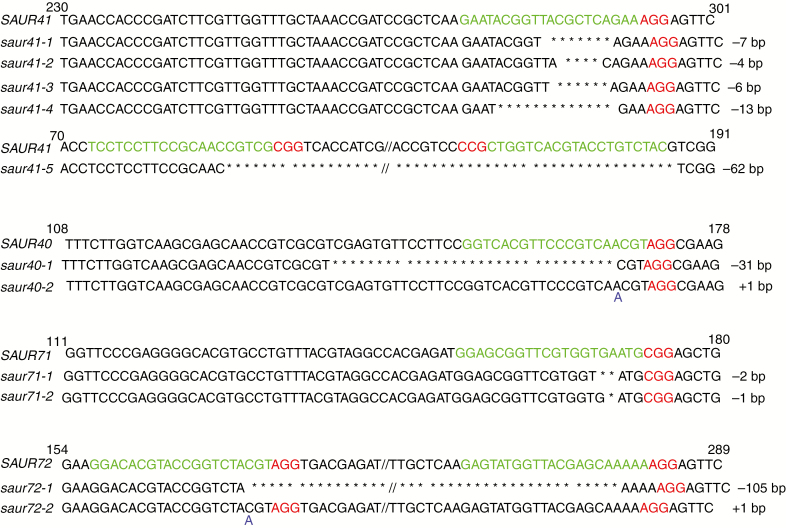
First, a set of single mutants was generated for each gene (Fig. 3). The impact of each mutation on the corresponding protein product was evaluated based on the integrity of the highly conserved approx. 60 amino acid SAUR domain (Ren and Gray, 2015). Mutations in saur40 completely deleted the SAUR domain; mutations in saur71 and saur72 deleted a very large part of the SAUR domain; and mutations in saur41-1 to saur41-4 deleted one-third of the SAUR domain and the entire C-terminus, while the saur41-5 mutation deleted the whole SAUR domain (Supplementary data Fig. S3). Subsequently, we crossed these single mutants and screened corresponding mutations to create double mutants, triple mutants and quadruple mutants. This allowed us to overcome potential functional redundancy among SAUR41 subfamily members and also to eliminate potential off-target effects from the CRISPR/Cas9 technique.
Fig. 3.
A set of single mutants for each SAUR41 family member was generated by CRISPR/Cas9 genomic editing. Protospacers (guide RNA targets) are marked in green and protospacer-adjacent motifs (PAMs) are marked in red. Heritable mutations were identified by Sanger sequencing.
In addition to the backcrosses, additional steps were taken to avoid potential off-target effects of the CRISPR/Cas9 technique. Transcriptome profiling studies (see below) provided both FPKM (fragments per kilobase of transcript sequence per millions base pairs sequenced) data as well as SNP (single nucleotide polymorphism) and InDel (insertion–deletion) data, which allowed us to confirm that there were no off-target knockouts of other SAUR genes during the mutation of SAUR41 genes. Furthermore, software to predict potential off-target knockouts (CCTop, https://crispr.cos.uni-heidelberg.de/) determined that the SAUR52 gene was the most likely candidate because it had only a 2 bp mismatch with the target of SAUR72. Using PAGE analysis, we did not detect any mutation in SAUR52 during the mutations of SAUR41 genes. From here on, the triple mutants saur41-1saur71-1saur72-1, saur41-1saur40-1saur71-2 and saur41-5saur40-1saur71-2 are abbreviated as saur41/71/72, saur41/40/71#1 and #2, while the quadruple mutants saur41-1saur40-1saur71-1saur72-1, saur41-1saur40-1saur71-2saur72-1, saur41-5saur40-1saur71-1saur72-1 and saur41-5saur40-1saur71-2saur72-1 are abbreviated as saur41/40/71/72#1 to #4, respectively.
Inactivation of SAUR41 subfamily genes leads to reduced and disordered cell expansion
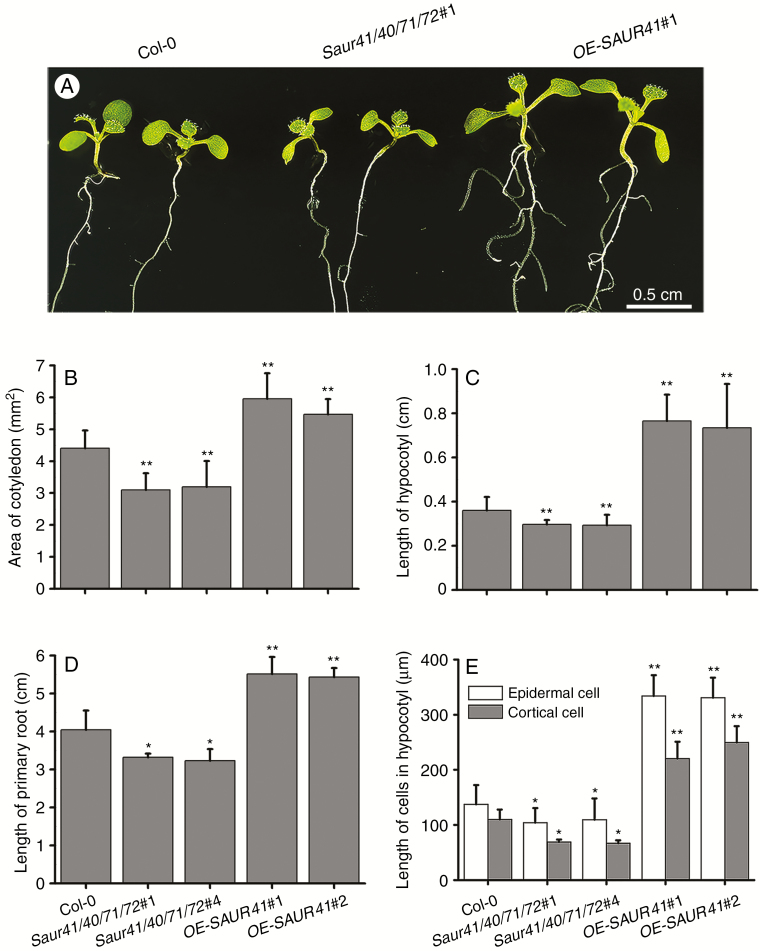
To characterize phenotypes of saur41 mutants, we first performed morphological analysis in comparison with the wild type and the SAUR41 overexpression line (35S::SAUR41-MYC lines, abbreviated as OE-SAUR41; Kong et al., 2013). In 7-day-old quadruple mutant seedlings, we found that the areas of cotyledons, and the lengths of hypocotyls and roots, were significantly reduced (Fig. 4A–D). The lengths of epidermal cells and cortical cells of hypocotyls were further measured across three genotypes, Col-0 (wild-type), saur41/40/71/72 and OE-SAUR41. The results showed that cell length correlated with organ length (Fig. 4E), indicating that the short hypocotyl phenotype in quadruple mutants should be the result of a reduction in cell elongation. In 10-day-old quadruple mutant seedlings, the lengths of cotyledon petioles were also significantly reduced (Supplementary data Fig. S4).
