Abstract
Purpose
Breast cancer (BC) is a heterogeneous disease consisting of various subtypes, with different prognostic and therapeutic outcomes. The amino acid transporter, SLC7A8, is overexpressed in oestrogen receptor-positive BC. However, the consequence of this overexpression, in terms of disease prognosis, is still obscure. This study aimed to evaluate the biological and prognostic value of SLC7A8 in BC with emphasis on the intrinsic molecular subtypes.
Methods
SLC7A8 was assessed at the genomic, using METABRIC data (n = 1980), and proteomic, using immunohistochemistry and TMA (n = 1562), levels in well-characterised primary BC cohorts. SLC7A8 expression was examined with clinicopathological parameters, molecular subtypes, and patient outcome.
Results
SLC7A8 mRNA and SLC7A8 protein expression were strongly associated with good prognostic features, including small tumour size, low tumour grade, and good Nottingham Prognostic Index (NPI) (all P < 0.05). Expression of SLC7A8 mRNA was higher in luminal tumours compared to other subtypes (P < 0.001). High expression of SLC7A8 mRNA and SLC7A8 protein was associated with good patient outcome (P ≤ 0.001) but only in the low proliferative ER+/luminal A tumours (P = 0.01). In multivariate analysis, SLC7A8 mRNA and SLC7A8 protein were independent factors for longer breast cancer specific survival (P = 0.01 and P = 0.03), respectively.
Conclusion
SLC7A8 appears to play a role in BC and is a marker for favourable prognosis in the most predominant, ER+ low proliferative/luminal A, BC subtype. Functional assessment is necessary to reveal the specific role played by SLC7A8 in ER+ BC.
Electronic supplementary material
The online version of this article (10.1007/s10549-020-05586-6) contains supplementary material, which is available to authorised users.
Keywords: SLC7A8, Breast cancer, Prognosis
Introduction
Many cancer cells alter their metabolism to provide energy and cellular building blocks required for their rapid proliferation. Amino acids, particularly glutamine and leucine, are essential for cancer cell growth, as they are critical for controlling protein translation and driving cell cycle progression through regulation of the mammalian target of rapamycin complex1 (mTORC1) pathway [1, 2]. This pivotal need for intracellular amino acids is reflected in the increased expression of amino acid transport systems in the majority of cancers, which is regulated by various transcription factors such as c-MYC, hormone receptors, and nutrient starvation responses [3–5].
Solute Carrier Family 7 Member 8 (SLC7A8) and Member 5 (SLC7A5) are sodium-independent amino acid exchangers (antiports), which transport small and large neutral amino acids, such as alanine, serine, threonine, cysteine, phenylalanine, tyrosine, leucine, and glutamine [6]. Both solute carriers require heterodimerisation with the heavy chain of SLC3A2 for their proper localisation in the plasma membrane [7–9].
Although the function of SLC7A8 and SLC7A5 are similar, the former displays relatively lower affinity for its substrates, glutamine, and serine. SLC7A5 has been extensively studied in a variety of cancers and it is regulated by the oncogene c-MYC [10–13]. We have previously described the potential utility of SLC7A5 as a poor prognostic factor for the highly proliferative breast cancer (BC) subtypes [14]. However, there is limited information whether SLC7A8 plays an equal role in BC. Previous studies showed that SLC7A8 is upregulated in ER+BC and it is controlled by oestrogen [4, 15]. Luo et al. also identified SLC7A8 as a novel progesterone target gene in uterine leiomyoma cells [16]. To our knowledge, the prognostic impact of SLC7A8 has not been studied.
In this study, we aimed to assess SLC7A8 gene copy number (CN) and mRNA expression, alongside SLC7A8 protein expression in large and well-characterised cohorts of BC to determine its clinicopathological and prognostic value with emphasis on the different molecular classes.
Material and methods
SLC7A8 genomic profiling
SLC7A8 gene copy number and gene expression were evaluated using the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort of invasive BC (n = 1980) [17]. In this study, DNA/RNA was isolated from fresh frozen samples and transcriptional profiling was acquired using the Illumina HT-12v3 platforms. Data were pre-processed and normalised as described previously [17]. Patients involved in the study who were Oestrogen Receptor-negative (ER-) and Lymph Node (LN)-positive received adjuvant chemotherapy, while ER+ and/or LN− patients did not receive adjuvant chemotherapy. Dichotomisation of SLC7A8 mRNA was achieved using X-tile (version 3.6.1, Yale University, USA), based on prediction of Breast Cancer Specific Survival (BCSS). SLC7A8 mRNA expression was associated with clinicopathological parameters, molecular subtypes and patient outcome.
The online dataset, Breast Cancer Gene-Expression Miner v4.0 (https://bcgenex.centregauducheau.fr), was used for external validation of SLC7A8 mRNA expression.
SLC7A8 protein expression
Immunhistochemistry for SLC7A8 was performed using a well-characterised cohort of early stage primary operable invasive BC patients aged ≤ 70 years. Patients presented at Nottingham City Hospital between 1989 and 2006. Patients were managed based on a uniform protocol. Clinical history, tumour characteristics, information on therapy, and outcomes are prospectively maintained. Outcome data included development and time to distant metastasis (DM) and BCSS.
Supplementary Table 1 summarises the clinicopathological parameters for the Nottingham and METABRIC series.
Western blotting
The specificity of anti-SLC7A8 primary antibody (HPA051950, Sigma-Aldrich, UK) was validated using Western blotting in BC lysates (American Type Culture Collection; Rockville, MD, USA) as previously described [18]. A single band for SLC7A8 was visualised at the correct predicted size (~ 58 KDa) (Supplementary Fig. 1).
Tissue arrays and Immunohistochemistry (IHC)
Tumour samples, 0.6 mm cores, were arrayed as previously described [14, 19]. Immunohistochemical staining was performed on 4 μm TMA sections using Novolink polymer detection system (Leica Biosystems, RE7150-K) as per the manufacturer’s instructions.
