Abstract
Long non-coding RNAs (lncRNAs) have crucial roles in lncRNAs in periodontal development and disorders of this tissue. A number of lncRNAs especially those regulating immune responses contribute in the pathophysiology of periodontitis. In the current case-control study, we assessed expression levels of two immune response-related lncRNAs namely the antisense non-coding RNA in the INK4 locus (ANRIL) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in gingival tissues and blood samples of patients with periodontitis and healthy subjects. Expression of ANRIL was significantly lower in peripheral blood of patients compared with controls (Posterior Beta RE = -1.734, P value = 0.035). However, when diving study participants based on their gender, no significant difference was found between patients and sex-matched controls. Expression of this lncRNA was not different between periodontitis tissues and normal tissues. Expression of MALAT1 was not different between samples obtained from cases and controls. Tissue or blood expressions of ANRIL or MALAT1 were not correlated with age of either patients or controls. There were significant correlations between expression levels of ANRIL and MALAT1 in gingival tissues both in cases (r = 0.62, P < 0.0001) and in controls (r = 0.37, P < 0.0001). However, blood levels of these lncRNAs were not correlated with each other either in cases or in controls. Most notably, there was no significant correlation between expression levels of these lncRNAs in gingival tissues and in the blood of study participants. The current study indicates dysregulation of ANRIL in the peripheral blood of patients with periodontitis in spite of its normal levels in gingival tissues which might reflect disturbance in systemic immune responses in these patients.
Keywords: Periodontitis, ANRIL, MALAT1, lncRNA
Highlights
-
•
Expression of ANRIL was significantly lower in peripheral blood of patients with periodontitis compared with controls.
-
•
Expression of ANRIL was not different between periodontitis tissues and normal tissues.
-
•
Expression of MALAT1 was not different between samples obtained from cases and controls.
-
•
Tissue or blood expressions of ANRIL or MALAT1 were not correlated with age.
1. Introduction
Periodontal disease is a common multifactorial disease that affects nearly all ages, with some groups being more susceptible. The main risk factors are smoking, poor dental care, systemic diseases such as diabetes, certain drugs, age and stress. Besides, genetic factors are regarded as contributing factors in its pathogenesis. Based on its high prevalence, periodontal disease is regarded as a public health problem [1]. The induction of inflammatory reaction to bacteria in the dental biofilm is the main pathogenic event in the periodontitis. Although certain microorganisms are linked with the progressive forms of this disorder; the existence of these microbes in persons with no sign of periodontitis implies that periodontitis is caused as the consequence of the inflammatory responses, not the sole existence of the microorganisms [2]. Regulation of immune responses is a complicated process in which several coding and non-coding genes participate [3]. Among non-coding RNAs whose roles in the pathogenesis of immune-related disorders have been identified are metastasis associated lung adenocarcinoma transcript 1 (MALAT1) [4,5] and antisense non-coding RNA in the INK4 locus (ANRIL) [6]. MALAT1 has been shown to be over-expressed patients with systemic lupus erythematous (SLE) patients [4]. Notably, MALAT1 knock down considerably decreased the expression of IL-21 in primary monocytes of these patients. Additional studies have shown that MALAT1 role in the pathogenesis of SLE is exerted through regulation of SIRT1 signaling [4], a pathway which is probably important in the reactive oxygen species homeostasis in the process of development of periodontitis [7]. Moreover, MALAT1 has been upregulated in response to lipopolysaccharide (LPS) [8]. Meanwhile, LPS has a pivotal role in the pathophysiology of periodontitis [9] in a way that severe periodontitis stimulates macrophage functions through this substance [10]. Expression of MALAT1 has been up-regulated in primary human gingival fibroblasts obtained from patients with periodontitis compared with controls. This lncRNA also enhances expression of inflammatory cytokines through sponging miR‐20a and releasing toll like receptor 4 [11].
ANRIL has a prominent role in the pathogenesis of immune-related disorders including coronary artery disease (CAD) [12], type 2 diabetes [13] and cancers [14] as revealed by genome wide association studies. Most notably, the genomic locus for this lncRNA has been identified as a risk locus for periodontitis by various research groups [[15], [16], [17]]. This lncRNA modulates immune response through interaction with the Yin Yang 1 protein [18], a transcription repressor which contribute in the regulation of immune reactions [19]. Certain polymorphisms within ANRIL have been associated with levels of C reactive protein (CRP) in CAD patients. Meanwhile, levels of this inflammation mediator have been correlated with severe periodontitis in these patients [20]. Moreover, expression of this lncRNA has been induced in gingival tissues following bacterial infection [21].