Fig. 4.
Inactivation of SAUR41 subfamily genes leads to reduced cell expansion. (A) Seven-day-old light-grown seedlings of the wild type, the saur41 quadruple mutants saur41/40/71/72 and the SAUR41 overexpression lines OE-SAUR41. (B) Statistical comparison of cotyledon areas in 7-day-old seedlings of three genotypes. (C) Statistical comparison of hypocotyl length in 7-day-old seedlings. (D) Statistical comparison of root length in 7-day-old seedlings. (E) Statistical comparison of epidermal cell and cortical cell length in 7-day-old hypocotyls. The quadruple mutants saur41-1sauir40-1saur71-1saur72-1 and saur41-5sauir40-1saur71-2saur72-1 are abbreviated as saur41/40/71/72#1 and saur41/40/71/72#4, while the 35S::SAUR41-MYC overexpression lines are abbreviated as OE-SAUR41#1 and #2. Each treatment contained approx. 30 seedlings or cells and was replicated three times. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
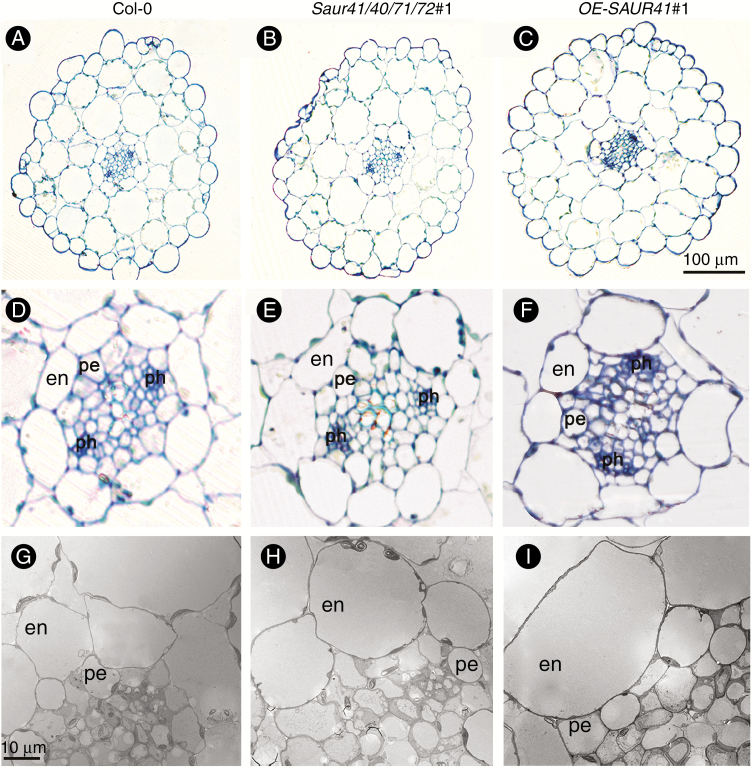
Optical sections from the hypocotyls of 6-day-old seedlings revealed heterogeneous cell size in the endodermis of both the quadruple mutants and the SAUR41 overexpression lines, with the shape of many endodermic cells changed from rectangular to circular (n = 6; Fig. 5A–F). Using TEM, we also found irregular arrangements of cell size and cell shape in the hypocotyl pericycles of saur41/40/71/72 and OE-SAUR41 (n = 4; Fig. 5G–I).
Fig. 5.
Both inactivation of SAUR41 genes and overexpression of SAUR41 led to irregular arrangement of cell size and shape in endodermis and pericycles of arabidopsis hypocotyls. (A) Optical section of hypocotyls of 6-day-old wild-type (Col-0) seedlings. (B) Optical section of the quadruple mutants saur41/40/71/72. (C) Optical section of the SAUR41 overexpression OE-SAUR41 lines. (D) Enlargement of an optical section of the wild type. (E) Enlargement of an optical section of the quadruple mutant. (F) Enlargement of an optical section of the SAUR41 overexpression line. (G) TEM of the wild type. (H) TEM of the quadruple mutant. (I) TEM of the SAUR41 overexpression line. en, endodermis; pe, pericycle cells; ph, phloem.