Stained TMA sections were scanned using high resolution digital images (NanoZoomer; Hamamatsu Photonics, Welwyn Garden City, UK), at × 20 magnification. Modified histochemical score (H-score) was applied to evaluate SLC7A8 immunostaining. This includes a semi-quantitative assessment of both the percentage and the intensity of stained cells [20]. Staining intensity was graded as: 0, negative; 1, weak; 2, medium; 3, strong and the percentage of the positively stained tumour cells was estimated subjectively. The final H-score was calculated multiplying the intensity (0–3) by the percentage of positive cells (0–100), producing a total range of 0–300. Dichotomisation of SLC7A8 protein expression was determined using X-tile software in predicting BCSS.
Immunhistochemical staining and dichotomisation of the other biomarkers included in this study were as per previous publications [14, 18–22]. ER and PgR positivity was defined as ≥ 1% staining. Immunoreactivity of HER2 was scored using standard HercepTest guidelines (Dako). Chromogenic in situ Hybridisation (CISH) was used to quantify HER2 gene amplification in borderline cases using the HER2 FISH pharmDx™ plus HER2 CISH pharmDx™ kit (Dako) and was assessed according to the American Society of Clinical Oncology guidelines. BC molecular subtypes were defined, based on tumour IHC profile and the Elston-Ellis [23] mitotic score as: ER+/HER2- Low Proliferation (mitotic score 1), ER+/HER2− high Proliferation (mitotic score 2 and 3), HER2− positive class: HER2+ regardless of ER status, Triple Negative (TN): ER−, PgR−, and HER2− [24].
Statistical analysis
SPSS 24.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied for statistical analysis. The Chi-square test was carried out for inter-relationships between categorical variables. One-way ANOVA with post hoc Tukey multiple comparison test and Pearson’s correlation coefficient was performed to analyse the association between continuous variables. Survival curves were examined by Kaplan–Meier with Log Rank test. Cox’s proportional hazard method was performed for multivariate analysis to identify the independent prognostic/predictive factors. P values were adjusted using Bonferroni correction for multiple testing, whenever applicable. A P value < 0.05 was considered significant. The study endpoints were 10-year BCSS or distant metastasis free survival (DMFS). This study complied with reporting recommendations for tumour marker prognostic studies (REMARK) criteria [25].
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ and the North West—Greater Manchester Central Research Ethics Committee under the title ‘Nottingham Health Science Biobank (NHSB)’ reference number 15/NW/0685.
Results
SLC7A8 in breast cancer
High SLC7A8 mRNA expression was observed in almost two-third (67%) of the METABRIC BC cases. A total of 90/1,980 (4.5%) of cases showed SLC7A8 copy number (CN) gain, whereas 45/1,980 (2.3%) cases showed a CN loss. A significant association was observed between SLC7A8 copy number variation (CNV) and SLC7A8 mRNA expression (P < 0.0001, Fig. 1a). There was a positive association between SLC7A8 CN gain and CN gain of the tumour suppressor gene, TP53 (P < 0.0001, Supplementary Table 2).
Fig. 1.
SLC7A8 mRNA expression and its association with copy number aberrations, clinicopathological parameters and molecular subtypes: a SLC7A8 and copy number aberrations, bSLC7A8 and tumour size, cSLC7A8 and tumour grade, dSLC7A8 and lymph node stage, eSLC7A8 and NPI, fSLC7A8 and PAM50 subtypes, gSLC7A8 and METABRIC Integrative Clusters. Pearson correlation was used for two variables and One-way ANOVA with post hoc tukey test for more than two variables
SLC7A8 protein expression was observed, predominantly in the cytoplasm of invasive BC cells, with expression levels varying from absent to high (Fig. 2a and b). Positive SLC7A8 protein expression (> 20 H-score) was observed in 177/1560 (11%) of cases.
Fig. 2.
SLC7A8 protein expression in invasive breast cancer cores. a Positive IHC expression, b negative IHC expression
SLC7A8 and clinicopathological parameters
High SLC7A8 mRNA expression was significantly associated with good prognostic parameters, including smaller tumour size (Fig. 1b, P = 0.007), lower tumour grade (Fig. 1c, P < 0.001), and good Nottingham Prognostic Index (NPI) (Fig. 1e, P < 0.001). These associations were confirmed using the Breast Cancer Gene-Expression Miner (Supplementary Fig. 2A–2C).
Similar associations were observed with SLC7A8 protein expression. Table 1 summarises the observed findings between high SLC7A8 protein and good prognostic factors, including small tumour size (P = 0.03), low tumour grade (P < 0.001), and good NPI (P < 0.001).
Table 1.
