Abstract
Alterations in HOXB genes expression in breast cancer have been described and related to therapy response and disease progression. However, due to breast cancer complexity and heterogeneity, added to the use of different technical approaches, the observed expression profiles are sometimes contradictory. Here, we provided the analyses of HOXB7, HOXB8 and HOXB9 expression profiles in cell lines extensively used in the literature addressing the putative role of HOXB genes in breast cancer (MCF7, BT474, SKBR3, MDA231 and MDA468) and representative of the clinical breast cancer molecular subtypes (Luminal A, Luminal B, HER2+ and Triple-negatives Claudin-low/Basal), compared to a normal breast model (MCF10A), using quantitative-PCR (qPCR). This technique allows a very sensitive quantification of gene expression and was performed using the fluorophore SYBR Green in order to obtain the expression levels relative to a reference gene, GAPDH in this case. We showed that HOXB7 is upregulated in all breast cancer cells analyzed, while HOXB8 and HOXB9 are significantly upregulated in MCF7 (Luminal A), BT474 (Luminal B) and MDA231 cells (Triple-negative Claudin-low). In addition, we found that the magnitude of the upregulation is highly subtype-specific, being the HER2+ cells the model with lowest HOXB7 upregulation, presenting very low or even null expression for HOXB8 and HOXB9, respectively. These results are analyzed in more detail in “HOX genes function in Breast Cancer development” [1] and are potentially relevant for a better understanding of the molecular heterogeneity of breast cancer, in addition to be a valuable tool assisting researchers in the choice of the most suitable cell models to perform functional assays concerning HOXB7, HOXB8 and HOXB9 genes.
Keywords: HOXB genes, Breast cancer, qPCR, Cell lines, Molecular classification
Specifications table
| Subject | Cancer Research |
| Specific subject area | Molecular genetics of breast cancer |
| Type of data | Gene expression profiles of HOXB7, HOXB8 and HOXB9 in breast cancer cell lines |
| How data were acquired | The qPCR runs were made in CFX 96™ Real-Time PCR Detection System (Bio-Rad). The data analyses were performed in CFX Manager 3.1 (Bio-Rad) and Prism 8 (GraphPad) |
| Data format | Raw and analyzed data [2] Histograms |
| Parameters for data collection | The first parameters analyzed were the quality of the duplicate samples’ amplification and negative controls, as well as the melting curve pattern for each primer pair in order to guarantee that only the specific PCR product was amplified. Secondly, we made the correct threshold positioning into the exponential phase of the amplification curve. |
| Description of data collection | The raw data were accessed by CFX Manager 3.1 (Bio-Rad) software and a threshold line was put into the exponential phase of the amplification curve generating a Cycle Threshold (CT) number for each sample. The CT numbers were transferred to an Excel file for analyses using the formula: RATIO = E target–(CT sample target gene)/E GAPDH–(CT sample GAPDH), in which “E” is the primer pair efficiency and GAPDH is the reference gene. These data are available on Mendeley Data [2]. The statistical analyses were made with Prism 8 (GraphPad) using the unpaired T test with Welch's correction generating the histograms displayed on Fig. 1A–D. P-values were considered statistically significant when P ≤ 0.05. Data are presented as the mean ± SD of at least three independent experiments. |
| Data source location | This study was conducted at i3S (Institute for Innovation and Health Research) Porto - Portugal 41° 10′ 30.008′' N, 8° 36′ 12.488′' W |
| Data accessibility | Repository name: Mendeley Data Data identification number: 10.17632/v77kmkzj88.3 Direct URL to data: http://dx.doi.org/10.17632/v77kmkzj88.3 |
| Related research article | S.A. de Bessa Garcia, M. Araújo, T. Pereira, J. Mouta, R. Freitas, HOX genes function in Breast Cancer development, Biochim Biophys Acta Rev Cancer, 2020, doi:10.1016/j.bbcan.2020.188358. Epub ahead of print. |
Value of the data
-
•
The HOXB7, HOXB8 and HOXB9 expression profiles in breast cancer are contradictory in the literature due to disease complexity and technical issues. The data presented here cover these two points by analyzing the expression of these genes in breast cancer cell lines (MCF7, BT474, SKBR3, MDA231 and MDA468) representative of the four molecular subtypes (Luminal A, Luminal B, HER2+, Triple-negatives Claudin-low/Basal) in comparison with a normal cell model (MCF10A), using a very sensitive quantitative technique, the qPCR.