Quadruple mutants exhibit disordered transcription of calcium and ABA signalling genes
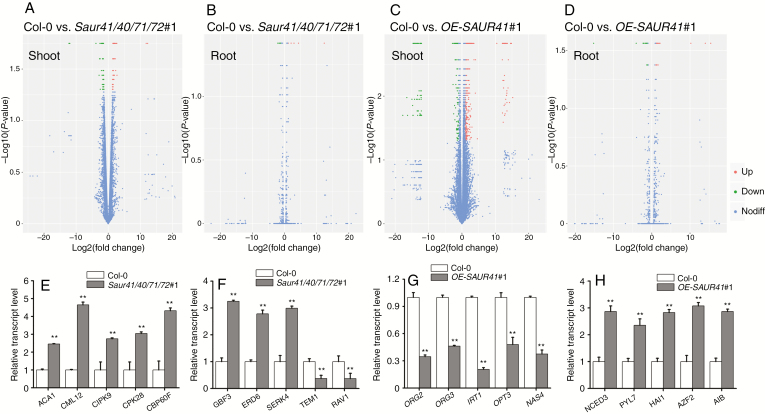
We used RNA-seq to profile differentially expressed genes in saur41/40/71/72 mutants and OE-SAUR41 overexpression lines as compared with the wild type. Shoot parts and root parts of 5-day-old seedlings were evaluated separately. Raw reads and processed data of RNA-seq experiments have been deposited in the GEO at NCBI. Information on differentially expressed genes including gene IDs, fold change and putative functions is given in Supplementary data Tables S2–S5. Venn diagrams (volcano plots) of wild-type shoot vs. mutant shoot, wild-type root vs. mutant root, wild-type shoot vs. overexpression shoot and wild-type root vs. overexpression root were graphically illustrated, and it seemed that shoot samples had far more differentially expressed genes than root samples (Fig. 6A–D). Four corresponding histograms for GO analysis (enrichments of cellular components, molecular function and biological processes) have been included in Supplementary data Figs S5 and S6. Considering that SAUR41 genes are induced by ABA (Fig. 2) to regulate cell expansion (Figs 4 and 5), among the significantly enriched biological processes, we chose certain gene sets related to ABA signalling/biosynthesis or ion homeostasis/signalling for qRT-PCR validation of gene transcription levels (Fig. 6E–H).
Fig. 6.
Transcriptome profiling analysis of saur41 quadruple mutants and SAUR41 overexpression lines. (A–D) Venn diagrams illustrating differentially expressed genes in wild-type shoot vs. mutant shoot (A), wild-type root vs. mutant root (B), wild-type shoot vs. overexpression shoot (C) and wild-type root vs. overexpression root (D). Shoot samples had far more differentially expressed genes than root samples. (E–H) In 5-day-old seedlings, qRT-PCR validation of differentially expressed genes identified by transcriptome analysis. For each gene, the expression level in Col-0 was set to l.0. (E) In the shoots of saur41/40/71/72, a set of calcium homeostasis/signalling genes are upregulated. (F) A set of ABA signalling-related genes are differentially expressed in the shoos of the quadruple mutants. (G) Overexpression of SAUR41 decreases the transcription of iron homeostasis genes in roots. (H) Overexpression of SAUR41 increases the transcription of ABA biosynthesis/signalling genes in shoots. Each treatment contained three biological replicates. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
In the shoot parts of saur41/40/71/72, a set of calcium homeostasis/signalling genes were upregulated as compared with the wild type; these included ACA1 (Autoinhibited Ca2+-ATPase 1), CML12/TOUCH3 (Calmodulin-Like 12), CIPK9 (CBL-Interacting Protein Kinase 9), CPK28 (Calcium-Dependent Protein Kinase28) and CBP60F (Calmodulin-Binding Proteins 60F) (Fig. 6E). In addition, a set of ABA signalling-related genes were differentially expressed in the shoot parts of the quadruple mutants: upregulated genes included GBF3 (G-box Binding Factor 3), encoding a positive transcription factor for ABA signalling in branch bud control and dehydration response (Gonzalez-Grandio et al., 2017; Ramegowda et al., 2017), ERD6 (Early Response To Dehydration 6), encoding a putative sucrose transporter, and BKK1/SERK4 (BRInsensitive1-Associated Kinase1-Like1/Somatic Embryogenesis Receptor-Like Kinase 4), encoding a component for cross-talk between ABA and BR and for stomatal patterning (Meng et al., 2015); downregulated genes included RAV1 (Related to ABA Insensitive3/Viviparous1) and TEM1 (Tempranillo 1), two negative transcription factor genes of ABA signalling (Feng et al., 2014) (Fig. 6F).
Overexpression of SAUR41 decreases transcription of iron homeostasis genes in roots and increases transcription of ABA biosynthesis/signalling genes in shoots
In roots of the SAUR41 overexpression seedlings, strikingly, of downregulated genes, approximately half were related to iron homeostasis; these included two transcription factor genes regulating the iron deficiency response (ORG2/AtHLH38 and ORG3/AtHLH39) (Yuan et al., 2008); iron transporter genes specific to the epidermis (IRT1) and phloem (OPT3), respectively; and a nicotianamine (NA) synthase gene (NAS4) that produces NA for the movement of iron via the NA–Fe complex (Fig. 6G).
In the shoot parts of the SAUR41 overexpression seedlings, a set of genes related to ABA biosynthesis and signalling had significantly changed their transcription levels; these included NCED3 (Nine-Cis-Epoxycarotenoid Dioxygenase3), a key gene for ABA biosynthesis which responds to leaf turgor decrease within minutes (Sussmilch et al., 2017), PYL7 (Pyrabactin Resistance 1-Like 7/Regulatory Components Of ABA Receptor), HAI1 (Highly ABA-Induced PP2C Family Clade A), AZF2 (Arabidopsis Zinc Finger Transcription Factor) and AIB (ABA-Inducible bHLH-type Transcription Factor) (Fig. 6H).