Clinicopathological associations of the SLC7A8 protein expression in breast cancer
| Parameter | SLC7A8 protein | χ2 (P value) | Adjusted P value | |
|---|---|---|---|---|
| Low n (%) |
High n (%) |
|||
| Tumour size | ||||
| < 2 cm | 756 (87.1) | 112 (12.9) | 7.03 (0.008) | 0.03 |
| ≥ 2 cm | 623 (91.3) | 59 (8.7) | ||
| Tumour grade | ||||
| 1 | 182 (82.7) | 38 (17.3) | 20.62 (0.00003) | 0.0002 |
| 2 | 491 (86.7) | 75 (13.3) | ||
| 3 | 704 (92.4) | 58 (7.6) | ||
| Lymph node stage | ||||
| 1 | 840 (87.9) | 116 (12.1) | 4.84 (0.08) | 0.16 |
| 2 | 400 (89.7) | 46 (10.3) | ||
| 3 | 136 (93.8) | 9 (6.2) | ||
| Nottingham Prognostic Index | ||||
| Good | 397 (83.8) | 77 (16.2) | 20.09 (0.00004) | 0.0002 |
| Moderate | 746 (90.6) | 77 (9.4) | ||
| Poor | 234 (93.2) | 17 (6.8) | ||
| IHC subtypes | ||||
| ER+/HER2− low proliferation | 706 (87.7) | 99 (12.3) | ||
| ER+/HER2− high proliferation | 230 (90.9) | 23 (9.1) | 5.67 | |
| Triple negative | 207 (88.5) | 27 (11.5) | (0.128) | 0.24 |
| HER2+ | 145 (93.5) | 10 (6.5) | ||
| Histological type | ||||
| Ductal (including mixed) | 1217 (89.6) | 142 (10.4) | ||
| Lobular | 88 (87.1) | 13 (12.9) | 15.93 | |
| Medullary | 23 (92.0) | 2 (8.0) | (0.01) | 0.03 |
| Miscellaneous | 7 (77.8) | 2 (22.2) | ||
| Special type | 41 (78.8) | 11 (21.2) | ||
Bold indicates the significant values
SLC7A8 expression in molecular BC subtypes
High expression of SLC7A8 mRNA was significantly associated with hormone receptor positive (ER+ and PgR+) and HER2− BC (all P < 0.001, Table 2). Likewise, SLC7A8 mRNA was highly expressed in non-triple negative (TN) compared with TN tumours (P < 0.001, Table 2). These results were validated using Breast Cancer Gene-Expression Miner (Supplementary Fig 2D–2G). Similarly, high SLC7A8 protein expression was associated with HER2 negative BC (P = 0.01) and although it was expressed primarily in hormone receptor positive tumours this did not reach significance (Table 2).
Table 2.
Expression of SLC7A8 in breast cancer and the expression of other molecular biomarkers
| SLC7A8 | ||||||||
|---|---|---|---|---|---|---|---|---|
| mRNA | protein | |||||||
| Low n (%) |
High n (%) |
χ2 (P value) | Adjusted P value |
Low n (%) |
High n (%) |
χ2 (P value) | Adjusted P value |
|
| ER | ||||||||
| Negative | 365 (77.2) | 108 (22.8) | 553.2 (2.5 × 10–122) | < 0.0001 | 296 (89.4) | 35 (10.6) | 0.09 (0.76) | 1.52 |
| Positive | 283 (18.9) | 1215 (81.1) | 1083 (88.8) | 136 (11.2) | ||||
| PR | ||||||||
| Negative | 453 (48.3) | 484 (51.7) | 193.6 (5.0 × 10–44) | < 0.0001 | 559 (91.0) | 55 (9.0) | 3.53 (0.06) | 0.30 |
| Positive | 195 (18.9) | 839 (81.1) | 807 (88.0) | 110 (12.0) | ||||
| HER2 | ||||||||
| Negative | 519 (30.1) | 1206 (69.9) | 48.7 (2.9 × 10–12) | < 0.0001 | 1174 (88.3) | 155 (11.7) | 8.82 (0.003) | 0.01 |
| Positive | 129 (52.4) | 117 (47.6) | 198 (95.2) | 10 (4.8) | ||||
| Triple negative | ||||||||
| No | 388 (23.5) | 1263 (76.5) | 405.07 (4.3 × 10–90) | < 0.0001 | 1169 (89.4) | 139 (10.6) | 0.56 (0.45) | 1.80 |
| Yes | 260 (81.3) | 60 (18.8) | 207 (87.7) | 29 (12.3) | ||||
| TP53 mutations | ||||||||
| Wild-type | 204 (28.5) | 512 (71.5) | 38.36 (4.6 × 10–9) | < 0.0001 | Not available | |||
| Mutation | 59 (59.6) | 40 (40.4) | ||||||
| p53 protein | ||||||||
| Negative | Not available | 860 (89.0) | 106 (11.0) | 0.24 (0.62) | 1.86 | |||
| Positive | 435 (89.9) | 49 (10.1) | ||||||
Bold indicates the significant values
Regarding the association of SLC7A8 CN and mRNA with the intrinsic (PAM50) subtypes, SLC7A8 CN gain was mainly observed in luminal B tumours (P < 0.001, Supplementary Table 2), whereas high mRNA expression was observed primarily in luminal A and B tumours and to lesser extent in HER2+BC (P < 0.001, Fig. 1f). In the METABRIC Integrative Clusters, high SLC7A8 mRNA expression was associated with clusters 7 and 8 which embrace ER+ tumours predominately of the luminal A intrinsic subtype (P < 0.001, Fig. 1g). Similar associations of SLC7A8 mRNA with the molecular subtypes were seen using Breast Cancer Gene-Expression Miner (Supplementary Fig. 2H).
Although the result of the association of SLC7A8 protein in the defined IHC subtypes were not nominally significant, it also showed higher SLC7A8 expression in the ER+ low proliferation tumours compared with the other subtypes (Table 1).
SLC7A8 expression and other associated markers
The correlations of SLC7A8 mRNA with other relevant genes were investigated using the METABRIC dataset (Table 3). These genes were selected based on previous publications showing a functional association between SLC7A8 and glutamine transport or metabolism [6, 26–28]. High SLC7A8 mRNA expression was significantly associated with enzymes involved in glutamine metabolism: glutaminase (GLS and GLS2;P < 0.001), which mediate the conversion of glutamine to glutamate. While the correlation with GLS was negative, it was positive with GLS2. There was also a positive association with the enzymes which mediate conversion of glutamine to proline, namely, ALDH4A1 and PRODH (P < 0.001). In contrast, some glutamine transporters were negatively correlated with SLC7A8 expression (P ≤ 0.009), including SLC1A5, SLC7A5, SLC7A6, SLC7A7, and SLC38A3, while others showed a positive association, including SLC7A9, SLC38A1, SLC38A2, and SLC38A7 (P ≤ 0.003). The associations between SLC7A8 and glutamine metabolic enzymes and transporters were primarily observed within luminal A tumours and to lesser extent in luminal B, HER2+ and TN subtypes. High SLC7A8 mRNA expression was associated with tumours which showed wild-type TP53 expression (P < 0.001, Table 3).
