Abstract
The family Coronavirtdae comprises a monogeneric group of 11 viruses which infect vertebrates. The main characteristics of the member viruses are: (i) Morphological: Enveloped pleomorphic particles typically 100 nm in diameter (range 60-220 nm), bearing about 20 nm long club-shaped surface projections, (ii) Structural: A single-stranded infectious molecule of genomic RNA of about (5-7) × 106 molecular weight. A phosphorylated nucleocapsid protein [mol.wt. (50-60) × 103] complexed with the genome as a helical ribonucleoprotein; a surface (peplomer) protein, associated with one or two glycosylated polypeptides [mol.wt. (90-180) × 103]; a transmembrane (matrix) protein, associated with one polypeptide which may be glycosylated to different degrees [mol.wt. (20-35) × 103]. (iii) Replicative: Production in infected cells of multiple 3′ coterminal sub genomic mRNAs extending for different lengths in the 5′ direction. Virions bud intracytoplasmically. (iv) Antigenic: 3 major antigens, each corresponding to one class of virion protein, (v) Biological: Predominantly restricted to infection of natural vertebrate hosts by horizontal transmission via the fecal/oral route. Responsible mainly for respiratory and gastrointestinal disorders.
Key Words: Coronavirus, Viral taxonomy
Since the second report of the Coronavirus Study Group in 1978 [1], considerable data, especially on the structure and replication of coronaviruses, have been published, and we feel a new report is justified. The Coronaviridae are a monogeneric family of pleomorphic, ether-labile, enveloped viruses. The virions have a diameter ranging from 60 to 220 nm and an average density in sucrose of 1.18 g/ml. They characteristically bear club-shaped surface projections about 20 nm in length from which the group derives its name (Latin corona, crown) [1]. The genomic RNA is an infectious single-stranded molecule which is capped and polyadenylated. The molecular weight is between 5 × 106 and 7 × 106, corresponding to about 15,000-20,000 nucleotides. There is no extensive sequence reiteration in the coronavirus genome. Coronavirions characteristically have three types of protein: a phosphorylated nucleocapsid protein [mol.wt. (50-60) × 103], complexed with the genome as a helical ribonucleoprotein (RNP); an N-glycosylated surface peplomer protein, associated with glycopolypeptides of (90-180) × 103 molecular weight, which is acylated and is responsible for virus attachment and cell-to-cell fusion (this protein may be removed by protease treatment); and a transmembrane matrix protein, associated with polypeptides of molecular weight (20-35) × 103 which have variable degrees of glycosylation. In the case of murine and bovine coronaviruses this polypeptide bears O-glycosidically linked oligosaccharides, and in the case of avian infectious bronchitis virus it bears N-glycosidically linked oligosaccharides.
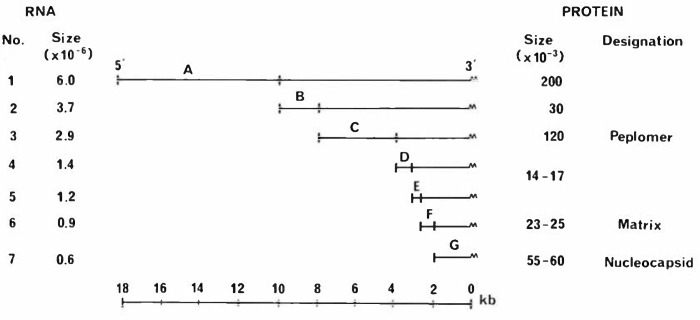
Most coronaviruses replicate in tissue culture within 12 h at 37°. Infection is often accompanied by cytopathic changes. There are conflicting reports as to whether a nuclear function is required for coronavirus replication. There are few data about the early events (adsorption, penetration, uncoating, etc.) involved in coronavirus replication. It is assumed that upon entering the cell the positive-stranded genome encodes protein(s) whose function is to replicate the genomic RNA and produce subgenomic mRNA. Recently, there have been reports of virus-specific RNA polymerases in coronavirus-infected cells, but the components of the enzyme have not been identified. Characteristic of coronavirus infection is the production of 3′ coterminal subgenomic RNAs which form a nested set extending in a 5′ direction. These RNAs are capped and polyadenylated. The replicative