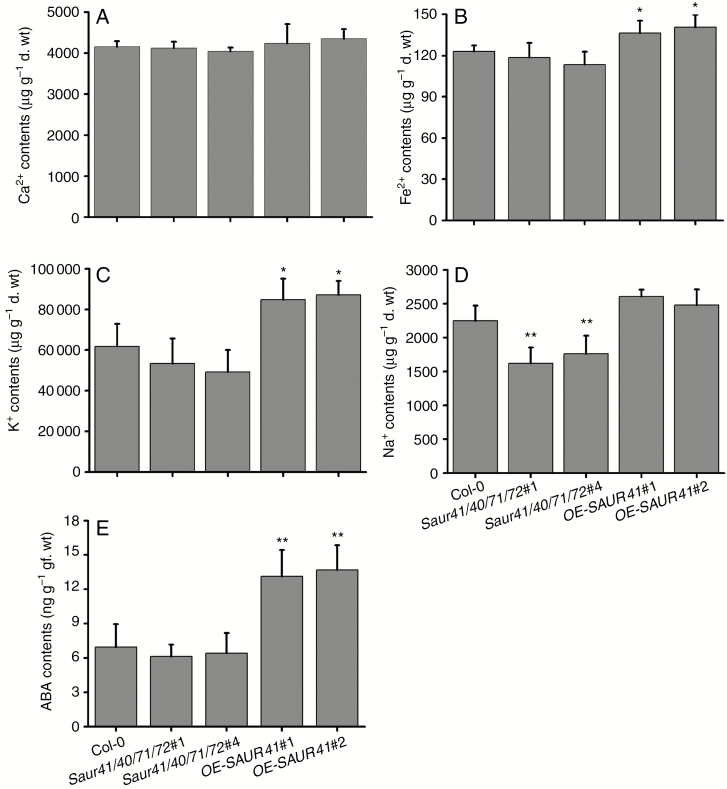
In light of the RNA-seq results, we measured metal ions (calcium, iron, sodium and potassium) and ABA contents in shoot parts of Col-0, saur41/40/71/72 and OE-SAUR41 seedlings. There were no differences on calcium content among the three genotypes (Fig. 7A), although calcium homeostasis/signalling genes were up-regulated in the quadruple mutants (Fig. 6E). This result indicated that the atomic emission spectrometry method used here to detect total calcium is unable to reveal the kinetic distribution of extracellular and cytosolic calcium. Interestingly, although the OE-SAUR41 seedlings showed a reduced expression of iron homeostasis genes in roots (Fig. 6G), they contained higher amounts of iron in shoots (Fig. 7B), indicating that SAUR41 overexpression may lead to an iron excess phenotype, resulting in a feedback effect. OE-SAUR41 shoots accumulated much more potassium (Fig. 7C), while the quadruple mutants had reduced contents of sodium (Fig. 7D). The OE-SAUR41 shoots showed increased expression of NCED3 (Fig. 6H); as expected, these shoots contained higher amounts of ABA than the wild type and the quadruple mutants (Fig. 7E).
Fig. 7.
Measurement of metal ion and ABA contents in 7-day-old seedling shoots. (A) There are no difference in calcium content among Col-0, saur41/40/71/72 and OE-SAUR41. (B) The SAUR41 overexpression lines have the highest levels of iron. (C) The SAUR41 overexpression lines accumulate much more potassium. (D) The saur41/40/71/72 mutants contain the lowest levels of sodium. (E) The SAUR41 overexpression lines have 2-fold increased ABA contents as compared with the wild type. Each treatment contained three biological replicates. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
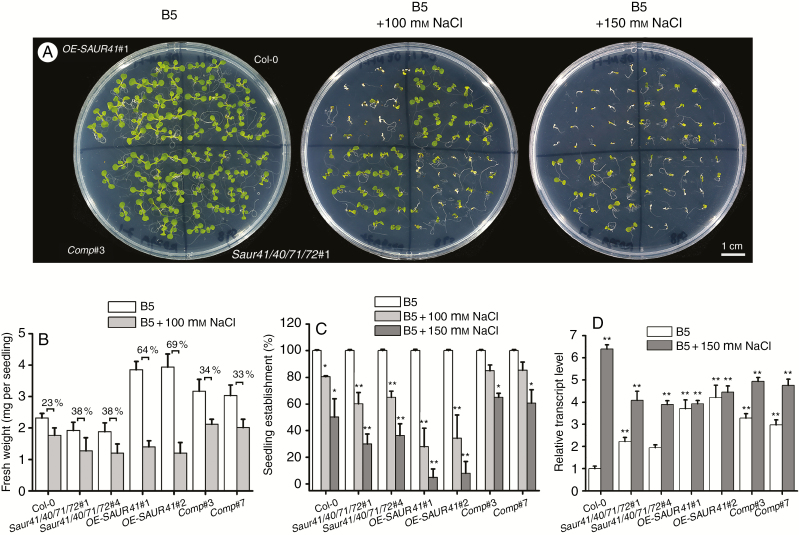
Both the quadruple mutants and the SAUR41 overexpression lines were hypersensitive to salt stress
As SAUR41 genes are ABA inducible (Fig. 2) and both the quadruple mutants and the SAUR41 overexpression lines exhibit altered expression of ABA biosynthesis and/or signalling genes (Fig. 6), we carried out abiotic stress response assays. After some primary investigations, we focused on salt stress in the present study. The results showed that both the quadruple mutants and the SAUR41 overexpression lines were hypersensitive to 100 and 150 mm NaCl stresses as compared with the wild type (Fig. 8A). The overexpression lines in control conditions had the longest and overexpanded hypocotyls, so they had much higher biomass than Col-0 in control conditions. However, under NaCl stress, the overexpression lines appeared to be most affected: they had the lowest seedling establishment rate and the most significant fresh weight reduction (Fig. 8B, C). Notably, in the quadruple mutants, specific expression of SAUR41 under the ABA-responsive RD29A (Responsive to Desiccation 29A) promoter rescued the inhibitory effect of salt on seedling establishment and growth (Fig. 8A–C). To carry out a rescue experiment for a quadruple mutant, the best approach is to introduce four transgenes into the mutant, but this will probably be quite difficult. As the SAUR41 genes were ABA inducible, here we drove SAUR41 expression by using the RD29A promoter (RA29A::SAUR41).
Fig. 8.
Both the quadruple mutants and the SAUR41 overexpression lines were hypersensitive to salt stress. (A) Seedling establishment and growth of the wild-type Col-0, quadruple mutants saur41/40/71/72, the SAUR41 overexpression OE-SAUR41 lines and the complementary lines Comp, grown on B5 media containing 100 or 150 mm NaCl for 7 d. The Comp lines were generated by specific expression of SAUR41-EGFP under the RD29A promoter (proRD29A::SAUR41-EGFP) in the quadruple mutant saur41/40/71/72#1. (B) Fresh weight of 7-day-old seedlings of four genotypes under salt stress. The labelled percentages are the rate of reduction of the fresh weight under salt stress. (C) Statistical comparison of percentage seedling establishment of four genotypes under salt stress. All the control treatments for the various lines had a 100 % seedling establishment rate. (D) qRT-PCR assay of the accumulation of SOS1 encoding an Na+/H+ antiporter and a salt tolerance determinant. Each treatment contained approx. 30 seedlings or approx. 90 seeds and was replicated three times. Error bars represent the s.d. **P < 0.01, *P < 0.05, Student’s t-test.
