Abstract
A reverse transcription-loop mediated isothermal amplification (RT-LAMP) assay was developed to detect the Potato virus S (PVS) in potato. Two sets of six novel primers that recognize the coat protein gene sequence of the PVS were designed and RT-LAMP assay was optimized for the parameters such as different concentrations of primers, MgSO4, betaine, dNTPs, Bst DNA polymerase, temperature and duration. The RT-LAMP was carried out under isothermal conditions without the thermal cycler using PVS infected leaf and tuber samples, LAMP specific primers with amplification at 65 °C for 60 min, and 80 °C for 5 min. The results were assessed by gel electrophoresis and visual observation of colour change using SYBR Green I dye. The detection limit of the developed RT-LAMP assay was determined and compared with a conventional reverse transcription-polymerase chain reaction (RT-PCR). RT-LAMP was found 100 times more sensitive than RT-PCR. The optimized RT-LAMP assay is robust, reliable, sensitive and convenient for the detection of the PVS in infected potato tubers including asymptomatic plants. No cross-reactions were observed with healthy plants and other potato viruses. The assay is economical and can be employed in large scale testing of potato plants against PVS under healthy seed potato production programme.
Keywords: Loop primer, Potato virus S, RT-LAMP, RT-PCR, Virus detection, SYBR green I
Introduction
Potato virus S (PVS, genus Carlavirus) is the most frequently found virus in potato crops worldwide. It has slightly flexuous filamentous particles 660 × 12 nm and single-stranded RNA. PVS is normally causing latent infection on a majority of the potato cultivars but sometimes transient leaf symptoms of faint mosaic or rugosity, vein deepening and leaf bronzing can be observed on the plants (Khurana 2004; Salama and El-Saghir 2017). The incidence of PVS ranges from 8.5 to 99.5% in Indian potato cultivars. However, tuber yield losses are reported from 5 to 20% in secondarily infected plants with PVS or more when associated with Potato virus X (PVX). PVS is highly contagious and spreads easily through seed cutting and even plant-to-plant contact (Khurana 2004; Kumar et al. 2017).
Accurate and efficient virus detection is essential for restricting the spread of PVS. Several serological and molecular methods have been developed for the detection of PVS (Agindotan et al. 2007; Kaushal et al. 2007; Bostan and Peker 2009; Lacomme et al. 2015; Kumar et al. 2017). However, these methods are time-consuming and require well-equipped laboratories with educated experts and frequent dealings with carcinogenic materials to visualize amplification. The PVS, which is transmitted by many species of aphids, has very low titres in the host plants, thus, necessitating the development of a highly sensitive and safe detection method to support seed potato certification schemes. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays are user friendly, time saver, highly sensitive, specific, simple and cost-effective (Bhat et al. 2013; Johnson et al. 2014; Jeevalatha et al. 2018; Kamala and Makeshkumar 2015; Banerjee et al. 2016; Keizerweerd et al. 2015; Zhao et al. 2015). The reactions can be easily visualized by the turbidity analysis, due to the production of a large amount of target DNA (Ju 2011; Nagamine et al. 2002). Besides, under a UV lamp, RT-LAMP products can be seen in the reaction tubes with the naked eye, if SYBR Green-like fluorescent dye is present in the reaction (Jeevalatha et al. 2018). The purpose of this study was to develop a visual detection method to diagnose PVS by RT-LAMP, based on its unique nucleotide sequences encoding a coat protein. The specificity, sensitivity, and rapidity of RT-LAMP were also assessed to optimize the detection of the PVS in potato leaves and tubers.
Materials and methods
Source of plant and viral materials
PVS infected leaf samples were collected from the virus pure culture facility of the Central Potato Research Institute, Shimla, India for optimization of RT-LAMP assay. The same samples were also used to evaluate the sensitivity and specificity of the RT-LAMP. For validation of the assay, leaf samples were randomly collected from the potato fields of Bihar, Himachal Pradesh, Madhya Pradesh, Punjab, Uttar Pradesh, and Gujarat states and frozen in − 80 °C until RNA extraction. Tubers harvested from infected potato plants and also stored tubers were used for validation. Virus-free potato leaf samples were used as negative control. PCR grade water was used as a control in place of the RNA template.
RNA extraction and RT-PCR
Total RNA was extracted from infected and healthy plant materials (leaves and tubers) using the Spectrum™ Plant Total RNA kit according to the manufacturer’s instructions (Sigma-Aldrich, Missouri, USA). The yield and quality of the RNA was evaluated in a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, USA). RT-PCR for comparative analysis with RT-LAMP was carried out using primers recognizing the targeted coat protein region and protocol described by Kumar et al. (2017) in the GeneAmp PCR system 9700 (Applied Biosystems). The amplified products were visualized by electrophoresis in 1% agarose gel stained with Ethidium bromide solution in 1× TAE buffer at 100 volts for 1 h alongside the DNA ladder under UV light.