structures from which they are produced have not been characterized, but it has been demonstrated that the negative-stranded template from which murine hepatitis virus mRNAs are copied is of genome length. UV inactivation studies indicate that coronavirus mRNAs are not produced by the processing of a larger RNA, although extensive sequence homologies have been detected at the 5′ ends of all murine hepatitis virus-specific subgenomic RNAs. For murine hepatitis virus, the mRNA function of each of the subgenomic viral RNAs has been demonstrated in vitro, and the mRNAs encoding each of the virion proteins, or its precursors, have been identified (fig. 1). Comparing the size of each mRNA with its translation product suggests that the expressed information lies within the 5′ sequences of each RNA which are not found in the next smallest RNA. For murine hepatitis virus, glycosylation of the peplomer protein is initiated cotranslationally in the rough endoplasmic reticulum, whereas the transmembrane protein is glycosylated posttranslationally in the Golgi apparatus. The infectious bronchitis virus matrix protein is glycosylated on the nascent polypeptide. After synthesis, genomic RNA and virion proteins are assembled at the rough endoplasmic reticulum and virions bud into cisternae, acquiring their lipid membranes from the cell. The virions are subsequently transported to and accumulate in Golgi and smooth-walled vesicles. There is an absence of budding from the plasmalemma. The mechanism of virus release has not been characterized.
Fig. 1.
Structure and expression of the MHV genome. The size and structural relationships of the MHV intracellular mRNAs arc shown. No difference between the genome RNA and mRNA 1 has yet been described. Each mRNA encodes only one protein, and the translation products of mRNAs 7, 6 and 3 have been identified as the intracellular precursors to the virion nucleocapsid, matrix and peplomer proteins, respectively. As the size of the translation product for each mRNA corresponds approximately to the coding potential of the 5′ sequences which are absent from the next smallest mRNA, it seems likely that only these regions are translated into protein.
The relationships between some coronaviruses have been studied by molecular and immunological methods, but the data are fragmentary. Molecular hybridization indicates extensive sequence homology (about 70%) between murine hepatitis virus strains, in particular within the gene encoding the nucleocapsid protein. This conclusion is supported by oligonucleotide fingerprinting of genomic RNAs and chymotryptic peptide fingerprinting of nucleocapsid proteins. Oligonucleotide fingerprinting of the genomic RNA of a number of infectious bronchitis virus strains indicates greater sequence divergence both between and within serotypes of this species. Coronaviruses contain 3 major antigens which can be distinguished by antibodies against virion subcomponents. Each antigen corresponds to one of the three types of coronavirus protein. Immunological studies with monoclonal antibodies, antisera directed against subcomponents prepared from purified virions, and immune electron microscopy indicate that the antigenic sites responsible for the induction of neutralizing antibodies are associated with the surface peplomer polypeptide(s). Studies on the antigenic relationships of coronaviruses present a complex pattern. Relationships have been determined by a wide variety of tests, mainly using polyvalent sera from naturally infected or hyperimmunized animals. These immunological studies indicate that there are two antigenic groups of mammalian coronaviruses and two antigenic groups of avian coronaviruses. One recent porcine isolate does not appear to fall into either mammalian group. Many viruses remain to be classified.