The transcription level and the stability of SOS1 (Salt Overly Sensitive 1) mRNA, encoding an Na+/H+ antiporter and a salt tolerance determinant, are very sensitive to salt tress (Shi et al., 2000; Deinlein et al., 2014). For qRT-PCR assay of SOS1 accumulation, 5-day-old seedlings were transferred into control medium or medium with 150 mm NaCl and grown for 2 d. The results showed that, under control conditions, the SAUR41 overexpression lines had the highest expression of SOS1, but, under NaCl stress, they failed to increase the expression of SOS1 further (Fig. 8D); this may one of the reasons why they were most sensitive to salt stress. The quadruple mutants were able to increase the expression of SOS1 under salt stress (Fig. 8D), although they were salt sensitive as compared with the wild type (Fig. 8A–C).
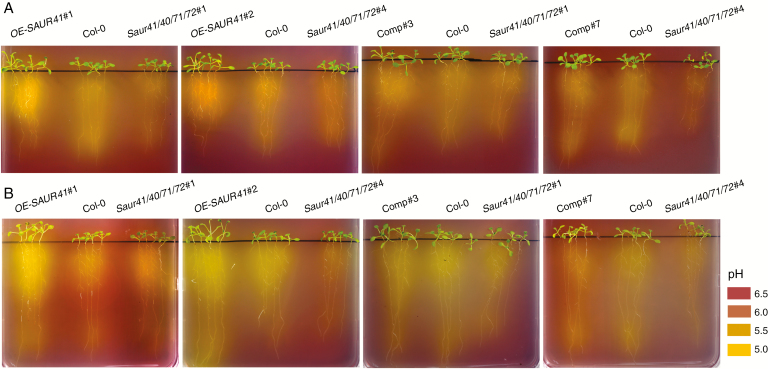
Mechanistically, SAUR19s activate plasma membrane H+-ATPases to induce apoplast acidification and cell expansion (Spartz et al., 2014). We finally performed rhizosphere acidification assays. Seedlings were grown on B5 medium with normal or one-fifth iron concentrations for 7 d and then transferred to plates containing the pH sensor bromocresol purple for 2 d. Extensive medium acidification was observed for the roots of OE-SAUR41, especially when the seedlings were from the one-fifth iron concentration medium (Fig. 9). In addition, overexpression of other members of the SAUR41 subfamily, including SAUR40, SAUR71 and SAUR72, also led to increased rhizosphere acidification (Supplementary data Fig. S7).
Fig. 9.
Overexpression of SAUR41 led to extensive rhizosphere acidification. Seedlings were grown on B5 medium with normal (A) or one-fifth (B) iron concentrations for 7 d and then transferred to B5 plates containing the pH sensor bromocresol purple (0.003 %) for 2 d. Extensive medium acidification was observed for the roots of OE-SAUR41, especially when the seedlings were from the one-fifth iron concentration medium. A pH colorimetric panel is illustrated.
DISCUSSION
Data mining on arabidopsis transcriptional profiles revealed that the SAUR41 subfamily genes are strikingly induced by the growth-inhibiting hormone ABA (Kodaira et al., 2011; reviewed in Ren and Gray, 2015). Using qRT-PCR assay, we found that the expression patterns of SAUR41 genes in arabidopsis seedlings were both overlapping and differential (Fig. 1). We verified that the transcription of SAUR41 genes was induced by ABA, but not by GA and BR (Fig. 2; Supplementary data Fig. S2), in contrast to most other arabidopsis SAUR genes, which are repressed by ABA and activated by GA and BR (Kodaira et al., 2011; reviewed in Ren and Gray, 2015). Thus, understanding the physiological roles of SAUR41 genes seems promising to identify new players in plant ABA pathways.
The protein sequences of SAUR41s are highly conserved within the subfamily but show significant divergence from other SAURs, largely in the N-terminus (Kong et al., 2013). Overexpression of SAUR41 genes led to cell expansion-related phenotypes (Kong et al., 2013; Qiu et al., 2013). Among the 79 SAURs of arabidopsis, approx. 20 have been overexpressed and found to promote cell expansion in young seedlings (see the Introduction), regardless of their diverse subcellular localization. In contrast, knockout studies on arabidopsis SAUR genes are less documented, but have begun to be carried out in recent years (van Mourik et al., 2017; Dong et al., 2019). Here, using the CRISPR/Cas9 genome editing technique developed recently, loss-of-function mutants were generated for the SAUR41 subfamily (Fig. 3), and some developmental and physiological phenotypes of quadruple mutants together with those of overexpression lines were characterized further (Figs 4–9). In those quadruple mutant seedlings, the lengths of hypocotyls, roots and cotyledon petioles and the areas of cotyledons were significantly reduced (Fig. 4; Supplementary data Fig. S4). In addition, thin and ultra-thin sections revealed that both in the quadruple mutants and in the SAUR41 overexpression lines, there existed irregular arrangements of cell size and cell shape in hypocotyl pericycles and endodermis (Fig. 5). Thus, based on both loss-of-function and gain-of-function investigations, we conclude that SAUR41s are positive regulators of cell expansion, similar to the other arabidopsis SAURs investigated in recent years, including SAUR19s (Franklin et al., 2011; Spartz et al., 2012), SAUR63s (Chae et al., 2012), SAUR36 (Stamm and Kumar, 2013), SAUR76–78 (Li et al., 2015), SAUR14/50/65 (Sun et al., 2016) and SAUR10s (van Mourik et al., 2017). Because these SAURs span three clades and exhibit divergent expression patterns, the cell expansion promotion function of these proteins should be related to or attributed to the highly conserved approx. 60 amino acid SAUR domain.