Although the role of lncRNAs in the pathophysiology of periodontitis has been uncovered [22], data regarding expression pattern of ANRIL or MALAT1 in gingival tissues or peripheral blood of patients with periodontitis is scarce. Thus, in the current study, we investigated expression of these lncRNAs in these two sets of samples obtained from patients with periodontitis and healthy subjects.
2. Material and methods
2.1. Enrolled individuals
Tissue samples were obtained during surgical procedure. Cases had the following criteria: chronic periodontitis (Stage II to IV) with at least two remaining periodontal pockets in each sextant after nonsurgical periodontal treatment, probing depth of 5 mm or greater, bleeding on probing (BOP), and at least 3 mm of attachment loss needing surgical periodontal treatment [23]. Moreover, they were older than 18 years and had at least 16 teeth. Exclusion criteria were smoking, systemic diseases, history of consumption of antibiotic or anti-inflammatory drugs 3 months prior to surgical procedures, pregnancy and breastfeeding. Diagnosis of periodontitis was based on the clinical and radiographic examinations performed by a periodontist. Control samples were obtained from BOP sites of patients who underwent crown lengthening. The sites were examined by a periodontist and sites with no BOP and less than 3 mm probing depths were included. The study protocol was approved by ethical committee of Shahid Beheshti University of Medical Sciences (Ethic Code: IR.SBMU.DRC.REC.1398.086).
2.2. Expression assays
Total RNA was extracted from both tissue and blood specimens by using Hybrid-RTM blood RNA extraction kit (GeneAll, Seoul, South Korea) according to the protocol provided by the company. The, cDNA was produced from RNA using the OneStep RT-PCR Series Kit (BioFact™, Seoul, South Korea). Relative expressions (RE) of ANRIL and MALAT1 were measured in all specimens using the RealQ Plus 2x PCR Master Mix Green Without ROX™ PCR Master Mix (Ampliqon, Odense, Denmark). Reactions were conducted in StepOnePlus™ RealTime PCR equipment (Applied Biosystems, Foster city, CA, USA) in duplicate. B2M gene was used as normalizer. Table 1 shows the sequences of primers and length of PCR products.
Table 1.
Sequences of primers and length of PCR products.
| lncRNA | Primer | Sequence | Product length |
|---|---|---|---|
| ANRIL | Forward Reverse |
TGCTCTATCCGCCAATCAGG GCGTGCAGCGGTTTAGGTTT |
108 bp |
| MALAT1 | Forward Reverse |
GACGGAGGTTGAGATGAAGC ATTCGGGGCTCTGTAGTCCT |
84 bp |
| B2M | Forward Reverse |
AGATGAGTATGCCTGCCGTG GCGGCATCTTCAAACCTCCA |
105 bp |
2.3. Statistical methods
Transcript quantities of ANRIL and MALAT1 were compared between periodontitis patients and healthy subjects using Bayesian regression model. The effects of independent variables were adjusted. Statistical analyses were performed in R 3.6.2 software, Rstan, ggplot 2 & non-parametric quantile regression packages. Bootstrap method and 100 iteration methods were used. Correlations between expressions of lncRNAs were evaluated through calculation of Spearman correlation coefficients.
3. Results
3.1. General characteristics of periodontitis patients and controls
The study included tissue samples from 30 patients with periodontitis (19 females, 11 males) and 30 controls (14 females, 16 males). We also gathered blood samples from 23 patients and 18 healthy controls. Table 2 summarizes the demographic data of periodontitis patients and controls.
Table 2.
