Abstract
Study Objectives
Sleep disturbances are common co-morbidities of epileptic disorders. Dravet syndrome (DS) is an intractable epilepsy accompanied by disturbed sleep. While there is evidence that daily sleep timing is disrupted in DS, the difficulty of chronically recording polysomnographic sleep from patients has left our understanding of the effect of DS on circadian sleep regulation incomplete. We aim to characterize circadian sleep regulation in a mouse model of DS.
Methods
Here we exploit long-term electrocorticographic recordings of sleep in a mouse model of DS in which one copy of the Scn1a gene is deleted. This model both genocopies and phenocopies the disease in humans. We test the hypothesis that the deletion of Scn1a in DS mice is associated with impaired circadian regulation of sleep.
Results
We find that DS mice show impairments in circadian sleep regulation, including a fragmented rhythm of non-rapid eye movement (NREM) sleep and an elongated circadian period of sleep. Next, we characterize re-entrainment of sleep stages and siesta following jet lag in the mouse. Strikingly, we find that re-entrainment of sleep following jet lag is normal in DS mice, in contrast to previous demonstrations of slowed re-entrainment of wheel-running activity. Finally, we report that DS mice are more likely to have an absent or altered daily “siesta”.
Conclusions
Our findings support the hypothesis that the circadian regulation of sleep is altered in DS and highlight the value of long-term chronic polysomnographic recording in studying the role of the circadian clock on sleep/wake cycles in pre-clinical models of disease.
Keywords: Dravet syndrome, circadian sleep regulation, epilepsy, sleep re-entrainment, siesta, jet lag, circadian rhythm, sleep timing
Statement of Significance.
Sleep disturbances are among the most common secondary symptoms of epileptic disorders. While there is evidence for a strong relationship between seizures, circadian rhythms, and sleep timing, performing long-term polysomnographic recordings to better understand this relationship is not feasible in patients. Here we use long-term electrocorticographic recordings to reveal impairments in the circadian regulation of sleep in a mouse model of Dravet syndrome, a severe form of childhood-onset epilepsy. In addition to furthering our understanding of sleep disturbances in this debilitating disorder, we propose an experimental framework in which sleep researchers can more thoroughly characterize the circadian regulation of sleep in pre-clinical models of disease.
Introduction
Sleep disturbance is common in epilepsy, and is associated with reduced seizure control, poor cognitive outcomes, and decreased quality of life [1]. The prevalence of nocturnal seizures in several forms of epilepsy [2], increased risk of seizure occurrence following sleep deprivation [3] and evidence that seizure propensity shows daily rhythms [4–9] highlights the need for a better understanding of the relationship between sleep, epilepsy, and the circadian clock.
Dravet syndrome (DS) is a severe form of treatment-resistant, childhood-onset epilepsy with a high mortality rate [10]. DS usually manifests at 6–9 months of age with fever and hyperthermia-induced seizures, and gradually progresses to include to spontaneous myoclonic, tonic–clonic, absence, and partial seizures [10, 11]. During this time, DS patients develop several other co-morbidities, including autism-like behaviors, ataxia, psychomotor regression, and high risk of sudden unexpected death (SUDEP). Sleep disruptions are reported by more than 70% of DS patients [12], and DS patients report more frequent nighttime awakenings and greater daytime sleepiness than patients with other forms of epilepsy [13]. Disturbances, including sleep-onset insomnia, difficulty maintaining sleep, and frequent nocturnal seizures are well-documented, and these disturbances are described as highly disruptive to quality of life for both patients and caregivers [14–16].
DS is caused by loss-of-function mutations in one allele of the SCN1A gene [17], which codes for the pore-forming alpha subunit of the NaV1.1 sodium channel [18]. NaV1.1 is widely expressed throughout the brain, primarily in GABAergic interneurons. Previous work in a mouse model of DS, in which the Scn1a gene is heterozygously deleted, has shown that the mutation causes a selective reduction in the excitability of inhibitory interneurons [19, 20]. Several characterizations of DS mice have revealed that, like human patients, they display both spontaneous and thermally induced seizures [19, 21], ataxia [22], autism-like cognitive and social deficits [23, 24], SUDEP [25], and sleep disturbances [26–28]. Specific heterozygous deletion of the Scn1a gene in forebrain interneurons recapitulates the epilepsy, SUDEP, cognitive impairment, autistic-like behaviors, and homeostatic sleep disruption observed in DS mice [23, 26, 28, 29]. This work has led to the unified hypothesis that hyperexcitability of different neural circuits due to the loss of functional NaV1.1 channels underlies the co-morbidities of DS [30].
The two-process model of sleep regulation proposes both homeostatic and circadian drives that influence the timing and consolidation of sleep [31]. While the neural circuitry underlying these sleep regulatory processes is multi-faceted, reliant on multiple neurotransmitter systems, and widely diffuse across the brain, many of the most important nodes in these circuits are GABAergic [32, 33]. In previous studies, we identified circadian behavioral deficits in DS mice including reduced circadian amplitude of wheel-running activity (WRA), elongated endogenous WRA period under conditions of constant darkness (DD), and increased time to re-entrainment of WRA to a new light–dark (LD) cycle [26]. These deficits were correlated with poor network synchrony and reduced photoresponsiveness in the GABAergic suprachiasmatic nucleus (SCN) of the hypothalamus, the master circadian clock in mammals and a critical component of daily sleep timing [34]. Later work showed that DS mice also lack normal homeostatic non-rapid eye movement (NREM) sleep rebound in response to sleep deprivation, as well as fragmented NREM sleep under baseline conditions [28]. This was associated with reduced excitability of GABAergic inhibitory neurons in the thalamic reticular nucleus (TRN), a brain region which contributes to maintaining the slow oscillations characteristic of NREM sleep and the homeostatic sleep response [2]. Based on these findings, we hypothesized that DS mice would also display deficits in the circadian regulation of sleep.
Here we leverage long-term electrocorticographic (ECoG) recordings to characterize the circadian regulation of sleep in DS mice. We hypothesized that DS mice would have disturbances in circadian sleep regulation due to reduced GABAergic tone in the SCN caused by the heterozygous deletion of the Scn1a gene. We report disturbances in circadian sleep behavior of DS mice including elongated circadian period of sleep, a fragmented rhythm of NREM sleep that was not correlated with epileptiform interictal activity and fragmented or absent siesta, a short bout of sleep under the control of the circadian clock. Interestingly, DS mice displayed no deficits in re-entrainment of sleep acrophase following jet lag, and similar total sleep time during each phase of the LD cycle when compared to wild type (WT) controls. These results highlight the value of coupling long-term sleep recordings with locomotor activity monitoring to more comprehensively disentangle the effects of environmental manipulations on different outputs of the circadian clock, and provide a novel approach to studying the circadian regulation of sleep in pre-clinical models of disease.
Methods
Animals and housing conditions
All experiments with animals were performed in accordance with animal protocols approved by the Office of Animal Welfare at the University of Washington. Mice with a heterozygous deletion of the Scn1a gene (Scn1a+/-), hereafter referred to as DS mice, were generated by targeted deletion of the last exon, encoding domain IV from the S3 to S6 segment and the entire C-terminal tail of NaV1.1 channels [19]. The mice used in this study were generated by crossing heterozygous mutant mice of C57BL/6 background with WT C57BL/6 mice (both males and females of each genotype), resulting in only WT or heterozygous Scn1a mutant offspring. Mice were genotyped as previously described [28].
