Abstract
The SARS-CoV-2 virus has spread around the world. At this time, there is no vaccine that can help people prevent the spread of coronavirus.
We are proposing amantadine as a drug that can be used to mitigate the effects of the virus.
It is demonstrated by docking models how amantadine can exert its action on Coronavirus viroporin E.
Keywords: COVID-19, Amantadine, Viroporin, Docking
Introduction
At the end of December 2019, a new virus was announced that was beginning to spread in an alarming way in the city of Wuhan in China.
SARS-CoV-2, the coronavirus that causes COronaVIrus Disease 2019 (COVID-19), comes from the Coronoviridae family and the symptoms it causes in people who contract it are: sore throat, cough, fever, difficulty breathing in severe cases. The World Health Organization has estimated a mortality rate of 3.4%.
It is known that the virus has spread practically all over the world and so far, several laboratories are looking to develop a vaccine. However, this process could take 12 to 18 months. For this reason, it is important that the effects of COVID-19 can be mitigated when it is acquired by people.
Recent studies showed that the drug chloroquine, used against malaria, showed positive effects that mitigate the symptoms of COVID-19. [1]
It has been determined that possibly the mechanism of action of chloroquine is to block the proton pump that prevents the decrease of the pH in the digestive tract, which is important for the metabolism of the plasmodium. [2]
Studies have also shown that amantadine, together with chloroquine, is able to enhance the effect against plasmodium falciparum, mainly in chloroquine-resistant strains. [3]
Amantadine has been used as an antiviral therapy against influenza A, the mechanism that has been proposed is to block the early stage of viral replication. When the viral particle enters the cell, an endosome is formed, which has an acidic pH of 5. The proton channel (viroporin) is formed by the transport protein M2, which carries protons into the virion. This proton pump is necessary to interrupt the macromolecular interactions that keep the integrity of the virion. Amantadine is able to cross the membrane of the endosome and breaks the hydrogen bridges formed by Ala 30 and Gly 34 [4] in that way it can capture protons preventing them from entering the virion, preventing the release of the viral nucleus into the cell. [5]
COVID-19 ion channel structure
Protein E is an integral membrane protein of 75 amino acids, from the amino acids 15 to 39 an alpha-helix structure is formed. The other amino acids in terms of their secondary structure form coils.
Studies have shown that a lack of E-protein attenuates damage in vivo in coronavirus-infected mice. [6]
Using the ProtScale software, Kyte-Doolittle scale [7] shows a region that is very hydrophobic which indicates that it is part of an intramembrane region. NMR studies show that a 5-alpha-helix (viroporin) channel is formed [8].
Hypothesis
Amantadine blocks the viroporine channel of COVID-19, preventing the release of the viral nucleus into the cell cytoplasm.
Methodology
The E protein in FASTA format was obtained from the GeneBank with access number: YP_009724392.
The software ProtScale was used to corroborate the hydrophobic region of the alpha helix of protein E.
The Tel-Aviv University PatchDock software was used, which is an algorithm for molecular docking [9]. The COVID-19 protein E with ID number in protein databank 5X29 and the amantadine molecule obtained from DrugBank.ca were sent to the server in PDB format.
The results obtained were analyzed in the CLCbio Main Workbench 20 program of Qiagen and PyMol.
Results
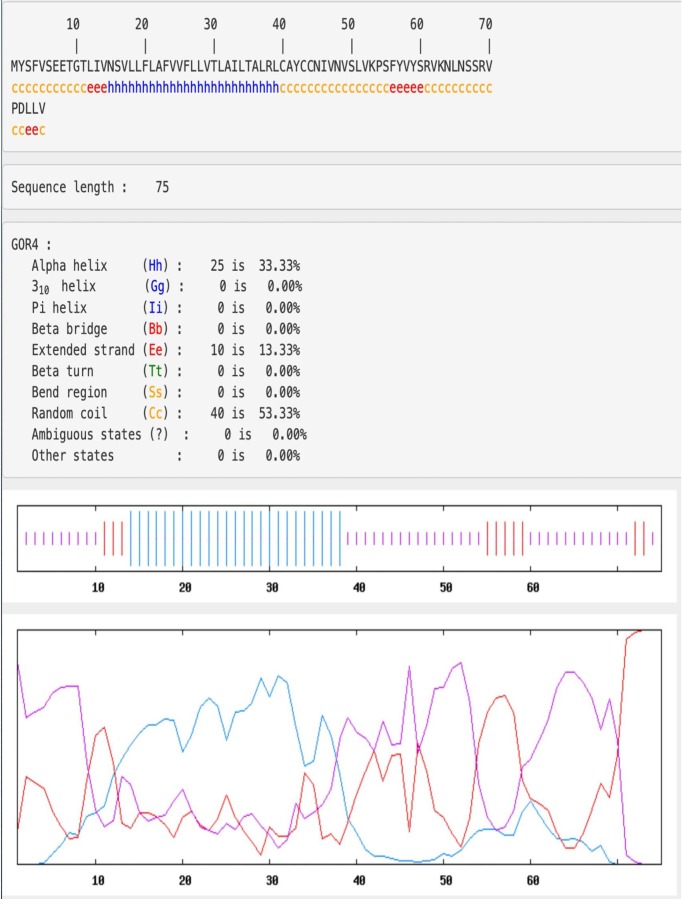
The Fig. 1 shows the primary structure of protein E in 1 letter code, the 75 amino acids are shown, where it is predicted that from the Met1 to Val14 amino acid has secondary coils structure. From the amino acid ASN15 to LEU39 a secondary alpha helix structure is formed. From amino acid Cys40 to Val75 form coils.
