Highlights
-
•
Volumetric MRI classification models can detect symptomatic Alzheimer disease.
-
•
Detection improved by using multiple regions instead of hippocampal volume alone.
-
•
Detection not improved by including age or controlling for age-related atrophy.
-
•
Most useful when frontotemporal dementia or non-neurodegenerative diseases possible.
Keywords: Alzheimer disease, Volumetrics, Diagnostic biomarkers, MRI, Aging
Abbreviations: AD, Alzheimer disease; Adj, Adjusted for age-related atrophy; AUC, Receiver operating characteristics area under the curve; CDR, Clinical dementia rating; CI, Confidence interval; CN, Cognitively Normal; HCV, Hippocampal Volume; MMSE, Mini Mental State Exam; SARA, Select Atrophied Regions in Alzheimer disease; SUVR RSF, Standard Uptake Value Ratio (Regional spread function applied)
Abstract
Introduction
Volumetric biomarkers for Alzheimer disease (AD) are attractive due to their wide availability and ease of administration, but have traditionally shown lower diagnostic accuracy than measures of neuropathological contributors to AD. Our purpose was to optimize the diagnostic specificity of structural MRIs for AD using quantitative, data-driven techniques.
Methods
This retrospective study assembled several non-overlapping cohorts (total n = 1287) with publicly available data and clinical patients from Barnes–Jewish Hospital (data gathered 1990–2018). The Normal Aging Cohort (n = 383) contained amyloid biomarker negative, cognitively normal (CN) participants, and provided a basis for determining age-related atrophy in other cohorts. The Training (n = 216) and Test (n = 109) Cohorts contained participants with symptomatic AD and CN controls. Classification models were developed in the Training Cohort and compared in the Test Cohort using the receiver operating characteristics areas under curve (AUCs). Additional model comparisons were done in the Clinical Cohort (n = 579), which contained patients who were diagnosed with dementia due to various etiologies in a tertiary care outpatient memory clinic.
Results
While the Normal Aging Cohort showed regional age-related atrophy, classification models were not improved by including age as a predictor or by using volumetrics adjusted for age-related atrophy. The optimal model used multiple regions (hippocampal volume, inferior lateral ventricle volume, amygdala volume, entorhinal thickness, and inferior parietal thickness) and was able to separate AD and CN controls in the Test Cohort with an AUC of 0.961. In the Clinical Cohort, this model separated AD from non-AD diagnoses with an AUC 0.820, an incrementally greater separation of the cohort than by hippocampal volume alone (AUC of 0.801, p = 0.06). Greatest separation was seen for AD vs. frontotemporal dementia and for AD vs. non-neurodegenerative diagnoses.
Conclusions
Volumetric biomarkers distinguished individuals with symptomatic AD from CN controls and other dementia types but were not improved by controlling for normal aging.
1. Introduction
Typical Alzheimer disease (AD)-specific biomarkers rely on in vivo detection and quantification of amyloid-β and tau, AD's hallmark proteins. These biomarkers are increasingly used to narrow the differential diagnosis and refine the treatment of symptomatic patients in clinical practice (Rabinovici et al., 2019) based upon their appropriate use criteria (Johnson et al., 2013; Shaw et al., 2018). Despite this increased use, histological confirmation (the diagnostic reference standard) currently confirms an AD diagnosis for only 83% of AD patients at autopsy (Beach et al., 2012). In clinical trials, biomarkers can improve accuracy, utility, and cost effectiveness of screening, and can assess response to investigational therapies (Jack et al., 2018; Sevigny et al., 2016; Sperling et al., 2014). Their value is expected to further increase as AD-modifying therapies are realized, creating a need and rationale for population-level screening and an influx of patients requiring timely diagnosis and treatment (Liu et al., 2017).
Established AD biomarkers require cerebrospinal fluid analyses or positron emission tomography (PET) imaging, which are limited by expense and inaccessibility. In addition, PET imaging exposes patients to radioactivity and lumbar punctures may cause back pain, headache, and bleeding (Duits et al., 2016). These issues highlight the need for accessible, AD-specific biomarkers that can be applied to broad clinical populations. The solution may lie in brain magnetic resonance imaging (MRI), which is already standard of care in the United States for diagnostic evaluation of patients with new cognitive complaints (Knopman et al., 2001).
Current MRI biomarkers match the high accuracy of PET and cerebrospinal fluid markers in separating AD from unimpaired individuals (Frisoni et al., 2010; Morris et al., 2016). However, MRI biomarkers cannot maintain the high accuracy of amyloid biomarkers in cohorts of patients with various causes of dementia (Ossenkoppele et al., 2018; Wollman and Prohovnik, 2003). One thing likely impairing MRI-based biomarkers is the confounding influence of age-related brain atrophy or other undetected co-pathologies attributed to aging (Fotenos et al., 2005). For example, the confounding influence of preclinical AD in cohorts of cognitively normal (CN) older adults for other neuroimaging and cognitive measures has been shown previously (Brier et al., 2014; Hassenstab et al., 2016; Jack et al., 2014).
Taking this into account, we sought to optimize the diagnostic specificity of structural MRIs for AD using quantitative, data-driven techniques. Specifically, we considered whether individually adjusting volumetric measures for age-related atrophy would improve volumetric-based AD biomarkers. We compared unadjusted AD classification models to those using normal aging curves generated from CN participants free from biomarker evidence of AD. These models were validated in a research cohort with biomarker-confirmed AD and CN individuals and in a large clinical cohort containing patients with various neurodegenerative dementing diseases (including AD) who underwent MRI as part of their diagnostic evaluation for the cause of dementia.
2. Methods
2.1. Participants
The 1287 participants in the Normal Aging (n = 383), Training (n = 216), Test (n = 109), and Clinical (n = 579) Cohorts were composed from research studies or clinical patient records (collected 1990–2018). All procedures in this retrospective study were HIPAA compliant and approved by the Washington University Institutional Review Board. Informed consent was waived for the Clinical Cohort and gained for all others. Those not in the Clinical Cohort were from open source datasets and have been reported in various previous publications; the analyses of this paper and the inclusion of the participants in the Clinical Cohort are unique. All participants are described in Table 1, and Supplemental Table S1 gives these demographics separated by data source and diagnosis. All participants met the inclusion criteria described in Supplemental Table S2, which at minimum included a clinical assessment.
Table 1.
Demographics.
| Normal aging cohort | Training cohort | Test cohort | Clinical cohort – defined diagnosis | Clinical cohort – uncertain diagnosis | |
|---|---|---|---|---|---|
| n | 383 | 216 | 109 | 426 | 153 |
| n by Data Source | |||||
| DIAN | 134 | 0 | 0 | 0 | 0 |
| OASIS | 249 | 136 | 77 | 0 | 0 |
| ADNI | 0 | 80 | 32 | 0 | 0 |
| MDC | 0 | 0 | 0 | 426 | 153 |
| Diagnosis (% with symptomatic AD) | 0 | 43.5 | 43.1 | 61.5 | NA |
| Age (median) | 18–88 (60) | 57–88 (75) | 57–86 (74) | 46–88 (73) | 55–87 (73) |
| Sex (% Men) | 35.8 | 49.1 | 52.3 | 48.1 | 49.0 |
| CDR [0,0.5,1,2,3] | 383,0,0,0,0 | 122,43,44,5,2 | 62,17,26,4,0 | 50,235,97,26,0* | 8,122,10,3,0* |
| MMSE (median) | 24-30 (30)* | 7-30 (28) | 9-30 (28) | 1-30 (20)* | 1-30 (21)* |
| APOE (% with an E4 allele) | 27.9 | 51.6* | 39.4 | NA | NA |
| Amyloid mean cortical SUVR rsf – Centiloid (median) | −9.34–19.0 (−0.880)* | −8.40–154 (14.0) | −14.0–142 (11.4) | NA | NA |
| Race (% Caucasian) | 91.2* | 90.3 | 79.8 | 86.9 | 84.3 |
| Education (years) (median) | 9–22 (16)* | 7–24 (16) | 8–22 (16) | Median completed college* | Median completed college* |
Table 1 presents the demographic information for all cohorts. The Clinical Cohort has been separated into those given either an AD or non-AD diagnosis vs. those whose diagnosis was uncertain (and thus were not used to measure model accuracy).