-
•
Researchers interested in the role of HOXB genes in breast cancer can use the data provided to pursue projects aiming to generate knowledge on the molecular networks involved in this highly heterogeneous disease as well as testing their value as therapeutic targets.
-
•
The HOXB7, HOXB8 an HOXB9 expression profiles in different breast cancer cell lines, and in a normal model, using sensitive quantitative techniques, are valuable tools shedding light into the most suitable cellular model to further address the function of these genes in vitro.
-
•
The cell lines analyzed in this work are extensively used in breast cancer research and allow comparisons with the basal expression profiles obtained by Hur et al. using reverse transcriptase PCR [3]. In addition, these cellular models were also used in studies in which HOXB7 expression was manipulated generating effects in the cells SKBR3 [4,7], MCF10A [5], MCF7 [6], BT474 [6], MDA231 [7], supporting their relevance for studies addressing the role of HOXB genes in breast cancer.
1. Data description
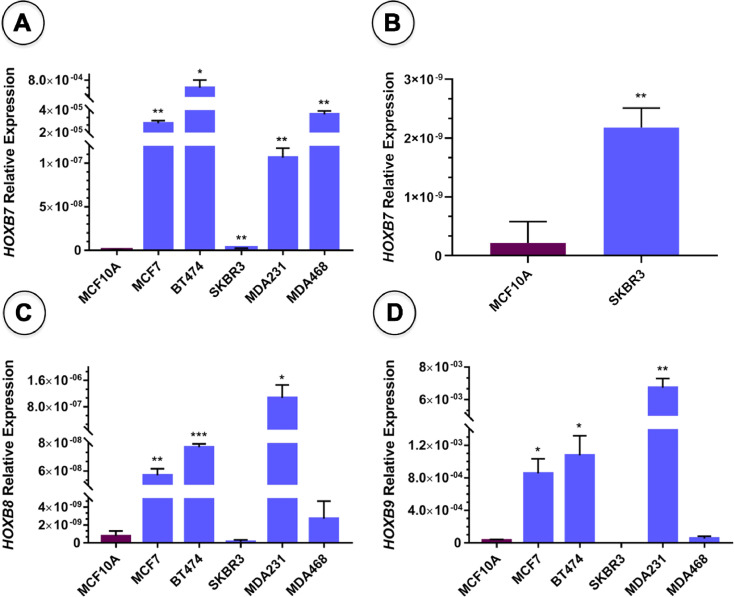
The total RNA extracted from one normal breast cell model (MCF10A) and five breast cancer cell lines (MCF7, BT474, SKBR3, MDA231 and MDA468) was reverse transcribed to cDNA and submitted to HOXB7, HOXB8 and HOXB9 relative expression analyses by qPCR. The respective raw and analyzed data are available on Mendley data (doi: 10.17632/v77kmkzj88.3) [2]. The data obtained after analyses show that HOXB7 tends to be upregulated in all breast cancer cell lines analyzed, when compared to the normal cell model (MCF10A) (Fig. 1A and B). However, its upregulation is subtype-dependent being particularly high in Luminal A and B cells (MCF7 and BT474, respectively) and Triple-negative Basal cells (MDA468). It is upregulated in moderate levels in Triple-negative Claudin-low cells (MDA231) and upregulated in low levels in HER2+ cells (SKBR3). In contrast, HOXB8 and HOXB9 are significantly upregulated in Triple-negative Claudin-low (MDA231) and Luminal cells (MCF7 and BT474) (Fig. 1C and D, respectively). Furthermore, these genes do not show expression differences in Triple-negative Basal cells (MDA468) when compared to MCF10A cells and HER2+ cells (SKBR3) present residual (HOXB8) or null expression (HOXB9).