ECoG recordings
Sleep was recorded as previously described [35]. Briefly, mice were anesthetized with isoflurane and placed into a stereotaxic device where isoflurane anesthesia continued throughout surgery. Each mouse was implanted with ECoG electrodes, consisting of dental screws (Pinnacle Technology, Lawrence, KS; No. 8209: 0.10-in.). A midline incision was made above the skull. Recording electrodes were screwed through cranial holes as follows: over the left frontal cortex (1.5 mm lateral and 2 mm anterior to bregma) and over the right parietal cortex (1.5 mm lateral and 2 mm posterior to bregma), a ground electrode was placed over the visual cortex (1.5 mm lateral and 4.0 mm posterior to bregma), and a reference electrode was placed over the cerebellum (1.5 mm lateral and 6.5 mm posterior to bregma). Electromyogram (EMG) signals were obtained by placing a pair of silver wires into the neck muscles. The screws were connected, through silver wires, to a common 6-pin connector compatible with the Pinnacle recording device. The screws and connector were fixed to the skull with dental cement. Mice were implanted at between 3 and 4 months of age to account for the long duration of circadian sleep experiments. After surgery, mice were housed in single recording cages under a 12:12 LD cycle.
Mice had a recovery period of 1 week and were then fitted with a preamplifier and tether, and connected to the Pinnacle Technology recording system, where they were allowed 1 day to acclimate before recording started. The ECoG and EMG signals were sampled at 400 Hz with low-pass filters of 80 Hz and 100 Hz, respectively.
Sleep scoring and processing
Raw ECoG/EMG data were automatically classified in 10-second bouts as either wake, NREM sleep or REM sleep using a previously described custom algorithm implemented in Python v2.7 [35]. Briefly, the power spectrums of the ECoG and EMG signals are calculated using Welch’s method with an overlap window of 50%. Then, the following features for each 10-second epoch of sleep are calculated: delta power, the sum of ECoG power at frequencies ranging between 0.5 and 4 Hz; theta power, the sum of ECoG power at frequencies ranging between 6 and 12 Hz; and EMG RMS, the mean square root of the raw EMG signal. If EMG RMS was higher than a visually determined threshold for each 24-hour recording session, typically set at 0.4 SDs above the mean, the epoch was classified as wake, otherwise it was classified as sleep. Next, sleep stage was classified as REM whenever the theta to delta power ratio during the epoch was higher than the average theta to delta ratio—over the whole 24-hour recording session—plus 1 SD. The remaining epochs were classified as NREM sleep. This scoring method was based on heuristics for manually scoring sleep in previous studies and validated by two independent experimenters that visually scored 10-second bins as previously described [36–38], with a minimum of 90% score agreement in both cases.
The 10-second scored epochs were further binned into 1- or 60-minute bins, for which we calculated the percentage of each sleep state. A 1- or 60-minute bin was classified as REM when more than 20% of the bin was spent in REM sleep. If REM sleep was lower than 20% during the bin, it was classified as either wake or NREM sleep, depending on which of the two brain states contributed to a higher percentage of the 1- or 60-minute bin. Hypnograms were generated with 10-second bins of sleep scores using a custom R code. NREM and REM intensities were calculated as the product of sleep stage percentage and either delta or theta power within each 1- or 60-minute bin. Statistical analysis was performed for both 1- and 60-minute bins and yielded similar results; 1-minute bins are reported for total sleep time, sleep re-entrainment, rhythm intradaily variability (IV), and circadian sleep period analyses, while 60-minute bin results are reported for daily sleep stage intensity scores under baseline conditions.
During long-term recording, the recording software was restarted every 24 hours to account for potential software or hardware failures, resulting in a loss of approximately 10–30 seconds of recording time each day. To account for this, missing data points were imputed after scoring by taking the mean of the values for delta, theta and sleep stage calls in the 60 seconds following and preceding the gap using a custom R code. This facilitated the combination of sleep scores into larger bins and enhanced the continuity of the recording.
Interictal spike quantification
Epileptiform interictal spikes were identified semi-automatically over a 24-hour recording session with the Sirenia Seizure software (Pinnacle Technology, Lawrence, KS) using a line length threshold [39] established for both ECoG channels, then confirmed visually by a manual scorer blind to experimental conditions. Spikes were then classified as occurring during either Wake, NREM or REM based on scores obtained through automated sleep classification.
Behavioral experiments and analysis
Our recordings were maintained in a ventilated, light-tight room under either a 12:12 LD cycle with 200-lux intensity or DD. To determine the effect of abrupt LD phase shifts on the phase of the circadian rhythm of sleep, we used a delay and advance “jetlag” protocol [26]. After mice were entrained to a 12:12 LD cycle, a delay jetlag was initiated by extending the light phase by 6 hours (i.e. delaying the time of lights off by 6 hours). The mice stayed under this new 12:12 LD cycle until they re-entrained to the new cycle. The advance jetlag was initiated by shortening the light phase by 6 hours (i.e. advancing the time of lights off by 6 hours). For each day of recording, acrophase was determined for both NREM and REM sleep using the El Temps software package (Dr. Toni Diéz-Noguera, Universidad de Barcelona), and time to re-entrainment was calculated as the number of days necessary for sleep stage acrophase to return to mean baseline levels within a 95% confidence interval.
To assess the endogenous circadian period of sleep, we released mice into DD for at least 14 days after they had synchronized to a 12:12 LD cycle. Free-running period of NREM, REM, and total sleep were assessed using 1-minute bins and Sokolove-Bushell periodogram analysis in El Temps and R version 3.1.2 [40]. Waveforms of total sleep in DD were generated using El Temps as previously described [26], and adjusted for the circadian period of each animal.
We defined the “siesta” as a brief (<2 hours) bout of sleep occurring primarily during the dark phase of the LD cycle that is visually discernible from the main bout and occurs with a regular onset each day. To compare the re-entrainment of the siesta and the main bout of sleep, we used total sleep (both NREM and REM), due to the brevity of the siesta and the limited number of REM bouts within it. We calculated the re-entrainment speed (RS) in minutes per day for both sleep bouts individually by extracting the slope of a line fitted to sleep offset for delays and onset for advances using actograms of sleep by three manual scorers blind to experimental conditions. RS was determined as the average slope calculated from the three manual scorers.
Wake after sleep onset (WASO) [41] was defined as the percentage of total time spent awake during the time of lights on in the chamber, as sleep onset occurred within minutes of lights on. WASO was calculated for each DS mouse during the same 24-hour recording period used for the interictal spike analysis.
Data analysis and statistics
Data are presented as mean ± SEM and typically analyzed using one-, two-, or three-way ANOVA followed by Tukey, or Student’s t-tests for comparisons in which type I error was not possible. Chi-squared tests were used to assess differences in the occurrence of siesta between genotypes. The 24-hour distributions of REM and NREM intensity were compared between genotypes using a two-sample Kolmogorov-Smirnov test, as the data were not normally distributed. Sleep stage bout durations from both genotypes were not normally distributed and were analyzed using a two-way analysis of variance (ANOVA) of the aligned rank transform of the data with the ARTool R package [42]. Occurrences of interictal spikes were analyzed using the Kruskal–Wallis test, and post hoc pairwise comparisons made with the Wilcoxon rank sum test. p < 0.05 was considered as statistically significant. IV index was calculated as previously described [43, 44]. Briefly, IV is the ratio of the mean square of the difference between all the successive hours on each day, and the same calculation around the overall mean:
where n is the total number of data points, xi is each individual data point, and is the mean of all the data points. An IV index of 0 represents a perfect sinusoid, while an IV index of 2 represents Gaussian noise. Interictal spiking frequency was correlated with sleep IV or WASO using Spearman’s rank correlation. All statistical tests were done with GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) or R version 3.1.2.