Transcriptome analysis demonstrated that, in the shoot parts of the quadruple mutants, the Ca2+ homeostasis gene ACA1 and certain Ca2+ signalling genes (CML12/TCH3, CIPK9, CPK28 and CBF60F) were upregulated (Fig. 6E). Additionally, GBF3, a transcription factor gene responsive to ABA, water deprivation and bud dormancy (Gonzalez-Grandio et al., 2017; Ramegowda et al., 2017), was also upregulated (Fig. 6F). CIPK9 is a CBL-interacting protein kinase that regulates potassium homeostasis under low potassium stress (Pandey et al., 2007; Liu et al., 2013). Calcium signalling is crucial for regulating ion homeostasis in plant stress responses to cell wall integrity, salt, K+ and pH (reviewed in Manishankar et al., 2018), and the integration of ABA and calcium signalling is essential for evoking substantial cellular responses to various stresses (reviewed in Edel and Kudla, 2016). Interestingly, most arabidopsis SAUR proteins, including SAUR71 and SAUR72, contain a potential calmodulin-binding motif, although no functional significance has been ascribed to this motif to date (reviewed in Ren and Gray, 2015).
In seedling shoots of the SAUR41 overexpression lines, the key ABA biosynthesis gene NCED3 was up-regulated (Fig. 6H), and ABA content was increased (Fig. 7E). As reported, the transcription of NCED3 is remarkably sensitive to the fluctuation of cell turgor (Sussmilch et al., 2017). Thus, it is likely that accelerated and premature cell expansion reduced the hydrostatic pressure of cells overexpressing SAUR41, thus inducing ABA biosynthesis. Alternatively, SAUR proteins have unknown pathways to induce ABA biosynthesis. Although the SAUR41 overexpression seedlings contained more ABA, these seedlings still exhibited increased cell expansion (Fig. 4; Supplementary data Fig. S4) and much higher potassium contents (Fig. 7C). These results may indicate that cell expansion induced by ectopic SAUR41 is epistatic to the putative inhibitory effects of ABA on cell expansion.
The SAUR41 overexpression lines were most sensitive to salt stress, whereas the quadruple mutants were moderately sensitive to salt stress (Fig. 8), indicating that a suitable expression level of SAUR41 genes is required for arabidopsis salt tolerance. Supporting this notion, context-dependent expression of SAUR41 under the ABA-responsive RD29A promoter in the quadruple mutants rescued the inhibitory effect of salt stress on seedling establishment (Fig. 8A–C). The RD29A promoter of arabidopsis contains two types of cis-acting elements for rapid (approx. 20 min) and slow (approx, 3 h) induction of gene expression under conditions of drought, high salt or low temperature (Yamaguchi-Shinozaki and Shinozaki, 1993, 1994). To improve plant stress tolerance, use of the strong constitutive 35S promoter resulted in severe side effects under normal growth conditions, whereas use of the stress-inducible RD29A promoter eliminated the side effects and provided an even greater tolerance than the 35S promoter (Kasuga et al., 1999).
Three lines of evidence may explain why the SAUR41 overexpression lines were those most unable to cope with salt stress. First, as reported, downregulation of most of the arabidopsis SAUR genes is involved in the inhibition of plant growth under salt stress (Kodaira et al., 2011). Constitutive expression of SAUR41 under a strong promoter (35S) may disturb the establishment of growth adaption under salt stress. Secondly, the SAUR41 overexpression lines failed to increase the expression of SOS1 under salt stress (Fig. 8D); enhanced expression of SOS1 is a key determinant of salt tolerance (Shi et al., 2000). Thirdly, overexpression of SAUR41 led to disordered cell size and shape in the endodermis (Fig. 5), and this may disrupt the functional relationship between root/hypocotyl/leaf anatomy and salt tolerance (reviewed in Munns and Tester, 2008). As regards the quadruple mutants, they were able to increase the expression of SOS1 under salt stress (Fig. 8D), and they contained the lowest contents of sodium under normal conditions (Fig. 7D), but they still displayed some weaknesses in salt tolerance (Fig. 8A–C). This could be explained in two ways. The quadruple mutants have the SAUR41 subfamily genes knocked out, so they have reduced cell expansion in cells expressing SAUR41 genes, which may lead to overinhibition under salt stress. In addition, they have disordered cell size and shape in the endodermis (Fig. 5).
The mechanisms whereby SAURs mediate cell expansion remain largely unknown. As reported, SAUR19s are thought to inhibit PP2C.D-type protein phosphatases to activate plasma membrane H+-ATPases and to induce apoplast acidification (Spartz et al., 2014). Similarly, it was found that SAUR40 and SAUR72 also inhibit PP2C.D1 in an in vitro phosphatase assay (Spartz et al., 2014). Plasma membrane H+-ATPases are powerhouses for plant physiology in regulation of cell expansion, solute uptake, phloem loading and tip growth (Palmgren 2001; Haruta et al., 2015; Falhof et al., 2016; Mangano et al., 2018). It has been suggested that arabidopsis roots cope with iron deficiency by promoting plasma membrane H+-ATPase 2 (AHA2)-mediated H+ release into the rhizosphere to enhance the dissolution of Fe3+ (reviewed in Jeong et al., 2017; Tsai and Schmidt, 2017). We found that overexpression of SAUR41 led to increased acidification of the medium (Fig. 9) and increased iron contents (Fig. 7B), which may be responsible for the reduced transcription of iron absorption and transport genes because of extensive bioavailability of iron (Fig. 6). Therefore, it is rational to assume that SAUR41s may inhibit PP2C.Ds to activate plasma membrane H+-ATPases. SAUR19s are peripheral membrane proteins, whereas SAUR41s are basically cytosolic proteins (Kong et al., 2013; Qiu et al., 2013). Nevertheless, the PP2C.D-type protein phosphatases are diverse in subcellular location, and different SAURs may have different PP2C.D partners (Ren et al., 2018; discussed in Stortenbeker and Bemer, 2018).
