Abstract
Schizaphis graminum Rondani (Hemiptera: Aphididae) and Sipha flava Forbes (Hemiptera: Aphididae) are two common pests of bioenergy grasses. Despite the fact that they are both considered generalists, they differ in their ability to colonize Panicum virgatum cultivars. For example, S. flava colonizes both P. virgatum cv. Summer and P. virgatum cv. Kanlow whereas S. graminum can only colonize Summer. To study the molecular responses of these aphids to these two switchgrass cultivars, we generated de novo transcriptome assemblies and compared the expression profiles of aphids feeding on both cultivars to profiles associated with feeding on a highly susceptible sorghum host and a starvation treatment. Transcriptome assemblies yielded 8,428 and 8,866 high-quality unigenes for S. graminum and S. flava, respectively. Overall, S. graminum responded strongly to all three treatments after 12 h with an upregulation of unigenes coding for detoxification enzymes while major transcriptional changes were not observed in S. flava until 24 h. Additionally, while the two aphids responded to the switchgrass feeding treatment by downregulating unigenes linked to growth and development, their responses to Summer and Kanlow diverged significantly. Schizaphis graminum upregulated more unigenes coding for stress-responsive enzymes in the Summer treatment compared to S. flava; however, many of these unigenes were actually downregulated in the Kanlow treatment. In contrast, S. flava appeared capable of overcoming host defenses by upregulating a larger number of unigenes coding for detoxification enzymes in the Kanlow treatment. Overall, these findings are consistent with previous studies on the interactions of these two cereal aphids to divergent switchgrass hosts.
Keywords: greenbug aphid, yellow sugarcane aphid, de novo transcriptome assembly, Kanlow, Summer
Considerable headway has been made towards understanding host resistant and susceptibility responses to aphids across a range of plant species (e.g., Gulsen et al. 2007, Gutsche et al. 2009, Endara and Coley 2011, Kerchev et al. 2012, Ramm et al. 2013, Louis and Shah 2015, Zust and Agrawal 2016, Donze-Reiner et al. 2017, Zust and Agrawal 2017). However, less is known about the molecular responses of insects resulting from biotic interactions, particularly those associated with plant resistance phenotypes (de la Paz Celorio-Mancera et al. 2013, Herde and Howe 2014, Eyres et al. 2016, Guidolin et al. 2018, Yates and Michel 2018). Previous studies with switchgrass (Panicum virgatum L.) cultivars and their interactions with the greenbug, Schizaphis graminum (Rondani) (Hemiptera: Aphididae), and yellow sugarcane aphid, Sipha flava (Forbes) (Hemiptera: Aphididae), have indicated differential levels of resistance within switchgrass populations to these two cereal aphids (Burd et al. 2012, Koch et al. 2014a, Koch et al. 2014b, Koch et al. 2015), making switchgrass an ideal system for studying the impacts of biotic stresses on aphid physiology and gene expression.
Schizaphis graminum and S. flava are primarily phloem feeders (Koch et al. 2015). During the feeding process, aphids secrete saliva into the plant and thereby modulate plant responses to herbivory (Miles 1999). Aphid salivary content and virulence can vary by biotype (Cooper et al. 2011, Nicholson and Puterka 2014). Schizaphis graminum saliva is known to contain toxins (Nicholson and Puterka 2014), and thus, herbivory by this aphid induces a strong defense response in Summer switchgrass (Donze-Reiner et al. 2017). Kanlow switchgrass plants (lowland ecotype) are not effective hosts for S. graminum, although S. graminum will attempt to feed on them (Koch et al. 2015). In contrast, S. flava readily establishes on Summer (upland ecotype) and Kanlow plants, although aphid performance is typically better on Summer plants (Koch et al. 2014b). Despite the fact that S. flava can grow and develop on Kanlow plants, Kanlow appeared to have antibiosis effects on both aphids, suggestive of physical barriers (i.e., callose) or feeding deterrents in phloem tissues (Koch et al. 2018). Collectively available data indicate that the genetics of the two switchgrass hosts differentially impacted aphid herbivory (Koch et al. 2018) and also potentially modulated the physiology of S. graminum and S. flava.
The host range for S. graminum includes many important cereals and grasses, including Agropyron (wheatgrass), Avena (oat), Hordeum (barley), Oryza (rice), Panicum (panicgrasses), Poa (bluegrasses), Sorghum (sorghum), Triticum (wheat), and Zea (maize) (Michels Jr. 1986, Blackman and Eastop 2000, Nuessly and Nagata 2005). In total, at least 70 Graminaceous species have been reported as suitable hosts (Michels Jr. 1986). Sipha flava is believed to be native to North America (Nuessly 2005) and has been recorded on approximately 60 plant species within diverse families such as Cyperaceae, Poaceae, and Commelinaceae (Kindler and Dalrymple 1999); including members of the genera Hordeum, Oryza, Panicum, Saccharum (sugarcane), Sorghum, Triticum, and Zea (Blackman and Eastop 2000, Nuessly 2005). In the United States, S. flava is considered an important pest of sugarcane, Saccharum officinarum L., and an occasional pest of small grains (Kindler and Dalrymple 1999). The wide host range of the two aphids and the antibiosis effects of Kanlow switchgrass on both aphids, but not in Summer switchgrass, indicated differential impacts on aphid metabolism when feeding on these two switchgrass cultivars with divergent defense responses. However, how the short-term biotic stress of feeding on plants with varying levels of resistance affects global gene expression in aphids and whether those responses differ from abiotic stress responses (i.e., starvation) are not known for either S. graminum or S. flava.
The aims of this study were to 1) generate de novo transcriptome assemblies of S. graminum and S. flava, 2) characterize gene expression of the two aphids stressed by starvation relative to aphids feeding on sorghum BCK60 plants, which are highly susceptible to both aphid species and present suitable controls for comparisons of resistant and susceptible interactions (Koch et al. 2014b), and 3) compare the transcriptomic profiles of starved aphids to aphids fed on switchgrasses with divergent defense responses to aphid herbivory. The overarching goal was to measure transcriptome-level observations that could be exploited to improve switchgrass germplasm with more durable resistance to aphid herbivory in the future.
Materials and Methods
Plant Material
Two switchgrass cultivars, ‘Kanlow’ and ‘Summer’, were used to explore the molecular responses of S. graminum and S. flava to feeding on switchgrass. In addition, the S. graminum-susceptible sorghum cultivar, ‘BCK60’, was used to compare switchgrass-aphid interactions with a well-documented host for both aphid species. In brief, switchgrass and sorghum plants were grown in SC-10 Super Cell Single Cell Cone-tainers (3.8 cm diameter × 21 cm deep) (Stuewe & Sons, Inc., Corvallis, OR) containing a Fafard growing media (Mix No. 3B) (Sun Gro Horticulture Distribution Inc., Agawam, MA). Plants were maintained in a greenhouse at 25 ± 4°C with the lighting augmented by LED lights (Pro 325, Lumigrow, Novato, CA) to produce a photoperiod of 16:8 (L:D) h and were grown to the V2 developmental stage (Moore et al. 1991). Plants were fertilized every 2 wk with a water-soluble fertilizer (20:10:20 N-P-K). After emergence, plants were thinned to one plant per cone-tainer.
Insect Colonies
Colonies for S. graminum (biotype I) and S. flava were obtained from Dr. John D. Burd, USDA-ARS in Stillwater, Oklahoma. The S. graminum colony was maintained in a plant growth chamber at 25 ± 2°C with a photoperiod of 16:8 (L:D) h, while the S. flava colony was kept in the greenhouse at 25 ± 4°C and a photoperiod of 16:8 (L:D) h within clear plastic cages (12.5 cm in diameter and ventilated with organdy fabric). Sipha flava was maintained in the greenhouse because it reproduces more efficiently under high-light intensity (Hentz and Nuessly 2004, Pallipparambil et al. 2014). Both aphid colonies used in these experiments were established from a single viviparous parthenogenetic female to limit variation from multiple genetic backgrounds and were maintained on a continuous supply of BCK60 sorghum plants.
Experimental Conditions and Sample Collection
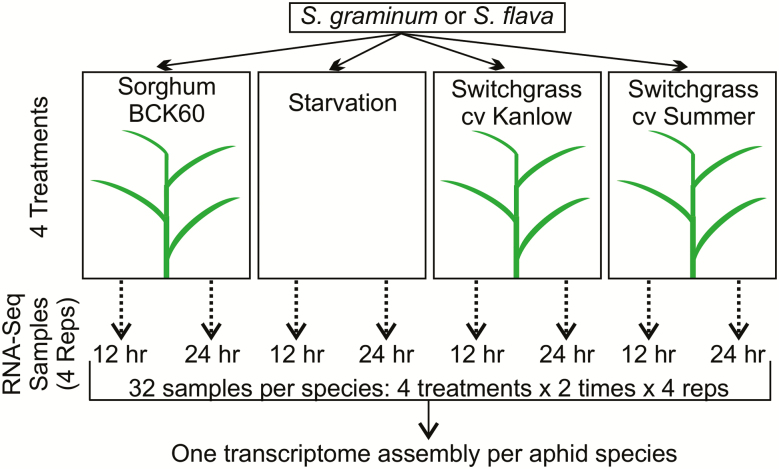
Twelve age-synchronized adult aphids were delicately transferred from BCK60 sorghum onto the youngest fully developed leaf of a single Kanlow, Summer, or BCK60 plant with a fine brush (Fig. 1). Aphids were transferred onto 12 plants per treatment and individual plants were caged with tubular plastic cages (4 cm diameter × 46 cm height) with vents covered with organdy fabric. Aphids were allowed to feed on plants for 12 or 24 h. Additionally, a fourth treatment group of aphids was starved for 12 or 24 h by caging the aphids within custom-built plastic Petri dish cages (8.9 × 2.5 cm) with two mesh windows (7 cm diameter) to allow for air circulation (Fig. 1). All experiments with S. graminum and S. flava were performed concurrently in the greenhouse so that the transcriptional responses of these two aphids to the various feeding treatments could be compared. Briefly, the greenhouse was maintained at 25 ± 4°C with the lighting augmented by LED lights (Pro 325, Lumigrow, Novato, CA) to produce a photoperiod of 16:8 (L:D) h. At 12 and 24 h after infestation, aphids were collected in 1.5 ml tubes, flash frozen in liquid nitrogen, and immediately transferred into a freezer at −80°C until samples could be processed. Aphids fed on three separate plants of the same cultivar were pooled to constitute one biological replicate of each treatment. Approximately 30 adult apterous aphids were collected for each replication, with four biological replications for each treatment.
Fig. 1.
Experimental design of feeding experiments. Each aphid species was subjected to four feeding treatments. Four biological replicates were collected after 12 and 24 h in each treatment.
RNA Extraction and Sequencing
Total RNA from 64 samples (2 timepoints [12 vs 24 h] × 4 treatments [starvation, Kanlow, Summer, and BCK60 sorghum] × 4 replicates × 2 aphid species; Fig. 1) were isolated and purified using a Qiagen RNeasy extraction kit (Qiagen, Valencia, CA) according to the manufacturer’s protocols. RNA quality was determined with a spectrophotometer (NanoDrop 1000, Wilmington, DE) and integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA was polyA-purified, reverse-transcribed, and barcoded using the TruSeq RNA-seq Library kit according to the manufacturer’s protocols (Illumina Inc, San Diego, CA). The individual samples were diluted to concentrations of 10 nM, multiplexed at a depth of five samples per lane, and sequenced on a single run on an Illumina HiSeq2500 system with paired-end 2x100-bp reads. All RNA-Seq library preparation, indexing, and sequencing were performed at the University of Nebraska DNA Sequencing Core at the University of Nebraska Medical Center (Omaha, NE) and all RNA extractions and sequencing libraries were prepared simultaneously to minimize any potential batch effects.