Results
DS mice show similar total amounts of sleep compared to WT mice under baseline conditions
We first sought to determine whether total sleep duration differed between WT and DS mice over the 24-hour day. We implanted WT and DS mice with ECoG/EMG electrodes and recorded continuously under 12:12 LD conditions. We then assessed total sleep time and intensity during the light and dark phases.
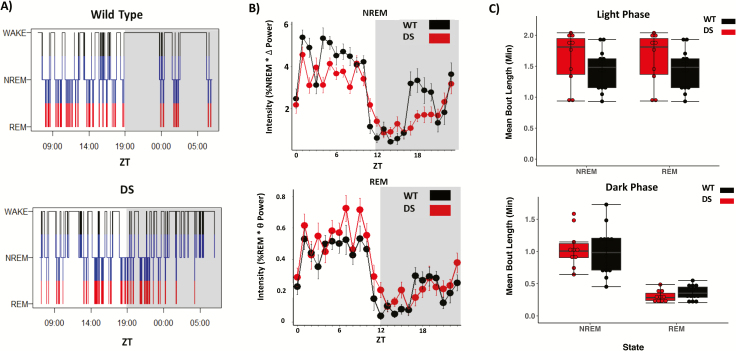
Sleep bouts of both genotypes synchronized to a 12:12 LD cycle. While representative hypnograms of 24 hours of baseline data (Figure 1A) reflect previously observed sleep fragmentation of DS mice [28], mean daily sleep time calculated from 48 consecutive hours of recording shown in Figure 1B reveal that WT and DS sleep time is comparable and highly consolidated during the light phase (two-way ANOVA: genotype [F1,44 = 0, p = 1]; light phase [F1,44 = 686.11, p < 0.001], n = 10 DS mice, 15 WT mice) (data not shown). We next generated daily waveforms of 1-hour bins of NREM and REM sleep intensity, defined as the product of the amount of time spent in each sleep stage per hour and the average delta or theta power, respectively (Figure 1B). The distribution of sleep intensity did not differ between genotypes for NREM (Kolmogorov-Smirnov test, p = 0.45) or REM (Kolmogorov-Smirnov test, p = 0.686). Finally, we found that, in agreement with previous reports [28], mean NREM and REM sleep bout duration is comparable between genotypes (Figure 1C), during both the light phase (two-way ANOVA of aligned rank transform data; no effect of sleep stage [F1,44 = 0, p = 1] or genotype [F1,44 = 3.42, p = 0.07] and no interaction [F1,44 = 0, p = 1]) and the dark phase (two-way ANOVA of aligned rank transform data; effect of sleep stage [F1,44 = 121.59, p < 0.001] but not genotype [F1,44 = 0.01, p = 0.92] and no interaction [F1,44 = 0.87, p = 0.35]).
Figure 1.
DS mice show similar sleep bout length compared to WT controls. (A) Representative hypnograms taken from a single day of sleep under a 12:12 LD cycle in wild type (top) and DS (bottom) mice. (B) Average waveforms 60-minute bins of non-rapid eye movement (NREM) and rapid-eye movement (REM) sleep intensity calculated from normalized power values in WT and DS mice. The distribution of sleep intensity did not differ between genotypes for NREM (Kolmogorov-Smirnov test, p = 0.45) or REM (Kolmogorov-Smirnov test, p = 0.686), and both WT and DS mice show similar total sleep time in both phases of the LD cycle (data not shown, two-way ANOVA, effect of LD phase [p < 0.001] but not genotype [p = 0.36]). (C) Average NREM and REM sleep bout lengths are comparable between genotypes, during both the light phase (two-way ANOVA of aligned rank transform data; no effect of sleep stage [F1,44 = 0, p > 0.05] or genotype [F1,44 = 3.42, p > 0.05], and no interaction [F1,44 = 0, p = 1]) and the dark phase (two-way ANOVA of aligned rank transform data; effect of sleep stage [F1,44 = 121.59, p < 0.001] but not genotype [F1,44 = 0.01, p > 0.05] and no interaction [F1,44 = 0.87, p = 0.35]).
DS mice display altered IV of NREM sleep that is not correlated with interictal activity
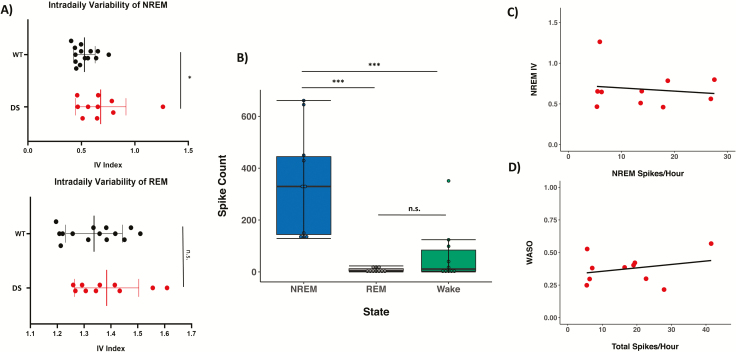
Previous work has demonstrated that DS mice display reduced circadian amplitude of locomotor activity under baseline 12:12 LD conditions [26], and highly fragmented NREM but not REM sleep [28]. We predicted that DS mice would also display a less stable daily rhythm of NREM and REM sleep and assessed the integrity of sleep rhythms using IV, a nonparametric index of rhythm fragmentation. We found that IV index (Figure 2A) was significantly higher in DS mice for NREM sleep (unpaired two-tailed t-test, p = 0.04), but not REM sleep (unpaired two-tailed t-test, p = 0.32), indicating that DS mice have a fragmented daily rhythm of NREM sleep under baseline conditions.
Figure 2.
Altered IV of NREM sleep is not correlated with interictal activity in DS mice. (A) IV, a nonparametric measure of daily rhythm fragmentation, is higher in DS mice than WT mice for NREM sleep (top panel; unpaired two-tailed t-test, p = 0.04), but not for REM sleep (bottom panel; unpaired two-tailed t-test, p = 0.32). (B) Interictal spiking occurs primarily in NREM sleep in DS mice (Kruskal–Wallis rank sum test, χ 2= 18.051, df = 2, p < 0.001), while some spikes were observed during wake and rarely during REM sleep (pairwise comparisons using Wilcoxon rank sum test; NREM-Wake, p = 0.001; NREM-REM, p < 0.001, REM-Wake, p = 0.27). Interictal spiking was not observed in WT mice. (C) Interictal spike frequency showed no significant correlation with intradaily variability of NREM sleep (Spearman’s rank correlation, rs = 0.19, p = 0.6). (D) Total interictal spike frequency showed no significant correlation with wake after sleep onset (Spearman’s rank correlation, rs = 0.19, p = 0.61).