A ‘*’ indicates missing data: 2 MMSEs, 124 Amyloids (all under age 45), and 6 Races from the Normal Aging Cohort; 1 APOE from the Training Cohort; 4 MMSEs, 18 CDRs, and 40 Educations from the Clinical Cohort – Defined Diagnosis; 1 MMSE, 10 CDRs, and 11 Educations from the Clinical Cohort – Uncertain Diagnosis
Abbreviations: AD – Alzheimer disease; ADNI – Alzheimer's Disease Neuroimaging Initiative (http://adni.loni.usc.edu); CDR – Clinical Dementia Rating; DIAN – Dominantly Inherited Alzheimer Network (https://dian.wustl.edu); MDC – Memory Diagnostic Center (http://memoryloss.wustl.edu/), soon to be available as OASIS-4; MMSE – Mini Mental State Exam; NA – Data Not Available; OASIS – Open Access Series of Imaging Studies – 3 (LaMontagne et al., 2019) (https://www.oasis-brains.org); SUVR rsf – Standard Uptake Value Ratio (Regional Spread Function applied)
The Normal Aging Cohort was restricted to CN participants who had a global Clinical Dementia Rating (CDR) of 0 (Morris, 1993) stable across longitudinal follow-up, and were free of substantial AD pathology as determined by a negative amyloid PET scan (defined in Section 2.2 Imaging). See Supplemental Table S2 for full inclusion criteria and Supplemental Figure S1 for distribution of ages. These participants were sourced from Open Access Series of Imaging Studies 3 (OASIS) and the Dominantly Inherited Alzheimer Network (DIAN). OASIS is an open-source dataset that is a retrospective compilation of data for >1000 participants. OASIS data was collected across several ongoing projects through the Knight Alzheimer Disease Research Center over the course of 30 years and includes CN controls and AD patients at various stages of impairment (LaMontagne et al., 2019). DIAN is a similar open source dataset that includes a greater number of younger controls due to its focus on dominantly inherited AD.
The Training and Test Cohorts contained CN and amyloid negative controls as well as cognitively impaired (CDR > 0) participants with a clinical diagnoses of AD and a PET scan indicating cerebral amyloidosis (full inclusion criteria in Supplemental Table S2). Participants were sourced from OASIS and the Alzheimer's Disease Neuroimaging Initiative (ADNI). ADNI is another open source dataset that includes several hundred healthy controls and AD patients at various stages of impairment from multiple sites across the United States. Two-thirds of the CN and symptomatic AD participants from these sources (not overlapping with the controls in the Normal Aging Cohort) were randomly assigned to the Training Cohort with the remaining one-third becoming the Test Cohort. Random assignment was done separately for the CN and AD participants to maintain equal distributions of AD diagnoses.
The Clinical Cohort drew from patients seen at the Washington University Memory Diagnostic Center outpatient clinic in Saint Louis, MO. Patients were split into symptomatic AD diagnoses and various non-AD diagnoses (including CN). See Supplemental Table S2 for full inclusion criteria. Patients listed separately as “Uncertain” (153/579) did not have an etiologic cause of dementia indicated; without this, they could not be used to test the classification models. The AD and non-AD groups were also split into groups of more specific diagnoses (Supplemental Table S3).
All participants underwent a clinical assessment conducted by experienced clinicians including a semi-structured interview with the participant and a knowledgeable collateral source as well as a thorough neurological examination (see Supplemental Table S2 for full inclusion criteria). A clinical diagnoses of dementia was considered at the conclusion of each assessment, integrating results from the clinical assessment and bedside measures of cognitive function (Day et al., 2017). Dementia severity was classified using the participant's CDR in accordance with established scoring rules (Morris, 1993). Etiologic diagnoses of dementia conformed to diagnostic criteria in use in clinical and research practices for AD (McKhann et al., 2011), dementia with Lewy bodies (McKeith et al., 2017), frontotemporal dementia (Rascovsky et al., 2011), and vascular cognitive impairment (Skrobot et al., 2018). See Supplemental Table S3 for a breakdown of specific diagnoses in the Clinical Cohort. Clinical diagnosis was made blinded to amyloid status in the OASIS participants, but not in the ADNI participants.
All CDRs and Mini Mental State Exams (MMSEs) (Folstein et al., 1975) used in this study occurred within a year of MRI, and those sourced from DIAN and ADNI all had a time difference of 0 days. Participants sourced from OASIS had an average time difference of 99 days and the Clinical Cohort had an average time difference of 118 days.
2.2. Imaging
All volumetric T1-weighted images underwent regional tissue segmentation with FreeSurfer 5.3 (freesurfer.net) using the Desikan-Killiany atlas (Desikan et al., 2006). Regional volumes (cortical and subcortical) underwent intracranial volume adjustment using a regression approach (Buckner et al., 2004), which fits a line to each region and the intracranial volume calculated by FreeSurfer. While studies typically fit this line to their entire cohort, we used the Normal Aging Cohort alone to mimic the conditions that would be used if the tool were to be implemented into clinical practice, enabling reproducibility at the single-subject level. Volumes after intracranial volume correction were summed across hemispheres and cortical thicknesses (not corrected for intracranial volume per standard practice) were averaged across hemispheres. For more specific imaging details see Supplemental Table S4.
Amyloid PET imaging used Florbetapir ([18F]-AV45) or [11C]-Pittsburgh compound B and was processed with an in-house pipeline using Freesurfer-derived regions (PET Unified Pipeline, github.com/ysu001/PUP) with a cerebellar cortex reference region. Partial volume correction in order to address signal spillover was done with a regional spread function (geometric transfer matrix) technique based on the scanner point spread function (determined at each imaging site) and the relative distance between regions (Su et al., 2015, 2013). We defined a negative amyloid PET scan as having a mean cortical standard uptake value ratio with regional spread function applied (SUVR rsf) < 1.42 (Centiloid < 16.4) for [11C]-Pittsburgh compound B-PET or SUVR rsf < 1.19 (Centiloid < 20.6) for Florbetapir-PET. The mean cortical SUVR rsf was defined as the average SUVR rsf from the precuneus, prefrontal cortex, gyrus rectus, and lateral temporal regions (Su et al., 2019). We used cutoffs determined individually for each tracer, as opposed to a unified centiloid cutoff, since these individually established cutoffs are likely more accurate due to the imperfect nature of harmonization procedures such as centiloid conversion (see Supplemental Table S5 for centiloid conversion details). Further imaging details varied by cohort (Supplemental Table S4).
2.3. Normal aging curves
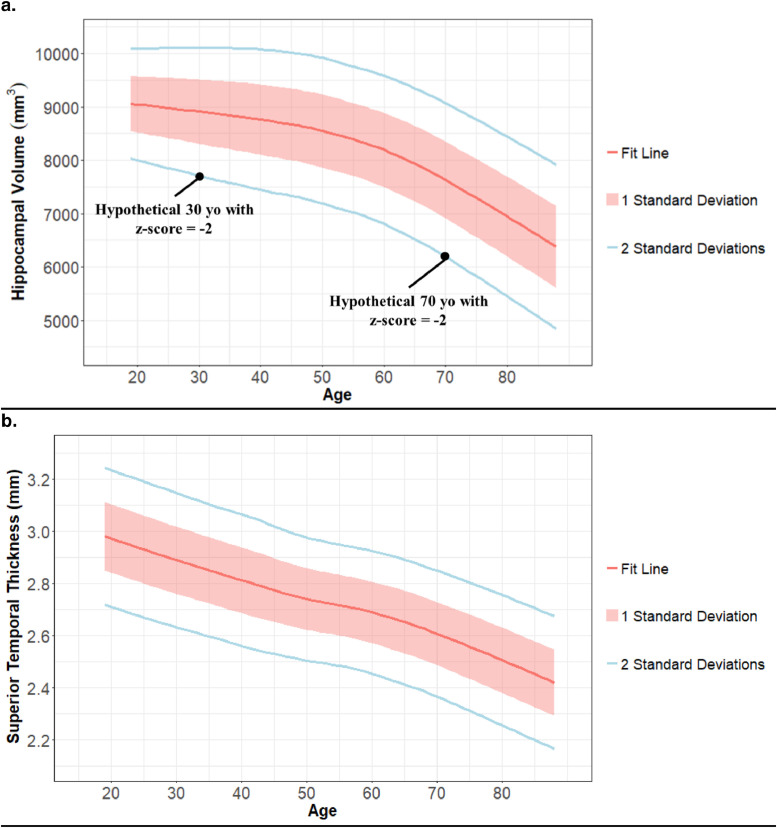
To describe age-related atrophy, normal aging curves were generated for each FreeSurfer region using the Normal Aging Cohort data. For each cortical thickness and intracranial-corrected volume at each age, the mean for each cortical thickness and intracranial-corrected volume was calculated using a locally weighted scatter-plot smoother regression and a smoothed sliding window of two years for standard deviation. The Training, Test, and Clinical Cohorts were then adjusted for age-related atrophy by transforming the volumes and cortical thicknesses into z-scores using these age-specific means and standard deviations (Fig. 1). The hemisphere-combined volumes and thicknesses, with volumes adjusted for intracranial volume, are referred to as ‘unadjusted for age-related atrophy’. The unadjusted dataset that has undergone the z-score adjusted described above is referred to as ‘adjusted for age-related atrophy’. This adjustment for age-related atrophy greatly reduced the correlation of volumetric data with age (examples in Supplemental Table S6).