De novo Assembly and Annotation of S. graminum and S. flava Transcriptomes
Reads for all biological replicates and treatments were used to construct separate de novo transcriptome assemblies for S. gramiunum and S. flava. Reads were quality filtered and k-mers (k = 25) were in silico normalized to 25X coverage prior to assembly to reduce memory requirements and to facilitate the assembly of transcripts derived from low abundance k-mers. Normalized reads were de novo assembled using Trinity (v 2.4.0) (Grabherr et al. 2011) with default parameters and coding regions were predicted using Transdecoder (https://github.com/TransDecoder). Pfam domain searches were used in conjunction with Transdecoder to facilitate the detection of open reading frames (ORFs). Initially, transcripts with complete ORFs (containing both start and stop codons) or, in the case of transcripts coding for partial ORFs, the mostly highly expressed isoform for each unigene were retained; however, in order to ensure that only the highest quality transcripts were included in the differential expression analysis and to facilitate comparisons between the transcriptional responses of S. graminum and S. flava, transcripts with transcripts per million (TPM) values less than 0.5 were removed from the assembly. Transcripts potentially derived from microbes, viruses, or plants were identified using BLASTp (Altschul et al. 1997) searches against the non-redundant protein database (NR; downloaded on 8 February 2017) in conjunction with MEGAN taxonomic classification (Huson et al. 2007) and were also removed from the assembly. Transcripts that had highest scoring BLASTp matches to proteins that are known to have been horizontally transferred from microbes into the genomes of other aphid species were not removed from the assembly as they had highest scoring BLASTp matches to aphid-derived proteins in NR.
High-quality transcripts were functionally annotated using the Trinotate pipeline (https://trinotate.github.io/). In brief, coding regions were annotated via BLASTp comparisons to the Swiss-Prot/UniProt database, Pfam domains (Bateman et al. 2004) were annotated using HMMER (Eddy 1998) and the Pfam A database, signal peptides were predicted with signalP (Petersen et al. 2011), and transmembrane regions were predicted using tmHMM (Krogh et al. 2001). Completeness was evaluated using BUSCO (Benchmarking Using Single Copy Orthologs) analysis (Simao et al. 2015) with the Insecta odb9 gene set available at https://busco.ezlab.org/datasets/insecta_odb9.tar.gz. Fully annotated transcripts derived from the most highly expressed isoform from each unigene for S. graminum and S. flava were deposited in NCBI’s Transcriptome Shotgun Assembly (TSA) database under the accessions GGMR00000000 and GGMS00000000, respectively, while raw RNA-Seq reads are available at NCBI’s Sequence Read Archive (SRA) under the accessions SRR6998954–SRR6998986 for S. graminum and SRR7009156–SRR7009190 for S. flava. Trinotate annotations for S. flava and S. graminum and other supplemental data are available at Ag Data Commons (https://data.nal.usda.gov/dataset/annotations-unigenes-assembled-schizaphis-graminum-and-sipha-flava).
Read Mapping and Differential Expression Analysis
The raw RNA-Seq reads were mapped back to the S. graminum and S. flava transcriptome assemblies using Bowtie2 (Langmead and Salzberg 2012), and transcripts were quantified using RSEM (Li and Dewey 2011). Prior to differential expression analysis, low abundance transcripts, defined as transcripts with counts per million (CPM) values less than one in all but two samples, were removed from the dataset. The program edgeR was used for differential expression analysis at the unigene level (Robinson et al. 2010). In brief, reads counts for each unigene were normalized using the trimmed mean of M-values (TMM) approach, tagwise dispersions were computed, and Fisher’s exact tests of pairwise comparisons were used to identify differentially expressed unigenes (DEUs) among all possible treatment combinations. Unigenes were flagged as differentially expressed if the false discovery rate (FDR) was less than 0.05 and the log2-fold change was ≥0.25 (https://data.nal.usda.gov/dataset/annotations-unigenes-assembled-schizaphis-graminum-and-sipha-flava). In most cases, differential expression was assessed relative to the susceptible BCK60 treatment, which is a well-established host for both S. graminum and S. flava. Additionally, discriminate analysis using principal components as covariates on normalized count data of all expressed unigenes was used to explore the relationships among the different feeding treatments. R (3.4.3) (Team 2017) was used for principal components analysis (PCA) using the ‘prcomp’ function and JMP12 was used for discriminant analysis (DA) using principal components that explained 80% of the total variance in the data as covariates. For S. graminum, the first four principal components were used for DA while the first six principal components were used for DA in S. flava.
Functional Analysis of DEUs
In order to facilitate comparisons of DEUs between S. graminum and S. flava, orthology analysis was performed using all-v-all BLASTp searches with single representative transcript isoform from each unigene as queries and the program OrthoFinder (Emms and Kelly 2015). The most abundant transcript for each unigene was selected for this analysis. Special emphasis was placed on the comparison of expression patterns of 1:1 orthologs shared between S. flava and S. graminum across the different feeding treatments. Additionally, in order to determine whether feeding treatments similarly impacted expression levels of unigenes assigned to core metabolic pathways in both aphids, DEUs were assigned to KEGG pathways using the KAAS server (Moriya et al. 2007) with BLASTp searches (single-directional best hit method) against a database consisting of annotated Acyrthosiphon pisum and Diuraphis noxia enzymes. Finally, GoSeq (Young et al. 2010) was used for gene ontology (GO) enrichment analyses. Unigenes were assigned to GO categories based on their highest scoring BLASTp match to the Swiss-Prot/UniProt database and basal terms were retrieved for all GO assignments using the ‘extract_GO_assignments_from_Trinotate_xls.pl’_script included with Trinotate (version r20140708). Enrichment was determined for unigenes that were either up- or downregulated in each treatment using the entire list of unigenes with detectable expression levels as a reference. All unigenes were weighted by length and nodes with less than five mapped terms were removed from the analysis. Enrichment was determined using the Wallenius approximation (‘pwf’ option) and categories with Benjamini–Hochberg adjusted P-values ≤ 0.05 were considered enriched.
Results
Sequencing, Assembly, and Annotation
For S. graminum (GC content ~ 35%), approximately 19.6 million normalized read pairs were initially assembled into 125,483 transcripts derived from 16,361 unigenes after first pass filtering to include only transcripts containing coding regions (Table 1). After filtering to remove transcripts with TPM ≤ 0.5, 38,420 transcripts derived from 8,428 unigenes with predicted coding regions remained. Median contig length and contig N50 for the filtered assembly were 2,482 nt and 3,564 nt, respectively, and 6,787 unigenes coded for full-length proteins containing both a start and stop codon (Table 1). Of the unigenes that coded for proteins, 6,592 (78.2%) had significant BLASTp matches to the Swiss-Prot database while 6,458 (76.6%) had at least one Pfam domain (Table 1). For S. flava (GC content ~36%), approximately 18.9 million normalized read pairs were assembled into 116,391 transcripts derived from 14,331 unigenes after first pass filtering to include only transcripts with coding potential (Table 1). After filtering to remove low abundance transcripts, 41,941 transcripts derived from 8,866 unigenes remained. For the filtered assembly, median contig length was 2,422 nt, contig N50 was 3,541 nt, and 7,159 coded for full-length proteins (Table 1). Additionally, 6,847 (77.3%) of the unigenes that coded for proteins had BLASTp matches to the Swiss-Prot database and 6,738 (76.0%) had at least one Pfam domain.
Table 1.
Transcriptome assembly and annotation metrics
| Metric | S. graminum | S. flava |
|---|---|---|
| Raw read pairs (M) | 988 | 1,103 |
| Initial assembly | ||
| Unigenes | 16,361 | 14,331 |
| Transcripts | 125,483 | 116,391 |
| Assembly length (Mb) | 37.8 | 36.5 |
| Assembly GC content | 34.89% | 36.41% |
| Median contig length (nt) | 1,436 | 1,633 |
| Average contig length (nt) | 1,813 | 2,011 |
| Contig N50 (nt) | 2,445 | 2,522 |
| Longest isoform only | ||
| Assembly length (Mb) | 31.3 | 30.7 |
| Median contig length (nt) | 1,538 | 1,766 |
| Average contig length (nt) | 1,914 | 2,139 |
| Contig N50 (nt) | 2,619 | 2,964 |
| Filtered assembly | ||
| Unigenes | 8,428 | 8,866 |
| Transcripts | 38,420 | 41,941 |
| Assembly length (Mb) | 24.1 | 25.9 |
| Median contig length (nt) | 2,482 | 2,422 |
| Average contig length (nt) | 2,863 | 2,861 |
| Contig N50 (nt) | 3,564 | 3,541 |
| Annotation | ||
| Full-length unigene | 6,787 (80.53%) | 7,159 (80.75%) |
| BLASTp match vs Swiss-Prot | 6,592 (78.20%) | 6,847 (77.30%) |
| PFAM domain | 6,458 (76.60%) | 6,738 (76.00%) |
| Predicted signal peptide | 1,219 (14.40%) | 1,347 (15.20%) |
| Predicted transmembrane domain | 2,720 (32.30%) | 3,090 (34.90%) |
Reads were normalized and assembled using Trinity with default parameters. Transcripts were collapsed into unigenes and annotations were performed using Trinotate. Percentages are expressed relative to the total number of unigenes with predicted coding regions.
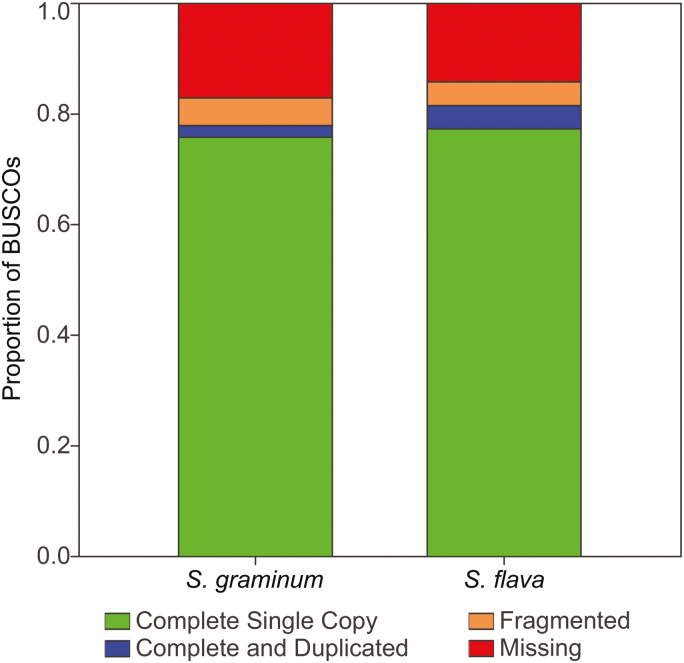
BUSCO analysis of the 8,428 high-quality unigenes for S. graminum indicated a fairly complete assembly of the transcriptome and a low rate of duplicated single copy orthologs with 75.8% identified as complete single copy orthologs in the assembly and 2.1% identified as complete and duplicated (Fig. 2). BUSCO results for S. flava were similar to those from S. graminum and indicated that 77.3% BUSCOs were present as complete single copies and 4.2% were complete and duplicated (Fig. 2). For both species, the recovery of complete and single copy orthologs from both the TPM filtered and unfiltered datasets were similar, indicating that removing low abundance transcripts from the datasets did not inadvertently remove transcripts coding for proteins with core metabolic functions. Instead, the percentage of complete and duplicated transcripts dropped from 7.1 to 2.1% in S. graminum and from 6.3 to 4.2% in S. flava and likely improved the quality of our assembly. Additional orthology analysis led to the detection of 4,502 1:1 orthologs (Table 2) that were shared between S. flava and S. graminum, the majority of which encoded enzymes linked to core metabolic pathways. However, many of the unigenes were not in 1:1 orthologous relationships, with 1,007 and 1,065 unigenes in S. graminum and S. flava assigned to 351 different orthogroups containing ‘many:many’ orthologs, as well as others that were assigned to ‘many:1’, ‘1:many’, ‘many:0’, ‘0:many’, ‘1:0’, and ‘0:1’ ortholog groups (Table 2). Overall, the distribution of unigenes coding for partial ORFs into the different ortholog categories did not significantly deviate from the distribution of unigenes coding for full-length ORFs for either aphid species (Table 2), suggesting that the presence of partial ORFs in our transcriptome assemblies did not inflate the abundance of ‘many:1’ or ‘many:0’ orthogroups in either transcriptome assembly.