It has been shown that interictal spiking events occur during NREM sleep in DS mice and these spikes are associated with an increased number of awakenings from NREM sleep [28]. We predicted that spiking frequency would also be positively correlated with NREM IV. We quantified the presence of interictal spikes during wake, NREM and REM sleep over a 24-hour recording session under baseline 12:12 LD conditions. In agreement with previous work, we found that interictal spiking occurs primarily during NREM sleep in DS mice (Kruskal–Wallis rank sum test, χ 2 = 18.051, df = 2, p < 0.001; Figure 2B). Some spikes were observed during wake, and rarely during REM sleep (pairwise comparisons using Wilcoxon rank sum test; NREM-Wake, p = 0.001; NREM-REM, p < 0.001, REM-Wake, p = 0.27). Spontaneous seizures were occasionally observed in DS mice during wake. Interictal spikes were not observed in WT mice. Interestingly, we found no significant correlation between NREM IV and interictal spike frequency during NREM sleep (Spearman’s rank correlation, rs = 0.19, p = 0.6; Figure 2C).
Previous reports have indicated that DS mice have highly fragmented sleep, and we predicted that WASO, a measure of sleep fragmentation commonly used in studies of human sleep, would also be positively correlated with interictal spiking activity in DS mice. Interestingly, we found no correlation between WASO and interictal spike frequency across behavioral states (Spearman’s rank correlation, rs = 0.19, p = 0.61; Figure 2D).
Sleep bouts take longer to re-entrain to phase advances than phase delays, and DS mice display normal re-entrainment
We previously demonstrated that DS mice take longer to re-entrain to abrupt phase advances and delays of the LD cycle simulating jet lag than WT mice, as measured by WRA [26]. We predicted that the acrophase of sleep stages of DS mice would also take longer to re-entrain to abrupt phase shifts of the LD cycle. DS and WT mice were implanted with ECoG/EMG electrodes, recorded from continuously for 3–5 days and subjected to either delay or advance jet lags (Figure 3). To our knowledge, continuous polysomnographic sleep recordings during single transient jet lags have not been performed in mice, so we first set out to determine the re-entrainment time of NREM and REM sleep stage acrophases following both phase delays and advances in WT mice.
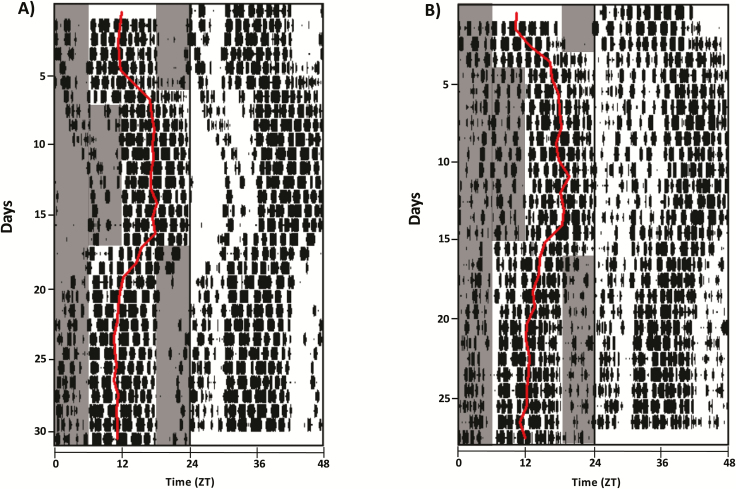
Figure 3.
Re-entrainment of WT and DS mice to delay and advance jet lags. (A) Representative double-plotted actogram of total sleep from a WT control mouse undergoing 30 days of continuous ECoG/EMG recording. Sleep acrophase for each day is overlaid in red on only one side of the actogram. All animals were monitored for 3–5 days under baseline conditions, and then subjected to either delay or advance jet lags. Shaded areas indicate periods of lights off, which are only indicated on the left side of the plots for easy visualization. (B) Same as (A) for one DS mouse.
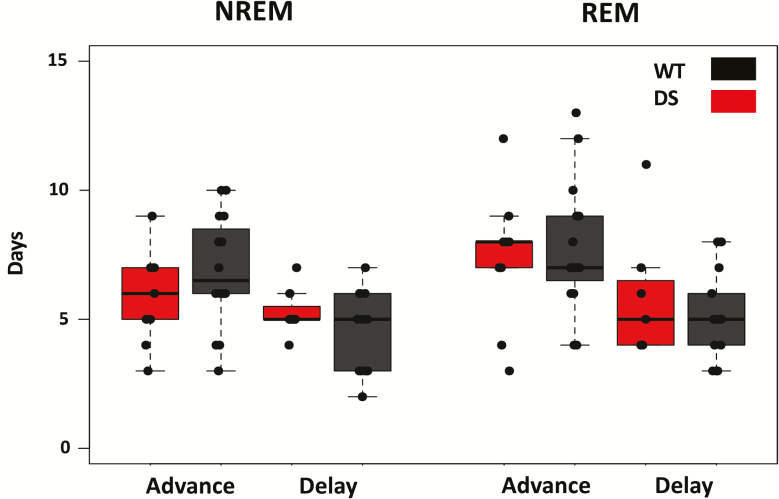
We find that following phase delays, WT NREM and REM sleep took an average of 4.5 ± 0.4 and 5.1 ± 5 days to re-entrain, respectively (n = 14). Following phase advances, NREM and REM re-entrained in 6.9 ± 0.5 and 7.8 ± 0.6 days, respectively (n = 16). Figure 3A shows a representative double-plotted actogram from one WT mouse, with acrophase overlaid in red on the left side. DS mice displayed comparable re-entrainment times for both delays (NREM = 5.3 ± 0.4 days, REM = 5.9 ± 1 days, n = 7) and advances (NREM = 5.9 ± 6 days, REM = 7.3 ± 0.9 days, n = 9). Three-way ANOVA with genotype, sleep stage and jet lag direction as factors reveals an effect of jet lag direction (F1,84 = 27.93, p < 0.001), where advances took longer to re-entrain to than delays, but no difference between genotypes (F1,84 = 0.003, p = 0.96) and no effect of the interaction (Figure 4). Interestingly, there was a trend for NREM sleep to re-entrain faster than REM sleep that reached the borderline of statistical significance (F1,84 = 3.9, p = 0.051).
Figure 4.
Sleep stage acrophase takes longer to re-entrain to phase advances than delays, and DS mice display normal re-entrainment when compared to WT littermates. Time to re-entrainment is calculated as the number of days necessary for sleep stage acrophase to return to mean baseline levels within a 95% confidence interval. Three-way ANOVA with genotype, sleep stage and jet lag direction as factors reveals a significant effect of jet lag condition (p < 0.001, F1,84 = 27.93), but no difference in re-entrainment time between sleep stages (p = 0.051, F1,84 = 3.9) or between genotypes (p = 0.96, F1,84 = 0.003), and no interactions. n = 14 WT and 7 DS mice for delays, 16 WT and 9 DS mice for advances.
These results indicate that in both WT and DS mice, sleep bouts take longer to re-entrain to advances than delays, as expected based on an abundance of previous work demonstrating the same result in mice using WRA. Furthermore, there was no significant difference in re-entrainment time between NREM and REM sleep stages, unlike previous demonstrations of this phenomenon from similar experiments in the rat [37]. Finally, we found that despite differences in re-entrainment times as measured by WRA [26], there was no difference in re-entrainment time of sleep bouts between WT and DS mice in our ECoG experiments.