Fig. 1.
Examples of region-specific atrophy observed in normal aging
Fig. 1 shows Hippocampal volume (1a) and superior temporal thickness (1b) as representatives of the normal aging curves used to adjust other participants’ volumes and cortical thicknesses for age-related atrophy. The red line displays the estimated average volumes. The red ribbon and blue line display the first and second standard deviations from that average, which is calculated locally. 1a additionally displays two black dots representing how two hypothetical participants at different ages could have different volumes but the same z-score after adjustment for age-related atrophy. Standard deviation is fairly consistent across the adult lifespan for both regions, but the averages suggest increasingly rapid atrophy at later ages for Hippocampal volume vs. a steady decline for superior temporal thickne.
2.4. Region selection for AD classification
Volumetric measures that optimally predicted symptomatic AD relative to CN controls were selected using the Training Cohort. For 1000 iterations, a random 50% sample of the Training Cohort was fit to a least absolute shrinkage and selection operator logistic regression. All regional volumes and cortical thicknesses, as measured by FreeSurfer, participant age, sex, and scanner strength (1.5 or 3T), were included as predictors (see Supplemental Fig. S2 for entire list). This regression minimizes the sum of squared errors and has a bound on the sum of the absolute values of the coefficients, which sets many coefficients to zero. The variables not set to zero within each iteration were recorded, determining the frequency each variable was selected. This process was done using data adjusted with the normal aging curves and separately using unadjusted data (volumes still corrected by intracranial volume). The final region set included regions selected in over half the iterations for both sets of data.
2.5. Development of classification models
All classification models in this study used a logistic regression model (R package ‘stats’) fit to the Training Cohort to predict an AD diagnosis. The Age model included only chronological age as a predictor. The Hippocampal Volume (HCV) model used only hippocampal volume. The Select Atrophied Regions in Alzheimer disease (SARA) model used the regions selected in the region selection process described in Section 2.4. The HCVadj and SARAadj models differ from HCV and SARA in that they use data that has undergone the z-score adjustment to remove age-related atrophy, as described in Section 2.3, while the HCV and SARA models use the unadjusted data (volumes still corrected by intracranial volume). The models HCV+Age, SARA+Age, HCVadj+Age, and SARAadj+Age added chronological age as an additional predictor. In this way, the HCVadj+Age and SARAadj+Age models used age as a risk factor for AD, and separately to determine age-specific means and standard deviations when normalizing age-related changes in brain volumes and cortical thicknesses.
Each models’ receiver operating characteristics area under the curve (AUCs) were compared using the Delong method (DeLong et al., 1988) with significance set to p < 0.003 (Bonferroni-corrected p < 0.05), and confidence intervals computed using 2000 stratified bootstrap replicates. Accuracy statistics, when reported, used thresholds determined by the maximal Youden's J statistic within the Training Cohort (Youden, 1950).
3. Results
3.1. Participants
Table 1 details the demographics for each cohort, while Supplemental Table S1 breaks down demographics by data source and AD/Non-AD diagnosis.
3.2. Normal aging curves
Fig. 1 displays the age-related atrophy observed in the Normal Aging cohort, which is free of biomarker evidence of AD. Graphs of other regions can be accessed at https://github.com/benzinger-icl/SARA.
3.3. Region selection for AD classification
The hippocampal volume, inferior lateral ventricle volume, amygdala volume, entorhinal cortical thickness, and inferior parietal cortical thickness were selected in over half of the iterations in both the adjusted and unadjusted data and were thus used in all multi-region (SARA) models. Unadjusted coefficients for all models are in Table S7. Age and nucleus accumbens were additionally selected when using data adjusted for age-related atrophy, while inferior parietal volume and banks of the superior temporal sulcus volume were selected only when using unadjusted data. Frequency of selection for all regions can be found in Supplemental Fig. S2.
3.4. Classification models: impact of adjusting for age-related atrophy
AUCs for each model within the Test and Clinical Cohorts are shown in Table 2; p-values for all comparisons are in Table 3. In the Test Cohort, no significant differences were found between models using adjusted and unadjusted data. In the Clinical Cohort, HCV+Age vs HCVadj+Age, and SARA+Age vs SARAadj+Age similarly showed no statistical difference in their AUCs, but HCV and SARA had higher AUCs than their counterparts HCVadj (0.801 vs 0.743, p < 0.001) and SARAadj (0.820 vs 0.764, p < 0.001). Thus, our adjustment for age-related atrophy did not improve classification ability within our cohorts and instead lowered classification ability in models that did not include age. Without reason to pursue the more complex processing required to adjust for age-related atrophy, further analyses were limited to unadjusted models.
Table 2.
AUCs for All Classification Models in Test and Clinical Cohorts.
| Model: | Test Cohort AUCs (95% CI) | Clinical Cohort AUCs (95% CI) |
|---|---|---|
| Age | 0.675 | 0.742 |
| (0.572–0.778) | (0.694–0.790) | |
| HCV | 0.944 | 0.801 |
| (0.902–0.987) | (0.756–0.846) | |
| SARA | 0.961 | 0.820 |
| (0.925–0.997) | (0.776–0.864) | |
| HCV + Age | 0.950 | 0.792 |
| (0.909–0.991) | (0.747–0.838) | |
| SARA + Age | 0.962 | 0.799 |
| (0.924–0.999) | (0.753–0.845) | |
| HCVadj | 0.948 | 0.743 |
| (0.905–0.992) | (0.693–0.792) | |
| SARAadj | 0.952 | 0.764 |
| (0.911–0.993) | (0.714–0.813) | |
| HCVadj + Age | 0.949 | 0.793 |
| (0.908–0.991) | (0.748–0.840) | |
| SARAadj + Age | 0.961 | 0.799 |
| (0.925–0.997) | (0.752–0.845) |
Table 2 displays each model's AUC (for AD vs. non-AD diagnoses) in the Test Cohort and Clinical Cohort along with its associated 95% CI. The AUC of an ROC plot (not displayed) gives a measure of model performance that does not depend on a specific cut-off or threshold. The various SARA models include hippocampal volume, inferior lateral ventricle volume, entorhinal thickness, amygdala volume, and inferior parietal thickness. X + Age indicates model X with age added as a covariate; Xadj indicates Model X using volumes and cortical thicknesses that have been adjusted for age-related atrophy.
Abbreviations: AD – Alzheimer disease; AUC – Receiver Operating Characteristics Area Under the Curve; CI – Confidence Interval; HCV – Hippocampal Volume; SARA – Select Atrophied Regions in Alzheimer disease
Table 3.
Comparisons of Classification Models’ AUCs.
| Test Cohort p-value | Clinical Cohort p-value | |
|---|---|---|
| Impact of adjusting for age-related atrophy | ||
| HCV vs HCVadj | 0.77 | <0.001 |
| SARA vs SARAadj | 0.32 | <0.001 |
| HCV+Age vs HCVadj+Age | 0.45 | 0.48 |
| SARA+Age vs SARAadj+Age | 0.87 | 0.88 |
| Impact of Age as a Predictor | ||
| HCV vs HCV+Age | 0.20 | 0.001 |
| SARA vs SARA+Age | 0.87 | <0.001 |
| HCV vs Age | <0.001 | 0.02 |
| SARA vs Age | <0.001 | 0.002 |
| Single vs Multi Region Model | ||
| HCV vs SARA | 0.18 | 0.06 |
Table 3 states the p-values for the Delong tests comparing AUCs in order to select the optimal classification model. Significant differences (after accounting for multiple comparisons) are bolded. The SARA models include hippocampal volume, inferior lateral ventricle volume, entorhinal thickness, amygdala volume, and inferior parietal thickness. X + Age indicates model X with age added as a covariate; Xadj indicates Model X using volumes and cortical thicknesses that have been adjusted for age-related atrophy.