Primer designing
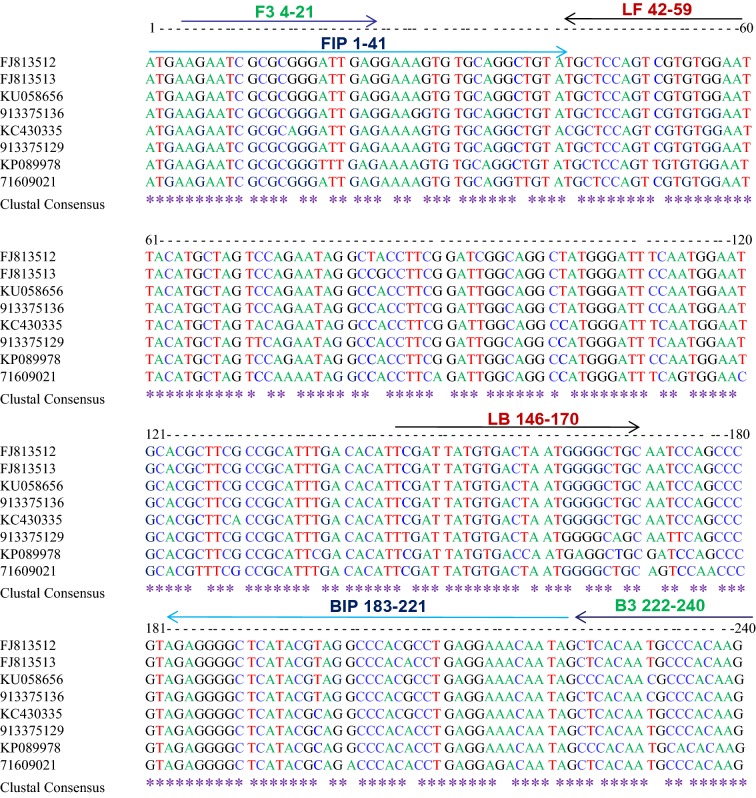
Two sets (A&B) of six oligonucleotide primers [F3, B3, FIP (F1c + F2), BIP (B1c + B2), LF and LB] that recognize a total of eight sequences of the coat protein gene of PVS were designed using Primer Explorer V4 software (http://primerexplorer.jp/e/) with default settings (Table 1). These primers were designed from the conserved region of complete coat protein gene sequences available in GenBank (FJ813512, FJ813513, KU058656, 913375136, KC430335, 913375129, KP089978 and 71609021) and their positions are shown in Fig. 1. All primers used here were of HPLC grade.
Table 1.
List of primers used for RT-LAMP assay
| Primer | Nucleotide sequence 5′–3′ | Length | Position in gene | Length of product (bp) |
|---|---|---|---|---|
| Primer set A | ||||
| PVS-LF | TTCCACACGACTGGAGCA | 18 | 567–584 | |
| PVS-LB | TCGATTATGTGACTAATGGGGCTGC | 25 | 671–695 | |
| PVS-F3 | AAGAATCGCGCGGGATTG | 18 | 526–546 | 237 |
| PVS-B3 | CTTGTGGGCATTGTGAGCT | 19 | 747–765 | |
| PVS-FIP | GCCGATCCGAAGGTAGCCTATGGAAAGTGTGCAGGCTGTA | 40 | 527–566 | 220 |
| PVS-BIP | CGCTTCGCCGCATTTGACACCCTACGTATGAGCCCCTCT | 39 | 708–668 | |
| Primer set B | ||||
| PVS-LF | TCGAGTCCAAGGGCACTG | 18 | 497–514 | |
| PVS-LB | AGTCGTGTGGAATTACATGCTAGTC | 25 | 573–597 | |
| PVS-F3 | CAAGCGTGAGTAGCTCTGTT | 20 | 437–456 | 219 |
| PVS-B3 | CGAAGCGTGCATTCCATTG | 19 | 637–655 | |
| PVS-FIP | CCTCAATCCCGCGCGATTCTTCGGGACTGTGGAGTTCCCAA | 41 | 450–490 | 196 |
| PVS-BIP | AAGTGTGCAGGCTGTATGCTCCTGCCGATCCGAAGGTAGC | 40 | 606–646 | |
Fig. 1.
Illustration showing locations and sequences of PVS CP primers used for RT-LAMP in the multiple sequence alignment of coat protein gene of PVS. Lines with the same colour indicate a primer set A
Optimization of RT-LAMP reaction
Initially, RT-LAMP assay was carried out in a total volume of 25 μl, the reaction mixture contained 1× ThermoPol reaction buffer (20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Tween 20), 1 μl of 100 pmol of FIP and BIP, 0.5 μl of 10 pmol F3 and B3, 1.0 μl of 100 pmol LF, LB, 8 mM MgSO4, 1.0 mM dNTPs, 0.8 M betaine hydrochloride, 8 U of Bst DNA polymerase (New England Biolabs), 0.5 μl of RevertAid M-MuLV reverse transcriptase (200 U/μl, Thermo Scientific, USA), 0.5 μl of RiboLock RNase Inhibitor (20 U/μl, Thermo Scientific, USA), DEPC-treated water and 2 µg total RNA. The reaction mixture was incubated at 65 °C for 60 min followed by denaturation at 80 °C for 5 min to inactivate the Bst DNA polymerase. RT-LAMP amplification products were analyzed and evaluated by direct visual observation of the reaction tube after the addition of SYBR green I (Sigma, St. Louis, USA) or under UV light (the reaction mixture becomes orange with no DNA amplification, whereas turns green with DNA amplification). Besides, RT-LAMP amplification products (3 μl) were also detected in gel electrophoresis in 2% agarose gel. A ladder-like amplification pattern was considered positive, while without a ladder-like band pattern was judged negative (Notomi et al. 2000).