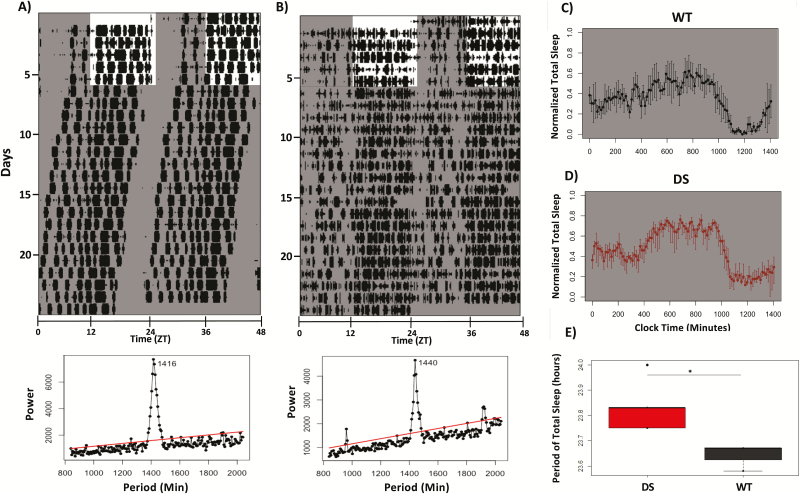
DS mice have an elongated circadian period of sleep
We predicted that like WRA, DS mice would display an elongated circadian period of sleep relative to WT controls. To test this prediction, we recorded sleep from WT and DS mice continuously for at least 14 days under conditions of constant darkness (Figure 5, A and B). We found that the circadian period of total sleep time in DD is significantly longer in DS (23.83 ± 0.04 hours, n = 5) than WT (23.64 ± 0.03 hours, n = 3) mice (two-tailed Student t-test, p = 0.01; Figure 5E). However, this difference was smaller than that measured from WRA [26]. To determine if there were any differences in the circadian period of NREM and REM sleep between and within genotypes, we also performed a two-way ANOVA with genotype and sleep stage as factors. While we found a strong effect of genotype (F1,17 = 19.96, p < 0.001), we found no effect of sleep stage (F1,17 = 0.15, p = 0.86) or the interaction (F1,17 = 0.5, p = 0.62; data not shown). These results suggest that, in agreement with previous findings [46], the rhythm of both NREM and REM sleep are under strong control of the circadian clock in free-running conditions.
Figure 5.
Elongated circadian period of sleep in DS mice under constant conditions. (A) Top panel: representative double-plotted actogram of total sleep bouts from a WT mouse. Shaded bars indicate times of lights off. Bottom panel: Sokolov-Bushell periodogram calculated for mouse above revealed an endogenous period of 23.6 hours. Red line indicates threshold of significance for the measured period. WT mice had a mean period of 23.64 hours. (B) Same as (A) for one DS mouse with period of 24 hours. Mean period for DS mice was 23.87 hours. (C) Mean waveform of sleep bouts under constant darkness for WT mice. (D) Same as (C) for DS mice. (E) DS mice have a significantly longer period of total sleep in constant darkness than WT mice (two-tailed Student t-test, p = 0.01, n = 3 WT mice, 5 DS mice).
DS mice are more likely to have an absent or fragmented daily “siesta”
The “siesta” is a brief bout of sleep during the active period commonly observed to occur in WT mice [46–50]. Recent work has demonstrated that the daily timing of the siesta is under control of vasoactive intestinal polypeptide (VIP) neurons in the SCN [50], suggesting it as an important output of the circadian system’s regulation of sleep. Our continuous ECoG/EMG recording paradigm offered a unique opportunity to directly assess how the timing of the siesta in mice responds to manipulations of the LD cycle, and to determine if the siesta is disrupted in DS mice.
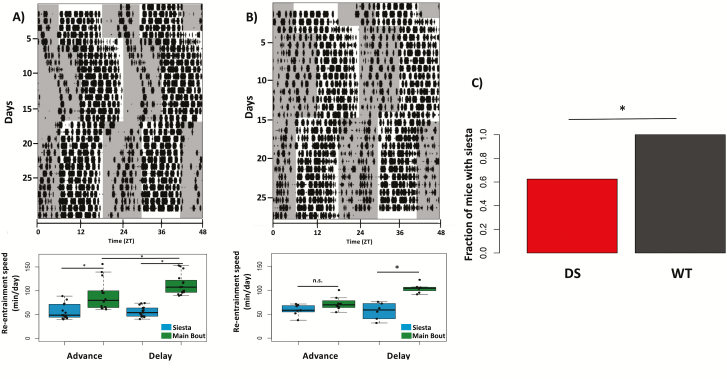
We first asked whether DS mice also display a consolidated siesta akin to that seen in WT mice (Figure 6). We found that DS mice were nearly 40% less likely to have a consolidated siesta than WT controls (Chi-squared test, χ 2(1) = 4.768, p = 0.02; Figure 6B, top left panel).We then set out to determine if there were any differences between the re-entrainment dynamics of the siesta and the main bout of sleep following phase shifts in WT mice. Here, we define the siesta as a brief (<2 hours) bout of sleep occurring primarily during the active period (dark phase) of the LD cycle that is visually discernible from the main bout and occurs with a regular onset each day. We first calculated the re-entrainment speed in minutes per day for both sleep bouts individually by extracting the slope of a line fitted to sleep offset for delays and onset for advances using actograms of sleep by manual scorers blind to experimental conditions. In WT mice, we find that re-entrainment of the siesta is slower than the primary bout of sleep for both advances and delays (two-way ANOVA: sleep bout [F1,54 = 65.88, p < 0.001], jet lag condition [F1,54 = 3.953, p = 0.052] and effect of the interaction [F1,54 = 4.023, p < 0.05], n = 14 mice for delays, 16 for advances; Figure 6A).
Figure 6.
Re-entrainment dynamics of primary sleep bout and siesta. (A) Top panel: Representative actogram of total sleep from WT mouse used for siesta scoring. Bottom panel: Re-entrainment speed (minutes/day) of the primary sleep bout and siesta, a daytime bout of sleep under control of the circadian clock, was calculated by extracting the slope of a line fitted to sleep offset for delays and onset for advances by a manual scorer blind to experimental conditions. In WT mice, re-entrainment of siesta is slower than the primary sleep bout (two-way ANOVA, effect of sleep bout [p < 0.001] and interaction with jet lag condition [p = 0.049], n = 14 mice for delays, 16 for advances). (B) Same as (A), for DS mouse. Tukey post hoc comparisons for DS mice revealed a significant difference in re-entrainment time between main bout and siesta following delay (p < 0.05) but not advance jet lags (p = 0.39). (C) DS mice are less likely to have a consolidated siesta than WT controls (Chi-squared test, χ 2(1) = 4.768, p = 0.02).
For those DS mice that did display a siesta, a two-way ANOVA with Tukey post hoc comparisons revealed a significant difference in RS between the siesta and main sleep bout following delays (p < 0.001), but not advances (p = 0.25; Figure 6B, bottom left panel). Therefore, we conclude that DS mice have a disrupted daily siesta relative to their WT counterparts.