Abbreviations: AUC – Receiver Operating Characteristics Area Under the Curve; HCV – Hippocampal Volume; SARA – Select Atrophied Regions in Alzheimer disease
3.5. Classification models: Impact of age as a predictor
In the Test Cohort, HCV and SARA models showed no statistical differences from their counterparts HCV+Age and SARA+Age (Table 3), but did outperform the Age model (0.944 vs 0.675, p < 0.001; and 0.961 vs 0.675, p < 0.001). In the Clinical Cohort, HCV and SARA had higher AUCs than their counterparts HCV+Age and SARA+Age (0.801 vs 0.792, p = 0.001 and 0.820 vs 0.799, p < 0.001), but only SARA maintained a significantly higher AUC than the Age model (0.820 vs 0.742, p = 0.002).
3.6. Classification models: Selecting a model
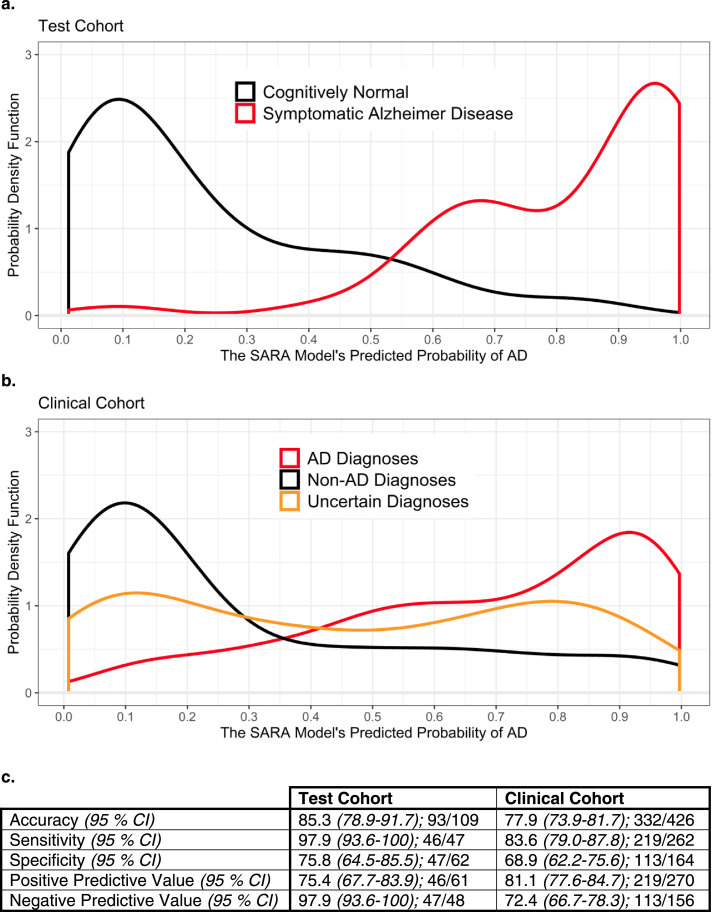
The AUCs of HCV and SARA were not significantly different from each other within the Test or Clinical Cohorts (Table 3), but SARA's AUC was numerically higher than HCV's in both cohorts (Table 2). SARA was selected as the optimal model for this reason in addition to being the only model significantly better than age alone in the Clinical Cohort. Fig. 2 provides more detail on the probabilities of AD predicted by SARA for the participants in the Test and Clinical Cohorts, as well as accuracy measures such as sensitivity and specificity. The x-axes represent the possible output from the SARA model, where a 1.00 indicates a predicted 100% probability of a symptomatic AD diagnosis. The y-axes indicate the probability density function, which is a smoothed histogram normalized to an area of 1 and allows comparison of different sized groups. An example of how to read Fig. 2a is to take the area under the curve of the CN line from x = 0 to 0.25. This is approximately 0.62, indicating that 62% of the CN Controls in the Test Cohort had a probability between 0 and 25%.
Fig. 2.
Distribution of predicted probabilities and accuracy statistics for the SARA model
Fig. 2a and b display the distribution of the SARA model's output for the Test Cohort and Clinical Cohort, respectively. Both 2a and 2b show good separation between the AD and non-AD groups. 2b has slightly less separation and additionally displays the Uncertain Diagnoses group – those that were unable to be classified into the AD or Non-AD groups. 2c displays more traditional diagnostic test measures for SARA using a cutoff of 0.381 (derived using the maximal Youden's J statistic in the Training Cohort) along with the 95% CIs.
Abbreviations: AD – Alzheimer disease; CI – Confidence Interval; SARA – Select Atrophied Regions in Alzheimer disease
3.7. Classification models: Specific diagnoses in the clinical cohort
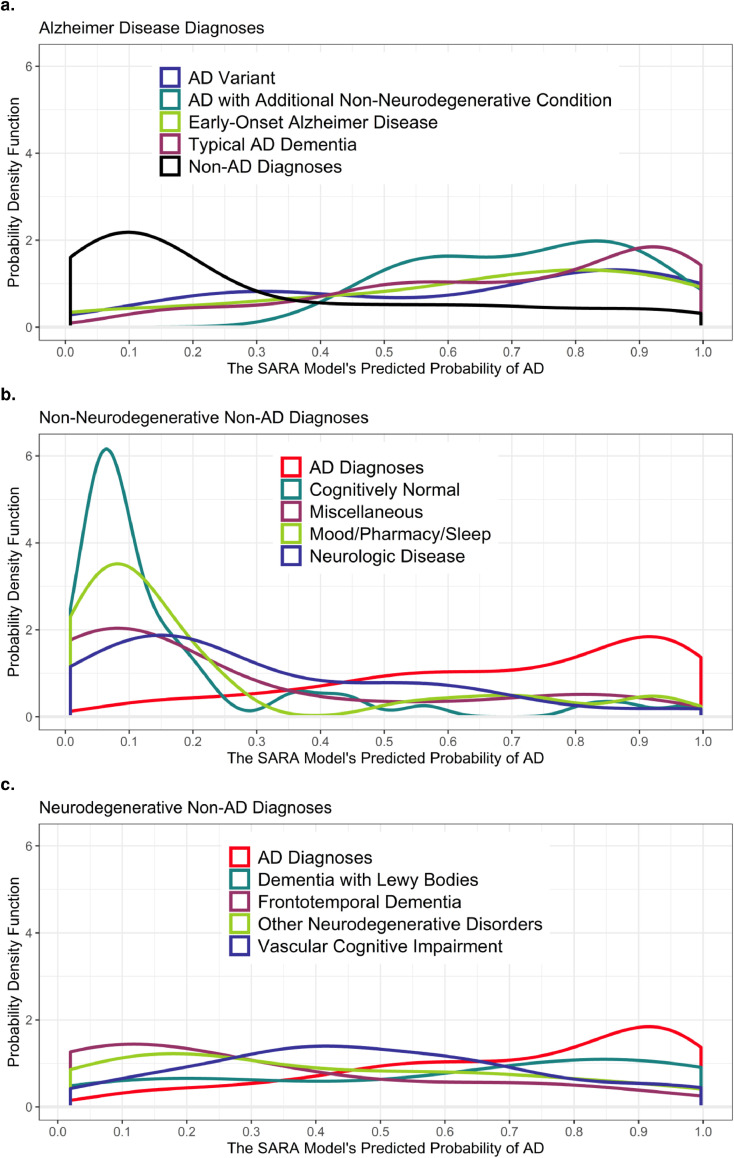
While models were compared using their ability to separate AD from non-AD diagnoses, the heterogeneity of the Clinical Cohort allowed us to examine more specific clinical diagnoses (groups defined in Supplemental Table S3). For non-AD diagnoses, these included other dementia types and non-dementia diagnoses that explained the cognitive complaints of the patient. For AD diagnoses, this included sub-groups of AD to test how the model behaved in atypical AD patients. Fig. 3 displays the unique probability density functions for each of the more specific etiologic diagnoses in the Clinical Cohort, with the AD and non-AD diagnoses groups included for comparison. The AUCs for each sub-group are in Supplemental Table S8, but small group sizes prevented robust statistical analyses. Thus, the following is qualitative rather than an assessment of p-values.
Fig. 3.
Distribution of Predicted Probabilities for the SARA Model for Specific Diagnoses in the Clinical Cohort
Figure 3 displays the distribution of the SARA model's output in the Clinical Cohort using more specific diagnoses than the AD and Non-AD binary from Figure 1b. Figure 3a displays the specific AD diagnoses along with the combined Non-AD Diagnoses line taken from Fig. 2b. Fig. 3b displays the specific Non-AD diagnoses that are non-neurodegenerative in nature and overlays the combined AD Diagnoses line. 3c displays the specific Non-AD diagnoses that are neurodegenerative in nature and overlays the combined AD Diagnoses line. Note the change in y-axes scale from Fig. 2 due to the tight distribution of CN patients in 3b.