The RT-LAMP assay was optimized by testing different incubation temperatures (60, 61, 62, 63 64 and 65 °C) and then with varying concentration of primers, MgSO4 (2, 4, 6 and 8 mM), dNTPs (0.25, 0.5, 0.75 and 1.0 mM), betaine (0.2, 0.4, 0.6 and 0.8 M) and Bst DNA polymerase (2, 4, 6 and 8 units). The values mentioned in parentheses represent the concentrations of MgSO4, dNTPs, betaine and Bst DNA polymerase in the 25 μl reaction mixture. To optimize the concentration of these four components, the experiments were conducted and best concentration was used one by one; (i) different concentration of MgSO4 with 0.8 M betaine, 1.0 mM dNTPs and 8 units of Bst DNA polymerase was used, (ii) different concentration of betaine with 6 mM MgSO4, 1.0 mM dNTPs and 8 units of Bst DNA polymerase, (iii) different concentration of dNTPs with 6 mM MgSO4, 0.6 M betaine and 8 units of Bst DNA polymerase, (iv) different concentration of Bst DNA polymerase with 6 mM MgSO4, 0.75 mM dNTPs and 0.6 M betaine. To determine the optimum reaction time, the RT-LAMP reaction was carried out at 65 °C for 30, 45, 60, 75 and 90 min. In all the experiments, 1 μl of 30 pmol of FIP and BIP, 0.5 μl of 10 pmol F3 and B3, 1.0 μl of 30 pmol LF and LB were used for optimization.
Sensitivity and specificity of RT-LAMP assay
The quality and quantity of the RNA may influence the results of RT-LAMP. To determine the sensitivity of RT-LAMP as compared to conventional RT-PCR, 10-fold serial dilutions of total RNA from infected potato leaves with PVS were used as the template. Detection limits were determined by the lowest input RNA concentration at which a positive result was obtained. The specificity analysis of optimized RT-LAMP was done by testing with other viruses viz., Tomato Leaf Curl New Delhi Virus (ToLCNDV), Potato Leafroll virus (PLRV), Potato virus Y (PVY), PVX, Potato virus A (PVA) and Potato virus M (PVM) along with a healthy sample. Furthermore, SYBR Green I dye was added to RT-LAMP products and positive reactions were directly detected by visual inspection. Similarly, the detection limit of the RT-LAMP and RT-PCR was approved by electrophoresis on 2.0% agarose gel.
Validation of optimized RT-LAMP assay
The optimized RT-LAMP assay was validated with infected leaf samples collected from potato growing fields in Bihar, Himachal Pradesh, Madhya Pradesh, Punjab, Uttar Pradesh, and Gujarat and virus free tissue culture raised mother plants as known healthy samples. Potato tubers harvested from infected plants and randomly selected tubers from store and asymptomatic plants were also used in the validation.
Results
Selection of specific primer set
Initially, the RT-LAMP for PVS detection was conducted with both the designed primers sets (A&B). Primer set A showed ladder-like bands on the agarose gel only for the PVS containing samples. The experiment was repeated and we confirmed positive reactions for primer set A, while primers set B do not show any ladder-like band/amplification (Fig. 2). For further studies, we have selected, primer set A and used for the optimization of RT-LAMP to detect the PVS.
Fig. 2.