The geographic distribution of many coronaviruses is known to extend over several continents and is probably worldwide. A seasonal incidence of infection occurs with some viruses, namely human coronavirus and transmissible gastroenteritis virus. Coronaviruses predominantly infect their natural vertebrate hosts. Biological vectors of coronaviruses have not been reported, and the natural hosts form the major reservoirs for further infection. In most cases, infection is by the fecal/oral route. Vertical intrauterine infection has been reported for infectious bronchitis virus and some murine hepatitis virus strains. Transmission from contaminated clothing and equipment is an important source of infection with infectious bronchitis virus and bovine coronavirus. Coronaviruses are associated with diseases of economic and clinical importance, predominantly respiratory and gastrointestinal disorders. Feline infectious peritonitis virus is responsible for peritonitis in cats, and hemagglutinating encephalomyelitis virus and some murine hepatitis virus strains are associated with encephalomyelitis. Diagnosis of coronavirus infection is initially clinical, and confirmation is most readily achieved by virus isolation and propagation in vitro and/or by a variety of immunological procedures [2].
At present the family Coronaviridae is recognized as 11 species, which are listed below. A number of recent isolates meet the morphological and, to some extent, the molecular criteria for inclusion in the group, but are as yet insufficiently characterized to be regarded as ‘possible’ family members. The question of speciation and whether the family should remain monogeneric will be considered by the Study Group in the near future. This report does not contain references to primary sources. Extensive bibliographies can be found in several recent reviews of coronavirus biology [3, 4, 5, 6]. The acronyms used throughout this report are defined in section 10.3.
1 Taxonomy
Intervirology 20: 181-189 (1983)
© 1983 S. Karger AG. Basel
0300-5526/83/0204-018182.75/0
Coronaviridae1
S.G. Siddell, R. Anderson, D. Cavanagh, K. Fujiwara, H.D. Klenk, M.R. Macnaughton, M. Pensaert, S.A. Stohlman, L. Sturman, B.A.M. van der Zeijst
Key Words. Coronavirus
Viral taxonomy
Summary. The family Coronaviridae comprises a monogeneric group of 11 viruses which infect vertebrates. The main characteristics of the member viruses are: (i) Morphological: Enveloped pleomorphic particles typically 100 nm in diameter (range 60-220 nm), bearing about 20 nm long club-shaped surface projections, (ii) Structural: A single-stranded infectious molecule of genomic RNA of about (5-7) × 106 molecular weight. A phosphorylated nucleocapsid protein [mol.wt. (50-60) × 103] complexed with the genome as a helical ribonucleoprotein; a surface (peplomer) protein, associated with one or two glycosylated polypeptides [mol.wt. (90-180) × 103]; a transmembrane (matrix) protein, associated with one polypeptide which may be glycosylated to different degrees [mol.wt. (20-35) × 103]. (iii) Replicative: Production in infected cells of multiple 3′ coterminal subgenomic mRNAs extending for different lengths in the 5′ direction. Virions bud intracytoplasmically. (iv) Antigenic: 3 major antigens, each corresponding to one class of virion protein. (v) Biological: Predominantly restricted to infection of natural vertebrate hosts by horizontal transmission via the fecal/oral route. Responsible mainly for respiratory and gastrointestinal disorders.
1 Third Report of the Coronavirus Study Group, Vertebrate Virus Subcommittee, International Committee on Taxonomy of Viruses (ICTV).
Address inquiries to: Dr. S. Siddell, Institute of Virology, University of Würzburg, Versbacher Strasse 7, D-8700 Würzburg (FRG)
Since the second report of the Coronavirus Study Group in 1978 [1], considerable data, especially on the structure and replication of coronaviruses, have been published, and we feel a new report is justified. The Coronaviridae are a monogeneric family of pleomorphic, ether-labile, enveloped viruses. The virions have a diameter ranging from 60 to 220 nm and an average density in sucrose of
Received: March 21, 1983
21.18 g/ml. They characteristically bear club-shaped surface projections about 20 nm in length from which the group derives its name (Latin corona, crown) [1]. The genomic RNA is an infectious single-stranded molecule which is capped and polyadenylated. The molecular weight is between 5 × 106 and 7 × 106, corresponding to about 15,000-20,000 nucleotides. There is no extensive sequence reiteration in the coronavirus genome. Coronavirions characteristically have three types of protein: a phosphorylated nucleocapsid protein [mol.wt. (50-60) × 103], complexed with the genome as a helical ribonucleoprotein (RNP); an N-glycosylated surface peplomer protein, associated with glycopolypeptides of (90-180) × 103 molecular weight, which is acylated and is responsible for virus attachment and cell-to-cell fusion (this protein may be removed by protease treatment); and a transmembrane matrix protein, associated with polypeptides of molecular weight (20-35) × 103 which have variable degrees of glycosylation. In the case of murine and bovine coronaviruses this polypeptide bears O-glycosidically linked oligosaccharides, and in the case of avian infectious bronchitis virus it bears N-glycosidically linked oligosaccharides.