Fig. 1.
Primary structure of protein E and the proposed region where alpha-helix are formed.
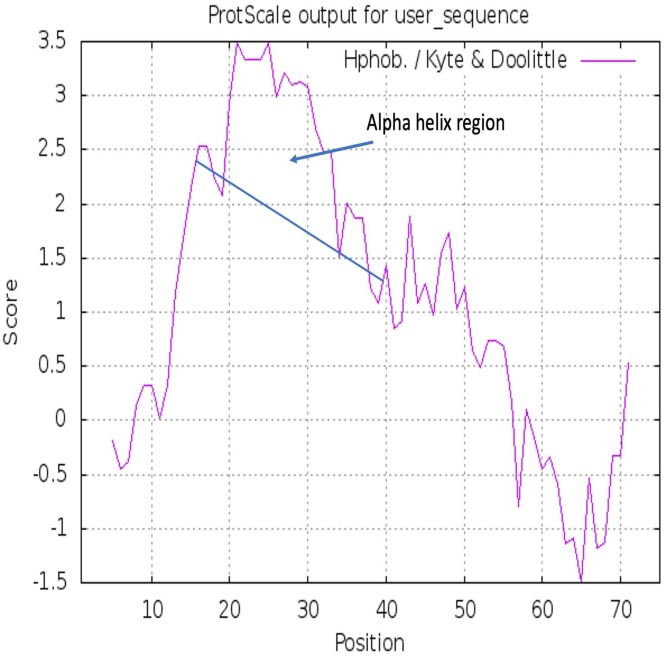
The Fig. 2 shows a hydrophobicity analysis using the Kyte-Doolittle scale, the degree of hydrophobicity shown from AA 15 to 39 is evident.
Fig. 2.
Hydrophobicity analysis using the Kyte-Doolittle scale.
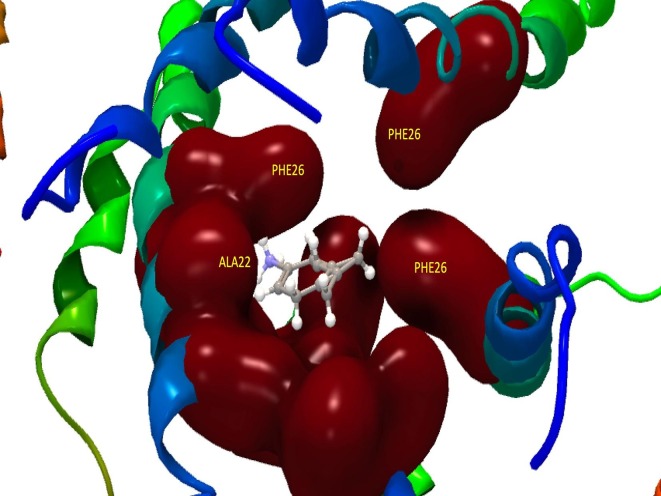
The Fig. 3 shows a model of protein E and amantadine where it is suggested that they could be interacting by hydrogen bridges with the amino acids PHE26 and ALA22.
Fig. 3.
Ribbons model of the channel formed by protein E and its possible interaction with amantadine in AA PHE26 and ALA22.
Fig. 4 shows a model where PHE26 and ALA22, which are part of the alpha-helices, could be forming hydrogen bridges with amantadine.
Fig. 4.
Ribbons model of the channel formed by the E protein and its possible interaction with amantadine in PHE26 and ALA22.