General data of periodontitis patients and controls.
| Patients | Healthy controls | ||
|---|---|---|---|
| Total Number of Tissues | 30 | 30 | |
| Sex | Female | 19 | 14 |
| Male | 11 | 16 | |
| Age (mean ± SD) | 41.28 ± 2.7 | 38.8 ± 1.9 | |
| Total Number of Blood Samples | 23 | 18 | |
| Sex | Female | 15 | 11 |
| Male | 8 | 7 | |
| Age (mean ± SD) | 39.65 ± 3.1 | 37.91 ± 2.8 | |
3.2. Expression assays
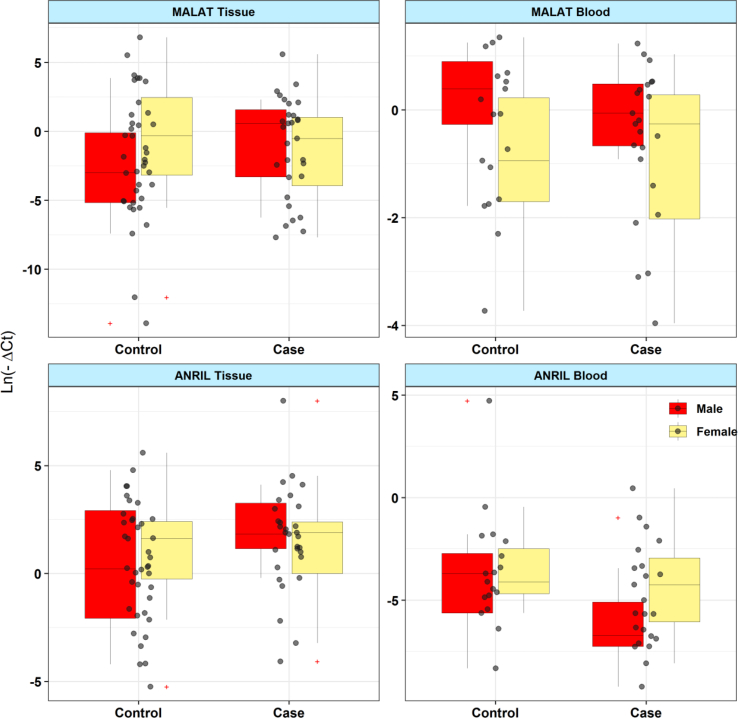
Fig. 1 shows relative expression (RE) of ANRIL and MALAT1 in tissue and blood samples obtained from patients and controls.
Fig. 1.
Relative expression (RE) of ANRIL and MALAT1 in tissue and blood samples obtained from patients and controls.
Expression of ANRIL was significantly lower in peripheral blood of patients compared with controls (Posterior Beta of RE = -1.734, P value = 0.035). However, when diving study participants based on their gender, no significant difference was found between patients and sex-matched controls (Posterior Beta of RE [95% CI] = -2.537 [-7.43, 2.59], P value = 0.043 for males; Posterior Beta of RE [95% CI] = -1.62 [-3.44, 0.45], P value = 0.209 for females). Expression of this lncRNA was not different between periodontitis tissues and normal tissues (Posterior Beta of RE = 0.549, P value = 0.12). This pattern was also seen among male subgroups (Posterior Beta of RE = 1.256, P value = 0.504) and among female subgroups (Posterior Beta of RE = 0.4, P value = 0.568). There was no correlation between tissue/blood levels of this lncRNA and age in either subgroups (Table 3).
Table 3.
Relative expression of ANRIL in tissues and blood specimens of periodontitis patients compared with controls (RE: relative expression, SE: standard error, CrI: credible interval).
| Tissue |
Blood |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior Beta of RE | SE | P-Value | 95% CrI for RE | Posterior Beta of RE | SE | P-Value | 95% CrI for RE |
| Total | Case/Control | 0.549 | 0.684 | 0.12 | [-0.71, 1.98] | −1.734 | 0.871 | 0.035 | [-3.46, -0.03] |
| Gender (F/M) | 0.195 | 0.673 | 0.522 | [-1.22, 1.42] | 1.282 | 0.815 | 0.482 | [-0.29, 2.92] | |
| Age (year) | 0.021 | 0.032 | 0.635 | [-0.04, 0.08] | 0.056 | 0.073 | 0.255 | [-0.08, 0.2] | |
| Male | Case/Control | 1.256 | 1.272 | 0.504 | [-1.07, 3.87] | −2.537 | 2.538 | 0.043 | [-7.43, 2.59] |
| Age (year) | 0.074 | 0.054 | 0.141 | [-0.03, 0.18] | 0.051 | 0.13 | 0.33 | [-0.19, 0.31] | |
| Female | Case/Control | 0.4 | 0.778 | 0.568 | [-1.06, 2.01] | −1.62 | 0.995 | 0.209 | [-3.44, 0.45] |
| Age (year) | −0.024 | 0.039 | 0.287 | [-0.11, 0.05] | 0.119 | 0.082 | 0.109 | [-0.04, 0.28] |
Expression of MALAT1 was not different between blood/gingival tissues of patients compared with controls (Posterior Beta of RE = 0.939, P value = 0.404 for tissues; Posterior Beta of RE = 0.055, P value = 0.605 for blood samples). Assessment of its expression levels in sex-based groups revealed no significant difference either between male patient and male controls (Posterior Beta of RE = 2.436, P value = 0.116 for tissues; Posterior Beta of RE = 0.595, P value = 0.64 for blood samples) or between female patients and female controls (Posterior Beta of RE = -0.161, P value = 0.88 for tissues; Posterior Beta of RE = 0.043, P value = 0.64 for blood samples) (Table 4).