Most coronaviruses replicate in tissue culture within 12 h at 37°. Infection is often accompanied by cytopathic changes. There are conflicting reports as to whether a nuclear function is required for coronavirus replication. There are few data about the early events (adsorption, penetration, uncoating, etc.) involved in coronavirus replication. It is assumed that upon entering the cell the positive-stranded genome encodes protein(s) whose function is to replicate the genomic RNA and produce subgenomic mRNA. Recently, there have been reports of virus-specific RNA polymerases in coronavirus-infected cells, but the components of the enzyme have not been identified. Characteristic of coronavirus infection is the production of 3′ coterminal subgenomic RNAs which form a nested set extending in a 5′ direction. These RNAs are capped and polyadenylated. The replicative structures from which they are produced have not been characterized, but it has been demonstrated that the negative-stranded template from which murine hepatitis virus mRNAs are copied is of genome length. UV inactivation studies indicate that coronavirus mRNAs are not produced by the processing of a larger RNA, although extensive sequence homologies have been detected at the 5′ ends of all murine hepatitis virus-specific subgenomic RNAs. For murine hepatitis virus, the mRNA function of each of the subgenomic viral RNAs has been demonstrated in vitro, and the mRNAs encoding each of the virion proteins, or its precursors, have been identified (fig. 1). Comparing the size of each mRNA with its translation product suggests that the expressed information lies within the 5′ sequences of each RNA which are not found in the next smallest RNA. For murine hepatitis virus, glycosylation of the peplomer protein is initiated cotranslationally in the rough endoplasmic reticulum, whereas the transmembrane protein is glycosylated posttranslationally in the Golgi apparatus. The infectious bronchitis virus matrix protein is glycosylated on the nascent polypeptide. After synthesis, genomic RNA and virion proteins are assembled at the rough endoplasmic reticulum and virions bud into cisternae, acquiring their lipid membranes from the cell. The virions are subsequently transported to and accumulate in Golgi and smooth-walled vesicles. There is an absence of budding from the plasmalemma. The mechanism of virus release has not been characterized.
Fig. 1. Structure and expression of the MHV genome. The size and structural relationships of the MHV intracellular mRNAs arc shown. No difference between the genome RNA and mRNA 1 has yet been described. Each mRNA encodes only one protein, and the translation products of mRNAs 7, 6 and 3 have been identified as the intracellular precursors to the virion nucleocapsid, matrix and peplomer proteins, respectively. As the size of the translation product for each mRNA corresponds approximately to the coding potential of the 5′ sequences which are absent from the next smallest mRNA, it seems likely that only these regions are translated into protein.
The relationships between some coronaviruses have been studied by molecular and immunological methods, but the data are fragmentary. Molecular hybridization indicates extensive sequence homology (about 70%) between murine hepatitis virus strains, in particular within the gene encoding the nucleocapsid protein. This conclusion is supported by oligonucleotide fingerprinting of genomic RNAs and chymotryptic peptide fingerprinting of nucleocapsid proteins. Oligonucleotide fingerprinting of the genomic RNA of a number of infectious bronchitis virus strains indicates greater sequence divergence both between and within serotypes of this species. Coronaviruses contain 3 major antigens which can be distinguished by antibodies against virion subcomponents. Each antigen corresponds to one of the three types of coronavirus protein. Immunological studies with monoclonal antibodies, antisera directed against subcomponents prepared from purified virions, and immune electron microscopy indicate that the antigenic sites responsible for the induction of neutralizing antibodies are associated with the surface peplomer polypeptide(s). Studies on the antigenic relationships of coronaviruses present a complex pattern. Relationships have been determined by a wide variety of tests, mainly using polyvalent sera from naturally infected or hyperimmunized animals. These immunological studies indicate that there are two antigenic groups of mammalian coronaviruses and two antigenic groups of avian coronaviruses. One recent porcine isolate does not appear to fall into either mammalian group. Many viruses remain to be classified.