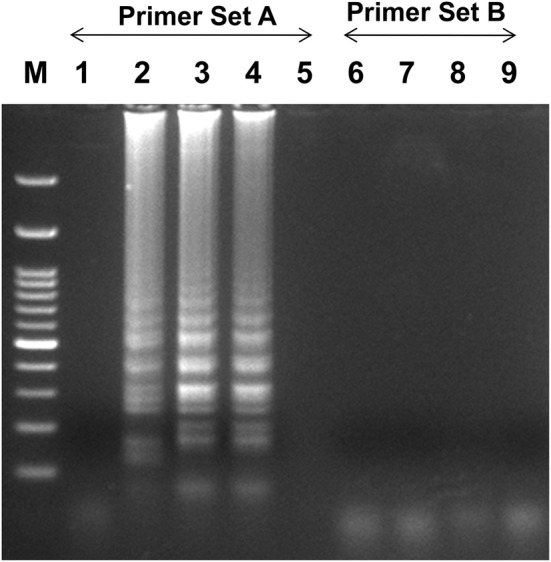
Optimization of RT-LAMP reaction for the detection of the PVS with two sets of primers (Set A&B). Agarose gel electrophoresis of the RT-LAMP products; M-100 bp ladder, lane 1-healthy control, lanes 2 to 4-PVS infected leaves with primer set A; lane 5-water control with primer set A, lanes 6 to 8- PVS infected leaves with primer set B, lane 9-water control with primer set B
Optimization of RT-LAMP reaction
The effect of temperatures, reaction time, concentrations of dNTP, primers, Bst DNA polymerase, Betaine, and MgSO4 was examined. The reactions were carried out using gradient PCR and temperature range was considered between 60 and 65 °C for 60 min followed by 5 min at 80 °C. The results have shown that all temperatures were capable of producing a ladder-like amplification in the RT-LAMP reaction (Fig. 3). However, we selected 65 °C for further studies. The concentrations of primers used in RT-LAMP also affect the amount of amplified products. To reduce the cost of primers, the concentration of different primer pairs was tested (results not shown). In these experiments, the concentration of primers viz., 1 μl of 30 pmol of FIP and BIP, 0.5 μl of 10 pmol F3 and B3, 1.0 μl of 30 pmol LF and LB were shown good amplification, and therefore, these concentrations were further used for RT-LAMP optimization. Then, different concentrations of MgSO4 (2, 4, 6 and 8 mM) were tested for the optimization. The 6 mM MgSO4 was observed best and below 4 mM MgSO4, no amplification was observed (results not shown). For further experiments, 6 mM MgSO4 was used along with an additional 2 mM MgSO4 present in the Thermo-Pol reaction buffer. Then, different concentrations of betaine (0.2, 0.4, 0.6 and 0.8 M) were tested and the RT-LAMP product was observed in all the concentrations of betaine. However, the amplified product was observed better at 0.6 M and 0.8 M betaine (results not shown) and for further optimization; we have used 0.6 M betaine. To see the effects of dNTPs concentration on RT-LAMP reaction, the concentration of dNTPs ranges from 0.25 to 1 mM were tested. The results show that at 0.75 mM and 1.0 mM, ladder-like DNA fragments were observed and 0.75 mM was selected for further studies (results not shown). Finally, Bst DNA polymerase concentration (2, 4, 6 and 8 units) was tested and only at 6 and 8 units in RT-LAMP has given clear amplification in gel electrophoresis. Less than 4 units of Bst DNA polymerase did not give amplification and it indicates that a minimum of 6 units of Bst DNA polymerase is required for RT-LAMP (results not shown). Simultaneously, the visual detection was observed in all the experiments after selection of suitable temperature and primer concentrations using SYBR green I dye under ambient light or UV light.
Fig. 3.
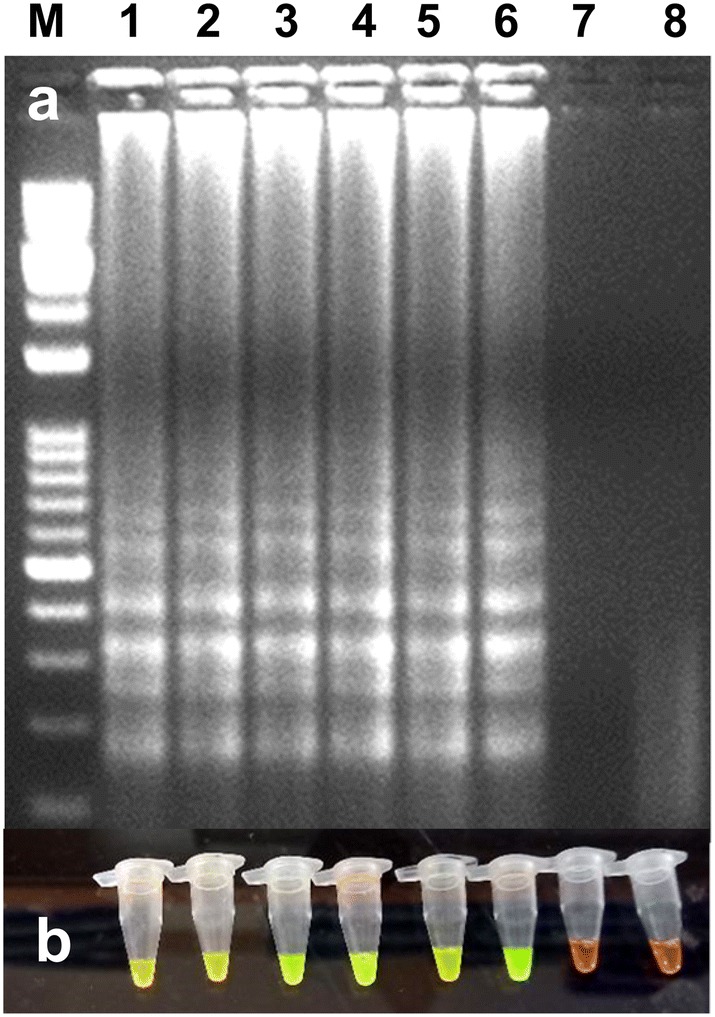
Optimization of RT-LAMP reaction for the detection of the PVS under different temperatures. a Agarose gel electrophoresis of the RT-LAMP products. M-100 bp ladder, lanes 1 to 6 (60, 61, 62, 63, 64 and 65 °C temperature, respectively), lane 7-water control and lane 8-healthy control. b Visualization of the RT-LAMP products under UV excitation at 365 nm. The tubes in (b) from left to right containing the RT-LAMP products and SYBR Green I are the same as those in lanes 1 to 8 in (a)