Table 4.
Relative expression of MALAT1 in tissues and blood specimens of periodontitis patients compared with controls (RE: relative expression, SE: standard error, CrI: credible interval).
| Tissue |
Blood |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior Beta of RE | SE | P-Value | 95% CrI for RE | Posterior Beta of RE | SE | P-Value | 95% CrI for RE |
| Total | Case/Control | 0.939 | 1.264 | 0.404 | [-1.6, 3.4] | 0.055 | 0.494 | 0.605 | [-0.89, 1.04] |
| Gender (F/M) | 1.651 | 1.067 | 0.038 | [-0.46, 3.64] | −0.463 | 0.376 | 0.091 | [-1.27, 0.24] | |
| Age (year) | −0.006 | 0.061 | 0.608 | [-0.12, 0.11] | −0.017 | 0.028 | 0.676 | [-0.07, 0.04] | |
| Male | Case/Control | 2.436 | 1.887 | 0.116 | [-1.33, 5.98] | 0.595 | 1.2 | 0.64 | [-1.67, 2.98] |
| Age (year) | −0.018 | 0.079 | 0.734 | [-0.17, 0.15] | −0.046 | 0.055 | 0.782 | [-0.15, 0.06] | |
| Female | Case/Control | −0.161 | 1.589 | 0.88 | [-3.2, 2.96] | 0.043 | 0.654 | 0.64 | [-1.26, 1.37] |
| Age (year) | −0.034 | 0.078 | 0.798 | [-0.18, 0.12] | −0.001 | 0.049 | 0.341 | [-0.1, 0.1] |
3.3. Correlation between expression levels of ANRIL and MALAT1 lncRNAs and age of enrolled individuals
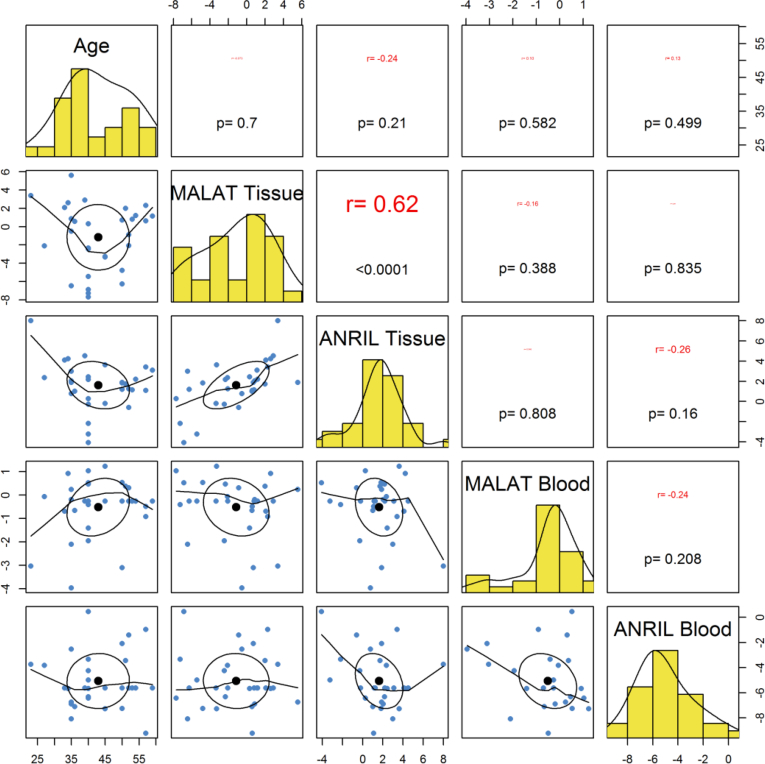
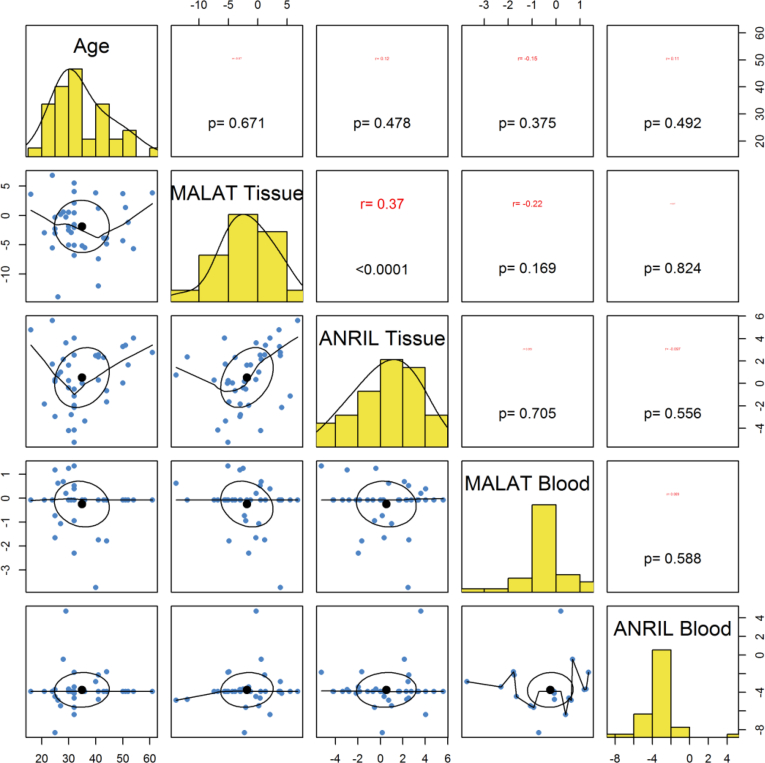
Tissue or blood expressions of ANRIL or MALAT1 were not correlated with age of either patients or controls (Fig. 2, Fig. 3). There were significant correlations between expression levels of ANRIL and MALAT1 in gingival tissues both in cases (r = 0.62, P < 0.0001) and in controls (r = 0.37, P < 0.0001). However, blood levels of these lncRNAs were not correlated with each other either in cases (r = -0.24, P = 0.208) or in controls (P = 0.588). Most notably, there was no significant correlation between expression levels of these lncRNAs in gingival tissues and in the blood of study participants (P = 0.556 for ANRIL and P = 0.388 for MALAT1) (Fig. 2, Fig. 3).
Fig. 2.
Correlation between expression levels of lncRNAs and age of periodontitis patients.
Fig. 3.
Correlation between expression levels of lncRNAs and age of controls.
4. Discussion