4The geographic distribution of many coronaviruses is known to extend over several continents and is probably worldwide. A seasonal incidence of infection occurs with some viruses, namely human coronavirus and transmissible gastroenteritis virus. Coronaviruses predominantly infect their natural vertebrate hosts. Biological vectors of coronaviruses have not been reported, and the natural hosts form the major reservoirs for further infection. In most cases, infection is by the fecal/oral route. Vertical intrauterine infection has been reported for infectious bronchitis virus and some murine hepatitis virus strains. Transmission from contaminated clothing and equipment is an important source of infection with infectious bronchitis virus and bovine coronavirus. Coronaviruses are associated with diseases of economic and clinical importance, predominantly respiratory and gastrointestinal disorders. Feline infectious peritonitis virus is responsible for peritonitis in cats, and hemagglutinating encephalomyelitis virus and some murine hepatitis virus strains are associated with encephalomyelitis. Diagnosis of coronavirus infection is initially clinical, and confirmation is most readily achieved by virus isolation and propagation in vitro and/or by a variety of immunological procedures [2].
At present the family Coronaviridae is recognized as 11 species, which are listed below. A number of recent isolates meet the morphological and, to some extent, the molecular criteria for inclusion in the group, but are as yet insufficiently characterized to be regarded as ‘possible’ family members. The question of speciation and whether the family should remain monogeneric will be considered by the Study Group in the near future. This report does not contain references to primary sources. Extensive bibliographies can be found in several recent reviews of coronavirus biology [3, 4, 5, 6]. The acronyms used throughout this report are defined in section 10.3.
51 Taxonomy
1.1 Family: Coronaviridae
1.1.1 Genus: Coronavirus.
1.1.2 Type species: Avian infectious bronchitis virus.
1.2 Taxonomic status: Family with one genus.
2 The virion
2.1 Chemical composition
2.1.1 Nucleic acid
2.1.1.1 RNA
2.1.1.2 Single-stranded
2.1.1.3 Linear
2.1.1.4 Unsegmented
2.1.1.5 Sedimentation coefficient: 50-70S.
2.1.1.6 Molecular weight:
IBV: (5.8-6.9) × 106
MHV: (5.4-6.0) × 106
HCV: (5.8-6.5) × 106
TGEV: 6.8 × 106
BCV: 6.8 × 106
2.1.1.10 Homology studies: High sequence homology among genomes of MHV (molecular hybridization, oligonucleotide fingerprinting). Greater divergence among IBV genomes (oligonucleotide fingerprinting).
2.1.1.11 Infectivity: Demonstrated for MHV, IBV, TGEV.
2.1.1.12 Other features: IBV, MHV, TGEV, BCV and HCV genomic RNA is polyadenylated. MHV genomic RNA is capped.
2.1.2 Proteins
2.1.2.2 Number of polypeptides:
6Nucleocapsid protein [mol.wt. (50-60) × 103]: Location demonstrated for IBV, MHV, HCV, TGEV, HEV, and BCV. Comparable protein shown for CCV. Phosphorylation demonstrated for IBV, MHV and BCV.
Surface (peplomer) protein: One or two glycosylated polypeptides [mol.wt. (90-180) × 103]. Location shown for IBV, MHV, HCV, TGEV, BCV, and HEV. Comparable protein reported for CCV. Primary sequence relationship shown for IBV and MHV polypeptides. Acylation shown for MHV and BCV polypeptide.