Abbreviations: AD – Alzheimer disease; CN – Cognitively Normal; SARA – Select Atrophied Regions in Alzheimer disease
Fig. 3a, with more detail in Supplemental Table S8, demonstrates that AD sub-groups have a high classification accuracy with only slightly lower predicted probabilities in the early-onset AD (<65) and AD variant (such as Posterior Cortical Atrophy) groups as compared to typical (amnestic, late onset) AD. Fig. 3b indicates SARA was good at distinguishing AD from the non-neurodegenerative diagnoses, including mood and sleep disorders; in total AD vs. non-neurodegenerative diagnoses had an AUC of 0.877 (95% Confidence Interval (CI): 0.833–0.922). Fig. 3c shows SARA was less able to separate AD from other neurodegenerative diagnoses and had a combined AUC of 0.719 (95% CI: 0.640–0.799). Only frontotemporal dementia participants (subtypes combined due to small n) approached the same level of separation from AD as the non-neurodegenerative diagnoses.
The impact of using the multi-region SARA over the simple HCV model also varied by diagnosis (Supplemental Table S8). SARA had only a marginally larger AUC than HCV for separating Typical AD from non-AD diagnoses (0.827 vs 0.819), but substantial improvements were seen for separating AD variants from non-AD diagnoses (0.795 vs 0.697).
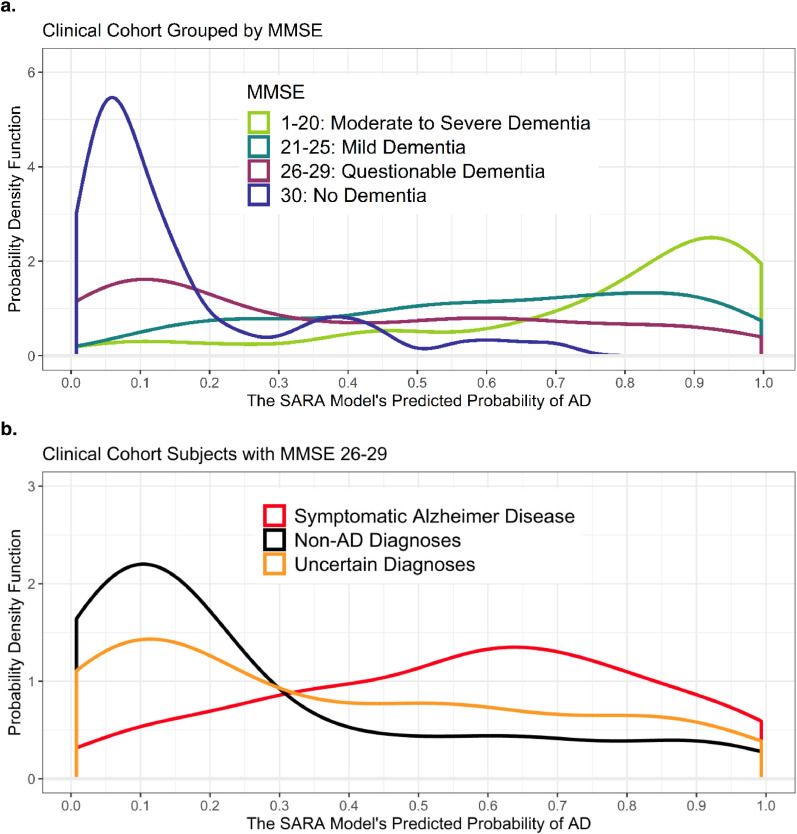
3.8. Classification models: MMSE and CDR in the clinical cohort
Fig. 4a shows the probability density plot of the Clinical Cohort separated by participants’ MMSE, while Supplemental Fig. S3a separates by CDR. MMSEs and CDRs were collected within one year of the MRI (average time difference of 118 days). The strong relationship between level of impairment and SARA's predicted probability of symptomatic AD reflects an alignment between an individual's level of impairment and atrophy in the regions used in SARA (atrophy indicated by a model output closer to one). Despite this strong relationship, SARA maintained a fairly high AUC (0.773) within the group of participants with MMSEs 26–29 (n = 154, Fig. 4b). This indicates the SARA model had good classification ability beyond predicting level of impairment. This was similarly true for participants with a global CDR of 0.5, which had an AUC of 0.782 (n = 235, Supplemental Fig. S3b). This pattern persisted even when considering only those whose MMSE and CDR occurred within 30 days of their MRI, with AUCs of 0.806 (n = 63) and 0.771 (n = 101), respectively.
Fig. 4.
MMSE Aligns, but is not equivalent, to predicted probability in SARA
Figure 4a displays the distributions of the SARA model's predicted probability of AD for all participants (including Uncertain Diagnoses) in the Clinical Cohort, grouped by MMSE score instead of by diagnosis. Note the change in y-axes scale due to the tight distribution of MMSE = 30 participants. 4b displays the distribution of SARA model's predicted probability of AD as in Fig. 2b, but only includes patients with MMSE scores of 26–29 (n = 154).
Abbreviations: AD – Alzheimer disease; AUC – Receiver Operating Characteristics Area Under the Curve; MMSE – Mini Mental State Exam; SARA – Select Atrophied Regions in Alzheimer disease
3.9. SARA as a possible clinical tool
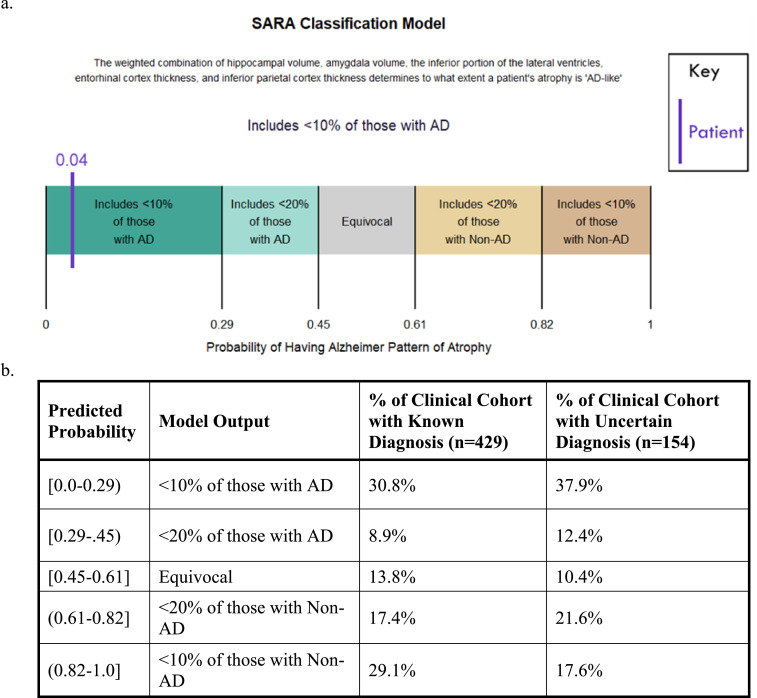
To create an example of how SARA could be used in clinical practice, we developed multiple thresholds reflecting 80% and 90% sensitivities and specificities. Fig. 5a shows the readout a clinician might be given for an individual patient and Fig. 5b lists the percent of the clinical cohort that fell into each category. Over half of the participants who had uncertain diagnoses were given a score within the 90% sensitivity/specificity ranges for AD or non-AD diagnoses, indicating it would be a valuable tool to support clinical decision making.
Fig. 5.
Possible Use of SARA in a Clinical Setting
Fig. 5a displays how the SARA model might be used by a clinician for a single patient, including a description of the model and multiple thresholds. The patient's specific probability of having AD as predicted by SARA is given (4%), as well as a statement reflecting that the sensitivity at that threshold is >90%, indicating both a measure of atrophy and the reliability of that measure. Fig. 5b displays the proportion of participants in the Clinical Cohort that fell into each bin of ranges of scores output from SARA. The next column shows the Clinical Cohort participants with uncertain diagnoses, with the distribution of scores suggesting that SARA would have helped provide a more certain diagnoses for the majority of the participants.