In the present study, we investigated expression of two lncRNAs in peripheral blood and affected gingival tissues of patients with periodontitis and healthy subjects. Expression of ANRIL was significantly lower in peripheral blood of patients compared with controls. However, expression of this lncRNA was not different between periodontitis tissues and normal tissues. This lncRNA has been recognized as a shared locus for both periodontitis and CAD [17]. Consistent with the mentioned results, a recent study demonstrated down-regulation of ANRIL in peripheral blood of CAD patients compared with healthy subjects [24]. Another study showed down-regulation of the main endothelial cell-associated transcript of ANRIL in CAD coronary arteries compared with non-CAD arteries, thus indicating the protective role for ANRIL against CAD [25]. Our present results also imply a protective role for this lncRNA against periodontitis, a disorder which has been linked with CAD in both genetic risks [17] and mechanisms [9]. Furthermore, the rs1333048 polymorphism within ANRIL has been associated with higher plasma levels of C reactive protein (CRP) in patients with periodontitis [26]. Notably, CRP is regarded a marker for CAD as well [27].
Expression of MALAT1 was not different between tissue or blood samples obtained from cases and controls. This lncRNA has been demonstrated to modulate immune responses [3]. Its silencing has enhanced expression of TNF-α and IL-6 cytokines [8]. In addition, MALAT1 has an inhibitory effect on NF-κB signaling, pathway that regulates several aspects of inflammatory responses [8]. This lncRNA also modulates macrophage activation to induce differentiation of M2 macrophages [28]. Notably, in silico analysis of miRNA and mRNA expression profiles has shown the role of MALAT1 in construction of the lncRNA-associated competing endogenous RNA network of periodontitis [22]. Our result was not consistent with the supposed role of this lncRNA in the pathogenesis of immune-related disorders. This might indicate different roles of this lncRNA in the pathogenesis of different immune-related disorders.