Transmembrane (matrix) protein: One polypeptide, which may be glycosylated to different degrees [(20-35) × 103 mol.wt.], reported for IBV, MHV, HCV, TGEV, CCV, BCV, and HEV. Location shown for IBV, MHV, HCV, and HEV. Glycosylated and nonglycosylated forms incorporated in MHV and IBV.
Others: Glycosylated and nonglycosylated envelope proteins reported sporadically for many coronaviruses. Relationships to other coronavirion polypeptides not established.
2.1.2.5 Enzymes: Protein kinase associated with MHV.
2.1.2.6 Other functional proteins: Hemagglutinin found in IBV, HCV-OC43, HEV, BCV, and MHV. Cell fusion activity associated with MHV peplomer protein.
2.1.3 Lipids: TGEV contains phospholipids and glycolipids resembling those of the host cell.
2.1.3.2 Other features: Fatty acids are covalently attached to MHV and BCV peplomer proteins.
2.1.4 Carbohydrates: The peplomer protein of MHV and IBV is N-glycosidically linked to complex and high-mannose oligosaccharides. The matrix protein of MHV is O-glycosidically linked to oligosaccharides. The 7matrix protein of IBV is N-glycosidically linked to complex and high mannose oligosaccharides.
2.2 Physicochemical properties
2.2.1 Density: Average density of 1.18 g/ml in sucrose, 1.23-1.24 g/ml in CsCl2; HEV has density of 1.17 g/ml in potassium tartrate.
2.2.2 Sedimentation coefficient: HCV, 380-400S; IBV, 330S; TGEV, 495S; FIPV, 400S.
2.2.4 Stability of infectivity
2.2.4.1 pH: Optimal stability of IBV, pH 6.0-6.5; TGEV, pH 6.5; MHV, pH 6.0-7.0. Conflicting data for more extreme conditions.
2.2.4.2 Heat: Rapid inactivation at 56°, moderate inactivation at 37°. Moderately stable at 4° in optimal suspending medium.
2.2.4.3 Lipid solvents: Chloroform- and ether-labile.
2.2.4.4 Radiation: IBV, MHV, TGEV, and HEV inactivated by UV radiation (30,000 erg/mm2).
2.2.4.5 Other agents: Agents capable of inactivation include SDS, sodium deoxycholate, formalin, ethanol (70%), KMNO4, β-propiolactone, hydroxylamine, and chlorohexidine.
2.3 Structure
2.3.1 Nucleocapsid: Genome and nucleocapsid protein associated as helical RNP.
2.3.2 Envelope: Lipid-containing envelope (host cell-derived) containing integral and peripheral viral proteins.
2.3.3 Cores: Electron-dense inner shell visible in thin sections. RNP core density: MHV, 1.27-1.28 g/ml in sucrose; HCV, 1.31 g/ml in CsCl2, sedimentation coefficient 180S.
2.4 Morphology
2.4.1 Overall shape: Pleomorphic, although roughly spherical.
2.4.2 Dimensions: 60-220 nm.
2.4.3 Surface projections: Usually characteristic club-shaped projections, about 20 nm long, widely spaced. IBV, HEV, and MHV can also have thin cone-shaped projections. BCV has two layers of projections.
2.4.4 Special features in thin sections: Inner and outer shells, sometimes separated by electron-lucent space.
2.4.5 Other features: Fragile attachment of projections to surface of virion, may be removed by protease treatment. Purified peplomer projections aggregate in aqueous environment. Purified transmembrane protein interacts specifically with genome RNA at 37°. Inner tongue-shaped membrane in IBV visible by negative staining.