Table 3.
Correlation of SLC7A8 expression with the expression of other related genes in the METABRIC data
| SLC7A8 mRNA expression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All cases (n = 1980) |
Luminal A (n = 368) |
Luminal B (n = 367) |
HER2+ (n = 110) | Triple negative (n = 150) |
||||||
| Correlation coefficient (P value) |
Adjusted P value | |||||||||
| Glutamine metabolism | ||||||||||
| GLS | − 0.10 (0.000009) | 0.0001 | 0.07 (0.05) | 0.40 | 0.05 (0.19) | 1.89 | 0.01 (0.85) | 3.84 | − 0.09 (0.08) | 1.52 |
| GLS2 | 0.25 (1.1 × 10–30) | < 0.0001 | 0.07 (0.04) | 0.36 | 0.01 (0.69) | 2.69 | 0.28 (0.000006) | 0.0001 | − 0.03 (0.53) | 1.06 |
| ALDH4A1 | 0.22 (3.1 × 10–23) | < 0.0001 | 0.11 (0.004) | 0.05 | 0.07 (0.09) | 1.08 | 0.13 (0.03) | 0.75 | 0.37 (3.1 × 10–12) | < 0.0001 |
| PRODH | 0.10 (0.000003) | 0.0001 | 0.16 (0.00001) | 0.0002 | 0.12 (0.005) | 0.09 | − 0.03 (0.56) | 4.96 | 0.13 (0.01) | 0.19 |
| PYCR1 | − 0.05 (0.01) | 0.08 | 0.09 (0.01) | 0.11 | 0.10 (0.02) | 0.32 | − 0.06 (0.33) | 4.90 | − 0.11 (0.05) | 0.55 |
| ALDH18A1 | − 0.004 (0.86) | 1.72 | 0.11 (0.003) | 0.04 | 0.11 (0.01) | 0.34 | 0.09 (0.16) | 3.12 | − 0.15 (0.004) | 0.15 |
| GLUL | 0.21 (3.8 × 10–21) | < 0.0001 | − 0.11 (0.002) | 0.03 | − 0.02 (0.58) | 3.25 | 0.01 (0.83) | 4.25 | 0.38 (3 × 10–13) | < 0.0001 |
| GLUD1 | 0.42 (1.7 × 10–84) | < 0.0001 | 0.26 (2.9 × 10–13) | < 0.0001 | 0.15 (0.001) | 0.02 | 0.27 (0.00001) | 0.0002 | 0.11 (0.05) | 0.70 |
| Glutamine/glutamate transporters | ||||||||||
| SLC1A5 | − 0.07 (0.001) | 0.009 | 0.06 (0.09) | 0.54 | 0.007 (0.88) | 1.76 | − 0.03 (0.62) | 4.34 | − 0.20 (0.0002) | 0.003 |
| SLC3A2 | − 0.08 (0.0003) | 0.003 | 0.01 (0.61) | 1.83 | 0.08 (0.06) | 1.17 | − 0.21 (0.001) | 0.01 | − 0.05 (0.35) | 2.04 |
| SLC6A19 | − 0.007 (0.74) | 2.96 | − 0.002 (0.95) | 1.90 | − 0.04 (0.37) | 3.22 | 0.03 (0.62) | 4.98 | 0.03 (0.51) | 1.59 |
| SLC7A5 | − 0.47 (1.3 × 10–112) | < 0.0001 | − 0.155 (0.00003) | 0.0005 | − 0.11 (0.02) | 0.60 | − 0.18 (0.004) | 0.48 | − 0.45 (3.3 × 10–18) | < 0.0001 |
| SLC7A6 | − 0.12 (2.6 × 10–8) | < 0.0001 | 0.07 (0.05) | 0.35 | 0.07 (0.09) | 1.54 | − 0.001 (0.98) | 1.98 | 0.10 (0.07) | 0.72 |
| SLC7A7 | − 0.25 (5.9 × 10–31) | < 0.0001 | − 0.21 (2.3 × 10–8) | < 0.0001 | − 0.14 (0.001) | 0.02 | − 0.33 (7.8 × 10–8) | < 0.0001 | 0.20 (0.0002) | 0.005 |
| SLC7A9 | 0.21 (1.2 × 10–20) | < 0.0001 | 1.33 (0.0003) | 0.005 | 0.15 (0.001) | 0.02 | 0.35 (1.7 × 10–8) | < 0.0001 | − 0.06 (0.25) | 1.92 |
| SLC38A1 | 0.25 (1.3 × 10–29) | < 0.0001 | 0.27 (8.4 × 10–14) | < 0.0001 | 0.22 (7.5 × 10–7) | < 0.0001 | 0.12 (0.05) | 0.84 | − 0.21 (0.00007) | 0.001 |
| SLC38A2 | 0.08 (0.0003) | 0.003 | 0.12 (0.001) | 0.01 | 0.02 (0.65) | 2.76 | − 0.003 (0.96) | 2.94 | 0.13 (0.01) | 0.13 |
| SLC38A3 | − 0.14 (3 × 10–11) | < 0.0001 | 0.05 (0.13) | 0.65 | 0.09 (0.04) | 0.84 | 0.19 (0.002) | 0.06 | − 0.19 (0.0003) | 0.06 |
| SLC38A5 | 0.02 (0.28) | 1.68 | 0.10 (0.006) | 0.07 | 0.03 (0.46) | 3.00 | 0.11 (0.06) | 2.08 | 0.13 (0.01) | 0.60 |
| SLC38A7 | 0.08 (0.0001) | 0.001 | 0.22 (5.9 × 10–10) | < 0.0001 | 0.14 (0.002) | 0.03 | 0.31 (7.1 × 10–7) | < 0.0001 | 0.26 (9.6 × 10–7) | < 0.0001 |
| SLC38A8 | − 0.05 (0.02) | 0.14 | − 0.07 (0.03) | 0.30 | − 0.06 (0.14) | 1.90 | − 0.07 (0.26) | 3.63 | 0.07 (0.19) | 1.75 |
| SLC7A11 | − 0.007 (0.75) | 2.25 | − 0.04 (0.22) | 0.88 | 0.05 (0.21) | 2.96 | 0.04 (0.44) | 5.04 | 0.05 (0.32) | 1.75 |
Bold indicates the significant values
SLC7A8 protein was significantly expressed with high GLS and GLS2 enzymes (P < 0.001 and P = 0.006, Table 4), respectively. High PRODH, ALDH18A1, and ALDH4A1 were expressed in breast tumours with high SLC7A8 expression (all P ≤ 0.004, Table 4). High SLC7A8 expression was associated with low levels of SLC7A5 (P < 0.03, Table 4), whereas paradoxical associations were observed with the other transporters, SLC38A2 and SLC7A11 (P ≤ 0.02, Table 4).