Besides, tissue or blood expressions of ANRIL or MALAT1 were not correlated with age of either patients or controls. Thus, if future studies reveal the biomarker role for ANRIL, as expression of this lncRNA is not influenced by the age, this biomarker can be used for follow-up of patients during long periods of time.
There were significant correlations between expression levels of ANRIL and MALAT1 in gingival tissues both in cases and in controls. However, blood levels of these lncRNAs were not correlated with each other either in cases or in controls. These observations may indicate the presence of tissue-specific regulatory mechanisms or interaction networks between these lncRNAs which should be explored in future studies. Consistent with this hypothesis, there was no significant correlation between expression levels of these lncRNAs in gingival tissues and in the blood of study participants.
The current study indicates dysregulation of ANRIL in the peripheral blood of patients with periodontitis in spite of its normal levels in gingival tissues which might reflect disturbance in systemic immune responses in these patients. Most notably, this finding is in line with the role of ANRIL in the pathogenesis of CAD, a disease which is genetically and mechanistically related with periodontitis.
CRediT authorship contribution statement
Leila Gholami: Data curation. Soudeh Ghafouri-Fard: Writing - original draft. Sara Mirzajani: Methodology. Shahram Arsang-Jang: Formal analysis, Data curation. Mohammad Taheri: Supervision. Zahra Dehbani: Formal analysis, Data curation. Safoora Dehghani: Methodology. Behzad Houshmand: Data curation. Reza Amid: Data curation. Arezou Sayad: Supervision. Bahareh Shams: Data curation.
Declaration of competing interest
The authors declare they have no conflict of interest.
Acknowledgement
The current study was supported by a grant from Shahid Beheshti University of Medical Sciences.
Contributor Information
Arezou Sayad, Email: ar.sayad@yahoo.com.
Bahareh Shams, Email: Baharshams2044@gmail.com.
References
- 1.Nazir M.A. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int. J. Health Sci. 2017;11(2):72. [PMC free article] [PubMed] [Google Scholar]
- 2.Cekici A., Kantarci A., Hasturk H., Van Dyke T.E. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol. 2000;64(1):57–80. doi: 10.1111/prd.12002. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadjicharalambous M.R., Lindsay M.A. Long non-coding RNAs and the innate immune response. Noncoding RNA. 2019;5(2):34. doi: 10.3390/ncrna5020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H., Liang N., Wang M., Fei Y., Sun J., Li Z. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget. 2017;8(44) doi: 10.18632/oncotarget.20490. 77400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghafouri-Fard S., Ashrafi Hafez A., Taheri M. Metastasis Associated Lung Adenocarcinoma Transcript 1: an update on expression pattern and functions in carcinogenesis. Exp. Mol. Pathol. 2020;112 doi: 10.1016/j.yexmp.2019.104330. [DOI] [PubMed] [Google Scholar]
- 6.Mirza A.H., Berthelsen C.H., Seemann S.E., Pan X., Frederiksen K.S., Vilien M. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7(1):39. doi: 10.1186/s13073-015-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C., Mo L., Niu Y., Li X., Zhou X., Xu X. The role of reactive oxygen species and autophagy in periodontitis and their potential linkage. Front. Physiol. 2017;8:439. doi: 10.3389/fphys.2017.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao G., Su Z., Song D., Mao Y., Mao X. The long noncoding RNA MALAT 1 regulates the lipopolysaccharide‐induced inflammatory response through its interaction with NF‐κB. FEBS Lett. 2016;590(17):2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 9.Liljestrand J.M., Paju S., Buhlin K., Persson G.R., Sarna S., Nieminen M.S. Lipopolysaccharide, a possible molecular mediator between periodontitis and coronary artery disease. J. Clin. Periodontol. 2017;44(8):784–792. doi: 10.1111/jcpe.12751. [DOI] [PubMed] [Google Scholar]