Sensitivity and specificity of RT-LAMP assay
Serially diluted (10−1 to 10−8 dilutions) total RNA from infected potato leaves with PVS was used to determine the sensitivity of optimized RT-LAMP assay. The optimized RT-LAMP assay was more sensitive than RT-PCR assay and it could detect up to 10−6 dilution in RT-LAMP, while in RT-PCR, virus detection was observed up to 10−4 (Fig. 4). Results were analyzed by gel electrophoresis on 2.0% agarose gel as well as by visual detection in the presence of SYBR Green I dye in PCR tubes. Hence, the optimized RT-LAMP assay was judged highly specific to PVS. The specificity of the optimized assay was also evaluated by performing the RT-LAMP assay with other potato viruses. No cross-reactivity was observed with ToLCNDV, PLRV, PVY, PVX, PVA and PVM infected samples with a healthy sample.
Fig. 4.
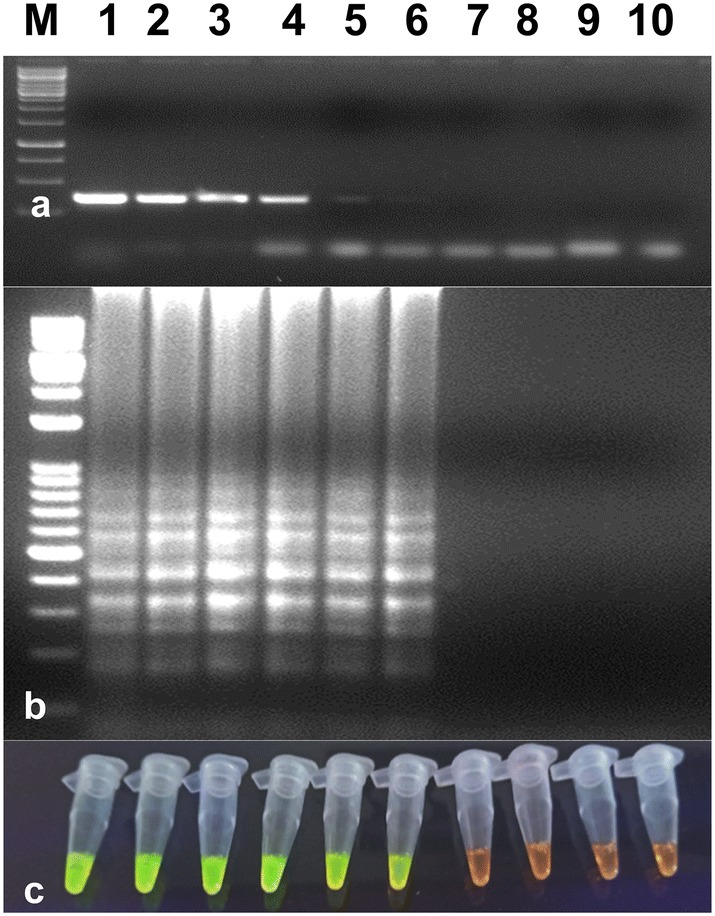
Comparison of the sensitivities of RT-LAMP and RT-PCR assays in the detection of the PVS. a Sensitivity of RT-PCR using a serial dilution of total RNA. M-1 kb ladder, lanes 1 to 8–10−1 to 10−8 dilutions of total RNA, lane 9-healthy control, lane 10-water control. b Sensitivity of RT-LAMP assay detected by agarose gel electrophoresis using a serial dilution of total RNA. The lanes in (b) from left to right containing the RT-LAMP products from the same dilution of total RNA as in lanes 1–8 in (a), lane 9-healthy control, lane 10-water control. c Sensitivity of RT-LAMP assay detected using UV excitation with the addition of SYBER Green I. The tubes in (c) from left to right containing the RT-LAMP products and SYBR Green I are the same as those in lanes 1 to 10 in (b)
Validation of optimized RT-LAMP assay
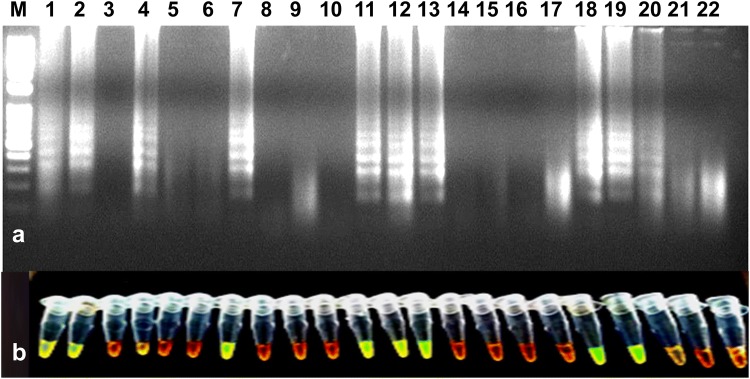
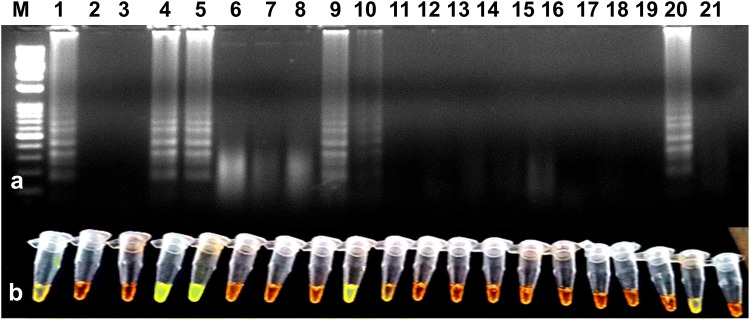
The developed RT-LAMP assay was successfully detected PVS in leaf samples collected (symptomatic and non-symptomatic) from potato fields. For the validation of RT-LAMP, a total of 19 PVS suspected leaf samples from different states along with known healthy samples were tested. Out of 19 samples, nine were found positive and remaining were negative (Fig. 5). The RT-LAMP was found enough sensitive as it could detect PVS in potato tubers kept in cold storage for a long time (Fig. 6). The infection of the virus was found higher in potato fields in Uttar Pradesh, Himachal Pradesh, followed by Punjab and Gujarat (detailed data on the distribution of PVS is not given).
Fig. 5.
Validation of RT-LAMP assay for the detection of the PVS using randomly collected leaf samples from potato fields. a Agarose gel electrophoresis of RT-LAMP products. M-1 kb ladder, lane 1-positive control, lanes 2 to 20-PVS suspected leaf samples, lane 21-water control, lane 22-healthy control. b Visualization of RT-LAMP products under UV excitation at 365 nm. The tubes in (b) from left to right containing the RT-LAMP products and SYBR Green I are the same as those in lanes 1–22 (a)
Fig. 6.