Fig. 1.
HOXB7, HOXB8 and HOXB9 basal mRNA expression levels analyzed by qPCR in five breast cancer cell lines representing distinct molecular subtypes (blue bars), in comparison with normal breast cells (MCF10A, purple bars). Statistical analyses by unpaired T test with Welch's correction. Y-axis depicts the ratios of HOXB expression relative to GAPDH expression. *p-value<0.05, **p-value<0.01 and ***p-value <0.001 (A) HOXB7 is significantly upregulated in all breast cancer cell lines analyzed. (B) Higher magnification evidencing HOXB7 upregulation in SKBR3 cells. (C) HOXB8 is significantly upregulated in MCF7, BT474 and MDA231 cells. (D) xHOXB9 is significantly upregulated in MCF7, BT474 and MDA231 cells and silenced in SKBR3 cells.
2. Experimental design, materials, and methods
2.1. Cell lines
Five cell lines, representing distinct breast cancer molecular subtypes, were grown in 1× Dulbecco's Modified Eagle Medium supplemented with 10% Fetal Bovine Serum and 1× antibiotic solution penicillin-streptomycin (pen-strep, Gibco™): MCF7 (Luminal A), BT474 (Luminal B), SKBR3 (HER2+), MDA231 (Triple-negative, Claudin-low) and MDA468 (Triple-negative, Basal). Immortalized cells derived from normal human mammary cells, MCF10A, were used as non-malignant control. The base medium for this cell line was DMEM/F12 with the following additives: human EGF (20 ng/mL), human insulin (40 µg/mL), cholera toxin (100 ng/mL), hydrocortisone (500 ng/mL), 5% of horse serum and 1× penicillin-streptomycin (pen-strep, Gibco™) [8].
All cells were maintained in a humidified atmosphere with 5% CO2 and at 37 °C. Cells were used in experiments when reached 70–80% of confluence. The cell lines are periodically authenticated by the Genomics Core Facility at i3s (University of Porto) using the PowerPlexⓇ 16 HS System (Promega Corporation, #DC2100). Detection of the amplified fragments was made with automated capillary electrophoresis using 3130 Genetic Analyzer (Applied Biosystems) and the assignment of genotypes was performed in GeneMapper software v5.0 (Applied Biosystems).
2.2. cDNA synthesis, quantitative reverse-transcriptase PCR (qPCR) and statistical analyses
Total RNA extraction, from at least three independent experiments, was performed by TRIzolⓇ (Invitrogen) method accordingly to manufacturer's instructions and adding one more wash with ethanol 75%. For cDNA synthesis, 800 ng of RNA was subjected to reverse transcription, using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, #4368814), following the manufacturer's instructions. The qPCRs reactions were performed in duplicates and carried out in the real-time thermal cycler CFX96™ (Bio-Rad) using the iTaq™ Universal SYBRⓇ Green Supermix (Bio-rad, #1725121). Each amplification reaction contained 2 µL of the respective cDNA diluted 1:4; 0.5 µL of the forward primer; 0.5 µL of the reverse primer; 5 µL of 2X SYBR Green and 2 µL of H2O DNase/RNase free, resulting in a 10 µL reaction. For each primer pair we prepared a mix containing all cited components, except the cDNA that was added in the correspondent well of the 96-well plate. The mix was prepared in sufficient quantity to the samples in analysis for a specific primer in the plate and distributed accordingly (8 µL mix/sample). The analyses of the qPCRs were performed using the method described by Schmittgen and Livak [9] and normalized with the glyceraldehyde-3-phosphate dehydrogenate (GAPDH) gene amplification. The raw data were obtained after the run using the CFX Manager 3.1 software (Bio-Rad). The amplification plot for each sample and negative controls, as well as the melting curve for each primer pair that were analyzed in order to guarantee the amplification of only one product and a threshold line was put into the exponential phase of the amplification curve generating a Cycle threshold number (CT) for each sample. The CTs were transferred to an Excel file and the expression differences obtained by the formula: Ratio= E target–(CT sample target gene)/E GAPDH–(CT sample GAPDH), where E refers to primer efficiencies previously determined.