3 Replication
83.1 Site of accumulation of viral proteins: Cytoplasm.
3.2 Nonstructural proteins: Not defined. Virus-specific nonvirion polypeptides for MHV.
3.3 Mode of nucleic acid replication
3.3.1 General account: Genomic RNA assumed to encode enzymes responsible for amplification of genome and production of subgenomic RNA. RNA-dependent RNA polymerase demonstrated in MHV- and TGEV-infected cells, but polypeptide components not identified. Subgenomic RNAs are capped and polyadenylated and form a 3′ coterminal nested set. Replicative structures not yet characterized. For MHV, negative-stranded 9template of genome length. UV inactivation studies on subgenomic RNA synthesis argue against extensive processing. Number of major subgenomic RNAs: IBV, 5; MHV, 6.
3.3.2 Effect of inhibitors: RNA synthesis is insensitive to actinomycin D, 5-iododeoxyuridine, 5-bromodeoxyuridine, 5-fiuorodeoxyuridine, cytosine arabinoside and aminopterin. Conflicting reports for IBV and MHV regarding α-amanitin. RNA synthesis is sensitive to 6-azauracil and virazole.
3.4 Site and mechanism of maturation: Matures in cytoplasm by budding through endoplasmic reticulum. No budding at plasmalemma.
3.5 Other features: For MHV, messenger function of the subgenomic RNAs has been demonstrated. RNAs 7, 6 and 3 encode the virion nucleocapsid, matrix and peplomer protein(s), or their precursors, respectively. Involvement of a nucleus or nuclear function in coronavirus replication is equivocal. MHV reported to replicate in enucleated cells. IBV (Beaudette) reported not to replicate in enucleated or UV-irradiated BHK21 cells.
4 Cooperative interactions: No data.
4.3 Phenotypic mixing: Reported between MHV and Friend leukemia virus.
5 Host range
5.1 Natural: Generally restricted to natural vertebrate host.
5.2 Experimental
5.2.1 In vivo: Generally specific for species of origin. Also,
HCV suckling mice,
(some strains) suckling hamsters
IBV suckling mice,
(some strains) suckling hamsters,
suckling rats,
newborn rabbits
MHV rats, hamsters, monkeys
TGEV dogs, foxes, cats
HEV suckling mice
BCV suckling mice, rats, hamsters
CCV pigs
10FIPV newborn mice, rats, hamsters, piglets
5.2.2 In vitro: Generally specific to organ cultures, primary and secondary cells or cell lines derived from species of origin. Also,
IBV first-passage monkey kidney cells: VERO, BHK, CHO (semipermissive)
CCV CFK, HRT 18
HCV first-passage monkey kidney cells: BSC1, AGMK, VERO
MHV L6, HTC, WI38, RN-2-2
BCV first-passage monkey kidney cells: HRT18, VERO, PK15, PK3, MA321
TGEV first-passage
canine kidney cells
6 Pathogenicity
6.1 Association with diseases: IBV-respiratory disease, nephritis and gonadal 11damage. MHV-acute hepatitis, encephalomyelitis and infantile diarrhea. FIPV-peritonitis and granulomatous inflammations in many organs. HCV-respiratory diseases. HEV-vomiting and wasting, encephalomyelitis. RCV-pneumonitis, rhinitis. SDAV-sialoadenitis, dacryoadenitis. TCV-enteritis. BCV, CCV, TGEV and PEDV associated with diarrhea.
6.2 Tissue tropism: IBV-respiratory tract, gonads, kidney. TCV-intestine, respiratory tract. BCV, CCV, PEDV-intestine. FIPV-peritoneum, lymphoid organs, liver, other organs. HCV-respiratory tract. MHV-liver, intestine, CNS, other organs. TGEV-intestine, respiratory tract. HEV-CNS, respiratory tract. RCV-respiratory tract, parotid gland. SDAV-salivary and lacrymal glands.
6.3 Cytopathology: Cellular vacuolation leading to cell disintegration, sometimes syncytium formation.
7 Geographical distribution: FIPV, HECV, HCV, IBV, MHV, TGEV, and HEV present over several continents, probably worldwide.