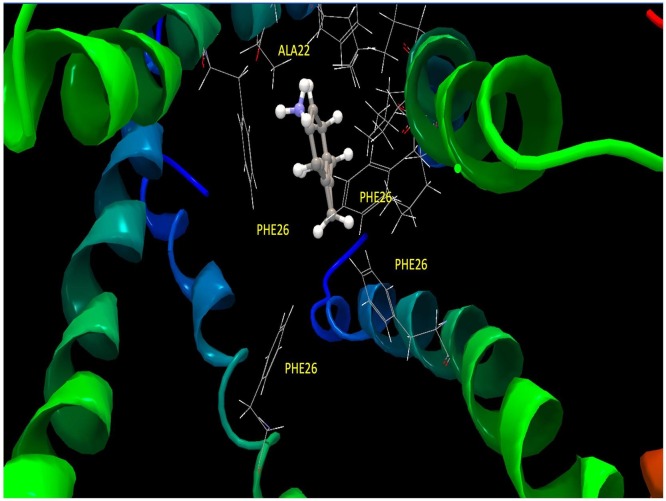
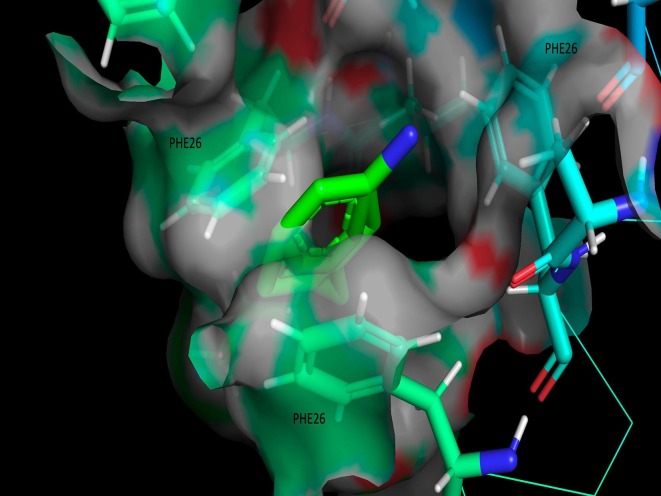
Fig. 5 shows the ligand site of amantadine, where it is shown that the PHE26 amino acids that are part of the alpha helix could be interacting with the ligand.
Fig. 5.
Region of the amantadine ligand-site.
Discussion
SARS-CoV-2 has spread rapidly around the world leaving thousands of people dead from lung problems.
That is the reason that laboratories have been experimenting to get a vaccine to help protect people from COVID-19.
However, the process of obtaining the vaccine can take more than 18 months.
Recently an article was published showing successful recovery of patients with COVID-19 when using chloroquine and azithromycin [1].
However, so far there are no models where the mode of action of chloroquine in the virus cycle could be suggested.
We are proposing a model where amantadine could enter the channel formed by the E-protein of the coronavirus, being able to break into the hydrogen bridges formed with water, in the same way as it does with the viroporin of the influenza virus [4].
A model has been proposed where amantadine is able to inhibit E-channel conductance in reconstituted lipid bilayers [10]. However, the primary sequence of COVID-19 has variations in its amino acids with respect to that analyzed by Torres et al.
We propose that the use of amantadine when the first symptoms of coronavirus disease occur could mitigate the effects of the disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Gautret P, Lagier JC, Parola P et al.Gautret P, Lagier JC, Parola P et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020105949. [DOI] [PMC free article] [PubMed]
- 2.Tang T., Xu W., Ma J. Inhibitory Mechanisms of DHA/CQ on pH and Iron Homeostasis of Erythrocytic Stage Growth of Plasmodium Falciparum. Molecules. 2019;24(10) doi: 10.3390/molecules24101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans S.G., Havlik IEvans S.G., Havlik I. Plasmodium falciparum: effects of amantadine, an antiviral, on chloroquine-resistant and -sensitive parasites in vitro and its influence on chloroquine activity. Biochem Pharmacol. 1993;45(5):1168–1170. doi: 10.1016/0006-2952(93)90264-w. [DOI] [PubMed] [Google Scholar]
- 4.Thomaston JL, Polizzi NF, Konstantinidi A et al.Thomaston JL, Polizzi NF, Konstantinidi A et al. Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters. J Am Chem Soc. 2018;140(45):15219-15226. [DOI] [PMC free article] [PubMed]
- 5.Wang C, Takeuchi K, Pinto LH et al.Wang C, Takeuchi K, Pinto LH et al. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67(9):5585-5594. [DOI] [PMC free article] [PubMed]
- 6.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML et al.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10(8):e1004320. [DOI] [PMC free article] [PubMed]
- 7.Kyte J., RFKyte Doolittle J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 8.Surya W., Li Y., Torres JSurya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta Biomembr. 2018;1860(6):1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneidman-Duhovny D, Inbar Y, Nussinov R et al.Schneidman-Duhovny D, Inbar Y, Nussinov R et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server issue):W363-7. [DOI] [PMC free article] [PubMed]
- 10.Torres J, Maheswari U, Parthasarathy K et al.Torres J, Maheswari U, Parthasarathy K et al. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16(9):2065-2071. [DOI] [PMC free article] [PubMed]