Validation of RT-LAMP assay for the detection of the PVS using randomly collected tuber samples from potato fields and cold storage. a Agarose gel electrophoresis of RT-LAMP products. M-1 kb ladder, lanes 1 to 19-PVS suspected potato tuber samples, lane 20-positive control, lane 21-water control. b Visualization of RT-LAMP products under UV excitation at 365 nm. The tubes in (b) from left to right containing the RT-LAMP products and SYBR Green I are the same as those in lanes 1–21 (a)
Discussion
At present, the testing facility for the detection of potato viruses is consistently improving especially in India due to well-established business in tissue culture raised plant and mini tubers production by several private as well as the Govt. sector. In this context, there was an urgent need to develop alternative methods that can visually detect the virus infection in less time for large scale testing. Moreover, to avoid the use of carcinogenic dye-like ethidium bromide in gel electrophoresis, an alternative option was required. Nowadays, RT-LAMP based methods have been employed for the detection of many plant pathogens including viruses (Ju 2011; Nie 2005; Jeevalatha et al. 2018). RT-LAMP assay is a prompt and highly efficient and can be performed under isothermal conditions (60–65 °C). More important is that amplification can be seen by the naked eye either as turbidity or due to colour change in the presence of a specific dye-like SYBR Green I, and therefore, the gel electrophoresis system is not required. Hence, we developed an RT-LAMP assay using RNA as a template for the detection of the PVS in leaves as well as tubers. The specificity of RT-LAMP is generally high because of the use of four primers that perceive six regions on the target gene. However, primer design is an important factor in RT-LAMP in RNA virus’s diagnosis (Boubourakas et al. 2009; Notomi et al. 2000; Wei et al. 2012; Jeevalatha et al. 2018). During the optimization of RT-LAMP assay, the good amplification was observed between 60 to 65 °C temperatures. Most of the literatures say that RT-LAMP reaction found successful between 60 and 65 °C temperature (Boubourakas et al. 2009; Kuan et al. 2010; Banerjee et al. 2016; Bhat et al. 2013; Jeevalatha et al. 2018) and any deviation from this temperature range results in lower detection. The reason behind this might be due to instability in the reaction or inactivation of the enzymes conferred by too low or high temperature (Fukuta et al. 2004; Wei et al. 2012; Jeong et al. 2015; Jeevalatha et al. 2018). We tested 0.2 to 0.8 M betaine and brighter amplification was found at 0.6 M and 0.8 M betaine concentration only, and therefore, 0.6 M betaine was selected in the final assay. Several findings are reported that 0.6–0.8 M betaine used in the optimization of LAMP assays (Banerjee et al. 2016; Keizerweerd et al. 2015). In this study, 0.75 mM dNTPs gave good amplification in gel electrophoresis and visual detection. Many researchers have used 0.6 to 3.2 mM dNTPs for LAMP (Banerjee et al. 2016; Wang et al. 2013; Bhat et al. 2013; Keizerweerd et al. 2015; Jeevalatha et al. 2018). Minimum concentration of 0.4 mM dNTPs was used in RT-LAMP to acquire positive results and for PVX detection, the optimum concentration was 1.0 to 1.2 mM dNTPs (Jeong et al. 2015). However, only 5 and 10 mM dNTPs concentration could detect