Table 4.
Association between SLC7A8 protein expression and other biomarkers
| SlC7A8 protein | |||
|---|---|---|---|
| Low, n (%) | High, n (%) | χ2 (P value) | |
| PRODH | |||
| Negative | 255 (89.8) | 29 (10.2) | 10.93 (0.001) |
| Positive | 46 (74.2) | 16 (25.8) | |
| GLS | |||
| Negative | 226 (92.6) | 18 (7.4) | 18.45 (0.00001) |
| Positive | 118 (77.6) | 34 (22.4) | |
| GLS2 | |||
| Negative | 171 (89.5) | 20 (10.5) | 7.41 (0.006) |
| Positive | 155 (79.5) | 40 (20.5) | |
| ALDH18A1 | |||
| Negative | 190 (91.8) | 17 (8.2) | 8.17 (0.004) |
| Positive | 157 (82.2) | 34 (17.8) | |
| ALDH4A1 | |||
| Negative | 185 (92.0) | 16 (8.0) | 11.19 (0.001) |
| Positive | 152 (80.4) | 37 (19.6) | |
| Positive | |||
| SLC7A5 | |||
| Negative | 1040 (88.1) | 140 (11.9) | 4.66 (0.03) |
| Positive | 253 (92.7) | 20 (7.3) | |
| SLC38A2 | |||
| Negative | 992 (89.5) | 116 (10.5) | 10.21 (0.001) |
| Positive | 89 (79.5) | 23 (20.5) | |
| SLC7A11 | |||
| Negative | 465 (92.1) | 40 (7.9) | 4.84 (0.02) |
| Positive | 580 (88.1) | 78 (11.9) | |
SLC7A8 expression and patient outcome
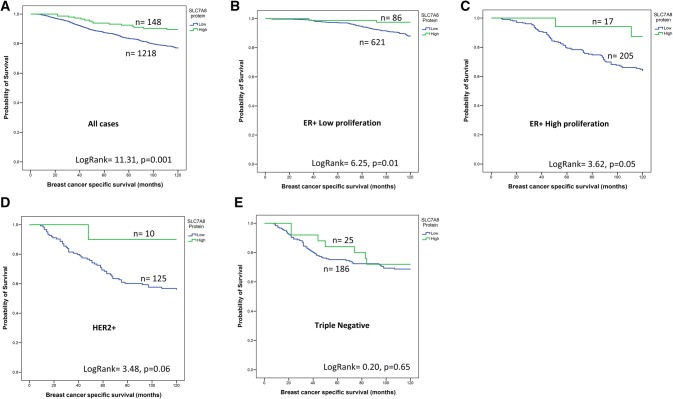
High expression of SLC7A8 mRNA and protein was associated with longer BCSS (P ≤ 0.001, Figs. 3a and 4a). While SLC7A8 mRNA expression was not predictive for BCSS in any specific molecular class (Fig. 3b–e), high expression of SLC7A8 protein was predictive of good survival in only the ER + low proliferation tumours (P = 0.01, Fig. 4b). There was no association between SLC7A8 protein and outcome in ER+ high proliferation, HER2+ or TN subtypes (Fig. 4c–e). Multivariate Cox regression analysis showed that SLC7A8 mRNA and SLC7A8 protein were predictors of longer BCSS independent of tumour size, grade, and lymph node stage (P = 0.01 and P = 0.03, Table 5) respectively.
Fig. 3.
SLC7A8 mRNA and breast cancer patient outcome. aSLC7A8 vs BCSS in all cases, bSLC7A8 vs BCSS in luminal A tumours, cSLC7A8 vs BCSS in Luminal B tumours, dSLC7A8 vs BCSS in HER2+ tumours, eSLC7A8 vs BCSS in Triple Negative tumours
Fig. 4.
SLC7A8 protein and breast cancer patient outcome. a SLC7A8 vs BCSS in all cases, b SLC7A8 vs BCSS of ER+—low proliferation tumours, c SLC7A8 vs BCSS of ER+—high Proliferation tumours, d SLC7A8 vs BCSS of HER2+ tumours, e SLC7A8 vs BCSS of Triple Negative tumours
Table 5.