Fig. 2.
BUSCO analysis of S. graminum and S. flava transcriptome assemblies. The proportion of BUSCOs identified as complete single copy (green), complete and duplicated (blue), fragmented (orange), and missing (red) in the S. graminum and S. flava transcriptome assemblies.
Table 2.
Orthogroup comparisons for S. graminum and S. flava
| Orthogroup class | No. of orthogroups | No. of S. graminum unigenes (complete) | No. of S. graminum unigenes (partial) | No. of S. flava unigenes (complete) | No. of S. flava unigenes (partial) |
|---|---|---|---|---|---|
| 1: 1 | 4,502 | 3,874 | 440 | 3,874 | 392 |
| Many: many | 351 | 630 | 202 | 683 | 203 |
| Many: 1 | 179 | 331 | 125 | 143 | 25 |
| 1: many | 167 | 155 | 16 | 380 | 79 |
| many: 0 | 6 | 81 | 12 | 0 | 0 |
| 0: many | 6 | 0 | 0 | 59 | 16 |
| 1: 0 | 2,287 | 1,719 | 843 | 0 | 0 |
| 0: 1 | 2,683 | 0 | 0 | 2,031 | 981 |
Orthologs were determined using reciprocal BLASTp searches and the program Orthofinder. Orthogroup ratios are presented as S. graminum: S. flava. Complete = complete ORFs; Partial = partial ORFs.
Analysis of KEGG Pathways and Unigenes Coding for Enzymes Linked to Digestion and/or Detoxification in S. graminum and S. flava
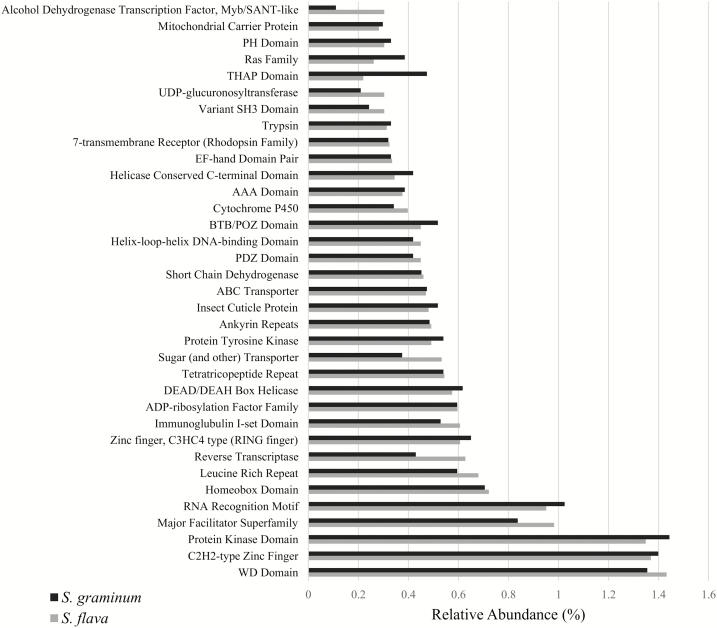
KEGG pathway analysis indicated a similar representation of unigenes coding for enzymes belonging to core metabolic pathways, and unigenes coding for all enzymes and proteins of all major metabolic pathways were readily identified from both aphid species. As in previous insect genome and transcriptome assemblies (Xie et al. 2012, Hull et al. 2013, Scully et al. 2013), the most common gene families in the transcriptome assemblies of S. flava and S. graminum were unigenes coding for structural proteins, proteins involved in signaling, transcription factors, and gene families linked to stress response and detoxification. In both assemblies, WD-domain containing unigenes were among the most abundant Pfam domains in the assembly. Additional abundant domains included protein kinases, C2H2-type zinc fingers, RNA recognition motifs, and major facilitator superfamilies (Fig. 3). Interestingly, both aphids coded for similar numbers of genes linked to digestion, detoxification and stress response, and plant host interactions, which were identified using highest scoring BLASTp matches to the Swiss-Prot database. Among others, these annotated unigenes included cytochrome P450s (CYP450s), ABC transporters, lipases, and glutathione-S-transferases (GSTs) (Table 3). Unigenes that were present in higher copy numbers in S. graminum relative to S. flava included β-glucuronidases, γ-glutamyltranspeptidases (GGTs), leucine-rich repeats (LRRs), cathepsin B cysteine proteinases, and maltases. Unigenes that were present in higher copy numbers in S. flava relative to S. graminum included B(0,+)-type amino acid transporters, lysosomal α-glucosidases, UDP-glucuronosyltransferases (UGTs), and vigilins. Some gene families were found exclusively in one of the aphid species and predominantly included transcription factors and transposable elements; however, eight unigenes containing glycoside hydrolase (GH) family 31 domains were found exclusively in S. flava (Table 3).
Fig. 3.
Relative abundances of the top 35 most abundant Pfam domains in the S. graminum and S. flava transcriptome assemblies. Occurrences of Pfam domains were counted only once for each protein and relative abundance is expressed relative to the total number of proteins in the transcriptome that had detectable Pfam domains. Pfam annotations for all unigenes are available at https://data.nal.usda.gov/dataset/annotations-unigenes-assembled- schizaphis-graminum-and-sipha-flava.
Table 3.
Copy number of unigenes related to digestion and stress response in S. graminum and S. flava based on BLASTp searches against the Swiss-Prot database
| Gene annotation | S. graminum | S. flava |
|---|---|---|
| Similar numbers in both species | ||
| Cytochrome P450 | 42 | 44 |
| Dehydrogenase/reductase SDR family | 17 | 14 |
| Glutathione S-transferase | 9 | 9 |
| Lipase | 17 | 13 |
| Carbonic anhydrase | 5 | 5 |
| Carboxylesterase | 15 | 13 |
| Esterase | 16 | 13 |
| Trehalase | 9 | 9 |
| Laccase | 4 | 4 |
| Peroxidase | 21 | 21 |
| ABC transporter | 22 | 22 |
| Heat shock protein | 28 | 28 |
| Facilitated trehalase transporter | 60 | 57 |
| Major facilitator superfamily | 23 | 20 |
| More abundant in S. graminum | ||
| Beta-glucuronidase | 8 | 3 |
| Gamma glutamyltranspeptidase | 15 | 4 |
| Cathepsin B cysteine proteinase | 15 | 8 |
| Glucose dehydrogenase | 37 | 26 |
| Peptide transporter | 10 | 3 |
| Phenol oxidase | 6 | 2 |
| Acetyl-CoA transporter | 51 | 13 |
| Beta-mannosidase | 4 | 1 |
| Leucine-rich repeat | 43 | 34 |
| Myrosinase | 8 | 3 |
| Maltase | 18 | 9 |
| Peroxidasin | 7 | 3 |
| Thiamine transporter | 13 | 9 |
| More abundant in S. flava | ||
| Lysosomal acid phosphatase | 3 | 9 |
| B(0,+)-type amino acid transporter | 4 | 8 |
| Lysosomal alpha-glucosidase | 3 | 8 |
| Vigilin | 1 | 12 |
| UDP-glucuronosyltransferase | 20 | 24 |
| Glycoside hydrolase family 31 | 0 | 8 |
Discriminate Analysis Reveals Similarities Between Aphids Exposed to Biotic and Abiotic Stresses
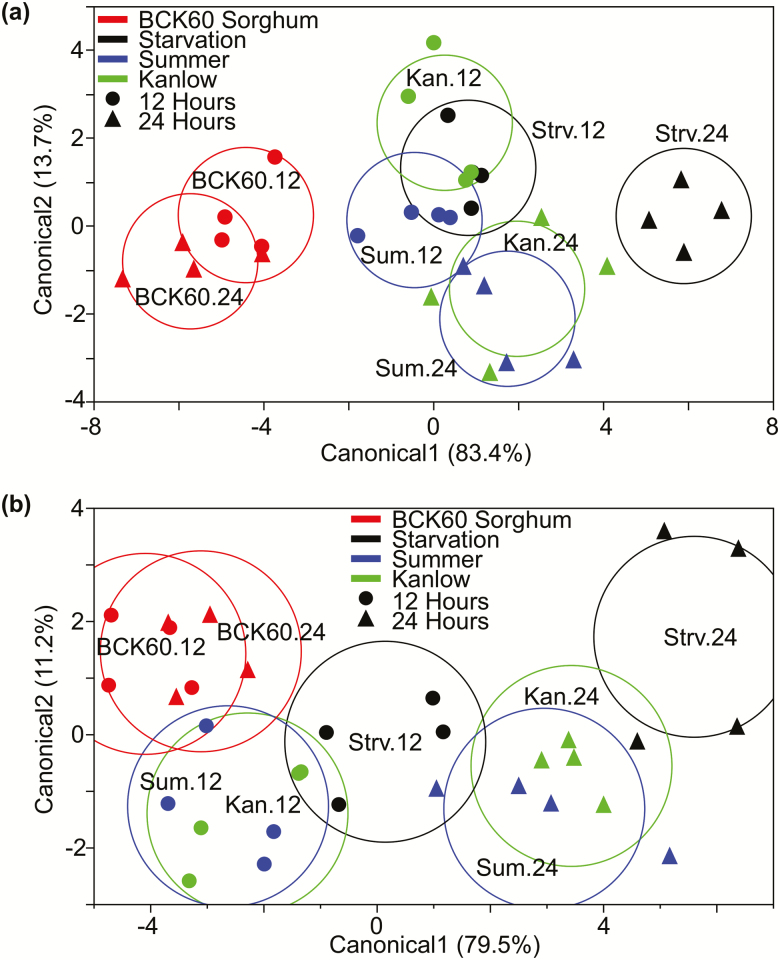
Canonical axis 1 in the discriminate analysis clearly separated the two switchgrass feeding treatments from the sorghum feeding treatment and the 24-h starvation treatment from all other treatments in S. graminum (Fig. 4a). Although there was considerable overlap between the Kanlow and Summer feeding treatments, the switchgrass samples were separated by time along canonical axis 2, with the 12- and 24-h sampling times each forming distinct clusters. The 12-h starvation treatment clustered strongly with both 12-h switchgrass feeding treatments. Similar relationships between the treatments were observed in the discriminate analysis for S. flava (Fig. 4b). For example, as was observed for S. graminum, the switchgrass treatments were mostly separable from the sorghum treatment and the 24-h starvation treatment was clearly separable from all other treatments along canonical axis 1. Additionally, there was significant overlap between the Kanlow and Summer treatments, but they were separated by time along canonical axis 1. Unlike S. graminum, however, the 12-h starvation treatment did not cluster strongly with the 12-h switchgrass feeding treatments and, instead, was positioned equidistantly between the 12- and 24-h switchgrass treatments along canonical axis 1 (Fig. 4b).
Fig. 4.
Discriminate analyses of (a) S. flava and (b) S. graminum transcriptional profiles. Discriminate analysis (DA) was performed using principal components as covariates using the ‘prcomp’ function in R and JMP12 for DA. Principal components that explained 80% of the total variance in the data were used as covariates in the DA analysis. Red indicates BCK60 samples, blue indicates Summer, green indicates Kanlow, and black indicates starvation. The 12 and 24 h timepoints are represented by circles and triangles, respectively. Colored circles represent the 95% confidence intervals around the true mean for each treatment.
Overall, the expression levels of 5,327 and 4,436 unigenes were impacted by the different feeding treatments in S. graminum and S. flava, respectively, and starvation had the strongest impacts on the expression profiles of both aphids, which altered the expression of 3,625 unigenes in S. graminum and 2,255 unigenes in S. flava relative to the BCK60 sorghum treatments over the 24-h time course. Expression levels of more unigenes were impacted by the different feeding treatments in S. graminum compared to S. flava and the 24-h timepoints had more significant impacts on global gene expression in both aphids relative to the 12-h timepoints in all feeding treatments. However, S. graminum seemed to mount an earlier and more robust stress response to starvation and feeding on Kanlow or Summer switchgrass in comparison to S. flava as more unigenes were differentially expressed at 12 h and expression levels were generally sustained through 24 h. In contrast, significant differential expression was not observed in S. flava until 24 h. Notably, it appeared that starvation and switchgrass feeding treatments did not similarly impact the expression levels of 1:1 orthologous unigenes in the two aphids. For example, 2,292 of the total 4,502 1:1 orthologs were differentially expressed in at least one aphid as a result of starvation, but only 46 (six downregulated and 40 upregulated) were impacted in both aphids at 12 h while 535 (251 downregulated and 284 upregulated) were differentially expressed in both aphids after 24 h of starvation. Similarly, 1,421 1:1 orthologs were differentially expressed when feeding on Kanlow while only 257 1:1 orthologs were impacted in both aphids after 24 h (117 downregulated and 140 upregulated), and of the 1,176 1:1 orthologs whose expression was impacted in the Summer treatment, only one was upregulated in both aphids at 12 h and 214 were shared at 24 h (70 downregulated and 144 upregulated).