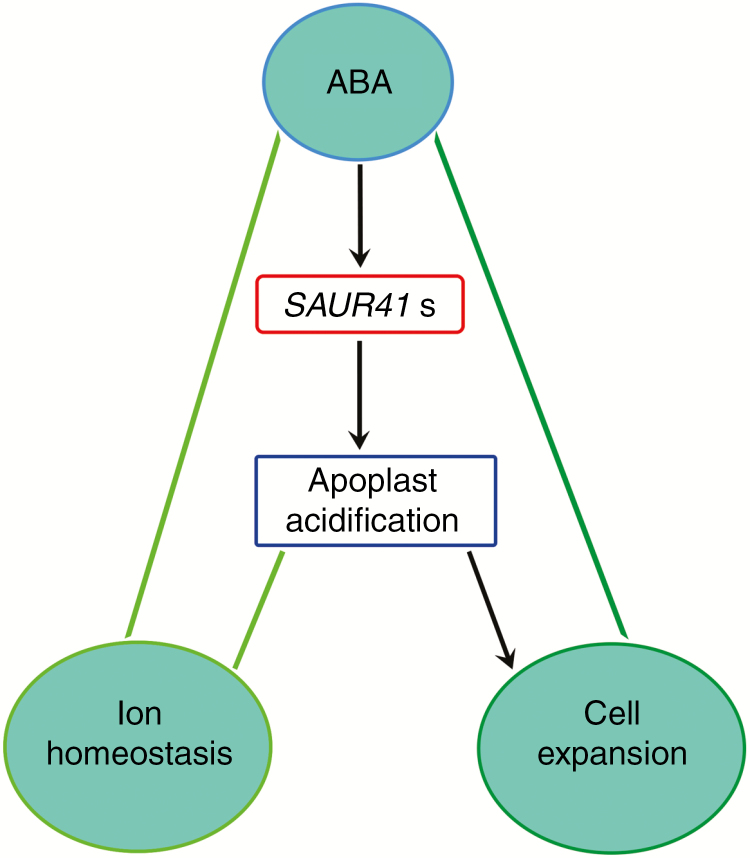
Taken together, we propose that besides the ABA-repressed SAUR genes (Kodaira et al., 2011), there exist ABA-induced SAUR genes (this study). Balanced expression of two types of SAUR genes may be necessary for salt tolerance. We suggest a working model for SAUR41 genes in which ABA induces the expression of SAUR41 genes to fine-tune seedling establishment and salt tolerance by modulating cell expansion and ion homeostasis (Fig. 10). Further investigation is needed to elucidate how the SAUR41 pathway interacts with the core ABA signalling pathways and how a plant cell can distinguish extracellular acidification required for ion transportation and cell expansion. In addition, further examination of cell size in single, double and triple mutants would help to reveal distinctive and/or overlapping roles of individual members of SAUR41 genes for cell size control. Considering that SAUR41 genes are ABA inducible and ABA is a key hormone in plant abiotic stress signalling, our work may provide new candidate genes for improvement of plant abiotic stress tolerance.
Fig. 10.
A working model for the function of SAUR41 genes. ABA induces the expression of SAUR41 genes to fine-tune seedling establishment and salt tolerance by modulating cell expansion and ion homeostasis.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: primer sequences used in the present study. Table S2: genes differentially expressed in root parts of the saur41 quadruple mutants. Table S3: genes differentially expressed in shoot parts of the saur41 quadruple mutants. Table S4: genes differentially expressed in root parts of the 35S::SAUR41-MYC seedlings. Table S5: genes differentially expressed in shoot parts of the 35S::SAUR41-MYC seedlings. Fig. S1: the design of guide RNAs targeting the coding sequences of SAUR41 genes for the CRISPR/Cas9 genome editing. Fig. S2: BR response of SAUR41 genes in 5-day-old arabidopsis seedlings as revealed by qRT-PCR. Fig. S3: predicted effects of each mutation on the corresponding protein product of SAUR41 genes. Fig. S4: cotyledon petiole lengths in single, double, triple and quadruple mutants of SAUR41 genes. Fig. S5: histograms for GO analysis of transcription profiling of the saur41 quadruple mutants. Fig. S6: histograms for GO analysis of transcription profiling of the SAUR41 overexpression lines. Fig. S7: overexpression of other SAUR41 genes increased medium acidification in arabidopsis rhizosphere.
FUNDING
This work was supported by the Fundamental Research Funds for the Central Universities (no. 2017QNA6003), the National Program on Key Basic Research Project (no. 2014CB541704) and the Natural Science Foundation of Zhejiang Province (no. LY18C020001).
ACKNOWLEDGEMENTS
We thank Dr Holger Puchta (Karlsruhe Institute of Technology, Germany) for the CRISPR/Cas9 vectors, and VIB-Ghent University for plant GATEWAY™ destination vectors.