SLC7A8 mRNA/ protein expression and patient outcome in the all breast cancer cases
| Parameter | SLC7A8 mRNA | SLC7A8 protein | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| SLC7A8 | 0.70 (0.52–0.94) | 0.01 | 0.57 (0.33–0.96) | 0.03 |
| Lymph node stage | 2.00 (1.56–2.55) | 2.4 × 10–8 | 1.89 (1.62–2.22) | 8.7 × 10–16 |
| Size | 1.48 (0.99–2.20) | 0.05 | 1.39 (1.08–1.78) | 0.009 |
| Grade | 1.41 (1.08–1.82) | 0.01 | 2.49 (1.97–3.16) | 2.8 × 10–14 |
Bold indicates the significant values
Likewise, high SLC7A8 protein expression was associated with longer distant metastases-free survival (DMFS) (P < 0.001, Supplementary Fig. 3A) within the ER+ low proliferation class (P = 0.003, Supplementary Fig. 3B) but not with other subtypes (Supplementary Fig. 3C, 2E). The relationship between high SLC7A8 mRNA expression and good patient outcome was verified using Breast Cancer Gene-Expression Miner (Supplementary Fig. 4A, 4B).
Discussion
BC represents a group of heterogeneous diseases that vary at the histopathological and molecular levels. These subtypes differ in their biology, clinical outcome, and response to therapy [29]. In addition, different BC subtypes showed disparity in their metabolic profiles and nutritional requirements. ER+/luminal tumours are the predominant BC subtype [30, 31] and characterised by having better prognosis and lower mortality rates as well as being targets for endocrine therapy [32]. In clinical practice, however, recognising patients who are likely to exhibit relapse or distant metastasis is challenging. Therefore, understanding the biology of BC is crucial in the pursuit of identifying targets for treatment and/or prognosis of BC patients particularly those with luminal tumours.
Altered metabolic pathways in human cancers are imperative to support cell proliferation and survival. Amino acid metabolism can vary substantially among BC subtypes, where TNBC display increased activity of amino acid consumption and metabolism compared with ER+ tumours [33, 34], suggesting that the latter subtype may allow for expression of lower levels of amino acid metabolic markers or express solute carriers that have lower affinity for their substrates. This study has revealed, for the first time, that SLC7A8 is a key amino acid transporter in the most predominant low proliferative ER+ tumours.
Unlike SLC7A5, SLC7A8 lacks studies that illustrate its prognostic role in human cancer. Data from Oncomine revealed a significant upregulation of SLC7A8 in several cancers, including breast, colorectal, head and neck, leukaemia, lymphoma, and melanoma [35]. However, this only has been validated at the mRNA level in a subset of breast tumours [15] and melanoma cell lines, which showed however more than five times increase in SLC7A5 expression compared to SLC7A8 [36]. Herein, we used large BC cohorts to reveal the significant associations between the high SLC7A8 expression, at mRNA and protein levels, and good prognostic clinicopathological parameters, including small tumour size, low tumour grade, and good NPI.
With respect to BC subtypes, the lowest levels of SLC7A8 mRNA were observed in the ER- and TNBC tumours, while SLC7A8 was higher in the ER+ subtypes which was more prominent in the luminal A tumours. These results were compatible with other studies which showed that SLC7A8 mRNA was expressed in the ER+, MCF7, cell line but not in the ER−, MDA-MB-231 cells [37]. Furthermore, we have shown that SLC7A8 was associated with better patient outcome and longer DMFS in the ER+ low proliferative tumours only and not in the other subtypes. Thakkar et al. also found that upregulation of SLC7A8 alongside GATA3 and MLPH significantly associated with longer relapse free survival in ER+ lymph node positive breast tumours [15]. These results may suggest that ER+ low proliferative tumours settle for the lower affinity transporter, SLC7A8, to satisfy their nutritional needs as they exhibit lower metabolic activity compared to the aggressive forms of BC.
The relationship between hormone receptor positivity and SLC7A8 indicate that hormone receptors have a possible role in stimulating SLC7A8 expression. It has been shown that 17β estradiol, in ER+ BC cells, regulates l-leucine uptake through SLC7A5 and SLC7A8 while no effect was observed in the ER- BC cells [4]. It has been further shown that SLC7A8 has oestrogen-dependent expression, in ER+ BC cells, and the existence of inhibitors of oestrogen signalling pathway (ICI182780 and tamoxifen) eliminates the oestrogen-induced upregulation of SLC7A8 [15]. Another study also revealed that progesterone significantly upregulates SLC7A8 mRNA and SLC7A8 protein expressions, in uterine leiomyoma tissues, and knockdown of SLC7A8 markedly increased leiomyoma cell proliferation [16].
P53 is a well-known tumour suppressor gene that responds to various stress signals through modulating other cellular processes, including cell cycle arrest and apoptosis. P53 also has a role in mediating other cellular mechanisms such as regulating metabolic pathways, including glutamine metabolism by inducing GLS2 expression [38]. Interestingly, this study showed a positive association between wild-type P53 and SLC7A8. This suggests that SLC7A8 may contribute to P53-dependent tumour suppression, which resulted in the presence of favourable prognosis and patient outcome in tumours expressing high SLC7A8. In addition, SLC7A8 was upregulated alongside two tumour suppressor genes, CEACAM1 and BMP2, in human colon cancer cells after exposure to anti-tumour agent [39].
We have previously showed that SLC7A5 is highly expressed in the aggressive BC subtypes and it was predictive of poor prognosis and poor patient outcome [14] while analysing SLC7A8 in the same cohort has resulted in opposite findings. Furthermore, this study showed that SLC7A8 at both mRNA and protein levels were mutually exclusive with SLC7A5 expression. It is noteworthy that although SLC7A5 and SLC7A8 have similar substrate selectivity and function, the latter has narrower tissue expression pattern and exhibits lower affinity to its substrates [6]. There is also evidence which suggests that the role of SLC7A8 is limited to equilibration of amino acids distribution across the cell membrane while SLC7A5 mediates the actual net of amino acid flux [40], which is required for further mTORC1 activation and cellular proliferation.
Furthermore, it seems that both solute carriers have contrasting effect in tumourigenesis and they could be a subject of different regulatory mechanisms, as SLC7A5 expression is induced by c-MYC while hormone receptors appear to control SLC7A8 expression [4, 14, 16].