Abiotic Starvation Stress had a Delayed Impact on Unigene Expression Levels in S. flava Compared to S. graminum
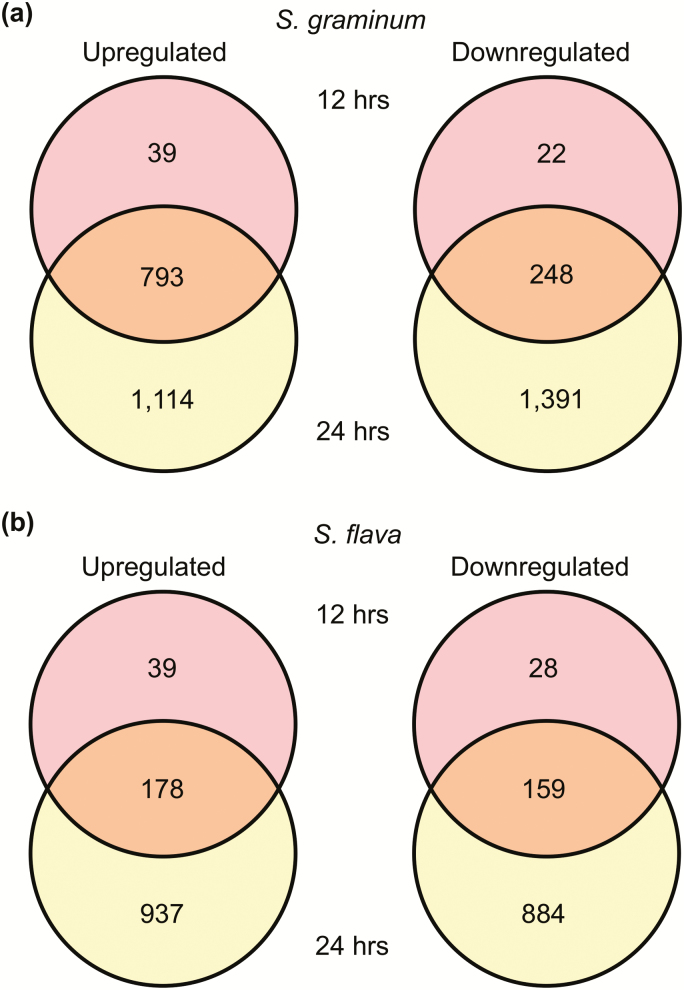
For S. graminum, 39 and 1,114 unigenes were exclusively upregulated in starved aphids relative to BCK60 sorghum at 12 and 24 h, respectively, whereas 22 and 1,391 were exclusively downregulated at these timepoints (Fig. 5a). Overall trends for S. flava were similar as 28 and 884 unigenes were downregulated exclusively at 12 and at 24 h while 39 and 997 were upregulated exclusively at these timepoints (Fig. 5b). The larger numbers of DEUs identified in S. graminum compared to S. flava suggest that the responses to starvation stress was differentially modulated in the two aphids at the transcriptomic level.
Fig. 5.
Venn diagrams of differentially expressed unigenes in starvation relative to BCK60 sorghum. (a) Up- and downregulated unigenes in S. graminum after 12 and 24 h of starvation. (b) Up- and downregulated unigenes at S. flava after 12 and 24 h of starvation.
GO enrichment analysis was performed to summarize the functional categories among the unigenes that were differentially expressed in response to starvation (Supp. Tables S1 and S2). More GO categories were commonly enriched between the two aphids when comparing the 12-h starvation treatment in S. graminum to the 24-h starvation treatment in S. flava, supporting the observation of a delayed response to starvation in S. flava. For example, only one GO category showed enrichment in the DEUs that were upregulated during starvation at 12 h in both aphids. In contrast, seven GO categories largely linked to lipid transport and carbohydrate catabolism were commonly enriched in upregulated DEUs at 12 h in starved S. graminum and after 24 h of starvation in S. flava. A similar trend was noted for the downregulated DEUs. No GO categories were enriched in the DEUs that were downregulated in S. flava at 12 h; however, 10 categories largely linked to mitosis and purine ribonucleoside metabolism that were downregulated in S. graminum at both 12 and 24 h timepoints were also enriched at 24 h in S. flava. Additional categories linked to DNA replication and cell cycle were downregulated in starved aphids of both species exclusively at 24 h (Supp. Table S1).
Several GO categories were enriched exclusively in either S. graminum or S. flava. In S. flava, 25 GO categories were enriched in the upregulated DEUs at both 12 and 24 h, which included unigenes coding for enzymes linked to carbohydrate transport and amino acid and nitrogen metabolism (Supp. Table S2). No GO categories were enriched in the downregulated DEUs. Nine categories linked to DNA replication and two linked to peptidase activity were enriched in downregulated DEUs in the starvation treatment at both 12 and 24 h in S. graminum while and 65 categories were enriched in upregulated DEUs at both 12 and 24 h. GO categories enriched in the upregulated DEUs were largely linked to detoxification, glycosyl compound metabolism, metabolism of purine nucleosides, vitamin transport, cation transport, and regulation of cholesterol storage (Supp. Table S2).
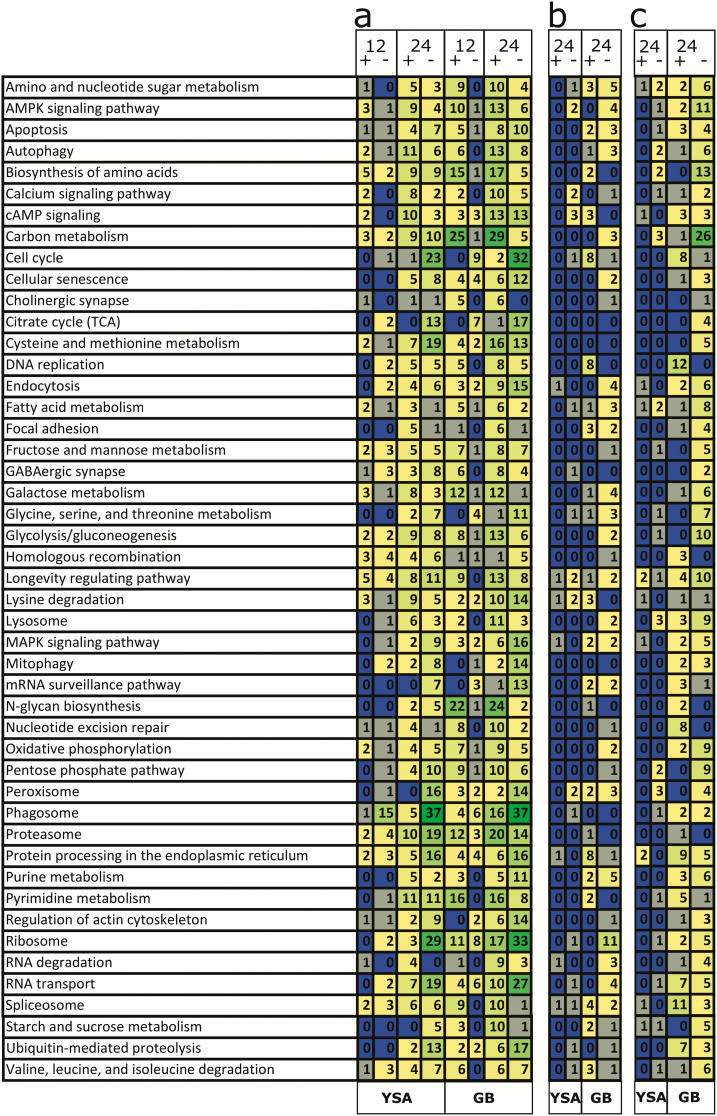
KEGG pathway analysis provided some additional insight into the types of metabolic processes that were impacted in both aphids by starvation (Fig. 6a) and again, this analysis showed that changes in gene expression were delayed in response to starvation in S. flava compared to S. graminum and that the expression levels of unigenes assigned to more metabolic pathways were impacted in S. graminum. For example, unigenes coding for ribosomal proteins, purine metabolism, biosynthesis of amino acids, and glycolysis were upregulated in starved S. graminum at both 12 and 24 h but were only upregulated in S. flava at 24 h. Similar trends were also noted for unigenes coding for enzymes involved in the following pathways: purine metabolism, biosynthesis of amino acids, glycolysis, peroxisome, apoptosis, endocytosis, and signaling pathways. In contrast, unigenes linked to lysine degradation were strongly upregulated in both aphids at 12 h and expression levels were sustained through the 24-h timepoint while unigenes linked to ubiquitin-mediated proteolysis and proteasome were downregulated strongly at 24 h in both S. graminum and S. flava. Of particular note, over 20 unigenes coding for enzymes linked to oxidative phosphorylation were strongly upregulated in S. graminum at both 12 and 24 h but were not similarly impacted in S. flava (Fig. 6a). Analysis of the downregulated DEUs largely mirrored the GO enrichment analysis in that unigenes coding for enzymes linked to cell cycle and DNA replication were downregulated earlier at 12 h in S. graminum and later at 24 h in S. flava (Fig. 6a). Similar patterns were noted for downregulated DEUs associated with pathways for the synthesis of N-glycans, focal adhesions, and mRNA surveillance. Notably, more pathway similarities between the two aphids were noted among the downregulated unigenes relative to the upregulated unigenes.
Fig. 6.
Representation of unigenes coding for enzymes assigned to KEGG pathways in (a) starvation, (b) Summer, and (c) Kanlow treatments. Unigenes were assigned to KEGG pathways based on highest scoring single-directional BLASTp matches to a database containing annotated Acyrthosiphon pisum and Diuraphis noxia enzymes. Numbers indicate the number of unigenes assigned to each pathway. Green indicates higher abundance pathways, yellow and gray indicate lower abundance pathways, and blue indicates pathways with 0 mapped unigenes. + designates upregulated unigenes, - indicates downregulated unigenes, GB = Schizaphis graminum, and YSA = Sipha flava. Because few differences were observed at the 12-h timepoint when S. flava was fed in either Summer or Kanlow, only the 24-h timepoints are shown for the switchgrass feeding treatments.
Finally, S. graminum seemed to mount an earlier stress response compared to S. flava based on expression patterns of unigenes related to detoxification. More unigenes annotated as carboxylesterases, CYPP450s, GSTs, heat shock proteins (HSPs), LRRs, cysteine proteinases, peroxidases, and UGTs (among others) were differentially expressed at both 12 and 24 h in S. graminum relative to S. flava (Supp. Table S3). In contrast, although induced earlier at 12 h in S. graminum, more unigenes annotated as alcohol dehydrogenases, sulfatases, and sulfate transporters were upregulated at 24 h in S. flava relative to S. graminum while more ABC transporter unigenes were downregulated in S. flava at this timepoint (Supp. Tables S3 and S4).