Abbreviations: AD – Alzheimer disease; SARA – Select Atrophied Regions in Alzheimer disease
4. Discussion
We demonstrated that volumetric models have excellent classification abilities that would aid in diagnosing symptomatic AD in various circumstances. We did observe region-specific atrophy even in our unique cohort of CN participants known to be without preclinical AD. However, controlling for this age-related atrophy did not improve classification. Doing so actually lowered accuracy within the Clinical Cohort if age was not included as an additional predictor. This reinforces the idea that age is a strong predictor of AD dementia and implies that these models require age or age-related atrophy to maintain the highest levels of accuracy. Thus, age-related atrophy may either convey increased risk for development of a neurodegenerative dementing illness, or, more likely, age-related atrophy may act as a proxy for age. Either way, total atrophy appears to be more predictive than atrophy specifically attributable to AD.
Our data-driven region selection approach, optimized to FreeSurfer, saw a specific pattern of atrophy in AD that overlapped with the medial temporal lobe regions reported in many previous papers. We evaluated if using these regions would improve classification of AD. While the single region HCV and multi-region SARA models did not show statistically different AUCs, other evidence suggested SARA was the stronger classifier. First, in both the Test and Clinical Cohorts, the value of the AUC was higher in SARA than HCV. Second, in the Clinical Cohort, it was only SARA that had an AUC statistically higher than the model using age alone, without any volumetric measures. Third, the pattern of higher AUCs in SARA than in HCV was seen for most specific diagnostic group within the Clinical Cohort.
Our results suggest SARA has the greatest diagnostic specificity when distinguishing AD from frontotemporal dementia or from non-neurodegenerative diagnoses (e.g. mood disorders, sleep disorders, CN individuals). The high performance in frontotemporal dementia is especially noteworthy. FDG-PET is recommended for patients with AD vs. frontotemporal dementia diagnoses (Silverman et al., 2001), but PET scans are limited in diagnostic sensitivity and by insurance coverage (Medicare will cover it, but often private insurance will not), availability, and cost. This AD vs. frontotemporal dementia differential is often considered, especially in younger patients, and highly available biomarkers would help identify the correct prognosis and treatment for these patients. The Clinical Cohort had the lowest diagnostic specificity when distinguishing AD from other neurodegenerative disorders. This was likely due to the disorders impacting overlapping regions and patients having co-incident diagnoses. Comorbidities increase with age and can include multiple neurodegenerative conditions, such as concurrent AD and dementia with Lewy bodies (Irwin and Hurtig, 2018).
Another way SARA reflects clinical reality is the correlation between atrophy and level of impairment measured by MMSE and global CDR. With this in mind, we evaluated the diagnostic utility of SARA beyond predicting impairment and found high classification ability in the Clinical Cohort even when limited to early symptomatic participants (CDR 0.5 or MMSE 26–29). These patients are also the ones for whom additional biomarkers would likely be most useful. These findings indicate SARA is not simply acting as a proxy for MMSE or CDR, but provides additional diagnostic information.
Strengths of this study include the large overall sample size of almost 1300 participants. We benefited from having research cohorts with participants diagnosed with the highest possible accuracy outside of postmortem testing, as well as a heterogeneous group of real-world patients seen at a dementia clinic. By using these cohorts in combination, we were able to demonstrate that our model approaches the sensitivity and specificity of amyloid PET and cerebrospinal fluid biomarkers. Strengths of the SARA algorithm include that it is fairly simple and transparent (unlike machine-learning algorithms), making it easier for doctors and patients to trust, will be freely available, uses MRI scans that are non-invasive and often already collected for dementia patients, and has been shown to work in both research and clinical populations. SARA and the Clinical Cohort's data will be made available online at https://github.com/benzinger-icl/SARA and https://www.oasis-brains.org/.
While our results indicate the potential usefulness of quantitative volumetric biomarkers, there are some limitations of this study. Though our cohorts had fairly good representation of African–Americans, the general lack racial and socioeconomic diversity may bias our models. Volumetric classification may be further improved if models, including the regions used, are optimized to specific non-AD diagnoses and/or incorporate longitudinal scanning. Our use of a single set of normal aging curves and a binary AD/non-AD prediction model was due to our limited numbers, despite surpassing the sample size of many neuroimaging studies. The threshold used in the reported accuracy statistics was based upon the Training Cohort and has not been optimized to a clinical setting. This optimization would need to be validated in a separate cohort, ideally with histopathologically-confirmed diagnoses. This confirmation was not available for the Clinical Cohort and misdiagnosis may have caused an under-estimation of model accuracy. An important question for future work to address is the overlap of AD and vascular disease, which we were unable to address due to the diagnostic difficulty and limited presence of vascular problems in our research cohorts.
5. Conclusions
In summary, we have shown that an MRI-based volumetric classification model can be used to separate AD from CN controls and other dementia types. Controlling for age-related atrophy did not improve our models, and lowered performance if age was not also included as a predictor. SARA may be useful as a first step for selecting symptomatic AD participants for entrance into clinical trials or as an adjunct to the diagnostic algorithm when a clinical differential diagnosis includes AD vs. frontotemporal dementia or non-neurodegenerative conditions.
Declaration of Competing Interest
GS Day is supported by a mentored career development award (K23AG064029), through additional research/grant support from the National Institutes of Health (P01AG03991, R56AG057195, U01AG057195) and from philanthropic support of the BJHF Willman Scholar Fund. Prior research has benefited from an in-kind gift of radiopharmaceuticals from Avid Radiopharmaceuticals. GS Day holds stock in ANI Pharmaceuticals, Inc., serves as a topic editor on dementia for DynaMed Plus (EBSCO Industries, Inc.), and has provided record review and expert medical testimony on legal cases pertaining to management of Wernicke. Amber Salter receives consulting fees for statistical reviews for the journal Circulation: Cardiovascular Imaging. Joy Snider does consulting with Eisai. Cyrus Raji does consulting for Brainreader ApS and Neurevolution LLC. Nupur Ghoshal has participated or is currently participating in clinical trials of anti-dementia drugs sponsored by the following companies: Bristol Myers Squibb, Eli Lilly/Avid Radiopharmaceuticals, Janssen Immunotherapy, Novartis, Pfizer, Wyeth, SNIFF (The Study of Nasal Insulin to Fight Forgetfulness) study, and A4 (The Anti-Amyloid Treatment in Asymptomatic Alzheimer's Disease) trial. Ghoshal receives research support from Tau Consortium and Association for Frontotemporal Dementia and is funded by the NIH. Michelle Miller-Thomas received payment as a site investigator for a multiple sclerosis imaging study sponsored by Biogen MA Inc. (completed May 2019), which is unrelated to this work. Miller-Thomas receives ongoing salary support from the Barnes Jewish Hospital Foundation to support her work on Neuroradiology Advanced Imaging, which is directly related to this manuscript. JC Morris is funded by NIH grants # P50AG005681; P01AG003991; P01AG026276 and UF1AG032438. Neither Dr. Morris nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company. Tammie Benzinger participated in clinical trials sponsored by Eli Lilly, Roche, and Biogen. Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly) provided doses of 18F-florbetapir, partial funding for 18F-florbetapir scanning, precursor for 18F-flortaucipir, and technology transfer for manufacturing of 18F-flortaucipir.
*DIAN Consortium:
Ricardo Allegri, Fatima Amtashar, Randy Bateman, Tammie Benzinger, Sarah Berman, Courtney Bodge, Susan Brandon, William (Bill) Brooks, Jill Buck, Virginia Buckles, Sochenda Chea, Jasmeer Chhatwal, Patricio Chrem, Helena Chui, Jake Cinco, Jack Clifford, Carlos Cruchaga, Mirelle D'Mello, Tamara Donahue, Jane Douglas, Noelia Edigo, Nilufer Erekin-Taner, Anne Fagan, Marty Farlow, Angela Farrar, Howard Feldman, Gigi Flynn, Nick Fox, Erin Franklin, Hisako Fujii, Cortaiga Gant, Samantha Gardener, Bernardino Ghetti, Alison Goate, Jill Goldman, Brian Gordon, Neill Graff-Radford, Julia Gray, Jenny Gurney, Jason Hassenstab, Mie Hirohara, David Holtzman, Russ Hornbeck, Siri Houeland DiBari, Takeshi Ikeuchi, Snezana Ikonomovic, Gina Jerome, Mathias Jucker, Celeste Karch, Kensaku Kasuga, Takeshi Kawarabayashi, William (Bill) Klunk, Robert Koeppe, Elke Kuder-Buletta, Christoph Laske, Jae-Hong Lee, Johannes Levin, Daniel Marcus, Ralph Martins, Neal Scott Mason, Colin Masters, Denise Maue-Dreyfus, Eric McDade, Lucy Montoya, Hiroshi Mori, John Morris, Akem Nagamatsu, Katie Neimeyer, James Noble, Joanne Norton, Richard Perrin, Marc Raichle, John Ringman, Jee Hoon Roh, Jee Hoon Roh, Stephen Salloway, Peter Schofield, Hiroyuki Shimada, Tomoyo Shiroto, Mikio Shoji, Wendy Sigurdson, Hamid Sohrabi, Paige Sparks, Kazushi Suzuki, Laura Swisher, Kevin Taddei, Jen Wang, Peter Wang, Mike Weiner, Mary Wolfsberger, Chengjie Xiong, Xiong Xu
Acknowledgments
Acknowledgments
Data collection and sharing for this project was supported by The Dominantly Inherited Alzheimer's Network (DIAN, U19AG032438) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), Partial support by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI).This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data were provided in part by OASIS-3: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50AG00561, P30NS09857781, P01AG026276, P01AG003991, R01AG043434, UL1TR000448, R01EB009352. Florbetapir F18 (18F-AV-45) doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
Funding information
This work was supported by the Knight Alzheimer Research Imaging (KARI) Program, Knight Alzheimer Disease Research Center (ADRC), ADRC Center Grant (NIH/NIA P50AG005681), the Charles and Joanne Knight Alzheimer Disease Research Center Support Fund, the Barnes-Jewish Hospital Foundation (BJHF) Adult Children's Study (ACS) NIH/NIA P01AG026276, support for image acquisition – CCIR/ICTS Human Imaging Unit (NIH/NCATS UL1TR000448), and support for imaging informatics – CNDA/XNAT through the Neuroimaging Informatics and Analysis Center (1P30NS098577) and R01 EB009352. These funding sources had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2020.102248.