Discussion
Here we show that DS mice have impairments in the circadian regulation of sleep including fragmented rhythm of NREM sleep, elongated endogenous period of sleep, and a disrupted daily siesta. These findings are consistent with our previous characterization of circadian wheel-running behavioral deficits in DS mice and suggest that a global heterozygous loss-of-function mutation in the NaV1.1 sodium channel is sufficient to cause disturbances in the circadian regulation of sleep. However, in contrast to our previous report, both NREM and REM sleep acrophase in DS mice re-entrain normally to phase shifts of the LD cycle simulating jet lag. Moreover, while the endogenous period of both WRA and sleep was significantly longer in DS mice, the magnitude of this difference is more pronounced in WRA than sleep [26]. This could be in part due to the fact that DS mice were previously documented to display less WRA in general than WT mice.
Despite these discrepancies, our results are consistent with reports of disturbed sleep in DS patients [14, 15]. We found that the endogenous period of both NREM and REM sleep is longer in DS mice than their WT counterparts. As noted previously [26], the elongation of circadian period in DS mice is striking, as period period changes are typically only seen in clock gene or clock-related gene mutants such as Clock [51], Rorβ [52], and CK1ε [53]. A similar phenomenon in DS patients could explain complaints of delayed sleep onset and difficulty maintaining sleep, as a longer endogenous period of sleep would result in a delayed phase of circadian entrainment. DS mice also showed rhythm fragmentation in NREM but not REM sleep. We hypothesize that disturbances to NREM sleep we report here and have reported previously [28] are caused by reduced excitability of GABAergic TRN neurons as a result of heterozygous deletion of Scn1a. Of note, recent work has demonstrated that another mouse model of DS displays hyperexcitability of TRN neurons due to compensation by calcium-activated SK potassium channels, leading to excessive burst firing and non-convulsive seizures [54]. However, the aforementioned DS mouse contains a human Scn1a mutation and is bred on a different genetic background than the DS model presented here, suggesting a number of different explanations for the discrepancies in TRN properties. Still, these results raise the possibility that cellular and/or network changes outside the TRN may contribute to the NREM sleep disturbances observed in DS. Interestingly, although interictal spiking during NREM sleep in DS mice has been associated with increased awakenings [28], we observed no correlation between interictal spiking frequency and both NREM IV and WASO. While we cannot completely rule out the contribution of epileptic activity to sleep disturbances in DS mice, our data suggest that epileptic activity identified at the recording sites in our experimental setup alone is unlikely to fully account for the fragmentation of sleep rhythms in DS. It is possible that the combination of epileptic activity and sleep rhythm fragmentation with a potentially independent etiology leads to the frequent nighttime awakenings reported by DS patients.
While most patients diagnosed with DS have a mutation in the SCN1A gene, these mutations exist on a spectrum, and the type and severity of symptoms seen in patients with DS and other SCN1A-related epilepsies can vary widely [18]. For example, a mouse model of generalized epilepsy with febrile seizures plus (GEFS+), which is the result of a different Scn1a mutation, was shown to have reduced NREM and REM sleep and normal homeostatic sleep rebound, in contrast to this and previous descriptions of sleep in the DS mouse studied here [27]. Continuing to characterize sleep and other co-morbidities in these different pre-clinical models of SCN1A-related epilepsies could be informative in helping to identify and more effectively address different manifestations of symptoms in patients.
The contradictions between the sleep phenotype found in this study and previously identified circadian behavioral phenotypes underscore the value of combining long-term ECoG/EMG recording and activity monitoring in disentangling different behavioral outputs of the circadian clock, particularly in pre-clinical models of disease. It is well-documented that DS mice are susceptible to thermally-induced seizures [19, 21], and that exercise such as wheel-running increases core body temperature. The risk of thermally induced seizures could deter DS mice from running on wheels. Furthermore, like many DS patients, DS mice show spasticity [23], which could reduce their ability to run on a wheel. Similarly, a seizure during WRA might cause premature termination, whereas a seizure before WRA might prevent initiation of a bout of running. Although it remains to be determined why it is clear that DS mice show reduced wheel-running and this could, in turn, affect their ability to achieve normal circadian phase resetting of WRA [26]. Therefore, existing circadian behavioral impairments in DS mice may appear exacerbated when measuring rest-activity cycles by WRA, as opposed to measuring home cage activity or ECoG/EMG.
There is evidence to suggest that the circadian deficits in DS mice are the result of reduced GABAergic neurotransmission caused by the loss-of-function mutation in the NaV1.1 channel, and thus impaired communication between SCN subregions [26]. The mutation reduces excitability of GABAergic neurons specifically [19], and GABA has been proposed as a critical component in coupling both SCN subregions [55], and individual SCN neuronal oscillators [55–57]. This network integrity is key to the maintenance of rhythmic sleep-wake behaviors of the type observed here [45]. Despite this, the possibility remains that neuronal damage sustained as a result of chronic seizures may also contribute to sleep phenotypes. The SCN projects to multiple known sleep-wake maintenance centers [45], as well as to neurons that drive endocrine, metabolic, and autonomic functions that affect sleep [34]. Depending on the frequency, severity and origin point of seizures, the SCN and its myriad downstream targets may be differentially affected. Though we are unaware of any evidence that seizures in DS directly affect the integrity of these areas, future work could incorporate histological evaluation of DS mouse brain tissue to determine if and how such centers are affected. This could also help to explain the high degree of inter-individual differences seen in circadian sleep behaviors in DS mice, such as rate of re-entrainment to jet lag (Figure 4). Although such an analysis is beyond the scope of the present study, previous histological work has confirmed a reduction in the expression of the NaV1.1 channel in GABAergic neurons in the SCN [26], TRN [28], hippocampus [19], cortex [19, 20, 24] and sleep-related hypothalamic nuclei [26, 27] of DS mice, suggesting the Scn1a+/- mutation is highly likely to contribute to the sleep phenotypes observed here. Of note, our previous study on WRA in DS mice used animals that were typically older than then ones used in the current study; some of the circadian deficits observed in WRA and not in the regulation of polysomnographically recorded sleep could be also due to the history of seizures that could presumably have cumulative neuronal damage in SCN targets. Future work on sleep-related co-morbidities of DS should employ region-specific targeting of the Scn1a+/- mutation to the SCN and its downstream targets—many of which also richly express the NaV1.1 channel [27]—in order to characterize their respective contributions to the phenotypes observed in the DS mouse.
While this study is not the first to employ polysomnographic recordings of this length in rodents [58], our results join a short but growing list of successes in using long-term ECoG/EMG recording to study the circadian regulation of sleep. A series of studies from our lab employed long-term continuous sleep recording in rats to demonstrate that under lighting conditions that cause internal circadian desynchrony, the daily rhythms of NREM and REM sleep become uncoupled [36], and that the daily timing of REM sleep specifically is associated with an oscillator in the dorsomedial subregion of the SCN [37]. In mice, long-term sleep recordings have been used to characterize temporal distributions and re-entrainment dynamics of sleep following chronic environmental circadian disruption in the form of serial advance jet lags over 3 months [59], as well as chronic sleep disturbances that occur under LD cycles that cause circadian misalignment [60], highlighting the value of this approach.