Biotic Stress Induced by Feeding on Switchgrass Plants Differentially Affected Expression Levels of Unigenes Related to Detoxification in S. flava and S. graminum
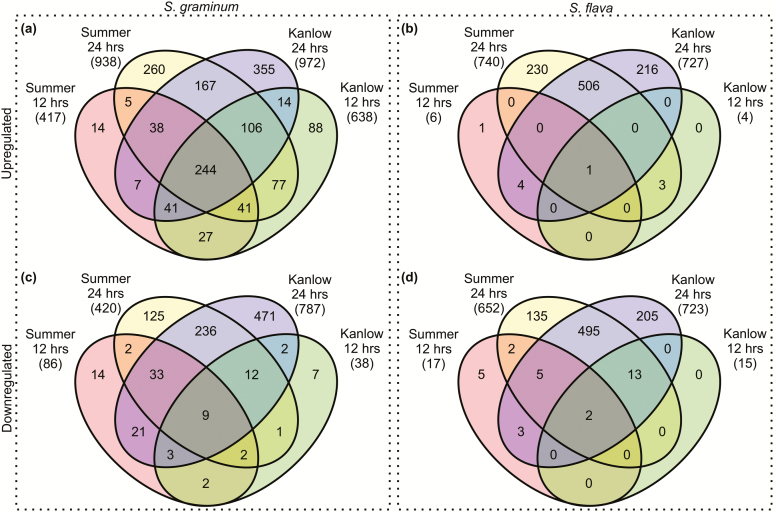
Switchgrass hosts differentially affected the transcriptomes of S. graminum and S. flava relative to BCK60 sorghum, and as in the starvation treatment, more pronounced impacts on global unigene expression were noted at 24 h in both aphids when they were fed in both hosts (Fig. 7). Although the expression levels of many DEUs were commonly impacted in aphids feeding on both switchgrass cultivars, several DEUs were exclusively impacted by feeding on either Kanlow or Summer (Fig. 7a and c), suggestive of specific host-induced effects on both aphid transcriptomes. Additionally, fewer numbers of DEUs were observed at 12 h in S. flava relative to S. graminum and more DEUs were identified in the Kanlow treatment in S. graminum relative to S. flava suggesting that S. graminum responds earlier to both hosts and mounts a more pronounced and complex transcriptional response to the resistant host (Fig. 7b and d).
Fig. 7.
Venn diagrams of differentially expressed unigenes in switchgrass treatments relative to BCK60 sorghum. Unigenes upregulated in S. graminum (a) and S. flava (b) as a result of feeding on Kanlow or Summer switchgrass after 12 and 24 h. Unigenes downregulated in S. graminum (c) and S. flava (d) as a result of feeding on Kanlow or Summer switchgrass after 12 and 24 h. Numbers in parenthesis indicate the total number of differentially expressed unigenes in each treatment.
The responses of the two aphid species to feeding in switchgrass also diverged significantly from each other based on the predicted unigene functions and signatures of stress responses. When comparing unigenes whose expression levels were impacted in both switchgrass cultivars, only a handful of GO terms and metabolic pathways were commonly enriched in both aphid species (Supp. Table S5). Of the over 30 GO categories that were enriched in downregulated DEUs in S. flava feeding from both switchgrass treatments and the over 100 different categories that were enriched in the upregulated DEUs in S. graminum from both switchgrass feeding treatments (Supp. Tables S5 and S6), only two GO terms were associated with DEUs upregulated at 24 h in both aphids and in both switchgrass treatments (small molecular metabolic process and extracellular region) and one GO term was associated with downregulated DEUs (mitotic cell cycle process) (Supp. Table S5). Likewise, few KEGG pathways were similarly impacted in unigenes that responded to both switchgrass feeding treatments in both aphid species. For example, DEUs encoding proteins for cell cycle, DNA replication, ubiquitin-mediated proteolysis, N-glycan metabolism, purine and pyrimidine metabolism, mitosis, RNA degradation, and splicesome were downregulated at 24 h while DEUs associated with the glycolysis and gluconeogenesis pathway were upregulated in both aphids after feeding on both switchgrass cultivars at this timepoint (Fig. 6b and c).
Despite the minor similarities between the common transcriptional responses of both aphids to both switchgrass cultivars, marked differences between the two aphids were noted to the GO and KEGG pathway assignments of the rest of the DEUs, especially with regards to those coding for proteins linked to digestion and stress response (Supp. Table S6). For example, S. graminum responded strongly to both switchgrass cultivars after 12 h of feeding by upregulating DEUs enriched for GO terms related to mitochondrial respiratory chain complex, monooxygenase activity, nucleoside biosynthesis, glycosyl compound biosynthesis, glycolytic processes, tetrapyrrole binding, oxidoreductase activity, xenobiotic metabolic processes, and heme binding (Supp. Table S6). All of the GO terms that were enriched in DEUs observed at the 12 h timepoint were also enriched at the 24 h timepoint. Likewise, unigenes assigned to KEGG pathways linked to oxidative phosphorylation, pentose phosphate pathway, ribosome, and starch and sucrose metabolism were upregulated at 12 h and sustained through 24 h in S. graminum (Fig. 6b and c). Additional upregulated pathways at 24 h included those related to lysosome, autophagy, peroxisome, and glycine, serine, and threonine metabolism (Fig. 6b and c). Finally, many unigenes coding for canonical stress, digestive, and detoxification enzymes were induced early at 12 h in S. graminum and were sustained through 24 h, including trypsins, papain family cysteine proteinases, CYP450s, HSP70s, peroxidases, UGTs, and LRRs (Supp. Tables S3 and S4). Several unigenes coding for chemosensory receptors were also upregulated exclusively at 12 h while upregulation of only one of these unigenes was sustained through 24 h (Supp. Table S7).
In contrast, only a few unigenes whose expression level was commonly impacted in both switchgrass treatments had expression changes at 12 h in S. flava and thus, no enriched GO terms were identified at this timepoint. The majority of the unigenes upregulated at 24 h in S. flava were assigned to GO terms related to carbohydrate transport, sulfur amino acid metabolic processes, and carboxylic acid transport while unigenes downregulated in both switchgrass feeding treatments were associated with microtubule-based processes, oligosaccharyltransferase, and cell cycle (Supp. Table S6). Likewise, unigenes assigned to major KEGG metabolic pathways were not strongly impacted at 12 h; however, unigenes assigned to KEGG pathways related to signaling, including AMPK signaling and MAPK signaling, were upregulated at 24 h in S. flava while those linked to the degradation of valine, leucine, and isoleucine, cysteine and methionine metabolism, glutathione metabolism, lysosome, and autophagy were downregulated (Fig. 6b and c). Additionally, in contrast to S. graminum, no unigenes related to digestion/nutrient acquisition and stress responses were upregulated at 12 h and fewer unigenes related to these processes were upregulated 24 h in S. flava in comparison to S. graminum (Supp. Tables S3 and S4). For example, only two unigenes coding for trypsins, one unigene coding for papain family cysteine proteinase, five unigenes coding for CYP450s, and three coding for HSP70s were upregulated in S. flava at 24 h. However, numbers of upregulated unigenes coding for LRRs, major facilitator superfamily transporters, UGTs, alcohol dehydrogenases, and melibiase in S. flava exceeded the numbers of those unigenes upregulated in S. graminum (Supp. Tables S3 and S4). Also in contrast to S. graminum, many unigenes linked to stress response and digestion were actually downregulated in S. flava at 24 h including unigenes coding for sugar transporters, GH family 1, peroxidase, HSP70, CYP450, UGT, and major facilitator transporters, among others (Supp. Tables S3 and S4). In addition, more unigenes coding for chemosensory related receptors were upregulated in S. flava compared to S. graminum (Supp. Table S7).
Feeding on the Susceptible Cultivar Summer was Associated With Upregulation of Stress-Related Unigenes in S. graminum at 24 h and Downregulation of These Unigenes in S. flava
Among the DEUs that were impacted exclusively in the Summer treatment, few were related to primary metabolic pathways and instead, coded for transcription factors, transporters, proteinases, detoxification enzymes, structural proteins, and molecules involved in signal transduction in both aphid species. For this reason, few pathway or GO enrichments were detected among the DEUs that were exclusively impacted in S. graminum or S. flava that had fed on Summer; however, GO terms associated with oligosaccharyltransferase, microtubule-based process, cell cycle, and intracellular part were exclusively enriched in the downregulated DEUs in the Summer treatment in S. graminum (Supp. Table S6). As in the comparisons described above, few similarities were noted between the two aphid species feeding on this cultivar. Schizaphis graminum responded early at 12 h by upregulating unigenes annotated as insect cuticle proteins, thioredoxin, chemosensory proteins, about half of which remained upregulated at 24 h, and unigenes associated with growth and development, including unigenes annotated as chitin-binding peritrophin A and juvenile hormone binding proteins (JHBPs) (Supp. Table S8). Additional unigenes related to digestion and stress response were induced at 24 h, in S. graminum including amino acid permease, sulfate transporter, ammonium transporter, GGTs, glutathione peroxidase, HSP70s, and UGT (Supp. Table S3). Few unigenes related to stress and digestion were downregulated at 24 h in S. graminum; however, expression levels of two of the unigenes coding chitin-binding peritrophin A proteins and three coding for JHBPs returned to similar levels as were observed in the BCK60 treatment after 24 h (Supp. Table S8).
In contrast, GO categories associated with monosaccharide metabolic processes and single organism catabolic processes were associated with upregulated DEUs in S. flava (Supp. Table S6). Additionally, no impacts to the expression levels of unigenes related to growth and development were observed at 12 h in S. flava while DEUs associated with detoxification, digestion, and stress response were downregulated at 12 h in S. flava feeding on Summer relative to BCK60 sorghum plants. These included unigenes annotated as ABC transporters, ammonium transporters, CYP450s, GMC oxidoreductases, lipases, HSP90s, sugar (and other) transporters, trypsins, insect cuticle proteins, and pheromone binding proteins (Supp. Tables S3, S4, S5, and S8). Downregulation of three of these DEUs was also observed through the 24-h timepoint in S. flava along with several other unigenes annotated as CYP450, GGT, LRR, and GH 1 (Supp. Tables S3 and S4). Unlike S. graminum, upregulated unigenes observed at 24 h included unigenes coding for different classes of digestive and stress-responsive proteins, such as major facilitator superfamily transporters, caspases, and lipases; however, like S. graminum, a single unigene coding for thioredoxin was also upregulated at 24 h (Supp. Tables S3 & S4). Additionally, three chemosensory genes were upregulated at 24 in S. flava (Supp. Table S7).
Feeding on Cultivar Kanlow was Associated With More Significant Changes to S. graminum Transcsriptomes Relative to S. flava
As in the Summer treatment, few DEUs that were exclusively impacted in the Kanlow treatment coded for enzymes assigned to primary metabolic pathways in S. flava and S. graminu (Fig. 7). Furthermore, no enriched GO terms were associated with DEUs exclusively from the Kanlow treatment in S. graminum or S. flava and the expression levels of few unigenes annotated as chemosensory proteins were exclusively impacted in the Kanlow treatment in either aphid species (Supp. Tables S6 and S7). Again, S. graminum transcriptomes responded earlier to feeding on Kanlow relative to S. flava as more DEUs were identified at 12 h, the majority of which were upregulated (Fig. 7). The 88 DEUs upregulated exclusively in S. graminum at 12 h in the Kanlow treatment were largely linked to stress response and included those annotated as antifungal peptides, CYP450s, lysosomal α-mannosidases, GMC oxidoreductases, LRRs, major facilitator superfamily proteins, papain cysteine proteinases, PBP/GOBP, short-chain dehydrogenases, and UGTs (Supp. Tables S3 and S4), among other functions. Few unigenes were downregulated exclusively in S. graminum in the Kanlow treatment at 12 h and all coded for putative transcription factors. Continued feeding on Kanlow plants resulted in the upregulation of additional unigenes related to plant interactions and stress responses at 24 h including 7-transmember chemosensory receptor, ABC transporters, peroxidases, LRRs, major facilitator superfamily transporters, torsins, sugar (and other) transporters, sulfatases, thioredoxins, insect cuticle proteins, and UGTs (Supp. Tables S3 and S4). Other upregulated unigenes included those annotated as a 7-transmemberane sweet taste receptor of 3 G-protein coupled receptor (GCPR) and several peptides involved in signaling (Supp. Tables S3, S4, and S7). In contrast to the Summer treatment, many more DEUs associated with stress responses and digestion were exclusively downregulated in the Kanlow treatment relative to BCK60 sorghum in S. graminum, potentially indicative of an inability to use Kanlow as a suitable host. These unigenes included those annotated as AKRs, CYP450s, GSTs, GMC oxidoreductases, LRRs, lipases, major facilitator transporters, short chain dehydrogenases, sugar (and other) transporters, amino acid transporters, and UGT (Supp. Tables S3 and S4). Other downregulated DEUs included those linked to MAPK signaling, cellular senescence, focal adhesion, endocytosis, TCA cycle, and autophagy (Fig. 6c).