This study further investigated the association of SLC7A8 expression with other solute carriers which involved in amino acid transport. While the majority of these transporters were negatively associated with SLC7A8, others were positively correlated, including SLC38A7 which showed consistent correlation in all BC subtypes. Positive associations with several glutamine metabolic enzymes were also detected, among these, GLS2 and glutamate dehydrogenase (GLUD1) which are associated with tumours of good prognosis and favourable patient outcome [21, 41]. Some variability in the expression of investigated markers across molecular subtypes was observed. For example, luminal A tumours were the main class which showed association between SLC7A8 and markers required for glutamine transport and metabolism. This could be attributed to the increased SLC7A8 expression and function in this particular BC subtype.
It is noteworthy that different transcriptional pathways such as c-MYC oncogenic transcription, hormone receptors and nutrient starvation responses regulate the expression of amino acid transporters in human cancers [27, 35]. This also applies to SLC7A5 and SLC7A8, as both belong to the system L transport family but their expression is controlled by different pathways. Furthermore, both solute carriers mediate the uptake of large neutral amino acids, with the latter showing decreased transporting capacity. However, SLC7A8 can also accept smaller neutral amino acids, such as glycine, alanine, serine, cysteine, and glutamine [42], which are known substrates of the key system A amino acid transporter, SLC1A5 that is highly expressed in TNBC [6, 43]. These statements indicate that SLC7A8 could be a key transporter in luminal A tumours through determining the actual net flux of not only the large but also the small neutral amino acids.
Conclusion
This study has revealed that the solute carrier SLC7A8 is an independent good prognostic marker in BC. Overexpression of SLC7A8 appears to have tumour suppressive characteristics especially in the low proliferative ER+ subtype, thus it could act as a potential prognostic factor. Functional assessment is necessary to reveal the specific role played by this amino acid transporter in the low proliferative ER+ tumours.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples.
Data availability
The dataset analysed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ and the North West—Greater Manchester Central Research Ethics Committee under the title ‘Nottingham Health Science Biobank (NHSB)’ reference number 15/NW/0685. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Release of data were also pseudoanonymised as per the UK Human Tissue Act regulations. This article does not contain any studies with animals performed by any of the authors.
Informed consent
All tissue samples from Nottingham used in this study were pseudoanonymised and collected prior to 1st September 2006; therefore under the UK Human Tissue Act informed patient consent was not needed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bond P. Regulation of mTORC1 by growth factors, energy status, amino acids and mechanical stimuli at a glance. J Int Soc Sports Nutr. 2016;13:8. doi: 10.1186/s12970-016-0118-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24(7):400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shennan DB, Thomson J, Gow IF, Travers MT, Barber MC. L-leucine transport in human breast cancer cells (MCF-7 and MDA-MB-231): kinetics, regulation by estrogen and molecular identity of the transporter. Biochem Biophys Acta. 2004;1664(2):206–216. doi: 10.1016/j.bbamem.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Hellsten SV, Tripathi R, Ceder MM, Fredriksson R. Nutritional stress induced by amino acid starvation results in changes for Slc38 transporters in immortalized hypothalamic neuronal cells and primary cortex cells. Front Mol Biosci. 2018;5:45. doi: 10.3389/fmolb.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhutia YD. Ganapathy V (2016) Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochem Biophys Acta. 1863;10:2531–2539. doi: 10.1016/j.bbamcr.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) J Biol Chem. 1998;273(37):23629–23632. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 8.Pineda M, Fernandez E, Torrents D, Estevez R, Lopez C, Camps M, Lloberas J, Zorzano A, Palacin M. Identification of a membrane protein, LAT-2, that Co-expresses with 4F2 heavy chain, an L-type amino acid transport activity with broad specificity for small and large zwitterionic amino acids. J Biol Chem. 1999;274(28):19738–19744. doi: 10.1074/jbc.274.28.19738. [DOI] [PubMed] [Google Scholar]
- 9.Rossier G, Meier C, Bauch C, Summa V, Sordat B, Verrey F, Kuhn LC. LAT2, a new basolateral 4F2hc/CD98-associated amino acid transporter of kidney and intestine. J Biol Chem. 1999;274(49):34948–34954. doi: 10.1074/jbc.274.49.34948. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi H, Ishii Y, Takayama T. Expression of L-type amino acid transporter 1 (LAT1) in esophageal carcinoma. J Surg Oncol. 2005;90(4):233–238. doi: 10.1002/jso.20257. [DOI] [PubMed] [Google Scholar]
- 11.Yoon JH, Kim IJ, Kim H, Kim HJ, Jeong MJ, Ahn SG, Kim SA, Lee CH, Choi BK, Kim JK, Jung KY, Lee S, Kanai Y, Endou H, Kim DK. Amino acid transport system L is differently expressed in human normal oral keratinocytes and human oral cancer cells. Cancer Lett. 2005;222(2):237–245. doi: 10.1016/j.canlet.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi K, Matsuo H, Kanai Y, Endou H, Hiroi S, Tominaga S, Mukai M, Ikeda E, Ozeki Y, Aida S, Kawai T. LAT1 expression in normal lung and in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Virchows Arch. 2006;448(2):142–150. doi: 10.1007/s00428-005-0063-7. [DOI] [PubMed] [Google Scholar]
- 13.Furuya M, Horiguchi J, Nakajima H, Kanai Y, Oyama T. Correlation of L-type amino acid transporter 1 and CD98 expression with triple negative breast cancer prognosis. Cancer Sci. 2012;103(2):382–389. doi: 10.1111/j.1349-7006.2011.02151.x. [DOI] [PubMed] [Google Scholar]