8 Transmission
8.1 Vertical Intrauterine for IBV, MHV and FIPV.
8.2 Horizontal: Probably all coronaviruses.
8.3 Vectors
8.3.1 Biological: None known.
8.3.2 Mechanical: HCV and HEV, airborne; IBV and BCV, contaminated material; others mainly oral/fecal route.
9 Antigenic properties
9.1 Number of distinct antigenic molecules in virion: Three for HCV, HEV, IBV, MHV, and TGEV. Each corresponds to one class of virion protein.
9.2 Antigen involved in neutralization: Surface peplomer.
9.3 Number of distinct nonstructural antigens: Not known.
9.4 Specificity of different antigens: Hemagglutinin (IBV, HCV, HEV, BCV, MHV) associated with peplomer protein. Also, virus attachment and cell-to-cell fusion activity.
9.5 Antigenic properties used for classification: Immunological reactivity determined by enzyme-linked immunosorbent assay or radioimmunoassay, immunofluorescence, immune electron microscopy, neutralization, hemagglutinin inhibition, Western blotting.
10 Classification
10.1 Definition of family Coronaviridae: Pleomorphic enveloped viruses, averaging 100 nm in diameter, bearing club-shaped projections about 20 nm long. The genome is one molecule of infectious RNA of about (5-7) × 106 molecular weight. Virions characteristically contain three (major) structural proteins: peplomer, matrix and nucleocapsid. Replication involves production 12of a 3′ coterminal nested set of subgenomic mRNAs. Virions bud intracellularly. The family is monogeneric. Serological relationships suggest 2 avian groups, each with one species (IBV and TCV), and 2 mammalian groups, comprised of HCV (229E), TGEV, CCV, and FIPV and of HCV (OC43), MHV, 13SDAV, RCV, HEV, and BCV [5]. IBV, MHV and HCV have many serotypes.
10.2 Genus Coronavirus
10.3 Type species:
IBV avian infectious bronchitis virus
Other species:
HCV human coronavirus
MHV murine hepatitis virus
BCV bovine coronavirus
TGEV transmissible gastroenteritis virus
HEV hemagglutinating encephalomyelitis virus
CCV canine coronavirus
FIPV feline infectious peritonitis virus
Possible species:
RCV rat (sialodacryoadenitis) coronavirus
TCV turkey coronavirus
PEDV porcine epidemic diarrhea virus
The classification of other isolates requires further information. In particular, further evidence is required to determine whether HECV, included in the previous report [1] as a coronavirus, is indeed a coronavirus. The problem of the characterization of HECV has been discussed in a recent review [7].
References
- 1.Tyrrell D.A.J., Alexander D.J., Almeida J.D., Cunningham C.H., Easterday B.C., Garwes D.J., Hierholzer J.C., Kapikian A., Macnaughton M.R., McIntosh K. Coronaviridae: second report. Intervirology. 1978;10:321–328. doi: 10.1159/000148996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohl E.H. Coronaviruses: diagnosis of infections Comp. Diagn. viral Dis. 1981;IV:301–328. [Google Scholar]
- 3.Siddell S.G., Wege H., ter Meulen V. The structure and replication of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:131–163. doi: 10.1007/978-3-642-68528-6_4. [DOI] [PubMed] [Google Scholar]
- 4.Siddell S.G., Wege. H., ter Meulen V. The biology of coronaviruses. J. gen. Virol. 1983;64:761–776. doi: 10.1099/0022-1317-64-4-761. [DOI] [PubMed] [Google Scholar]
- 5.Sturman L.S., Holmes K.V. The molecular biology of coronaviruses. Adv. Virus Res. 28 doi: 10.1016/S0065-3527(08)60721-6. (in press 1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wege H., Siddell S., ter Meulen V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- 7.Macnaughton M.R., Davies H.A. Human enteric coronaviruses. Archs Virol. 1981;70:301–313. doi: 10.1007/BF01320245. [DOI] [PMC free article] [PubMed] [Google Scholar]