In contrast to S. graminum, no DEUs were exclusively up or downregulated in S. flava after 12 h feeding on Kanlow plants (Fig. 7b and d) and comparatively fewer DEUs were downregulated in the Kanlow treatment in S. flava relative to S. graminum. The majority of the upregulated DEUs at 24 h were largely associated with stress responses and included those encoding carbonic anhydrase, hemopexin, LRR, UGT, and lipase (Supp. Table S3). In contrast to previous treatments, fewer unigenes coding for stress-related proteins were downregulated in this treatment at 24 h. Among the downregulated DEUs included unigenes annotated as GMC oxidoreductases, trypsins, CYP450s, insect cuticle proteins, insect pheromone binding proteins, JHBPs, DNA mismatch repair proteins (mutS), neurotransmitter-gated ion-channel, GH 18 chitinase, elongation factor-Tu, and UGT (Supp. Tables S3, S4, S7, and S8).
Starvation and Feeding on Switchgrass Trigger Shared Transcriptomic Responses
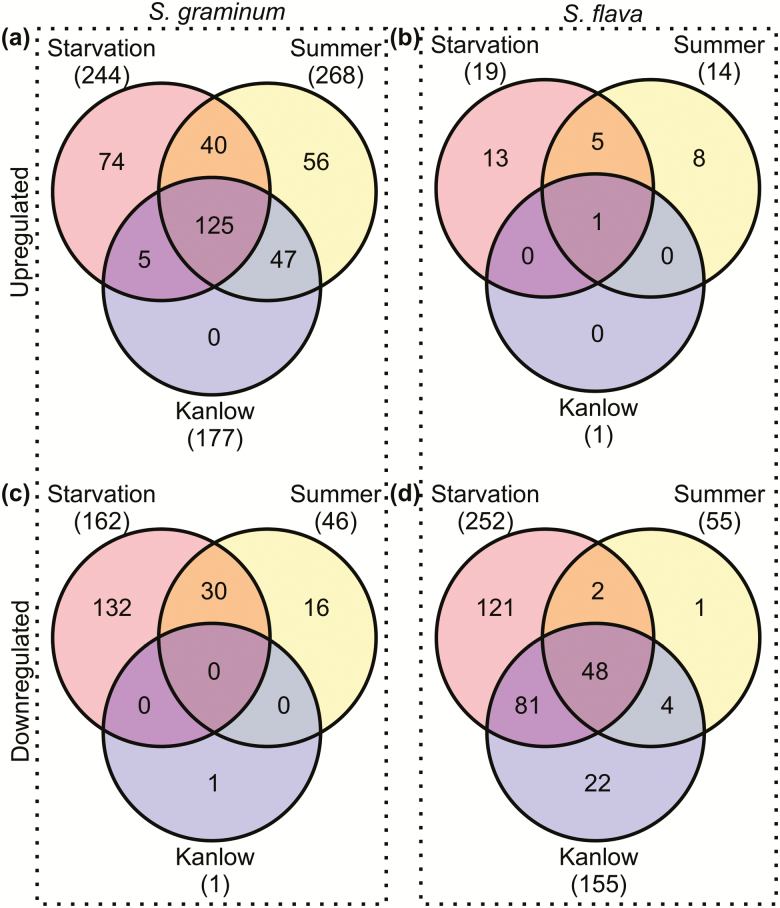
The transcriptomes obtained from starved aphids across the two harvest timepoints were compared to the transcriptomes obtained from aphids feeding on Kanlow, which is a poor host for S. graminum and a suboptimal host for S. flava, and Summer, which is a suitable host for both aphids (Koch et al. 2014a, Koch et al. 2014b, Koch et al. 2015) to identify similarities and differences between abiotic stress (starvation) and biotic stress (feeding on switchgrass) responses (Fig. 8). Overall, starvation, and feeding on Summer, and Kanlow induced the upregulation of 1,946, 1,027 and 1,205 DEUs, and the downregulation of 1,661, 460, and 799 DEUs in S. graminum, respectively (Fig. 8a and c). Of the DEUs, 725 upregulated and 311 downregulated DEUs were shared between the three treatments regardless of timepoint, indicating common transcriptional responses to starvation and feeding on switchgrass in S. graminum. However, 191 unigenes were exclusively upregulated in both the starvation and Summer treatments, respectively, while 121 were exclusively downregulated. Interestingly, more unigenes were commonly and exclusively differentially expressed in the Kanlow and starvation treatments relative to the Summer-starvation comparison with 323 upregulated and 312 downregulated (Fig. 8a and c).
Fig. 8.
Venn diagrams contrasting starvation and feeding on switchgrass relative to BCK60 sorghum. Shared and unique unigenes upregulated in S. graminum (a) and S. flava (b) as a result of starvation, feeding on Kanlow, or feeding on Summer. Shared and unique unigenes downregulated in S. graminum (c) and S. flava (d) as a result of starvation, feeding on Kanlow, or feeding on Summer. Numbers in parenthesis indicate the total number of differentially expressed unigenes in each treatment.
Overall, many of the unigenes that were upregulated in both the Summer and starvation treatments were linked to stress response. These included unigenes coding for α-amylases, lipases, melibiases, trypsins, cysteine proteinases, ABC transporters, peroxidases, CYP450s, major facilitator superfamily transporters, UGTs, and immuglobulin domain proteins (Supp. Tables S3 and S4). Several unigenes were also downregulated in both Summer and starvation, including HSP70s, LRRs, major facilitator superfamily transporters, immunoglobulin domain proteins, and sugar (and other) transporters (Supp. Tables S3 and S4). In contrast, more unigenes related to stress response and digestion were commonly impacted in the Kanlow-starvation comparison relative to the Summer-starvation comparison in S. graminum (Supp. Tables S3 and S4). For example, unigenes coding for enzymes linked to oxidative phosphorylation and lysosomes were strongly upregulated in both the starvation and Kanlow treatments (Fig. 6a and b). Other upregulated unigenes with predicted roles in stress response included ABC transporters, AKRs, alcohol dehydrogenases, amino acid permeases, animal haem peroxidases, carboxylesterases, CYP450s, glutathione peroxidases, GSTs, GH 1s, super oxide dismutases, LRRs, major facilitator superfamily transporters, melibiases, GH 30s, cysteine proteinases, PBP/GOBPs, short-chain dehydrogenases, trehalases, trypsins, and UGTs (Supp. Tables S3, S4, S7, and S8). Furthermore, large numbers of chemosensory related genes were also upregulated at 24 h in both starved S. graminum and those fed on Kanlow (Supp. Table S7). Interestingly, large numbers of unigenes potentially linked with digestion and stress responses were downregulated both the Kanlow and starvation treatments (Supp. Tables S3 and S4). Predominant gene families that were downregulated included HSP70s, LRRs, major facilitator superfamily transporters, ubiquitin-related proteins, immunoglobulin domain proteins, GH family 1 enzymes, and trypsins (Supp. Tables S3 and S4).
Although several unigenes were commonly differentially expressed between the starvation and switchgrass treatments in S. flava, these numbers were lower in comparison to S. graminum. Overall, starvation resulted in upregulation of 1,154 DEUs and downregulation of 1,071 DEUs (Fig. 8b and d), of which 473 and 488 were upregulated and downregulated in all three treatments, respectively, regardless of time. Only 118 upregulated and 71 downregulated DEUs were exclusively shared between the Summer and starvation treatments and like S. graminum, more DEUs were exclusively shared in Kanlow-starvation comparison relative to the Summer-starvation comparison in S. flava with 149 upregulated and 162 downregulated DEUs (Fig. 8b and d). In the Summer-starvation comparison, fewer stress and digestive-related DEUs were upregulated at 24 h in S. flava relative to S. graminum, which included trehalase, trypsin, cysteine proteinase, peroxidases, major facilitator superfamily transporters, UGTs, and immunoglobulin domain protein, while downregulated unigenes at this timepoint included GST, HSP70s, and thioredoxin (Supp. Tables S3 and S4). Like S. graminum, a more diverse response to Kanlow and starvation was also observed in S. flava in terms of the different families of digestive and stress-related unigenes whose expression levels were impacted in these treatments.
In contrast, however, more stress-related unigenes were upregulated in S. graminum in the Kanlow treatment at 24 h relative to S. flava, including unigenes annotated as amylases, amino acid permeases, carboxylesterases, CYP450s, ferritin-like proteins, GGTs, GSTs, lipases, melibiases, papain family cysteine proteinases, PBP/GOBPs, GPCR chemoreceptors, sugar (and other) transporters, transmembrane amino acid transporter proteins, trehalases, thiolases, trypsins, UGTs, and ubiquitin-related proteins (Supp. Tables S3, S4, and S7). Similar numbers of animal haem peroxidases and ABC transporters were upregulated in both S. graminum and S. flava at 24 h in the Kanlow treatment (Supp. Tables S3 and S4), but significantly fewer unigenes that were related to digestion and stress response were downregulated in the Kanlow and starvation treatments in S. flava relative to S. graminum.
Discussion
Intuitively, starvation and feeding on switchgrass induced large-scale transcriptional responses in both S. graminum and S. flava, with S. graminum showing a more rapid response to abiotic and biotic stresses relative to S. flava. Despite this difference, unigenes coding for enzymes with similar functions were impacted in the starvation treatment in both aphids when comparing DEUs at 12 h in S. graminum with DEUs at 24 h in S. flava. Many of the downregulated unigenes were linked to cell division, DNA replication, cell membrane biosynthesis, mitosis, and ribonucleoside metabolism, indicating that starvation likely impacted growth and development of both aphids. Interestingly, many of these same unigenes were also downregulated in aphids fed on both Kanlow and Summer, especially in S. graminum, indicating that growth and development were likely also impacted in these treatments and that these processes may be more strongly impacted in S. graminum compared to S. flava. In contrast, unigenes that were upregulated in both aphids in response to starvation and feeding in switchgrass were largely associated with carbohydrate biosynthesis, lipid metabolism, lysine degradation, and other unigenes related to energy production and ATP metabolism. In general, this stress-induced upregulation of genes coding for basic energy components, such as glycogen synthase, is consistent with previous reports in aphids (Silva et al. 2012, Enders et al. 2014). Notably, Enders et al. (2014) also documented an upregulation of carbohydrate biosynthetic genes including those coding for 1,4 α-glucan-branching enzyme-like, glycogen synthase, and glycogenin in starved soybean aphids, Aphis glycines (Matsumura) (Hempitera: Aphididae). Possibly, upregulation of unigenes associated with carbohydrate biosynthesis and lipid metabolism may be an attempt to mitigate stress, at least temporarily, by accessing and mobilizing energy stores (Silva et al. 2012, Enders et al. 2014). Alternatively, the upregulation of these unigenes in aphids that had fed on both switchgrass cultivars could indicate that anti-feeding or anti-nutritional mechanisms play a role in defense against herbivory in switchgrass. Indeed, transcriptional analysis of Summer switchgrass infested with S. graminum revealed that infested plants downregulate genes coding for primary metabolic enzymes, which may effectively function to starve aphids of nutrients, and upregulate defensive components, which could act as antifeedants (Donze-Reiner et al. 2017). Furthermore, trehalose concentrations were higher in infested plants, which regulates leaf senescence and programmed degradation of cellular components. This is an important mechanism of resistance in Arabidopsis to Myzus persicae (Sulzer) (Hempitera: Aphididae) feeding (Singh et al. 2011). In this case, premature leaf senescence results in the export of nutrients out of the senescing leaf, effectively starving aphids and limiting their growth (Pegadaraju et al. 2005, Louis and Shah 2015).