PLRV in RT-LAMP (Almasi et al. 2013) and the fact may be that dNTPs requirements can vary due to target genes in RT-LAMP assay (Jeong et al. 2015). The 6 mM MgSO4 was observed best and used in the final assay in addition to 2 mM MgSO4 present in the ThermoPol reaction buffer. Similar studies reported earlier the optimal MgSO4 range between 4 and 8 mM. Optimization of Mg2+ concentration is crucial as it influences primer annealing and enzyme activities in RT-LAMP assay (Banerjee et al. 2016; Keizerweerd et al. 2015; Nie 2005; Wei et al. 2012; Jeevalatha et al. 2018). In the case of Bst DNA polymerase requirement, with the application of 6 and 8 U, we could see clear cut amplification in gel electrophoresis and visual detection. Similar findings reported by Almasi et al. (2013), Nie (2005) and Jeevalatha et al. (2018), where they observed poor amplification with 2 U of Bst DNA polymerase and had to raise enzyme concentration to 8 U to get good amplification. The developed RT-LAMP assay for the detection of the PVS was found 100 times more sensitive than RT-PCR (Fig. 4). Similar findings reported that because of high efficiency, the RT-LAMP was 10 to 1000 times more sensitive than conventional RT-PCR (Ju 2011; Kuan et al. 2010; Banerjee et al. 2016; Wei et al. 2012; Zhao et al. 2015). However, in the case of RT-LAMP assays for Sorghum mosaic virus and Sugarcane mosaic virus, the conventional RT-PCR was more sensitive compared to RT-LAMP (Keizerweerd et al. 2015). In our studies, we found that extreme care is required during reaction set up as a minor mistake or ill handling may give the false results quite evident from the laddering in non-template reactions. Therefore, to avoid the wrong testing, the laboratory set up should be cleaned and careful handling is needed during RT-LAMP assay. However, non-specific amplification can be seen in the case of primer dimers which gives false-positive results too (Jeevalatha et al. 2018). In addition, the concentration of dNTPs, primers, Mg2+, betaine and Bst DNA polymerase must be optimized to avoid non-specific amplification (Wang et al. 2013, Jeevalatha et al. 2018). The optimized RT-LAMP assay was found effective in the detection of PVS in potato leaves from asymptomatic plants and potato tubers from storage. Hence, it could be a better option for a large scale testing of PVS under a healthy seed production system. Since the virus mainly spreads through seed tubers, the successful application of the developed RT-LAMP assay in potato tubers further adds value to the study. The assay was sensitive, specific and robust for routine testing of potato plants under healthy seed potato production.
Acknowledgements
This study was supported by the Indian Council of Agricultural Research, New Delhi, India.
Authors’ contributions
RK conceptualization. RK and PK did the analytical work of processing samples for RNA extraction, cDNA synthesis, and PCR assays. RK, JA, PK and SS methodology. RK, PK and JA Data analysis. SS validation. RK, SS, and SKC helped in preparing and editing the MS.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest by the co-authors.
Ethical clearance
No human subjects were used in the writing of the manuscripts.