The statistical differences between breast cancer cells and MCF10A normal cells were determined by unpaired T test with Welch's correction using the Prism8 software (GraphPad). P-values were considered statistically significant when P ≤ 0.05. Data are presented as the mean ± SD of at least three independent experiments. The primers sequences are described in Table 1.
Table 1.
HOXB7, HOXB8, HOXB9 and GAPDH primer sequences, product length and annealing temperature. Primer sequences for HOXB7 and HOXB9 were obtained, respectively, from references [10] and [11]. HOXB8 and GAPDH primers were designed using primer3 online software [12]. Bp, base pair; FW, forward primer; RV, reverse primer.
| Sequence (5′-3′) | Product bp | Annealing temperature | ||
|---|---|---|---|---|
| HOXB7 | FW | TACCCCTGGATGCGAAGCTC | 171 | 60° |
| RV | AATCTTGATCTGTCTTTCCGTGA | |||
| HOXB8 | FW | GACAGGTCAAAATCTGGTTCC | 111 | |
| RV | GCTTCTGTTTCTCCAGCTCCT | |||
| HOXB9 | FW | CTACGGTCCCTGGTGAGGTA | 198 | |
| RV | TAATCAAAGACCCGGCTACG | |||
| GAPDH | FW | ACTGGCGTCTTCACCACCAT | 142 | |
| RV | TCTTGAGGCTGTTGTCATACTTC |
Acknowledgments
Acknowledgments
This work was supported by North Portugal Regional Operational Program (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) and by the European Regional Development Fund (ERDF) through the COMPETE – Operational Competitiveness Programme and POPH – Operational Human Potential Programme (NORTE-01-0145-FEDER-000029); and national funds through FCT–Foundation for Science and Technology (POCI-01-0145-FEDER-030562).
Conflict of Interest
The authors declare no competing financial interests or personal relationships that could influence the work reported in this paper.
References
- 1.de Bessa Garcia S.A. HOX genes function in breast cancer development. Biochim. Biophys. Acta Rev. Cancer. 2020 doi: 10.1016/j.bbcan.2020.188358. [DOI] [PubMed] [Google Scholar]
- 2.de Bessa Garcia S.A., Araújo M., Freitas R. Dataset of HOXB7, HOXB8 and HOXB9 RNA expression in breast cancer cell lines. Mendeley Data. 2020;(V3) doi: 10.17632/v77kmkzj88.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hur H. Analysis of HOX gene expression patterns in human breast cancer. Mol. Biotechnol. 2014;56(1):64–71. doi: 10.1007/s12033-013-9682-4. [DOI] [PubMed] [Google Scholar]
- 4.Care A. Transduction of the SKBR3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16(25):3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 5.Wu X. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66(19):9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 6.Jin K. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc. Natl. Acad. Sci. U.S.A. 2012;109(8):2736–2741. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S. HOXB7 promotes malignant progression by activating the TGF beta signaling pathway. Cancer Res. 2015;75(4):709–719. doi: 10.1158/0008-5472.CAN-14-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soule H.D. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086. [PubMed] [Google Scholar]
- 9.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 10.Ma R. HOXB7-S3 inhibits the proliferation and invasion of MCF-7 human breast cancer cells. Mol. Med. Rep. 2015;12(4):4901–4908. doi: 10.3892/mmr.2015.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha B. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279(19):3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Untergasser A. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]