Both aphids also responded to starvation and feeding in switchgrass by altering the expression levels of unigenes with roles in xenobiotic metabolism and stress response, which were broadly induced in both aphids among the three treatments. Overall, unigenes coding for CYP450s, proteinases, ABC transporters, peroxidases, HSP70s, LRRs, UGTS, and GH family 1 enzymes were among those whose expression levels were commonly impacted, which is in accordance with previous studies on molecular stress responses of aphids. The expression levels of similar copy numbers of unigenes derived from each family were impacted across the three treatments in the two aphid species; however, more unigenes annotated as major facilitator superfamily transporters, alcohol dehydrogenases, α-amylases, amino acid permeases, peroxidases, GH family 1 enzymes, GH family 35 enzymes, folate carriers, sulfate transporters, thioredoxins, trehalases, JHBPs, and lipases were differentially expressed in S. flava. In contrast, the expression levels more unigenes annotated as ecdysteroid kinase, GGTs, lipocalins, and papain family cysteine proteinases were impacted in S. graminum across the three treatments. Furthermore, induction of these unigenes was generally more rapid in S. graminum with upregulation of unigenes linked to xenobiotic metabolism occurring after 12 h. However, for S. flava, significant induction of unigenes coding for xenobiotic enzymes often did not occur until 24 h. In addition, many more stress-related genes were often upregulated in S. graminum in the starvation and switchgrass treatments compared to S. flava while unigenes linked to xenobiotic metabolism were often downregulated in S. flava. The only exception was in the Kanlow treatment where the number of stress-related genes that were downregulated in S. graminum after 24 h exceeded those that were upregulated in this time period. The downregulation of fewer DEUs associated with detoxification and stress response in S. flava may help this aphid persist on this switchgrass cultivar.
Xenobiotic metabolism of plant defensive compounds typically occurs in three phases (Xu et al. 2005, Li et al. 2007) and CYP450s play a crucial role in oxidation–reduction of metabolites in phase I. Phase II enzymes, which conjugate the products of phase I reactions to increase their solubility and prepare them for eventual excretion include sulfotransferases, include UGTs, GSTs, and carboxylesterases (Xu et al. 2005), while phase III enzymes include ATP-binding cassette (ABC) transporters that function as efflux transporters to facilitate the removal of the conjugated compounds from the cell (Xu et al. 2005). All three classes of enzymes have been previously implicated in insect responses to plant defensive compounds and stress responses (Schuler 1996, Despres et al. 2007, Bansal et al. 2014). Accordingly, unigenes coding for phase I CYP450s and phase III ABC transporters were among the most commonly impacted in this study while unigenes coding for phase II enzymes were impacted a lesser extent in both aphids. The expression levels of the majority of these unigenes were impacted by both starvation and feeding in switchgrass and did not seem to respond exclusively to abiotic or biotic stresses. For example, of the 21 DEUs coding for CYP450s in S. flava, the majority (14) were differentially expressed in the starvation treatment and at least one of the switchgrass treatments. Unigenes coding for one CYP450 were induced in the Kanlow and Summer treatments whereas three unigenes each were differentially expressed exclusively in the Summer and Kanlow treatments. Similar trends were noted in the 27 DEUs identified in S. graminum, which included seven that were differentially expressed exclusively in the starvation treatment and one that was differentially expressed exclusively in the Kanlow treatment. Likewise, many of the UGTs, carboxylesterases, and ABC transporters were differentially expressed in the starvation treatment; however, one UGT was specifically upregulated in S. flava in the Kanlow treatment after 24 h and one ABC transporter was upregulated in exclusively in the Kanlow and Summer treatments in S. graminum after 24 h. This finding indicates that many of the unigenes coding for proteins involved in stress response identified in the transcriptomes of S. flava and S. graminum may not be inducible by specific host plants and instead, may respond to general biotic and abiotic stresses. Accordingly, it remains unclear to what extent insect-mediated detoxification of plant metabolites might play in S. flava’s ability to colonize and feed on Kanlow plants; however, these xenobiotic metabolic enzymes often perform multiple biological functions and act on multiple endogenous and exogenous substrates (Li et al. 2004). Due to the promiscuity of CYP450s, it is possible that unigenes coding for detoxification enzymes that are upregulated in both the starvation and switchgrass treatments could play roles in detoxification of plant defensive compounds, even though the majority do not appear to be specifically induced or downregulated in response to switchgrass.
Another mechanism plants employ to protect against herbivory is the deployment of proteinase inhibitors (PIs). PIs are small proteins that are induced in plants in response to insect injury and may interfere with the digestive process of insects (Habib and Fazili 2007). In response, insects may circumvent the deleterious effects of PIs by elevating expression levels of unigenes coding proteinases (Bown et al. 1997) or by expressing different types of proteinases whose activity is not impaired by the PIs (Jongsma and Bolter 1997). Moreover, aphid salivary proteinases may play an important role in establishing a compatible interaction by suppressing plant defense responses (Furch et al. 2015). Previously, Donze-Reiner et al. (2017) documented a significant induction of PIs at 5 and 10 days after infestation (DAI) in Summer plants upon S. graminum herbivory. Although no studies have evaluated PIs in Kanlow in response to aphid herbivory, it is possible that PIs contribute the antibiosis impacts on the two aphids in this cultivar. Despite their potential importance in overcoming plant defense responses, unigenes coding for trypsins were broadly differentially expressed across the three treatments in both aphids and none were induced specifically by feeding on switchgrass, with the exception of one trypsin that was upregulated at 24 h in the Summer treatment in S. flava. Similar trends were noticed for the expression profiles of the cysteine proteinases and only one cysteine proteinase that was exclusively upregulated in the Summer treatment in S. graminum was identified. Overall, more unigenes coding for trypsins and cysteine proteinases were upregulated in both switchgrass treatments in S. graminum (11 and nine unigenes, respectively) compared to S. flava (two and one unigene, respectively). While this observation suggests that many proteinases coded by S. flava and S. graminum are not specifically induced by feeding in switchgrass and may instead be induced as a general stress response mechanism, the PIs previously detected in the cultivar Summer were not induced until 5 or 10 DAI and it is unknown if PIs are induced earlier in response to aphid herbivory.
None of the other classes of unigenes coding for stress-responsive enzymes seemed to be induced specifically by the switchgrass treatments, although common genes that were triggered by both starvation and switchgrass in the two aphids included peroxidases and HSPs. HSPs respond not only to temperature-induced stresses, but to a variety of other abiotic and biotic stresses, such as exposure to pathogens, starvation, UV light, cuticular damage, and dehydration (Feder and Hofmann 1999, Zhao and Jones 2012). The broad upregulation of unigenes coding for these proteins in this study confirms their diverse role in stress responses. Specific roles of HSPs in mitigating stress caused by starvation or feeding in switchgrass are not known; however, ingestion of plant defensive compounds during feeding could cause oxidative damage to critical structural proteins or enzymes and HSPs could help quickly identify and repair misfolded or damaged proteins (Zhao and Jones 2012). Alternatively, HSPs could also mitigate oxidative stress caused by an upregulation of unigenes coding for CYP450s and other detoxification enzymes in stressed aphids. Peroxidases were also broadly induced across treatments and are often found in aphid saliva where they are capable of oxidizing plant defensive compounds including alkaloids and terpenoids (Urbanska et al. 1998). In previous studies, these enzymes were released into the diets regardless of whether toxins were present (Miles and Peng 1989), indicating that many of these canonical defensive enzymes are not directly inducible by the occurrence of specific toxins in the diet. Similar trends were noted for other unigenes associated with stress response; however, more unigenes annotated as major facilitator superfamily transporters, alcohol dehydrogenases, α-amylases, amino acid permeases, GH family 1 enzymes, GH family 35 enzymes, folate carriers, sulfate transporters, thioredoxins, trehalases, and lipases were differentially expressed in S. flava while more GGTs and lipocalins were impacted in S. graminum across the three treatments. The broad upregulation of large numbers of defensive and stress-responsive genes or unigenes belonging to diverse gene families in response to a variety of different stresses may be a strategy of some generalist species (Govind et al. 2010).
One other major transcriptional difference that was detected between the two aphids in response to both the starvation and switchgrass feeding treatments was the upregulation of unigenes encoding proteins required for oxidative phosphorylation in S. graminum after 12 and 24 h in both the starvation and switchgrass feeding treatments. Induction of unigenes coding for similar enzymes was not observed in S. flava in any of the three treatments. Plant defensive compounds have the ability to interfere with oxidative phosphorylation (Berenbaum 1988), and upregulation of these unigenes may be an attempt to overcome this inhibition. Alternatively, oxidative phosphorylation and ATP may be required to sustain activity levels of stress-responsive enzymes (du Rand et al. 2015). For example, an upregulation of genes coding for enzymes involved in oxidative phosphorylation was also observed previously in Asian longhorned beetle, Anoplophora glabripennis (Motschulsky), when feeding in a preferred host (Acer saccharum; sugar maple) compared to a nutrient-rich artificial diet (Mason et al. 2016). In this system, it was hypothesized that insects needed to expend more energy to digest woody tissue and detoxify defensive compounds produced by the host tree. In this study, S. graminum upregulated a larger number of unigenes coding for detoxification enzymes in comparison to S. flava, and it is plausible that S. graminum required more energy to sustain the activity levels of these enzymes. Furthermore, the downregulation of unigenes coding for detoxification enzymes after 24 h in the Kanlow treatment could be reflective of an inability to obtain sufficient nutrients from this plant to generate sufficient ATP to sustain a stress response.
Overall, changes at the transcriptome-level provide data consistent with previous studies on the interactions of these cereal aphids to divergent switchgrass hosts (Koch et al. 2015, Donze-Reiner et al. 2017, Koch et al. 2018), and the overlap between the starvation response to feeding on the two switchgrass cultivars was largely consistent with the feeding behaviors observed for S. graminum and S. flava on Summer and Kanlow switchgrass (Koch et al. 2018). Notably, S. graminum spent more time probing and less time feeding in both Summer and Kanlow switchgrass compared to S. flava, an observation that was more pronounced when it was feeding in Kanlow. The reduced time spent feeding could explain why there were more transcriptional similarities between the two switchgrass treatments and the starvation treatment in S. graminum relative to S. flava. In contrast, comparisons of S. flava transcriptomes from the different treatments indicate this aphid’s ability to feed on either switchgrass, potentially through a sustained upregulation of stress-related genes. It is also possible that S. flava is not as readily detected as S. graminum by its switchgrass host, or that S. flava is better able to detoxify/sequester switchgrass produced antifeedants.
This work provides a comprehensive analysis of the transcriptional changes in two aphids belonging to different taxonomic subfamilies in response to feeding on two switchgrass cultivars. Indeed, a comprehensive understanding of the molecular interactions between switchgrasses cultivars that are being developed as bioenergy feedstocks and cereal aphids can provide important clues about the resistance mechanisms involved in these interactions, which can be valuable to developing and improving integrated pest management strategies. Additionally, understanding the molecular, physiological and behavioral responses of aphids to those resistance mechanisms could potentially lead to more sustainable pest management strategies by providing foresight into possible insect countermeasures to host resistance.
Supplementary Material
Acknowledgments
This research was funded by U.S. Department of Agriculture, National Institute of Food and Agriculture (2011-67009-30096), and by U.S. Department of Agriculture, Agricultural Research Service (3042-21000-030-00D and 3020-43000-032-00D). The U.S. Department of Agriculture, Agricultural Research Service, is an equal opportunity/affirmative action employer and all agency services are available without discrimination. Mention of commercial products and organizations in this manuscript is solely to provide specific information. It does not constitute endorsement by USDA-ARS over other products and organizations not mentioned. The University of Nebraska DNA Sequencing Core receives partial support form the U.S. National Institute of Health, National Institute for General Medical Studies (P20GM10327 and 1P30GM110768-01) as well as The Fred & Pamela Buffet Cancer Center Support Grant – P30CA036727. This publication’s contents are the sole responsibility of the authors and do not necessarily represent the official views of the NIH or NIGMS.