LITERATURE CITED
- Arsuffi G, Braybrook SA. 2017. Acid growth: an ongoing trip. Journal of Experimental Botany 69: 137–146. [DOI] [PubMed] [Google Scholar]
- Bechtold N. 1993. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Compte Rendu de l’Academie des Sciences Paris, Sciences de la Vie 316: 1194–1199. [Google Scholar]
- Bemer M, van Mourik H, Muino JM, Ferrandiz C, Kaufmann K, Angenent GC. 2017. Fruitfull controls SAUR10 expression and regulates Arabidopsis growth and architecture. Journal of Experimental Botany 68: 3391–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, et al. 2012. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. The Plant Journal 71: 684–697. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Deinlein U, Strphan AB, Horie T, Luo W, Xu G, Schroeder JI. 2014. Plant salt-tolerance mechanisms. Trends in Plant Science 19: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Sun N, Yang J, et al. 2019. TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. The Plant Cell 31: 1155–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel KH, Kudla J. 2016. Integration of calcium and ABA signaling. Current Opinion in Plant Biology 33: 83–91. [DOI] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J, Pedersen JT, Fuglsang AT, Palmgren M. 2016. Plasma membrane H+-ATPase regulation in the center of plant physiology. Molecular Plant 9: 323–337. [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H. 2014. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. The Plant Journal 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Favero DS, Le KN, Neff MM. 2017. Brassinosteroid signaling converges with Suppressor of Phytochrome B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. The Plant Journal 89: 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF. 2014. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. The Plant Journal 80: 654–668. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, et al. 2011. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences, USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Grandio E, Pajoro A, Franco-Zorrilla JM, Tarancon C, Immink RG, Cubas P. 2017. Abscisic acid signaling is controlled by a branched1/HD-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences, USA 114: e245–e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Jiang Q, Hu Z, Sun X, Fan S, Zhang H. 2017. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. The Crop Journal 6: 181–190. [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin responsive gene expression – genes, promoters and regulatory factor. Plant Molecular Biology 49: 373–385. [PubMed] [Google Scholar]
- Haruta M, Gray WM, Sussman MR. 2015. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Current Opinion in Plant Biology 28: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Li Z, Waadt R, Schroeder JI. 2017. Snapshot: abscisic acid signaling. Cell 171: 1708–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SL, Hsieh HL, Jauh GY. 2018. SMALL AUXIN UP RNA62/75 are required for the translation of transcripts essential for pollen tube growth. Plant Physiology 178: 626–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS. 2013. SAUR36, a SMALL AUXIN UP RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiology 161: 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes & Development 24: 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Merkovich A, Clyne M, Connolly EL. 2017. Directing iron transport in dicots: regulation of iron acquisition and translocation. Current Opinion in Plant Biology 39: 106–113. [DOI] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ. 2009. SAUR39, a SMALL AUXIN-UP RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiology 151: 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. Gateway™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1999. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LS, et al. 2011. Arabidopsis Cys2/His2 Zinc-Finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology 157: 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Zhu Y, Gao C, et al. 2013. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant & Cell Physiology 54: 609–621. [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. 2004. Microarray expression analyses of arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. The Plant Cell 16: 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZG, Chen HW, Li QT, et al. 2015. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Scientific Reports 5: 12477. doi: 10.1038/srep12477 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH. 2013. A protein kinase, Calcineurin B-Like Protein-Interacting Protein Kinase9, interacts with Calcium Sensor Calcineurin B-Like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiology 161: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Chen X, Liu C, et al. 2017. MeSAUR1, encoded by a SMALL AUXIN-UP RNA gene, acts as a transcription regulator to positively regulate ADP-glucose pyrophosphorylase small subunit1A gene in Cassava. Frontiers in Plant Science 8: 1315. doi: 10.3389/fpls.2017.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Pacheco JM, Marino-Buslje C, Estevez JM. 2018. How does pH fit in with oscillating polar growth? Trends in Plant Science 23: 479–489. [DOI] [PubMed] [Google Scholar]
- Manishankar P, Wang N, Koster P, Alatar AA, Kudla J. 2018. Calcium signaling during salt stress and in the regulation of ion homeostasis. Journal of Experimental Botany 69: 4215–4226. [DOI] [PubMed] [Google Scholar]
- Markakis MN, Boron AK, Van Loock B, et al. 2013. Characterization of a SMALL AUXIN-UP RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS One 8: e82596. doi: 10.1371/journal.pone.0082596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, et al. 2015. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Current Biology 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mourik H, van Dijk ADJ, Stortenbeker N, Angenent GC, Bemer M. 2017. Divergent regulation of Arabidopsis SAUR genes: a focus on the SAUR10-clade. BMC Plant Biology 17: 245. doi: 10.1186/s12870-017-1210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. 2010. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nature Protocols 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Biology 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S. 2007. CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in arabidopsis. Cell Research 17: 411–421. [DOI] [PubMed] [Google Scholar]
- Qiu T, Chen Y, Li M, et al. 2013. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of SMALL AUXIN UP RNA genes: potential implications. Plant Signaling & Behavior 8: e25283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda V, Gill US, Sivalingam PN, et al. 2017. GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Scientific Reports 7: 9148. doi: 10.1038/s41598-017-09542-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM. 2015. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Molecular Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Park MY, Spartz AK, Wong JH, Gray WM. 2018. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genetics 14: e1007455. doi: 10.1371/journal.pgen.1007455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Lin W, Zhu Y, et al. 2010. The Gypsy insulator of Drosophila melanogaster, together with its binding protein Suppressor of Hairy-Wing, facilitate high and precise expression of transgenes in Arabidopsis thaliana. Genetics 185: 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. 2000. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putativeNa+/H+ antiporter. Proceedings of the National Academy of Sciences, USA 97: 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Mila I, Liu M, et al. 2018. The RIN-regulated Small Auxin-Up RNA SAUR 69 is involved in the unripe-to-ripe phase transition of tomato fruit via enhancement of the sensitivity to ethylene. New Phytologist 222: 820–836 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, et al. 2012The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. The Plant Journal 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, et al. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. The Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lor VS, Ren H, et al. 2017. Constitutive expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiology 173: 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P, Kumar PP. 2013. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Reports 32: 759–769. [DOI] [PubMed] [Google Scholar]
- Stortenbeker N, Bemer M. 2018. The SAUR gene family: the plant’s toolbox for adaptation of growth and development. Journal of Experimental Botany 70: 17–27 [DOI] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, et al. 2016. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proceedings of the National Academy of Sciences, USA 113: 6071–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussmilch FC, Brodribb TJ, McAdam SAM. 2017. Up-regulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. Journal of Experimental Botany 68: 2913–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Sepahi M, Arendall B, Bohnert HJ. 1995. Enhancement of seed germination in high salinity by engineering mannitol expression in Arabidopsis thaliana. Plant, Cell & Environment 18: 801–806. [Google Scholar]
- Tsai HH, Schmidt W. 2017. Mobilization of iron by plant-borne coumarins. Trends in Plant Science 22: 538–548. [DOI] [PubMed] [Google Scholar]
- Wild M, Daviere JM, Regnault T, et al. 2016. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Developmental Cell 37: 190–200. [DOI] [PubMed] [Google Scholar]
- Xu YX, Xiao MZ, Liu Y, Fu JL, He Y, Jiang DA. 2017. The SMALL AUXIN-UP RNA OsSAUR45 affects auxin synthesis and transport in rice. Plant Molecular Biology 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. Characterization of the expression of a desiccation responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular and General Genetics 236: 331–340. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jin H, Chen Y, et al. 2012. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytologist 193: 81–95. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, et al. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research 18: 385–397. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Zhao T, Kermode AR. 2012. A conifer ABI3-interacting protein plays important roles during key transitions of the plant life cycle. Plant Physiology 161: 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.