In addition to the identification of circadian sleep regulation deficits in DS, our study offers a thorough characterization of re-entrainment dynamics of sleep following a single phase shift of the LD cycle simulating jet lag in WT mice, as well as, to our knowledge, the first direct observation of the effects of LD cycle manipulation on the daily siesta. Interestingly, we found no significant difference in re-entrainment time between NREM and REM sleep stages. This is in contrast to results from similar experiments in the rat, which demonstrated that REM sleep takes longer to re-entrain to delay jet lags than NREM, and that the timing of REM is associated with the activity of the dorsomedial SCN [37]. A recent study found that under 20-hour LD cycles causing circadian desynchrony in the mouse, REM sleep increases during the dark phase during misaligned days (coinciding with the subjective day), suggesting that REM sleep timing is regulated primarily by the ~24-hour circadian clock, and is dissociated from the LD cycle [60]. This is in agreement with our previous finding in rats that under 22-hour LD circadian forced desynchrony, REM is dissociated from LD cycle and displays an endogenous period of approximately 25 hours [36]. Future work seeking to reconcile these findings on the role of the circadian clock in the timing of REM sleep in mice could combine continuous sleep recording and a serial jet lag protocol as used by Casiraghi et al. [61] to examine both re-entrainment dynamics and the resulting period of sleep stages.
Both our protocol and Casiraghi et al.’s serial jet lag paradigm could also be useful in further characterizing the mechanisms underlying both the timing and the re-entrainment dynamics of the siesta, which recent evidence suggests is under control of VIP neurons in the ventral SCN [50]. We previously demonstrated reduced signal propagation from the VIP-rich ventral SCN to the dorsal SCN resulting in poor network connectivity in DS mice [26], which could help to explain the disturbed siesta phenotype reported here. However, NREM sleep rhythm fragmentation was observed in DS mice, and because NREM sleep comprises a sizable portion of the daily siesta, it is possible that the absence or fragmentation of siesta in DS mice is an epiphenomenon of NREM fragmentation in general. Still, the difference in re-entrainment dynamics of siesta, but not the main sleep bout, between WT and DS mice that display a consolidated siesta, as well as the complete absence of siesta in some animals, suggest that while NREM fragmentation likely contributes, the siesta phenotype cannot be explained by this fragmentation alone. Interestingly, a recent study using a mouse model of Angelman syndrome with a maternal deletion of the Ube3A gene, which is expressed in the SCN, found that the daily siesta is lost while circadian rhythmicity remains intact [48], suggesting the siesta may also be regulated by the sleep homeostat. Yet another report found that mice with a deficiency in serotonin synthesis exhibit both abnormal circadian behavior and a complete lack of siesta [49]. Taken together, these data suggest a combination of circadian and homeostatic components are at play in timing the siesta and highlight the need for further study of its regulation.
Long-term sleep experiments of the kind performed here and in previous studies also offer a framework in which to study diseases with circadian and sleep-related co-morbidities where long-term polysomnographic studies are not possible in patients. Additionally, these approaches would be invaluable in determining both efficacy and mechanisms of interventions to improve sleep hygiene, and in turn, the core symptoms of these diseases. Pharmacological imposition of sleep improved both circadian and cognitive outcomes in a mouse model of Huntington’s disease [62, 63]. Non-pharmacological behavioral interventions using circadian entraining cues to improve the temporal organization of sleep-wake cycles have also proven effective in treating disease. For example, social rhythm therapy has shown success in improving both sleep hygiene and symptoms in patients with mood disorders [64], and temporally restricting feeding has been shown to reduce seizure frequency and severity in a rat model of pilocarpine-induced epilepsy [65]. Combining these interventions with continuous sleep monitoring can offer greater insight into how sleep timing and quality is affected and provide clues into how these changes may result in improvement of disease symptoms.
Acknowledgments
We thank Franck Kalume for guidance in quantifying epileptiform interictal events, Gideon Dunster for help with analyzing actograms, Tenley Weil for assistance in manual sleep scoring, and all members of the de la Iglesia lab for thoughtful discussions in performing this study.
Conflict of interest statement. None declared.
Funding
This work was supported by US National Institutes of Health Research Grant R01 NS094211 to H.O.D., US National Institutes of Health Research Grant R01 NS025704 to W.A.C. and a Washington Research Foundation Innovation Graduate Fellowship in Neuroengineering to R.S.
References
- 1. Bazil CW. Epilepsy and sleep disturbance. Epilepsy Behav. 2003;4(Suppl 2):S39–S45. [DOI] [PubMed] [Google Scholar]
- 2. Steriade M. Sleep, epilepsy and thalamic reticular inhibitory neurons. Trends Neurosci. 2005;28(6):317–324. [DOI] [PubMed] [Google Scholar]
- 3. Malow BA. Sleep deprivation and epilepsy. Epilepsy Curr. 2004;4(5):193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shouse MN, et al. . Circadian rhythm, sleep, and epilepsy. J Clin Neurophysiol. 1996;13(1):32–50. [DOI] [PubMed] [Google Scholar]
- 5. Hofstra WA, et al. . The circadian rhythm and its interaction with human epilepsy: a review of literature. Sleep Med Rev. 2009;13(6):413–420. [DOI] [PubMed] [Google Scholar]
- 6. Cho CH. Molecular mechanism of circadian rhythmicity of seizures in temporal lobe epilepsy. Front Cell Neurosci. 2012;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirzoev A, et al. . Circadian profiles of focal epileptic seizures: a need for reappraisal. Seizure. 2012;21(6):412–416. [DOI] [PubMed] [Google Scholar]
- 8. Sánchez Fernández I, et al. . Clinical evolution of seizures: distribution across time of day and sleep/wakefulness cycle. J Neurol. 2013;260(2):549–557. [DOI] [PubMed] [Google Scholar]
- 9. Karoly PJ, et al. . The circadian profile of epilepsy improves seizure forecasting. Brain. 2017;140(8):2169–2182. [DOI] [PubMed] [Google Scholar]
- 10. Dravet C, et al. . Severe myoclonic epilepsy in infancy: Dravet syndrome. Adv Neurol. 2005;95:71–102. [PubMed] [Google Scholar]
- 11. Dravet C. The core Dravet syndrome phenotype. Epilepsia. 2011;52(Suppl 2):3–9. [DOI] [PubMed] [Google Scholar]
- 12. Licheni SH, et al. . Sleep problems in Dravet syndrome: a modifiable comorbidity. Dev Med Child Neurol. 2018;60(2):192–198. [DOI] [PubMed] [Google Scholar]
- 13. Schoonjans AS, et al. . More daytime sleepiness and worse quality of sleep in patients with Dravet Syndrome compared to other epilepsy patients. Eur J Paediatr Neurol. 2019;23(1):61–69. [DOI] [PubMed] [Google Scholar]
- 14. Nolan K, et al. . Coping with a child with Dravet syndrome: insights from families. J Child Neurol. 2008;23(6):690–694. [DOI] [PubMed] [Google Scholar]
- 15. Villas N, et al. . Dravet syndrome: characteristics, comorbidities, and caregiver concerns. Epilepsy Behav. 2017;74:81–86. [DOI] [PubMed] [Google Scholar]
- 16. Losito E, et al. . Age-related “Sleep/nocturnal” tonic and tonic clonic seizure clusters are underdiagnosed in patients with Dravet Syndrome. Epilepsy Behav. 2017;74:33–40. [DOI] [PubMed] [Google Scholar]
- 17. Claes L, et al. . De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68(6):1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Catterall WA, et al. . NaV1.1 channels and epilepsy. J Physiol. 2010;588(Pt 11):1849–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu FH, et al. . Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9(9):1142–1149. [DOI] [PubMed] [Google Scholar]