References Cited
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R., Mian M. A., Mittapalli O., and Michel A. P.. 2014. RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean. BMC Genomics 15: 972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., . et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum M. 1988. Allelochemicals in insect-microbe-plant interactions; agents provocateurs in the coevolutionary arms race, pp. 97–123. InBarbosa P. and Letourneau D. K. (eds.), Novel aspects of insect-plant interactions. John Wiley & Sons Ltd, New York, NY. [Google Scholar]
- Blackman R. L., and Eastop V. F.. 2000. Aphids on the world’s crops, an identification and information guide, 2nd ed. John Wiley & Sons Ltd, West Sussex, United Kingdom. [Google Scholar]
- Bown D. P., Wilkinson H. S., and Gatehouse J. A.. 1997. Differentially regulated inhibitor-sensitive and insensitive protease genes from the phytophagous insect pest, Helicoverpa armigera, are members of complex multigene families. Insect Biochem. Mol. Biol. 27: 625–638. [DOI] [PubMed] [Google Scholar]
- Burd J. D., Prasifka J. R., and Bradshaw J. D.. 2012. Establishment and host effects of cereal aphids on switchgrass (Panicum virgatum L.) cultivars. Southwest. Entomol. 37: 115–122. [Google Scholar]
- Cooper W. R., Dillwith J. W., and Puterka G. J.. 2011. Comparisons of salivary proteins from five aphid (Hemiptera: Aphididae) species. Environ. Entomol. 40: 151–156. [DOI] [PubMed] [Google Scholar]
- Després L., David J. P., and Gallet C.. 2007. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 22: 298–307. [DOI] [PubMed] [Google Scholar]
- Donze-Reiner T., Palmer N. A., Scully E. D., Prochaska T. J., Koch K. G., Heng-Moss T., Bradshaw J. D., Twigg P., Amundsen K., Sattler S. E., . et al. 2017. Transcriptional analysis of defense mechanisms in upland tetraploid switchgrass to greenbugs. BMC Plant Biol. 17: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics. 14: 755–763. [DOI] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endara M.-J., and Coley P. D.. 2011. The resource availability hypothesis revisited: a meta-analysis. Funct. Ecol. 25: 389–398. [Google Scholar]
- Enders L. S., Bickel R. D., Brisson J. A., Heng-Moss T. M., Siegfried B. D., Zera A. J., and Miller N. J.. 2014. Abiotic and biotic stressors causing equivalent mortality induce highly variable transcriptional responses in the soybean aphid. G3 (Bethesda). 5: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyres I., Jaquiéry J., Sugio A., Duvaux L., Gharbi K., Zhou J. J., Legeai F., Nelson M., Simon J. C., Smadja C. M., . et al. 2016. Differential gene expression according to race and host plant in the pea aphid. Mol. Ecol. 25: 4197–4215. [DOI] [PubMed] [Google Scholar]
- Feder M. E., and Hofmann G. E.. 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61: 243–282. [DOI] [PubMed] [Google Scholar]
- Furch A. C., van Bel A. J., and Will T.. 2015. Aphid salivary proteases are capable of degrading sieve-tube proteins. J. Exp. Bot. 66: 533–539. [DOI] [PubMed] [Google Scholar]
- Govind G., Mittapalli O., Griebel T., Allmann S., Böcker S., and Baldwin I. T.. 2010. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One 5: e8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., . et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin A. S., Cataldi T. R., Labate C. A., Francis F., and Cônsoli F. L.. 2018. Spiroplasma affects host aphid proteomics feeding on two nutritional resources. Sci. Rep. 8: 2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsen O., Shearman R. C., Heng-Moss T. M., Mutlu N., Lee D. J., and Sarath G.. 2007. Peroxidase gene polymorphism in buffalograss and other grasses. Crop Sci. 47: 767. [Google Scholar]
- Gutsche A., Heng-Moss T., Sarath G., Twigg P., Xia Y., Lu G., and Mornhinweg D.. 2009. Gene expression profiling of tolerant barley in response to Diuraphis noxia (Hemiptera: Aphididae) feeding. Bull. Entomol. Res. 99: 163–173. [DOI] [PubMed] [Google Scholar]
- Habib H., and Fazili K. M.. 2007. Plant protease inhibitors: a defense strategy in plants. Biotechnol. Mol. Biol. Rev. 2: 68–85. [Google Scholar]
- Hentz M., and Nuessly G.. 2004. Development, longevity, and fecundity of Sipha flava (Homoptera: Aphididae) feeding on Sorghum bicolor. Environ. Entomol. 33: 546–553. [Google Scholar]
- Herde M., and Howe G. A.. 2014. Host plant-specific remodeling of midgut physiology in the generalist insect herbivore Trichoplusia ni. Insect Biochem. Mol. Biol. 50: 58–67. [DOI] [PubMed] [Google Scholar]
- Hull J. J., Geib S. M., Fabrick J. A., and Brent C. S.. 2013. Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome. PLoS One 8: e55105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J., and Schuster S. C.. 2007. MEGAN analysis of metagenomic data. Genome Res. 17: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M. A., and Bolter C.. 1997. The adaptation of insects to plant protease inhibitors. J. Insect Physiol. 43: 885–895. [DOI] [PubMed] [Google Scholar]
- Kerchev P. I., Fenton B., Foyer C. H., and Hancock R. D.. 2012. Plant responses to insect herbivory: interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant. Cell Environ. 35: 441–453. [DOI] [PubMed] [Google Scholar]
- Kindler S. D., and Dalrymple R. L.. 1999. Relative susceptibility of cereals and pasture grasses to the yellow sugarcane aphid (Homoptera: Aphididae). J. Agric. Urban Entomol. 16: 113–122. [Google Scholar]
- Koch K. G., Bradshaw J. D., Heng-Moss T. M., and Sarath G.. 2014a. Categories of resistance to greenbug and yellow sugarcane aphid (Hemiptera: Aphididae) in three tetraploid switchgrass populations. Bioenerg. Res. 7: 909–918. [Google Scholar]
- Koch K. G., Fithian R., Heng-Moss T. M., Bradshaw J. D., Sarath G., and Spilker C.. 2014b. Evaluation of tetraploid switchgrass (Poales: Poaceae) populations for host suitability and differential resistance to four cereal aphids. J. Econ. Entomol. 107: 424–431. [DOI] [PubMed] [Google Scholar]
- Koch K. G., Palmer N., Stamm M., Bradshaw J. D., Blankenship E., Baird L. M., Sarath G., and Heng-Moss T. M.. 2015. Characterization of greenbug feeding behavior and aphid (Hemiptera: Aphididae) host preference in relation to resistant and susceptible tetraploid switchgrass populations. Bioenerg. Res. 8: 165–174. [Google Scholar]
- Koch K. G., Donze-Reiner T., Baird L. M., Louis J., Amundsen K., Sarath G., Bradshaw J. D., and Heng-Moss T.. 2018. Evaluation of greenbug and yellow sugarcane aphid feeding behavior on resistant and susceptible switchgrass cultivars. Bioenerg. Res. 11: 480–490. [Google Scholar]
- Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C. N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Baudry J., Berenbaum M. R., and Schuler M. A.. 2004. Structural and functional divergence of insect CYP6B proteins: from specialist to generalist cytochrome P450. Proc. Natl. Acad. Sci. U. S. A. 101: 2939–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Schuler M. A., and Berenbaum M. R.. 2007. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52: 231–253. [DOI] [PubMed] [Google Scholar]
- Louis J., and Shah J.. 2015. Plant defence against aphids: the PAD4 signalling nexus. J. Exp. Bot. 66: 449–454. [DOI] [PubMed] [Google Scholar]
- Mason C. J., Scully E. D., Geib S. M., and Hoover K.. 2016. Contrasting diets reveal metabolic plasticity in the tree-killing beetle, Anoplophora glabripennis (Cerambycidae: Lamiinae). Sci. Rep. 6: 33813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels G. J., Jr 1986. Graminaceous North American host plants of the greenbug with notes on biotypes. Southwest. Entomol. 11: 55–66. [Google Scholar]
- Miles P. W. 1999. Aphid saliva. Biol. Rev. 74: 41–85. [Google Scholar]
- Miles P. W., and Peng Z.. 1989. Studies on the salivary physiology of plant bugs - detoxification of phytochemicals by the salivary peroxidase of aphids. J. Insect. Physiol. 35: 865–872. [DOI] [PubMed] [Google Scholar]
- Moore K. J., Moser L. E., Vogel K. P., Waller S. S., Johnson B. E., and Pedersen J. F.. 1991. Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83: 1073–1077. [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., and Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35: W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S. J., and Puterka G. J.. 2014. Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum Rondani) biotypes. J. Proteomics 105: 186–203. [DOI] [PubMed] [Google Scholar]
- Nuessly G. S. 2005. Yellow sugarcane aphid, Sipha flava (Forbes) (Insecta: Hemiptera: Aphididae), featured creatures, EENY354. University of Florida/Institute of Food and Agricultural Sciences Extension. [Google Scholar]
- Nuessly G. S., and Nagata R. T.. 2005. Greenbug, Schizaphis graminum (Rondani) (Insecta: Hemiptera: Aphididae), Featured Creatures. University of Florida/Institute of Food and Agricultural Sciences Extension. [Google Scholar]
- Pallipparambil G. R., Cha G., and Gray M. E.. 2014. A comparative life-table analysis of Sipha flava (Hemiptera: Aphididae) on two biofuel hosts, Miscanthus x giganteus and Saccharum spp. J. Econ. Entomol. 107: 1069–1075. [DOI] [PubMed] [Google Scholar]
- de la Paz Celorio-Mancera M., Wheat C. W., Vogel H., Soderlind L., Janz N., and Nylin S.. 2013. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Mol. Ecol. 22: 4884–4895. [DOI] [PubMed] [Google Scholar]
- Pegadaraju V., Knepper C., Reese J., and Shah J.. 2005. Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol. 139: 1927–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., and Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Ramm C., Saathoff A., Donze T., Heng-Moss T., Baxendale F., Twigg P., Baird L., and Amundsen K.. 2013. Expression profiling of four defense-related buffalograss transcripts in response to chinch bug (Hemiptera: Blissidae) feeding. J. Econ. Entomol. 106: 2568–2576. [DOI] [PubMed] [Google Scholar]
- du Rand E. E., Smit S., Beukes M., Apostolides Z., Pirk C. W., and Nicolson S. W.. 2015. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci. Rep. 5: 11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M. A. 1996. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 112: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Hoover K., Carlson J. E., Tien M., and Geib S. M.. 2013. Midgut transcriptome profiling of Anoplophora glabripennis, a lignocellulose degrading cerambycid beetle. BMC Genomics 14: 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. X., Jander G., Samaniego H., Ramsey J. S., and Figueroa C. C.. 2012. Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: Aphididae) I: a transcriptomic survey. PLoS One 7: e36366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Singh V., Louis J., Ayre B. G., Reese J. C., Pegadaraju V., and Shah J.. 2011. TREHALOSE PHOSPHATE SYNTHASE11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J. 67: 94–104. [DOI] [PubMed] [Google Scholar]
- Team, R. C 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN3-900051-07-0 https://www. R-project. org [Google Scholar]
- Urbanska A., Tjallingii W. F., Dixon A. F. G., and Leszczynski B.. 1998. Phenol oxidising enzymes in the grain aphid’s saliva. Entomol. Exp. Appl. 86: 197–203. [Google Scholar]
- Xie W., Lei Y., Fu W., Yang Z., Zhu X., Guo Z., Wu Q., Wang S., Xu B., Zhou X., . et al. 2012. Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut. Int. J. Biol. Sci. 8: 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Li C. Y., and Kong A. N.. 2005. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 28: 249–268. [DOI] [PubMed] [Google Scholar]
- Yates A. D., and Michel A.. 2018. Mechanisms of aphid adaptation to host plant resistance. Curr. Opin. Insect Sci. 26: 41–49. [DOI] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., and Oshlack A.. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., and Jones W. A.. 2012. Expression of heat shock protein genes in insect stress responses. Isj-Invert. Surviv. J. 9: 93–101. [Google Scholar]
- Züst T., and Agrawal A. A.. 2016. Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2: 15206. [DOI] [PubMed] [Google Scholar]
- Züst T., and Agrawal A. A.. 2017. Plant chemical defense indirectly mediates aphid performance via interactions with tending ants. Ecology 98: 601–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.