Appendix. Supplementary materials
References
- Beach T.G., Monsell S.E., Phillips L.E., Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J. Neuropathol. Exp. Neurol. 2012;71:266–273. doi: 10.1097/NEN.0b013e31824b211b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier M.R., Thomas J.B., Fagan A.M., Hassenstab J., Holtzman D.M., Benzinger T.L., Morris J.C., Ances B.M. Functional connectivity and graph theory in preclinical Alzheimer's disease. Neurobiol. Aging. 2014;35:757–768. doi: 10.1016/j.neurobiolaging.2013.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R.L., Head D., Parker J., Fotenos A.F., Marcus D.S., Morris J.C., Snyder A.Z. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Day G.S., Lim T.S., Hassenstab J., Goate A.M., Grant E.A., Roe C.M., Cairns N.J., Morris J.C. Differentiating cognitive impairment due to corticobasal degeneration and Alzheimer disease. Neurology. 2017;88:1273–1281. doi: 10.1212/WNL.0000000000003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Desikan R.S., Ségonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Buckner R.L., Dale A.M., Maguire R.P., Hyman B.T., Albert M.S., Killiany R.J. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Duits F.H., Martinez-Lage P., Paquet C., Engelborghs S., Lleó A., Hausner L., Molinuevo J.L., Stomrud E., Farotti L., Ramakers I.H., Tsolaki M., Skarsgård C., Åstrand R., Wallin A., Vyhnalek M., Holmber-Clausen M., Forlenza O.V, Ghezzi L., Ingelsson M., Hoff E.I., Roks G., de Mendonça A., Papma J.M., Izagirre A., Taga M., Struyfs H., Alcolea D.A., Frölich L., Balasa M., Minthon L., Twisk J.W., Persson S., Zetterberg H., van der Flier W.M., Teunissen C.E., Scheltens P., Blennow K. Performance and complications of lumbar puncture in memory clinics: results of the multicenter lumbar puncture feasibility study. Alzheimer's Dement. 2016;12:154–163. doi: 10.1016/j.jalz.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fotenos A.F., Snyder A.Z., Girton E., Morris J.C., Buckner R.L. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology. 2005;64:1032–1039. doi: 10.1212/01.WNL.0000154530.72969.11. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassenstab J., Chasse R., Grabow P., Benzinger T.L., Fagan A.M., Xiong C., Jasielec M.S., Grant E.A., Morris J.C. Certified normal: Alzheimer's disease biomarkers and normative estimates of cognitive functioning. Neurobiol. Aging. 2016;43:23–33. doi: 10.1016/j.neurobiolaging.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D.J., Hurtig H.I. The contribution of Tau, amyloid-beta and alpha-synuclein pathology to dementia in Lewy body disorders. J. Alzheimer's Dis. Park. 2018;08:139–148. doi: 10.4172/2161-0460.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J.H., Liu E., Molinuevo J.L., Montine T.J., Phelps C.H., Rankin K.P., Rowe C.C., Scheltens P., Siemers E., Snyder H.M., Sperling R.A., Elliott C., Masliah E., Ryan L., Silverberg N. NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimer's Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Wiste H.J., Weigand S.D., Rocca W.A., Knopman D.S., Mielke M.M., Lowe V.J., Senjem M.L., Gunter J.L., Preboske G.M., Pankratz V.S., Vemuri P., Petersen R.C. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: a cross-sectional study. Lancet Neurol. 2014;13:997–1005. doi: 10.1016/S1474-4422(14)70194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.A., Minoshima S., Bohnen N.I., Donohoe K.J., Foster N.L., Herscovitch P., Karlawish J.H., Rowe C.C., Carrillo M.C., Hartley D.M., Hedrick S., Pappas V., Thies W.H. Appropriate use criteria for amyloid PET: a report of the amyloid imaging task force, the society of nuclear medicine and molecular imaging, and the Alzheimer's association. Alzheimer's Dement. 2013;9:E1–E16. doi: 10.1016/j.jalz.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopman D.S., DeKosky S.T., Cummings J.L., Chui H., Corey-Bloom J., Relkin N., Small G.W., Miller B., Stevens J.C. Practice parameter: diagnosis of dementia (an evidence-based review): report of the quality standards subcommittee of the American academy of neurology. Neurology. 2001;56:1143–1153. doi: 10.1212/WNL.56.9.1143. [DOI] [PubMed] [Google Scholar]
- LaMontagne P.J., Benzinger T.L.S., Morris J.C., Keefe S., Hornbeck R., Xiong C., Grant E., Hassenstab J., Moulder K., Vlassenko A., Raichle M.E., Cruchaga C., Marcus D. OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. medRxiv 2019. 2019;12:13. doi: 10.1101/2019.12.13.19014902. 19014902. [DOI] [Google Scholar]
- Liu J., Hlavka J., Hillestad R., Mattke S. Assessing the preparedness of the U.S. Health care system infrastructure for an Alzheimer's treatment, assessing the preparedness of the U.S. Health care system infrastructure for an Alzheimer's treatment. RAND Corp. 2017 doi: 10.7249/RR2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeith I.G., Boeve B.F., Dickson D.W., Halliday G., Taylor J.-P., Weintraub D., Aarsland D., Galvin J., Attems J., Ballard C.G., Bayston A., Beach T.G., Blanc F., Bohnen N.I., Bonanni L., Bras J., Brundin P., Burn D., Chen-Plotkin A., Duda J.E., El-Agnaf O., Feldman H., Ferman T.J., Ffytche D., Fujishiro H., Galasko D., Goldman J.G., Gomperts S.N., Graff-Radford N.R., Honig L.S., Iranzo A., Kantarci K., Kaufer D., Kukull W., Lee V.M., Leverenz J.B., Lewis S., Lippa C., Lunde A., Masellis M., Masliah E., McLean P., Mollenhauer B., Montine T.J., Moreno E., Mori E., Murray M., O'Brien J.T., Orimo S., Postuma R.B., Ramaswamy S., Ross O.A., Salmon D.P., Singleton A., Taylor A., Thomas A., Tiraboschi P., Toledo J.B., Trojanowski J.Q., Tsuang D., Walker Z., Yamada M., Kosaka K. Diagnosis and management of dementia with Lewy bodies. Neurology. 2017;89:88–100. doi: 10.1212/WNL.0000000000004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, E., Chalkidou, A., Hammers, A., Peacock, J., Summers, J., Keevil, S., 2016. Diagnostic accuracy of 18 F amyloid PET tracers for the diagnosis of Alzheimer's disease : a systematic review and meta-analysis374–385. 10.1007/s00259-015-3228-x. [DOI] [PMC free article] [PubMed]