- 10.Pussinen P.J., Vilkuna-Rautiainen T., Alfthan G., Palosuo T., Jauhiainen M., Sundvall J. Severe periodontitis enhances macrophage activation via increased serum lipopolysaccharide. Arterioscler. Thromb. Vasc. Biol. 2004;24(11):2174–2180. doi: 10.1161/01.ATV.0000145979.82184.9f. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Wang M., Song L., Wang X., Lai W., Jiang S. Lnc RNA MALAT 1 regulates inflammatory cytokine production in lipopolysaccharide‐stimulated human gingival fibroblasts through sponging miR‐20a and activating TLR 4 pathway. J. Periodontal. Res. 2019 doi: 10.1111/jre.12700. [DOI] [PubMed] [Google Scholar]
- 12.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taheri M., Ghafouri-Fard S. Antisense non-coding RNA in the INK4 locus (ANRIL) in human cancers. Int. J. Canc. Manag. 2018;11(10) [Google Scholar]
- 15.Schaefer A.S., Bochenek G., Manke T., Nothnagel M., Graetz C., Thien A. Validation of reported genetic risk factors for periodontitis in a large-scale replication study. J. Clin. Periodontol. 2013;40(6):563–572. doi: 10.1111/jcpe.12092. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer A.S., Richter G.M., Dommisch H., Reinartz M., Nothnagel M., Noack B. CDKN2BAS is associated with periodontitis in different European populations and is activated by bacterial infection. J. Med. Genet. 2011;48(1):38–47. doi: 10.1136/jmg.2010.078998. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer A.S., Richter G.M., Groessner-Schreiber B., Noack B., Nothnagel M., El Mokhtari N.E. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5(2) doi: 10.1371/journal.pgen.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X., Han X., Wittfeldt A., Sun J., Liu C., Wang X. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016;13(1):98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J., He Y., Chen J., Zeng Z., Yang B., Ou Q. A critical role of transcription factor YY1 in rheumatoid arthritis by regulation of interleukin-6. J. Autoimmun. 2017;77:67–75. doi: 10.1016/j.jaut.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Reichert S., Seitter L., Schaller H.-G., Schlitt A., Schulz S. ANRIL polymorphisms (rs1333049 and rs3217992) in relation to plasma CRP levels among in-patients with CHD. Cytokine. 2020;127 doi: 10.1016/j.cyto.2019.154932. 154932. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer A.S., Richter G.M., Dommisch H., Reinartz M., Nothnagel M., Noack B. CDKN2BAS is associated with periodontitis in different European populations and is activated by bacterial infection. J. Med. Genet. 2011;48(1):38–47. doi: 10.1136/jmg.2010.078998. [DOI] [PubMed] [Google Scholar]
- 22.Li S., Liu X., Li H., Pan H., Acharya A., Deng Y. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J. Periodontal. Res. 2018;53(4):495–505. doi: 10.1111/jre.12539. [DOI] [PubMed] [Google Scholar]
- 23.Sayad A., Taheri M., Sadeghpour S., Omrani M.D., Shams B., Mirzajani S. Exploring the role of long non-coding RNAs in periodontitis. Meta Gene. 2020;24 [Google Scholar]
- 24.Ebadi N., Ghafouri-Fard S., Taheri M., Arsang-Jang S., Omrani M.D. Expression analysis of inflammatory response-associated genes in coronary artery disease. Arch. Physiol. Biochem. 2020:1–7. doi: 10.1080/13813455.2019.1708953. [DOI] [PubMed] [Google Scholar]
- 25.Cho H., Shen G.Q., Wang X., Wang F., Archacki S., Li Y. Long noncoding RNA ANRIL regulates endothelial cell activities associated with coronary artery disease by up-regulating CLIP1, EZR, and LYVE1 genes. J. Biol. Chem. 2019;294(11):3881–3898. doi: 10.1074/jbc.RA118.005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teeuw W.J., Laine M.L., Bizzarro S., Loos B.G. A lead ANRIL polymorphism is associated with elevated CRP levels in periodontitis: a pilot case-control study. PloS One. 2015;10(9) doi: 10.1371/journal.pone.0137335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auer J., Berent R., Lassnig E., Eber B. C-reactive protein and coronary artery disease. Jpn. Heart J. 2002;43(6):607–619. doi: 10.1536/jhj.43.607. [DOI] [PubMed] [Google Scholar]
- 28.Cui H., Banerjee S., Guo S., Xie N., Ge J., Jiang D. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI insight. 2019;4(4) doi: 10.1172/jci.insight.124522. [DOI] [PMC free article] [PubMed] [Google Scholar]