References
- Agindotan BO, Shiel PJ, Berger PH. Simultaneous detection of potato viruses, PLRV, PVS, PVX, and PVY from dormant potato tubers by TaqMan real-time RT-PCR. J Virol Methods. 2007;142:1–9. doi: 10.1016/j.jviromet.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Almasi MA, Jafary H, Moradi A, Zand N, Ojaghkandi MA, Aghaei S. Detection of coat protein gene of the Potato leafroll virus by reverse transcription loop-mediated isothermal amplification. J Plant Pathol Microb. 2013;4:156. [Google Scholar]
- Banerjee A, Roy S, Sharma SK, Dutta SK, Chandra S, Ngachan SV. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for rapid diagnosis of Chilli veinal mottle virus. Arch Virol. 2016;161:1957–1961. doi: 10.1007/s00705-016-2850-7. [DOI] [PubMed] [Google Scholar]
- Bhat AI, Siljo A, Deeshma KP. Rapid detection of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) by loop-mediated isothermal amplification (LAMP) J Virol Methods. 2013;193:190–196. doi: 10.1016/j.jviromet.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Bostan H, Peker PK. The feasibility of tetraplex RT-PCR in the determination of PVS, PLRV, PVX and PVY from dormant potato tubers. Afr J Biotechnol. 2009;17:4043–4047. [Google Scholar]
- Boubourakas IN, Fukuta S, Kyriakopoulou PE. Sensitive and rapid detection of Peach latent mosaic viroid by the reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2009;160:63–68. doi: 10.1016/j.jviromet.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Fukuta S, Ohishi K, Yoshida K, Mizukami Y, Ishida A, Kanbe M. Development of immune-capture reverse transcription loop-mediated isothermal amplification for the detection of Tomato spotted wilt virus from chrysanthemum. J Virol Methods. 2004;121:49–55. doi: 10.1016/j.jviromet.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Jeevalatha A, Kaundal P, Kumar R, Raigond B, Kumar R, Sharma S, Chakrabarti SK. Optimized loop-mediated isothermal amplification assay for Tomato leaf curl New Delhi virus-[potato] detection in potato leaves and tubers. Eur J Plant Pathol. 2018;150:565–573. doi: 10.1007/s10658-017-1300-z. [DOI] [Google Scholar]
- Jeong J, Cho SY, Lee WH, Lee KJ, Ju HJ. Development of a rapid detection method for Potato virus Y by reverse transcription loop-mediated isothermal amplification. Plant Pathol J. 2015;31:219–225. doi: 10.5423/PPJ.OA.03.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AMA, Dasgupta I, Sai Gopal DVR. Development of loop-mediated isothermal amplification and SYBR green real-time PCR methods for the detection of Citrus yellow mosaic badnavirus in citrus species. J Virol Methods. 2014;203:9–14. doi: 10.1016/j.jviromet.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Ju HJ. Simple and rapid detection of Potato leafroll virus by reverse transcription loop-mediated isothermal amplification. Plant Pathol J. 2011;27:385–389. doi: 10.5423/PPJ.2011.27.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamala S, Makeshkumar T. Rapid and sensitive detection of Dasheen mosaic virus infecting elephant foot yam by reverse transcription loop-mediated isothermal amplification of coat protein gene. J Virol Methods. 2015;222:106–109. doi: 10.1016/j.jviromet.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Kaushal N, Gawande SJ, Kaundal P, Garg ID. Reverse transcriptase-polymerase chain reaction (RT-PCR) based detection of PVS and PVS strain by using degenerate primers. Potato J. 2007;34:85–86. [Google Scholar]
- Keizerweerd AT, Chandra A, Grisham MP. Development of a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of Sugarcane mosaic virus and Sorghum mosaic virus in sugarcane. J Virol Methods. 2015;212:23–29. doi: 10.1016/j.jviromet.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Khurana SMP. Potato viruses and their management. In: Naqvi SAMH, editor. Diseases of fruits and vegetables: diagnosis and management. Dordrecht: Kluwer Academic; 2004. pp. 389–440. [Google Scholar]
- Kuan CP, Wu MT, Lu YL, Huang HC. Rapid detection of Squash leaf curl virus by loop-mediated isothermal amplification. J Virol Methods. 2010;169:61–65. doi: 10.1016/j.jviromet.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Kumar R, Jeevalatha A, Raigond B, Kumar R, Sharma S, Nagesh M. A multiplex RT-PCR assay for simultaneous detection of five viruses in potato. J Plant Pathol. 2017;99:37–45. [Google Scholar]
- Lacomme C, Holmes R, Evans F. Molecular and serological methods for diagnosis of viruses in potato tubers. PVX, PLRV and PVS by RT-PCR in potato leaf and tuber. Australas Plant Dis Notes. 2015;1:41–46. doi: 10.1007/978-1-4939-2620-6_13. [DOI] [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Nie X. Reverse transcription loop-mediated isothermal amplification of DNA for the detection of Potato virus Y. Plant Dis. 2005;89:605–610. doi: 10.1094/PD-89-0605. [DOI] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama M, El-Saghir J. Incidence, serological and molecular characterization of Potato virus S from commercial potato in Egypt. J Virol Sci. 2017;1:67–75. [Google Scholar]
- Wang X, Teng D, Guan Q, Tian F, Wang J. Detection of roundup ready soybean by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Food Control. 2013;29:213–220. doi: 10.1016/j.foodcont.2012.06.007. [DOI] [Google Scholar]
- Wei QW, Yu C, Zhang SY, Yang CY, Miriam K, Zhang WN, Dou DL, Tao XR. One-step detection of Bean pod mottle virus in soybean seeds by the reverse transcription loop-mediated isothermal amplification. Virol J. 2012;9:187–193. doi: 10.1186/1743-422X-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LM, Li G, Gao Y, Zhu YR, Liu J, Zhu XP. Reverse transcription loop-mediated isothermal amplification assay for detecting Tomato chlorosis virus. J Virol Methods. 2015;213:93–97. doi: 10.1016/j.jviromet.2014.11.013. [DOI] [PubMed] [Google Scholar]