- Morris J.C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Ossenkoppele R., Rabinovici G.D., Smith R., Cho H., Schöll M., Strandberg O., Palmqvist S., Mattsson N., Janelidze S., Santillo A., Ohlsson T., Jögi J., Tsai R., La Joie R., Kramer J.H., Boxer A.L., Gorno-Tempini M.-L., Miller B.L., Choi J.Y., Ryu Y.H., Lyoo C.H., Hansson O. Discriminative Accuracy of [18 F]flortaucipir positron emission tomography for Alzheimer disease vs other neurodegenerative disorders. JAMA. 2018;320:1151. doi: 10.1001/jama.2018.12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici G.D., Gatsonis C., Apgar C., Chaudhary K., Gareen I., Hanna L., Hendrix J.A., Hillner B.E., Olson C., Lesman-Segev O.H., Romanoff J., Siegel B.A., Whitmer R.A., Carrillo M.C. Association of amyloid positron emission tomography with subsequent change in clinical management among medicare beneficiaries with mild cognitive impairment or dementia. JAMA. 2019;321:1286. doi: 10.1001/jama.2019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascovsky K., Hodges J.R., Knopman D.S., Mendez M.F., Kramer J.H., Neuhaus J., van Swieten J.C., Seelaar H., Dopper E.G., Onyike C.U., Hillis A.E., Josephs K.A., Boeve B.F., Kertesz A., Seeley W.W., Rankin K.P., Johnson J.K., Gorno-Tempini M.-L., Rosen H., Prioleau-Latham C.E., Lee A., Kipps C.M., Lillo P., Piguet O., Rohrer J.D., Rossor M.N., Warren J.D., Fox N.C., Galasko D., Salmon D.P., Black S.E., Mesulam M., Weintraub S., Dickerson B.C., Diehl-Schmid J., Pasquier F., Deramecourt V., Lebert F., Pijnenburg Y.A.L., Chow T.W., Manes F., Grafman J., Cappa S.F., Freedman M., Grossman M., Miller B.L. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y., O'Gorman J., Qian F., Arastu M., Li M., Chollate S., Brennan M.S., Quintero-Monzon O., Scannevin R.H., Arnold H.M., Engber T., Rhodes K., Ferrero J., Hang Y., Mikulskis A., Grimm J., Hock C., Nitsch R.M., Sandrock A. The antibody aducanumab reduces Aβ plaques in Alzheimer's disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- Shaw L.M., Arias J., Blennow K., Galasko D., Molinuevo J.L., Salloway S., Schindler S.E., Carrillo M.C., Hendrix J.A., Ross A., Illes J., Ramus C., Fifer S. Appropriate use criteria for lumbar puncture and cerebrospinal fluid testing in the diagnosis of Alzheimer's disease. Alzheimer's Dement. 2018;14:1505–1521. doi: 10.1016/j.jalz.2018.07.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman D.H.S., Small G.W., Chang C.Y., Lu C.S., de Aburto M.A.K., Chen W., Czernin J., Rapoport S.I., Pietrini P., Alexander G.E., Schapiro M.B., Jagust W., Hoffman J.M., Welsh-Bohmer K.A., Alavi A., Clark C.M., Salmon E., de Leon M.J., Mielke R., Cummings J.L., Kowell A.P., Gambhir S.S., Hoh C.K., Phelps M.E. Positron Emission Tomography in Evaluation of Dementia. JAMA. 2001;286:2120. doi: 10.1001/jama.286.17.2120. [DOI] [PubMed] [Google Scholar]
- Skrobot O.A., Black S.E., Chen C., DeCarli C., Erkinjuntti T., Ford G.A., Kalaria R.N., O'Brien J., Pantoni L., Pasquier F., Roman G.C., Wallin A., Sachdev P., Skoog I., Ben-Shlomo Y., Passmore A.P., Love S., Kehoe P.G., Taragano F.E., Kril J., Cavalieri M., Jellinger K.A., Kovacs G.G., Engelborghs S., Lafosse C., Bertolucci P.H., Brucki S., Caramelli P., de Toledo Ferraz Alves T.C., Bocti C., Fulop T., Hogan D.B., Hsiung G.R., Kirk A., Leach L., Robillard A., Sahlas D.J., Guo Q., Tian J., Hokkanen L., Jokinen H., Benisty S., Deramecourt V., Hauw J., Lenoir H., Tsatali M., Tsolaki M., Sundar U., Coen R.F., Korczyn A.D., Altieri M., Baldereschi M., Caltagirone C., Caravaglios G., Di Carlo A., Di Piero V., Gainotti G., Galluzzi S., Logroscino G., Mecocci P., Moretti D.V., Padovani A., Fukui T., Ihara M., Mizuno T., Kim S.Y., Akinyemi R., Baiyewu O., Ogunniyi A., Szczudlik A., Bastos-Leite A.J., Firmino H., Massano J., Verdelho A., Kruglov L.S., Ikram M.K., Kandiah N., Arana E., Barroso-Ribal J., Calatayud T., Cruz-Jentoft A.J., López-Pousa S., Martinez-Lage P., Mataro M., Börjesson-Hanson A., Englund E., Laukka E.J., Qiu C., Viitanen M., Biessels G.J., de Leeuw F.-E., den Heijer T., Exalto L.G., Kappelle L.J., Prins N.D., Richard E., Schmand B., van den Berg E., van der Flier W.M., Bilgic B., Allan L.M., Archer J., Attems J., Bayer A., Blackburn D., Brayne C., Bullock R., Connelly P.J., Farrant A., Fish M., Harkness K., Ince P.G., Langhorne P., Mann J., Matthews F.E., Mayer P., Pendlebury S.T., Perneczky R., Peters R., Smithard D., Stephan B.C., Swartz J.E., Todd S., Werring D.J., Wijayasiri S.N., Wilcock G., Zamboni G., Au R., Borson S., Bozoki A., Browndyke J.N., Corrada M.M., Crane P.K., Diniz B.S., Etcher L., Fillit H., Greenberg S.M., Grinberg L.T., Hurt S.W., Lamar M., Mielke M., Ott B.R., Perry G., Powers W.J., Ramos-Estebanez C., Reed B., Roberts R.O., Romero J.R., Saykin A.J., Seshadri S., Silbert L., Stern Y., Zarow C. Progress toward standardized diagnosis of vascular cognitive impairment: guidelines from the vascular impairment of cognition classification consensus study. Alzheimer's Dement. 2018;14:280–292. doi: 10.1016/j.jalz.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Sperling R.A., Rentz D.M., Johnson K.A., Karlawish J.H., Donohue M., Salmon D.P., Aisen P.S. The A4 study: stopping AD before symptoms begin? Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3007941. 228fs13-228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Blazey T.M., Snyder A.Z., Raichle M.E., Marcus D.S., Ances B.M., Bateman R.J., Cairns N.J., Aldea P., Cash L., Christensen J.J., Friedrichsen K.A., Hornbeck R.C., Farrar A.M., Owen C.J., Mayeux R., Brickman A.M., Klunk W.E., Price J.C., Thompson P.M., Ghetti B., Saykin A.J., Sperling R.A., Johnson K.A., Schofield P.R., Buckles V., Morris J.C., Benzinger T.L. Partial volume correction in quantitative amyloid imaging. Neuroimage. 2015;107:55–64. doi: 10.1016/j.neuroimage.2014.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., D'Angelo G.M., Vlassenko A.G., Zhou G., Snyder A.Z., Marcus D.S., Blazey T.M., Christensen J.J., Vora S., Morris J.C., Mintun M.A., Benzinger T.L. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One. 2013;8:e73377. doi: 10.1371/journal.pone.0073377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Flores S., Wang G., Hornbeck R.C., Speidel B., Joseph-Mathurin N., Vlassenko A.G., Gordon B.A., Koeppe R.A., Klunk W.E., Jack C.R., Farlow M.R., Salloway S., Snider B.J., Berman S.B., Roberson E.D., Brosch J., Jimenez-Velazques I., van Dyck C.H., Galasko D., Yuan S.H., Jayadev S., Honig L.S., Gauthier S., Hsiung G.-Y.R., Masellis M., Brooks W.S., Fulham M., Clarnette R., Masters C.L., Wallon D., Hannequin D., Dubois B., Pariente J., Sanchez-Valle R., Mummery C., Ringman J.M., Bottlaender M., Klein G., Milosavljevic-Ristic S., McDade E., Xiong C., Morris J.C., Bateman R.J., Benzinger T.L. Comparison of Pittsburgh compound B and florbetapir in cross-sectional and longitudinal studies. Alzheimer's dement. diagnosis. Assess. Dis. Monit. 2019;11:180–190. doi: 10.1016/j.dadm.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman D.E., Prohovnik I. Sensitivity and specificity of neuroimaging for the diagnosis of Alzheimer's disease. Dialogues Clin. Neurosci. 2003;5:89–99. [PMC free article] [PubMed] [Google Scholar]
- Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO. 2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.