- 20. Ogiwara I, et al. . Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27(22):5903–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oakley JC, et al. . Temperature- and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc Natl Acad Sci U S A. 2009;106(10):3994–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalume F, et al. . Reduced sodium current in Purkinje neurons from Nav1.1 mutant mice: implications for ataxia in severe myoclonic epilepsy in infancy. J Neurosci. 2007;27(41):11065–11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han S, et al. . Autistic-like behaviour in Scn1a+/- mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489(7416):385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tatsukawa T, et al. . Impairments in social novelty recognition and spatial memory in mice with conditional deletion of Scn1a in parvalbumin-expressing cells. Neurobiol Dis. 2018;112:24–34. [DOI] [PubMed] [Google Scholar]
- 25. Kalume F, et al. . Sudden unexpected death in a mouse model of Dravet syndrome. J Clin Invest. 2013;123(4):1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han S, et al. . NaV1.1 channels are critical for intercellular communication in the suprachiasmatic nucleus and for normal circadian rhythms. Proc Nat Acad Sci U S A. 2012b;109:E368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papale LA, et al. . Altered sleep regulation in a mouse model of SCN1A-derived genetic epilepsy with febrile seizures plus (GEFS+). Epilepsia. 2013;54(4):625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalume F, et al. . Sleep impairment and reduced interneuron excitability in a mouse model of Dravet Syndrome. Neurobiol Dis. 2015;77:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheah CS, et al. . Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A. 2012;109(36):14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Catterall WA. Dravet syndrome: a Sodium Channel Interneuronopathy. Curr Opin Physiol. 2018;2:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borbély AA, et al. . Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14(6):557–568. [DOI] [PubMed] [Google Scholar]
- 32. Luppi PH, et al. . Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med Rev. 2017;32:85–94. [DOI] [PubMed] [Google Scholar]
- 33. Scammell TE, et al. . Neural Circuitry of Wakefulness and Sleep. Neuron. 2017;93(4):747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mistlberger RE. Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus. Brain Res Brain Res Rev. 2005;49(3):429–454. [DOI] [PubMed] [Google Scholar]
- 35. Hsu YA, et al. . The dorsal medial habenula minimally impacts circadian regulation of locomotor activity and sleep. J Biol Rhythms. 2017;32(5):444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cambras T, et al. . Circadian desynchronization of core body temperature and sleep stages in the rat. Proc Natl Acad Sci U S A. 2007;104(18):7634–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee ML, et al. . Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus. Curr Biol. 2009;19(10):848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee ML, et al. . Fragmentation of rapid eye movement and nonrapid eye movement sleep without total sleep loss impairs hippocampus-dependent fear memory consolidation. Sleep. 2016;39(11):2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esteller R, et al. Line length: an efficient feature for seizure onset detection. Engineering in Medicine and Biology Society, 2001. Proceedings of the 23rd Annual International Conference of the IEEE. 2. 1707–1710 vol.2. doi:10.1109/IEMBS.2001.1020545.
- 40. Sokolove PG, et al. . The chi square periodogram: its utility for analysis of circadian rhythms. J Theor Biol. 1978;72(1):131–160. [DOI] [PubMed] [Google Scholar]
- 41. Sudbrack-Oliveira P, et al. . Sleep architecture in adults with epilepsy: a systematic review. Sleep Med. 2019;53:22–27. [DOI] [PubMed] [Google Scholar]
- 42. Wobbrock JO, et al. . The Aligned Rank Transform for nonparametric factorial analyses using only ANOVA procedures. In: Proceedings of the ACM Conference on Human Factors in Computing Systems; May 7–12, 2011; Vancouver, British Columbia, Canada. ACM Press, pp. 143–146. [Google Scholar]
- 43. Witting W, et al. . Alterations in the circadian rest-activity rhythm in aging and Alzheimer’s disease. Biol Psychiatry. 1990;27(6):563–572. [DOI] [PubMed] [Google Scholar]
- 44. Duhart JM, et al. . Circadian alterations in a murine model of hypothalamic glioma. Front Physiol. 2017;8:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Albers HE, et al. . The dynamics of GABA signaling: revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front Neuroendocrinol. 2017;44:35–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tankersley CG, et al. . Circadian rhythm variation in activity, body temperature, and heart rate between C3H/HeJ and C57BL/6J inbred strains. J Appl Physiol (1985). 2002;92(2):870–877. [DOI] [PubMed] [Google Scholar]
- 47. Stowie AC, et al. . Longitudinal study of changes in daily activity rhythms over the lifespan in individual male and female C57BL/6J Mice. J Biol Rhythms. 2015;30(6):563–568. [DOI] [PubMed] [Google Scholar]
- 48. Ehlen JC, et al. . Maternal Ube3a loss disrupts sleep homeostasis but leaves circadian rhythmicity largely intact. J Neurosci. 2015;35(40):13587–13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whitney MS, et al. . Adult brain serotonin deficiency causes hyperactivity, circadian disruption, and elimination of siestas. J Neurosci. 2016;36(38):9828–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Collins B, et al. . A Population of VIPergic Clock Neurons in the Suprachiasmatic Nucleus Consolidate Daily Siesta Sleep. In: Presentation at Society for Research on Biological Rhythms, May 2018; Amelia Island, FL.
- 51. Vitaterna MH, et al. . Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. André E, et al. . Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17(14):3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meng QJ, et al. . Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58(1):78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ritter-Makinson S, et al. . Augmented reticular thalamic bursting and seizures in Scn1a-Dravet syndrome. Cell Rep. 2019;26(1):54–64.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Albus H, et al. . A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol. 2005;15(10):886–893. [DOI] [PubMed] [Google Scholar]
- 56. Liu C, et al. . GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25(1):123–128. [DOI] [PubMed] [Google Scholar]
- 57. Welsh DK, et al. . Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Richardson GS, et al. . Circadian rhythms of sleep and wakefulness in mice: analysis using long-term automated recording of sleep. Am J Physiol. 1985;248(3 Pt 2):R320–R330. [DOI] [PubMed] [Google Scholar]
- 59. Brager AJ, et al. . Sleep loss and the inflammatory response in mice under chronic environmental circadian disruption. PLoS One. 2013;8(5):e63752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hasan S, et al. . Effects of circadian misalignment on sleep in mice. Sci Rep. 2018;8(1):15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Casiraghi L, et al. . Effects of chronic forced circadian desynchronization on body weight and metabolism in male mice. Physiol Rep. 2016;4(8):e12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pallier PN, et al. . Pharmacological imposition of sleep slows cognitive decline and reverses dysregulation of circadian gene expression in a transgenic mouse model of Huntington’s disease. J Neurosci. 2007;27(29):7869–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pallier PN, et al. . Management of sleep/wake cycles improves cognitive function in a transgenic mouse model of Huntington’s disease. Brain Res. 2009;1279:90–98. [DOI] [PubMed] [Google Scholar]
- 64. Haynes PL, et al. . Social rhythm therapies for mood disorders: an update. Curr Psychiatry Rep. 2016;18(8):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Landgrave-Gómez J, et al. . Anticonvulsant effect of time-restricted feeding in a pilocarpine-induced seizure model: metabolic and epigenetic implications. Front Cell Neurosci. 2016;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]