- 14.El Ansari R, Craze ML, Miligy I, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018;20(1):21. doi: 10.1186/s13058-018-0946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakkar A, Raj H, Ravishankar MB, Balakrishnan A, Padigaru M. High expression of three-gene signature improves prediction of relapse-free survival in estrogen receptor-positive and node-positive breast tumors. Biomark insights. 2015;10:103–112. doi: 10.4137/bmi.S30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X, Yin P, Reierstad S, Ishikawa H, Lin Z, Pavone ME, Zhao H, Marsh EE, Bulun SE. Progesterone and mifepristone regulate L-type amino acid transporter 2 and 4F2 heavy chain expression in uterine leiomyoma cells. J Clin Endocrinol Metab. 2009;94(11):4533–4539. doi: 10.1210/jc.2009-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craze ML, Cheung H, Jewa N, Coimbra ND, Soria D, El-Ansari R, Aleskandarany MA, Cheng KW, Diez-Rodriguez M, Nolan CC. MYC regulation of Glutamine-Proline regulatory axis is key in Luminal B breast cancer. Br J Cancer. 2017;118(2):258–265. doi: 10.1038/bjc.2017.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116(3):340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 20.McCarty KS, Jr, McCarty KS., Sr Histochemical approaches to steroid receptor analyses. Semin Diagn Pathol. 1984;1(4):297–308. [PubMed] [Google Scholar]
- 21.Craze ML, El-Ansari R, Aleskandarany MA, Cheng KW, Alfarsi L, Masisi B, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. Glutamate dehydrogenase (GLUD1) expression in breast cancer. Breast Cancer Res Treat. 2019;174(1):79–91. doi: 10.1007/s10549-018-5060-z. [DOI] [PubMed] [Google Scholar]
- 22.Green AR, Aleskandarany MA, Agarwal D, Elsheikh S, Nolan CC, Diez-Rodriguez M, Macmillan RD, Ball GR, Caldas C, Madhusudan S, Ellis IO, Rakha EA. MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br J Cancer. 2016;114(8):917–928. doi: 10.1038/bjc.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elston CW, Ellis IO (2002) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19; 403–410. Histopathology 41 (3a):151–152, discussion 152–153 [PubMed]
- 24.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S, Cardoso F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 25.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93(4):387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pochini L, Scalise M, Galluccio M, Indiveri C. Membrane transporters for the special amino acid glutamine: structure/function relationships and relevance to human health. Front Chem. 2014;2:61. doi: 10.3389/fchem.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Ansari R, McIntyre A, Craze ML, Ellis IO, Rakha EA, Green AR. Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology. 2018;72(2):183–190. doi: 10.1111/his.13334. [DOI] [PubMed] [Google Scholar]
- 28.Cha YJ, Kim ES, Koo JS. Amino acid transporters and glutamine metabolism in breast cancer. Int J Mol Sci. 2018;19(3):907. doi: 10.3390/ijms19030907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 30.Rakha EA, El-Sayed ME, Green AR, Paish EC, Powe DG, Gee J, Nicholson RI, Lee AH, Robertson JF, Ellis IO. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J Clin Oncol. 2007;25(30):4772–4778. doi: 10.1200/jco.2007.12.2747. [DOI] [PubMed] [Google Scholar]
- 31.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32(5):617–628. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ring BZ, Seitz RS, Beck R, Shasteen WJ, Tarr SM, Cheang MC, Yoder BJ, Budd GT, Nielsen TO, Hicks DG, Estopinal NC, Ross DT. Novel prognostic immunohistochemical biomarker panel for estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(19):3039–3047. doi: 10.1200/jco.2006.05.6564. [DOI] [PubMed] [Google Scholar]
- 33.Kanaan YM, Sampey BP, Beyene D, Esnakula AK, Naab TJ, Ricks-Santi LJ, Dasi S, Day A, Blackman KW, Frederick W, Copeland RL, Sr, Gabrielson E, Dewitty RL., Jr Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genomics Proteom. 2014;11(6):279–294. [PubMed] [Google Scholar]
- 34.Cao MD, Lamichhane S, Lundgren S, Bofin A, Fjosne H, Giskeodegard GF, Bathen TF. Metabolic characterization of triple negative breast cancer. BMC Cancer. 2014;14:941. doi: 10.1186/1471-2407-14-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res. 2015;5(4):1281–1294. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Beaumont KA, Otte NJ, Font J, Bailey CG, van Geldermalsen M, Sharp DM, Tiffen JC, Ryan RM, Jormakka M, Haass NK, Rasko JE, Holst J. Targeting glutamine transport to suppress melanoma cell growth. Int J Cancer. 2014;135(5):1060–1071. doi: 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- 37.Shennan DB, Thomson J, Barber MC, Travers MT. Functional and molecular characteristics of system L in human breast cancer cells. Biochem Biophys Acta. 2003;1611(1–2):81–90. doi: 10.1016/s0005-2736(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bermudez-Soto MJ, Larrosa M, Garcia-Cantalejo JM, Espin JC, Tomas-Barberan FA, Garcia-Conesa MT. Up-regulation of tumor suppressor carcinoembryonic antigen-related cell adhesion molecule 1 in human colon cancer Caco-2 cells following repetitive exposure to dietary levels of a polyphenol-rich chokeberry juice. J Nutr Biochem. 2007;18(4):259–271. doi: 10.1016/j.jnutbio.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Meier C, Ristic Z, Klauser S, Verrey F. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 2002;21(4):580–589. doi: 10.1093/emboj/21.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Zhang C, Lin M, Zhu W, Liang Y, Hong X, Zhao Y, Young KH, Hu W, Feng Z. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget. 2014;5(9):2635–2647. doi: 10.18632/oncotarget.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Amo EM, Urtti A, Yliperttula M. Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur J Pharm Sci. 2008;35(3):161–174. doi: 10.1016/j.ejps.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 43.van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O'Toole SA, Rasko JE, Holst J. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35(24):3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset analysed during the current study are available from the corresponding author on reasonable request.